The ability of bacteria to respond to a multitude of environmental signals and integrate these signals to trigger adaptive responses provides a successful strategy for survival in rapidly changing environments. In many cases integration can be achieved via the interlinking of different regulatory circuits in which various master regulators respond to different environmental cues. However, in some systems an economy of scale can be achieved if individual regulatory proteins are able to respond to more than one input signal. In this review we consider a particular example where multiple signals are integrated by a regulatory protein complex to finely tune transcriptional regulation of nitrogen fixation in free-living diazotrophic bacteria.
The ability to fix atmospheric nitrogen to ammonia by using the enzyme nitrogenase enables diazotrophs not only to survive but also to proliferate under conditions of extreme fixed-nitrogen deprivation. This strategy, however, incurs an energetic penalty since nitrogenase consumes 16 mol of ATP per mol of dinitrogen fixed in vitro, and the energy cost increases to up to 40 mol of ATP per mol in vivo (44). The irreversible inactivation of nitrogenase by oxygen also imposes physiological constraints on diazotrophs such that they may only be able to utilize the enzyme under anaerobic conditions or, alternatively, they may employ elegant physiological strategies to protect the enzyme from damage by oxygen under aerobic conditions. In addition to its oxygen sensitivity, nitrogenase is a kinetically slow enzyme (97). Growth in the absence of fixed nitrogen requires a high concentration of the enzyme, and some diazotrophs accumulate 10 to 20% of the total cell protein as nitrogenase under nitrogen-fixing conditions. Nitrogen fixation thus provides an opportunistic strategy to colonize nitrogen-deficient environments, but the cellular commitment in terms of protein synthesis and ATP consumption is appropriate only under specific environmental conditions. The oxygen concentration and the availability of fixed nitrogen are therefore important factors in the regulation of nitrogenase biosynthesis. These signals are integrated to provide transcriptional control of nif gene expression in free-living diazotrophs.
In diazotrophic representatives of the Proteobacteria, nif gene transcription is mostly dependent on the alternative sigma factor σ54, which recognizes promoters with consensus sequences located at positions −24 and −12 (9, 16). Transcription initiation by the σ54-RNA polymerase holoenzyme requires a specific class of transcriptional activator, which binds to enhancer-like elements upstream of σ54-dependent promoters and through ATP hydrolysis catalyzes conformational changes in σ54 which enable the holoenzyme to undergo the transition to the transcriptionally competent open promoter complex. σ54-dependent activators, which are also known as enhancer binding proteins (EBPs) (72, 95), belong to the AAA+ superfamily of ATPases that transform energy into mechanical function to remodel their substrates (75, 103). Activation of σ54-dependent nif gene transcription requires NifA, a conserved EBP that regulates genes necessary for the synthesis of molybdenum nitrogenase in proteobacteria.
Activation of transcription by NifA is regulated in response to oxygen and fixed nitrogen. In many diazotrophic members of the alpha and beta subgroups of the Proteobacteria, NifA activity is apparently directly responsive to the oxygen status, and conserved cysteine residues have been implicated in the response (32, 33). There is also evidence that the activity of these NifA proteins is regulated in response to fixed nitrogen. However, in some nitrogen-fixing organisms, particularly members of the gamma subgroup of the Proteobacteria, the activity of NifA is not intrinsically oxygen or nitrogen responsive, and a partner protein, NifL, is required to modulate NifA activity directly in response to oxygen and fixed nitrogen (24, 89).
Initial sequencing of the nifL-nifA operon suggested that these genes could encode a two-component regulatory system. Although NifA does not contain an archetypal response regulator receiver-like domain, the C-terminal region of NifL shows homology to the histidine protein kinases (HPKs) (12, 26). Moreover, Azotobacter vinelandii NifL contains a conserved histidine residue found in the transmitter domains of histidine kinases, suggesting that this NifL might employ a classical phosphoryl transfer mechanism to communicate environmental signals to NifA. However, replacement of this conserved histidine by a number of other amino acids does not disable signal transduction (101). Furthermore, NifL is competent to inhibit NifA in vitro in the absence of ATP, and signal transduction requires stoichiometric protein-protein interactions between the two regulatory proteins (6, 43, 57, 69, 87). Current evidence suggests that NifL controls the activity of NifA by a relatively stable protein-protein interaction that is modulated by redox changes, ligand binding, and interactions with other signal transduction proteins and membrane components.
NifL homologues are found in various enterobacteria, including Klebsiella pneumoniae (51), Klebsiella oxytoca (53), and Pantoea agglomerans (92), and in the aerobic diazotrophs A. vinelandii (12, 81) and Pseudomonas stutzeri (23). Database searches have suggested that the NifL-NifA system is also present in the plant pathogen Erwinia chrysanthemi and in the methane-oxidizing bacterium Methylococcus capsulatus strain Bath. A NifL-NifA system has also been reported in Azoarcus sp. strain BH72, a diazotrophic grass endophyte belonging to the beta subgroup of the Proteobacteria (29). A sequence (PY07698) encoding a NifL-NifA fusion protein in the genome of the rodent malaria parasite Plasmodium yoelii yoelii (18) is probably an artifact due to contamination with Methylococcus DNA sequences. In this review we focus on the NifL-NifA systems of K. pneumoniae and A. vinelandii, which are the most well-studied systems and provide examples of sophisticated multidomain signaling interactions that are responsive to environmental cues. The NifL and NifA proteins of K. pneumoniae are designated Kp NifL and Kp NifA, respectively, and the NifL and NifA proteins of A. vinelandii are designated Av NifL and Av NifA, respectively.
DOMAIN STRUCTURES
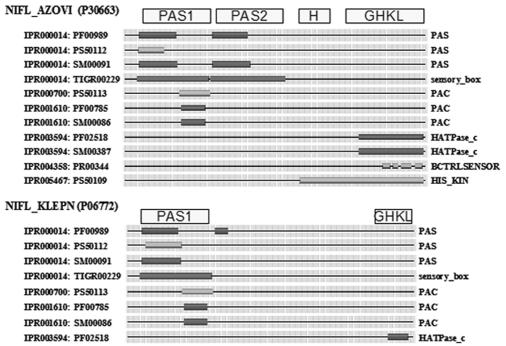
Av NifL contains at least three discrete domains. The N-terminal region has a sensing function, and the C terminus exhibits homology to transmitter domains of the HPKs. Separating these regions is a glutamine-rich hydrophilic sequence representing a Q linker (102). Kp NifL has a domain structure that is more difficult to define, but it also has an N-terminal sensory region and a C-terminal region required for signal transmission. The domain annotation for Av NifL and Kp NifL is shown in Fig. 1. One or more PAS domains are found in the N-terminal region (104). Although limited homology has been observed among PAS domains at the sequence level, structural studies have demonstrated that various cofactors, including heme, flavin adenine dinucleotide (FAD), flavin mononucleotide, and 4-hydroxycinnamic acid, are retained by a common α/β fold (96). The NifL sequences of aerobic diazotrophs (e.g., A. vinelandii) possess two PAS domains, which we designate PAS1 and PAS2, whereas the NifL sequences of bacteria which fix nitrogen under anaerobic conditions (e.g., K. pneumoniae) have only a single PAS domain, PAS1 (Fig. 1). The PAS1 domains of both Kp NifL and Av NifL have been shown to contain FAD as a prosthetic group (45, 88) and are required for the redox-sensing functions of these proteins, as discussed below. Like wild-type Av NifL, the isolated PAS1 domain is tetrameric and contains ∼1 mol of FAD per monomer (40-42). However, the function of the Av NifL PAS2 domain is unknown. Truncated derivatives of Av NifL lacking the PAS1 domain are not responsive to the redox status but are competent to signal the response to fixed nitrogen, demonstrating that redox and nitrogen sensing are discrete functions of the protein (Table 1) (93).
FIG. 1.
Graphic view of the domain structure of Av NifL (NIFL_AZOVI) and Kp NifL (NIFL_KLEPN) from the INTERPRO server (http://www.ebi.ac.uk/interpro/). INTERPRO assignments and database cross-references are indicated on the left, and the corresponding domains are indicated on the right. Domain designations used in this paper are above the diagram. PAS domains are subdivided into PAS and PAC motifs in some databases (78), but both of these motifs form an integral part of a single structural domain. The term PAS domain in this paper refers to the single structural fold that encompasses both the PAS and PAC motifs.
TABLE 1.
Properties of truncated derivatives of Av NifL
| Fragment | Domain(s) | FADa | Association stateb | ATP-ADP bindingc | NifA interactiond | GlnK interactione | ADP responsef | Nitrogen response
|
Oxygen response
|
||
|---|---|---|---|---|---|---|---|---|---|---|---|
| In vivog | In vitroh | In vivog | In vitroi | ||||||||
| Wild-type NifL | PAS1, PAS2, H, GHKL | + | Tetramer | + | + | + | + | + | + | + | + |
| 1-140 | PAS1 | + | Tetramer | NDj | ND | ND | ND | ND | ND | ND | ND |
| 1-284 | PAS1, PAS2 | + | Tetramer | − | − | − | − | ND | − | ND | − |
| 147-519 | PAS2, H, GHKL | − | Mostly dimer | + | + | + | + | + | + | − | − |
| 360-519 | GHKL | − | Monomer | + | + | + | + | ND | − | ND | − |
Derived from limited proteolysis data (93).
Complex formation measured by coaffinity chromatography in the presence of Mg-ADP (69).
Ability to bind GlnK in the presence of 2-oxoglutarate and ATP, determined by pull down assays and surface plasmon resonance (60).
Ability to inhibit open promoter complex formation by NifA in vitro in the presence of ADP (31, 93).
Inhibition of open promoter complex formation by NifA in vitro in the presence of ADP, 2-oxoglutarate, and GlnK (60, 63).
Inhibition of open promoter complex formation by NifA in vitro under oxidizing conditions (45, 93).
ND, not determined.
As noted above, the sequence of Av NifL suggested homology to the transmitter domains of members of the HPK family. Structural studies of representatives of the HPK family have revealed a C-terminal catalytic domain, required for ATP binding and trans phosphorylation, and a dimerization domain (H domain) containing the conserved histidine residue required for phosphotransfer to the response regulator (94). The catalytic kinase domain belongs to the GHKL (HATPase_c) superfamily of ATPases, which includes DNA gyrase B, Hsp90, SpoIIAB, and MutL (27). Av NifL contains conserved residues corresponding to the N, G1, F, and G2 boxes that constitute the ATP binding domain of the GHKL superfamily, and this domain of Av NifL has been demonstrated to bind adenosine nucleotides (93) (see below). However, the corresponding region of Kp NifL is less homologous to the GHKL superfamily and apparently does not bind nucleotides (56). Structural predictions also indicate that the region of Av NifL located between the PAS2 and GHKL domains may constitute an antiparallel helix bundle, similar to the structures of the dimerization (H) domains of the HPKs EnvZ and CheA.
The domain architecture of diazotrophic NifA proteins is more uniform than that of NifL proteins. The N-terminal region of NifA proteins constitutes a GAF domain, a ubiquitous sensory module found in signaling proteins in all phyla (1). GAF domains have a three-dimensional structure similar to that of PAS domains (47) and are known to bind small molecules, including cyclic nucleotides (50) and formate (48). Truncated NifA proteins lacking the N-terminal GAF domain have altered regulatory properties and, in the case of Av NifA and Kp NifA, have altered responses to NifL (8, 25). The central domain of NifA proteins is highly conserved and comprises the AAA+ module required for nucleotide hydrolysis and σ54 interaction that is common to all EBPs (72, 95). AAA+ proteins are oligomeric and commonly form a hexameric ring-like structure in which nucleotide is bound between adjacent protomers (99). Oligomerization of EBPs promotes ATP hydrolysis, which in turn couples changes in protomer structure to binding interactions with σ54 (17, 19, 103). A conserved GAFTGA motif in the AAA+ domain that is unique to EBPs is critical for transcriptional activation (100). This region constitutes an interaction surface that binds σ54 and couples the energy of ATP hydrolysis to open complex formation (15). As is the case for all EBPs, the C-terminal domain of NifA contains a helix-turn-helix motif required for recognition of the enhancer-like, upstream activator sequences (70). Mutations in the recognition helix of Kp NifA that influence transcriptional activation occur at residues that exhibit chemical shifts in the nuclear magnetic resonance spectra of the C-terminal domain upon DNA binding, directly demonstrating that the helix-turn-helix motif interacts with the enhancer-like sequences (82).
LIGAND BINDING AND THE NifL-NifA INTERACTION
Since NifL proteins have a domain structure similar to that of the HPKs, it was expected that these proteins might bind adenosine nucleotides. Av NifL apparently does not hydrolyze ATP, exhibit autophosphorylation, or phosphotransfer to NifA. However, the C-terminal GHKL domain of Av NifL binds ATP and ADP (93). The binding of these nucleotides influences the conformation of the C-terminal region, which is particularly susceptible to trypsin cleavage in the absence of nucleotide. The affinity of Av NifL for ADP (apparent Kd, ∼13 μM) is approximately 10-fold higher than that of ATP (93). The presence of ADP also influences the activity of NifL in vitro, strongly stimulating the ability of Av NifL to inhibit the activity of Av NifA (31, 45). The ability of ADP to act as an effector of NifL activity is reflected by the increased stability of the protein complexes formed between Av NifL and Av NifA in the presence of this ligand (69). Removal of the nucleotide causes dissociation of the complex. Neither the N-terminal region of Av NifL (69) nor the GAF domain of Av NifA (8, 58) is essential for the interaction. Protein footprinting experiments indicate that complex formation alters the protease sensitivity of the Q linker region in Av NifL and the GAF domain and linker region between the GAF and AAA+ domains of Av NifA (68). Overall, these experiments suggest that the binding of adenosine nucleotides to the GHKL domain of Av NifL influences the conformation of the transmitter region and facilitates interaction with Av NifA.
Expression of K. pneumoniae nifL and nifA genes is coupled at the translational level, and immunoprecipitation experiments have demonstrated that Kp NifL and Kp NifA form a protein-protein complex in vivo (34, 35, 43). However, the requirements for complex formation appear to be different from those of the A. vinelandii NifL-NifA system (Table 2). As mentioned above, the C-terminal region of Kp NifL is less homologous to the HPKs than Av NifL is, and it lacks conserved residues required for nucleotide binding in the GHKL superfamily. In contrast to Av NifL, the presence of adenosine nucleotides is not specifically required for the inhibition of Kp NifA activity by Kp NifL (57). Although neither ATP hydrolysis (10) nor kinase activities (87) have been reported previously, Kp NifL has recently been shown to exhibit ATP binding and ATPase activity when it is purified from cultures grown under conditions of nitrogen sufficiency (56). In contrast, Kp NifL purified from cultures grown under conditions of nitrogen deficiency is catalytically inactive. Although both of these forms are competent to inhibit Kp NifA in vitro, an increase in inhibitory activity is observed in the presence of adenosine nucleotides. Surprisingly however, nucleotide binding to Kp NifL is apparently associated with the PAS1 domain and not with the C-terminal GKHL-like domain (56). The sequences required for the interaction between Kp NifL and Kp NifA also appear to be different than the sequences required in the A. vinelandii NifL-NifA system. Unlike the isolated C-terminal kinase-like domain of Av NifL, the C-terminal domain of Kp NifL is sufficient to inhibit Kp NifA both in vivo and in vitro (74) (Table 2).
TABLE 2.
Properties of truncated derivatives of Kp NifL
| Fragment | Domain(s) | FADa | Association stateb | ATP bindingc | ATP hydrolysisd | Inhibition of NifAe |
|---|---|---|---|---|---|---|
| Wild-type NifL | PAS1, GHKL? | + | NDf | + | + | + |
| 1-113 | − | ND | + | ND | − | |
| 1-242 | PAS1 | + | ND | + | + | − |
| 136-495 | GHKL? | − | ND | ND | ND | + |
| 266-495 | GHKL? | − | Dimeric | ND | ND | + |
| 306-495 | GHKL? | ND | Dimeric | − | − | + |
Wild-type Kp NifL is highly insoluble (7) and is highly aggregated even when it is expressed as an MBP fusion protein (74). The association states of truncated derivatives were measured by analytical gel filtration (74).
Measured by filter binding with labeled ATP and affinity chromatography on ATP-agarose. This activity was detected only when NifL was purified from cells grown under nitrogen-sufficient conditions (56).
Determined by hydrolysis of labeled ATP. This activity was detected only when NifL was purified from cells grown under nitrogen-sufficient conditions (56).
Wild-type Kp NifL is competent to inhibit Kp NifA in response to fixed nitrogen and oxygen in vivo (46, 66). When expressed from a multicopy vector, wild-type Kp NifL and some deletion derivatives are competent to inhibit Kp NifA even under anaerobic, nitrogen-limiting conditions in vivo. These deletion derivatives are also competent to inhibit transcriptional activation by NifA in vitro (74).
ND, not determined.
Although the response of Av NifL to adenosine nucleotides might provide a mechanism for sensing the energy charge, the physiological relevance of nucleotide binding is unclear, since the intracellular concentrations of ATP and ADP are far greater than the association constants of Av NifL for these nucleotides. Potentially, the affinity of Av NifL for nucleotides could be altered in response to environmental cues, as postulated for Kp NifL (56). Alternatively, other ligands may influence the NifL-NifA interaction. It has recently been shown that 2-oxoglutarate is an allosteric effector of the A. vinelandii NifL-NifA system that counteracts the inhibitory function of the ADP-bound form of Av NifL (63). 2-Oxoglutarate has been implicated as a key metabolic signal of the carbon status, but the concentration of this metabolite also reflects the nitrogen status. The response of the A. vinelandii NifL-NifA system to 2-oxoglutarate in vitro is within the physiological range, which extends from ∼100 μM under conditions of carbon limitation (excess of fixed nitrogen) to ∼1 mM under conditions of nitrogen deficiency (excess of carbon) (85, 90). Thus, at relatively low 2-oxoglutarate concentrations, the ADP-bound form of NifL is competent to inhibit NifA activity, but at high 2-oxoglutarate levels, NifA is not responsive to NifL in the presence of ADP (63). Isothermal titration calorimetry experiments have demonstrated that 2-oxoglutarate binds to Av NifA but not to Av NifL (61). The GAF domain of Av NifA exhibits stoichiometric binding of 2-oxoglutarate with a dissociation constant of 60 μM, and binding is not observed with a truncated form of Av NifA lacking the GAF domain. The interaction of 2-oxoglutarate with the GAF domain may induce conformational changes in Av NifA which render it resistant to Av NifL, since the binding of 2-oxoglutarate alters the susceptibility of this domain to digestion with trypsin (61). In contrast, the GAF domain of K. pneumoniae NifA apparently does not bind 2-oxoglutarate, emphasizing the mechanistic differences between the A. vinelandii and K. pneumoniae NifL-NifA systems.
NITROGEN REGULATION
Nitrogen fixation is essential for growth only under conditions of fixed-nitrogen deficiency, and thus the activity of NifA is stringently controlled by NifL in response to the fixed-nitrogen status. Key information related to the intracellular nitrogen and the carbon status is communicated by the signal transduction protein PII, which integrates these signals and transmits the information globally to various receptors to control nitrogen assimilation (in this case the NifL-NifA system). PII-like proteins constitute a highly conserved family of signaling proteins found in all three kingdoms of life (2, 76). Many bacteria possess more than one PII-like protein; these proteins have common functions but perform discrete physiological roles. Enteric bacteria express two PII paralogues, designated GlnB and GlnK, that are both subject to reversible covalent modification by the uridylyltransferase/uridylyl-removing enzyme, the product of glnD (2). The activity of this enzyme is regulated by the intracellular level of glutamine, a key signal of the nitrogen status. Under conditions of nitrogen deprivation, when the intracellular level of glutamine is relatively low, GlnD uridylylates the PII proteins. Conversely, at high glutamine concentrations, the PII proteins are deuridylylated by GlnD. The site of covalent modification is a conserved tyrosine residue (Tyr 51) which is located at the tip of a surface-exposed loop (the T loop) required for the interaction of PII proteins with their receptors. The activity of PII is controlled not only by covalent modification but also by the binding of the ligands ATP and 2-oxoglutarate (76). As with Av NifA, the interaction with 2-oxoglutarate enables PII proteins to respond allosterically to a signal of central carbon metabolism. PII proteins are trimeric and contain three binding sites for 2-oxoglutarate and ATP.
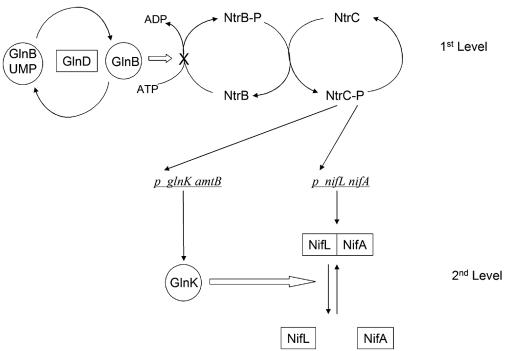
The primary physiological role of GlnB in enteric bacteria is to control nitrogen assimilation under conditions of relative nitrogen sufficiency, whereas GlnK mediates nitrogen control under conditions of nitrogen starvation (5, 13). This enables nitrogen regulatory cascades in which the PII paralogues operate at different levels. Such a cascade mediates nitrogen regulation of nitrogen fixation in K. pneumoniae. By modulating the activity of the NtrB-NtrC two-component regulatory system, GlnB controls the phosphorylation state of the EBP, NtrC, a key activator of nitrogen-regulated genes. This first level of the cascade ensures that activation of the glnK and nifLA promoters is determined by the phosphorylation state of NtrC. Both of these promoters possess relatively weak enhancers, and a high level of NtrC-P is required for their activation (5, 67). The first level of nitrogen control, therefore, governs the expression of NifL, NifA, and GlnK, the major players at the second level of the cascade (39) (Fig. 2). Although glnB mutations influence the first step of the cascade by altering the level of NtrC-P, they do not influence the fixed-nitrogen response of the Kp NifL and Kp NifA proteins per se, which occurs at the second level of the cascade. However, in glnK mutants, Kp NifA activity is constitutively inactivated by Kp NifL, indicating that in the second step of the cascade, GlnK is required to prevent Kp NifL from inhibiting Kp NifA under conditions of nitrogen starvation (38, 49). Although there is no biochemical evidence, these results suggest that GlnK could bind to either Kp NifL or Kp NifA to prevent interaction between the partners under nitrogen-limiting conditions (Fig. 2). The ability of GlnK to relieve inhibition by Kp NifL under conditions of nitrogen starvation does not require covalent modification of GlnK, since normal regulation by NifL is observed in glnD mutants and with a mutant form of GlnK, GlnK-Y51N, which cannot be uridylylated by GlnD (28, 38). The specificity of the interaction between GlnK and the K. pneumoniae nif-specific regulatory proteins has been investigated by comparing the T-loop regions of Escherichia coli GlnB and GlnK. Substitution of residues 43 and 54 of GlnB by the analogous residues in GlnK enables GlnB to relieve NifL inhibition of Kp NifA (3). Expression of GlnB on a multicopy plasmid also relieves NifL inhibition (4), which is congruent with the finding that specific physiological roles played by GlnB and GlnK are dependent upon expression levels rather than discrete functions (13).
FIG. 2.
Cascade regulation of nif genes in K. pneumoniae in response to the fixed nitrogen status. Under nitrogen-limiting conditions, GlnB is uridylylated and NtrB phosphorylates NtrC, leading to activation of transcription of the glnK, amtB, and nifLA operons. Expression of GlnK prevents Kp NifL from inhibiting Kp NifA, leading to activation of nif transcription. Under nitrogen-sufficient conditions, GlnD deuridylylates GlnB, which is then competent to activate the phosphatase activity of NtrB; this limits the availability of NtrC-P and prevents expression of nifLA and glnK amtB. Following ammonium upshift GlnK may be sequestered by AmtB, as discussed in the text.
In A. vinelandii similar components required for nitrogen regulation are present, but the cascade appears to have become disconnected and the mechanism of regulation is somewhat different. Although this organism possesses a bona fide NtrB-NtrC two-component regulatory system which regulates nitrate assimilation, NtrC is not required to activate transcription from the nifLA promoter (98). This promoter is active under conditions of nitrogen excess and, surprisingly, is not dependent on rpoN even though there is a potential σ54 interaction site appropriately positioned upstream of the transcription start site (12, 81). Another unusual feature of A. vinelandii is the presence of only a single gene encoding a PII-like protein, designated glnK, which is located in an operon with amtB and is transcribed constitutively (20, 65). Therefore, both the nifLA and glnK amtB operons are transcribed under conditions of nitrogen sufficiency, and thus the first level of the nitrogen regulatory cascade present in enteric bacteria is absent in A. vinelandii. The glnK gene is apparently essential in A. vinelandii (65), and GlnK is uridylylated by a homologue of GlnD previously known as NfrX (20). Mutants with mutations in glnD which decrease uridylyltransferase activity are unable to fix nitrogen. However, the Nif− phenotype of these strains can be suppressed by secondary mutations that inactivate NifL (20, 21). This suggests that the uridylylation function of GlnD is necessary to prevent Av NifL from inhibiting Av NifA in A. vinelandii, in contrast to enteric bacteria, in which uridylylation of GlnK is not required. Since A. vinelandii glnK is an essential gene, it has not been possible to examine the phenotype of strains with glnK null mutations. However, a strain expressing a mutant form of GlnK with a mutation in the T loop, GlnK-Y51F, which prevents uridylylation by GlnD, is stable providing that the strain contains a secondary mutation (gln-71) that also prevents the adenylylation of glutamine synthetase (86). The secondary mutation is necessary because the nonuridylylated form of GlnK results in constitutive adenylylation of glutamine synthetase, thus preventing ammonia assimilation (20). Strains containing the glnK-Y51F and gln-71 mutations are impaired for nitrogen fixation, and Av NifA is inactivated by Av NifL even under nitrogen-fixing conditions. As is the case for the glnD mutations, this phenotype is suppressed by insertion mutations in nifL (86). Thus, uridylylation of GlnK is necessary in order to prevent inhibition by Av NifL under conditions of fixed-nitrogen deficiency.
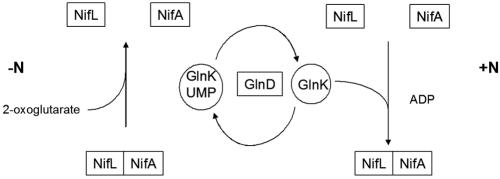
The A. vinelandii nifLA system responds to nitrogen regulation when it is introduced into E. coli (93), which allows comparisons between the responses of the K. pneumoniae and A. vinelandi NifL-NifA systems to well-characterized nitrogen-regulatory mutations. In contrast to the response of the K. pneumoniae system in E. coli, neither GlnK nor GlnB is required to relieve inhibition by Av NifL under nitrogen-limiting conditions. Moreover, in double glnB ntrC mutants or in triple glnB glnK ntrC mutants, which do not express the PII proteins, the activity of Av NifA is not regulated by Av NifL in response to the nitrogen source (85). This suggests that in contrast to the K. pneumoniae NifL-NifA system, the PII proteins are required to activate the inhibitory function of Av NifL rather than prevent inhibition (Fig. 3). This conclusion is strongly supported by the results of biochemical experiments performed with purified components. The inhibitory function of Av NifL is activated by the nonuridylylated form of E. coli GlnB but not by the fully uridylylated form (GlnB-UMP) (63). Likewise, the nonmodified form of A. vinelandii GlnK (Av GlnK) is competent to activate the inhibitory function of Av NifL in the presence of 2-oxoglutarate and ATP. However, when fully uridylylyated by the A. vinelandii GlnD protein, Av GlnK does not activate Av NifL (63). This is fully consistent with the in vivo data and demonstrates that covalent modification of Av GlnK by GlnD is necessary to prevent Av GlnK from activating the inhibitory function of Av NifL.
FIG. 3.
Nitrogen source regulation of nif gene transcription in A. vinelandii. Under conditions of fixed-nitrogen limitation (−N), GlnK is mainly uridylylated and not competent to interact with NifL. Under these conditions, binding of 2-oxoglutarate to the GAF domain of Av NifA relieves inhibition by Av NifL, freeing NifA to activate transcription. Under conditions of fixed-nitrogen sufficiency (+N), GlnD deuridylylates GlnK, which interacts with NifL, promoting formation of the NifL-NifA complex.
Potentially, Av GlnK could interact with either Av NifL or Av NifA to modulate their activities. Interactions between Av GlnK and Av NifL, but not between Av GlnK and Av NifA, have been detected by coaffinity assays, surface plasmon resonance experiments, and yeast two-hybrid assays (13, 86). The in vitro interaction of Av GlnK with Av NifL is abolished by a mutation, E44C, in the T loop of GlnK and also, as expected, when Av GlnK is fully uridylylated. Similar to the interactions of the E. coli PII proteins with their receptors, the binary interaction is dependent upon Mg2+, ATP, and 2-oxoglutarate (60). The role of Av GlnK in communicating the nitrogen status to Av NifL may be analogous to the role of E. coli GlnB in signaling to the histidine kinase NtrB, since in both cases the interactions have been localized to the C-terminal GHKL domain of the receptors (60, 77). However, the response of the interaction to 2-oxoglutarate appears to be different in each case. This ligand is an allosteric effector of the E. coli GlnB-NtrB interaction but not the Av GlnK-Av NifL interaction (60). This difference may arise from the discrete ligand binding properties of E. coli GlnB and Av GlnK. As mentioned above, GlnB binds a single molecule of 2-oxoglutarate with high affinity, and the binding of subsequent molecules is inhibited by anticooperativity. However, isothermal titration calorimetry experiments have suggested that unlike E. coli GlnB, Av GlnK does not exhibit negative cooperativity in the binding of 2-oxoglutarate (60). The involvement of 2-oxoglutarate in the Av GlnK-Av NifL interaction is also complicated by the binding of this ligand to the GAF domain of Av NifA. Protease footprinting experiments have suggested that the interaction with Av GlnK promotes the formation of a GlnK-NifL-NifA ternary complex (61). Whereas the binding of 2-oxoglutarate to the GAF domain of NifA appears to favor dissociation of the binary A. vinelandii NifL-NifA complex, the presence of nonmodified GlnK favors the ternary interaction, even at high 2-oxoglutarate concentrations.
The mechanism by which GlnK communicates the nitrogen status to the A. vinelandii NifL-NifA system is thus clearly very different from that observed in K. pneumoniae. In A. vinelandii, Av GlnK is required to activate the inhibitory function of Av NifL, whereas in K. pneumoniae GlnK is required to prevent Kp NifL from inhibiting Kp NifA. K. pneumoniae GlnK could interact either with Kp NifL or Kp NifA to promote dissociation of the complex. In contrast, the nonmodified form of Av GlnK promotes association of a ternary complex. Covalent modification of Av GlnK by GlnD modulates the interaction with Av NifL in response to the N status, whereas in K. pneumoniae the signal for nitrogen starvation is provided by activation of glnK expression by NtrC-P, and modification of GlnK is not required to transmit the signal (compare Fig. 2 and 3).
REDOX-OXYGEN SENSING
The need to reconcile the oxygen sensitivity of nitrogenase with strictly aerobic metabolism in A. vinelandii has necessitated the evolution of various protection mechanisms to ensure that nitrogenase is not damaged by oxygen (44, 73, 79). However, when respiration is unable to cope with excess oxygen, it is necessary to ensure that synthesis of nitrogenase is prevented. Likewise in K. pneumoniae, which is capable of fixing nitrogen only under anaerobic conditions, it is important to prevent nif transcription in response to aerobiosis. It is well established that NifL inhibits NifA in response to the external oxygen concentration, but the first clue to a potential mechanism for redox sensing was the demonstration that Av NifL is a flavoprotein with FAD as a prosthetic group (45). Spectroscopy of purified Av NifL revealed a characteristic flavin spectrum with absorption maxima at 360 and 445 nm and shoulders at 420 and 470 nm indicative of a protein-bound flavin moiety. While the oxidized form of Av NifL is competent to inhibit transcriptional activation by Av NifA, this inhibition is reversed when the flavin is reduced with sodium dithionite (45). These observations demonstrate that Av NifL is a redox-sensitive regulatory protein. Av NifL binds FAD in the N-terminal PAS1 domain, which has sequence similarity with the corresponding PAS domain in the E. coli flavoprotein Aer, a signal transducer for aerotaxis (11, 83). The N terminus of Kp NifL also contains bound FAD (88), although in this case redox-mediated control of NifA activity has not been demonstrated in vitro, as truncated forms of NifL lacking the N-terminal domain inhibit NifA activity. However, the biochemical experiments to date with the K. pneumoniae system have not included GlnK, which may be required to promote dissociation of Kp NifL and Kp NifA under reducing conditions.
The FAD moiety in Av NifL can be reduced with a variety of enzymes as electron donors in a two-electron reduction with a redox potential of ∼225 mV at pH 8 (64). Oxygen is a potential physiological oxidant as Av NifL is rapidly oxidized upon exposure to air, which yields hydrogen peroxide as a product (62). However, the physiological electron donor to Av NifL in A. vinelandii is unknown. The redox potential of the reduced FAD-oxidized FAD couple in Kp NifL is ∼275 mV at pH 8, and the reduced form of the protein also oxidizes rapidly in the presence of air (55). Although there is no evidence that Kp NifL contains Fe or an iron-sulfur cluster, iron is required in the culture medium to prevent Kp NifL from inhibiting Kp NifA even under anaerobic, nitrogen-limiting conditions (87). This suggests that an iron-containing protein could be required to signal the redox status to NifL by acting as an electron donor. One candidate iron-containing protein that acts as a general oxygen sensor is the global regulator Fnr, which contains an oxygen-labile [4Fe-4S] cluster (52). Analysis of the K. pneumoniae NifL-NifA system in fnr mutants of E. coli and K. pneumoniae indicated that Fnr is required to maintain Kp NifL in a noninhibitory state under anaerobic, nitrogen-limiting conditions (36). Since Fnr is unlikely to be a direct electron donor to Kp NifL, the physiological electron donor is likely to be an electron transport component encoded by a gene(s) that is subject to transcriptional activation by Fnr under anaerobic conditions. K. pneumoniae strains having mutations in either fdnG (encoding formate dehydrogenase N) or nuoCD (encoding NADH:ubiquinone oxidoreductase) show reduced nif gene activation, but the mutations do not influence the expression levels of Kp NifL and Kp NifA (37). This suggests that in the absence of these membrane-bound oxidoreductases, NifL is maintained in an inhibitory form under anaerobic conditions, although it is not obvious why both mutations apparently influence electron donation to Kp NifL. The fdn operon is subject to Fnr control in E. coli (59), whereas the nuo operon is regulated by ArcA (14). It will therefore be of interest to determine whether Fnr is involved in regulation of the corresponding K. pneumoniae operons. It has been proposed that Kp NifL is reduced by electrons from the reduced quinone pool since the quinone derivatives dimethyl naphthoquinol and menadiol are able to reduce the flavin moiety of Kp NifL in the absence of an electron mediator in vitro (37).
DYNAMIC LOCALIZATION
Translational coupling of the nifLA operon in K. pneumoniae ensures that the two encoded proteins are produced in stoichiometric amounts (34). Alterations in the ratio of Kp NifL to Kp NifA disrupt bona fide regulation, demonstrating that the stoichiometry is important for effective signal transduction (35). Under anaerobic nitrogen-limiting conditions, a high proportion of Kp NifL partitions to the membrane, whereas NifL is found primarily in the cytoplasm when cultures are grown aerobically or under conditions of nitrogen sufficiency (54). In contrast, Kp NifA remains primarily in the cytoplasm under all conditions tested. The spatial separation of NifL and NifA under derepressing conditions suggests that membrane association plays a significant role in releasing Kp NifA from inhibition by Kp NifL. It seems likely that reduction of Kp NifL promotes membrane association, since NifL is found mainly in the cytoplasm in fnr, nuoCD, and fdnG mutant strains in which redox sensing by Kp NifL is disabled (37, 54). Likewise, membrane association is not observed in glnK mutants which are unable to signal the nitrogen status (54). Thus, both the presence of GlnK and reduction of NifL appear to be necessary for membrane association. The sequestration of GlnK by AmtB upon ammonium upshift may be a factor in promoting the release of NifL from the membrane (22, 54). The oxidation of the flavin moiety may also release NifL to the cytoplasm. However, the precise factors involved in promoting association and dissociation of NifL with the membrane remain to be determined.
MECHANISM OF NIFL INHIBITION
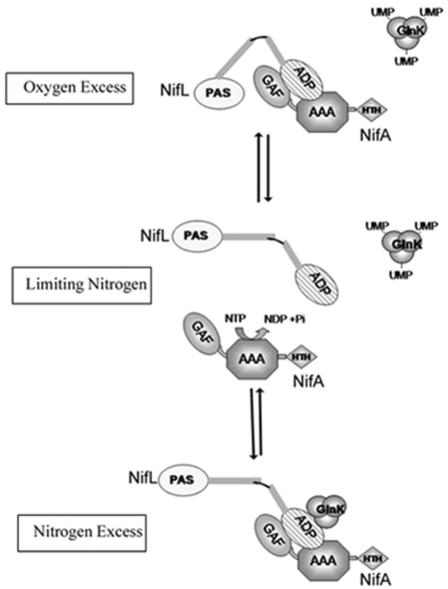
There are several stages at which NifL could inhibit transcriptional activation by NifA through the formation of a protein-protein complex. These stages could potentially involve prevention of binding to enhancers by the C-terminal DNA binding domain, inhibition of the functions of the AAA+ domain, which include oligomerization, nucleotide binding hydrolysis, and interaction with σ54 (103), and remodeling of the GAF domain to induce intramolecular repression of the AAA+ domain (91). Since inhibition by NifL is specific to NifA and NifL apparently does not control transcriptional activation by other EBPs, it is likely that there are surface residues in NifA that are specific to the interaction with NifL. Mutations in Av NifA that confer resistance to inhibition by Av NifL have been isolated from a library of random PCR-generated nifA mutants. Mutations conferring resistance are located in both the GAF and AAA+ domains of Av NifA, implying that both of these domains are involved in the response (84). Some NifA mutants appear to discriminate between the forms of NifL present in response to different environmental conditions. One of the mutations in the AAA+ domain, NifA-Y254N, is sensitive to Av NifL under aerobic growth conditions but is resistant to inhibition under conditions of nitrogen excess. This mutation may disfavor formation of the ternary complex with GlnK but remains sensitive to the binary complex formed with oxidized NifL (Fig. 4).
FIG. 4.
Model showing potential interactions between Av NifL and Av NifA in response to environmental cues. Only the PAS1 and ADP binding domains of NifL are shown (open and cross-hatched ovals, respectively). The three domains of Av NifA are labeled GAF, AAA, and HTH. Under nitrogen-limiting conditions GlnK is uridylylated, and provided that the flavin moiety in Av NifL is reduced, Av NifA is free to activate transcription, catalyzed by ATP hydrolysis (center diagram). However, when Av NifL is oxidized, the NifL-NifA binary complex is formed, perhaps promoted by conformational changes mediated via the PAS domain. Formation of the complex sequesters Av NifA, preventing transcriptional activation. Under nitrogen-excess conditions, when GlnK is in the noncovalently modified form, it interacts with the C-terminal ADP binding domain of Av NifL to promote formation of a ternary complex in which the activity of Av NifA is also inhibited.
As mentioned above, nucleotide hydrolysis catalyzed by the central domain of EBPs is necessary to drive conformational changes that enable σ54-RNA polymerase to form the open promoter complex. The ATPase activity of Av NifA is inhibited by Av NifL (8, 31), suggesting that NifL inhibits steps required either for nucleotide binding (e.g., assembly of AAA+ domain protomers) or for nucleotide hydrolysis. A major function of NifL is therefore to inhibit catalysis by NifA and thus prevent transcriptional activation. However, in a truncated form of Av NifA lacking the GAF domain, nucleotide hydrolysis is not strongly inhibited by Av NifL, even though the ability of NifA to activate open complex formation is inhibited (8). Similarly, transcriptional activation by the isolated central domain of Kp NifA is inhibited by Kp NifL in the absence of inhibition of the ATPase activity of Kp NifA (10). This suggests that NifL may inhibit access of σ54 to interaction surfaces in the AAA+ domain of NifA (e.g., the GAFTGA motif). Thus, NifL may be able to inhibit two discrete functions of the AAA+ domain, nucleotide hydrolysis and interaction with σ54. Since inhibition of nucleotide hydrolysis apparently requires the GAF domain, it appears that this domain may control the ATPase activity of the AAA+ domain in response to NifL by interdomain repression (8). Thus, as suggested by the mutations conferring resistance to NifL, both the GAF and AAA+ domains of Av NifA may contact Av NifL. There is also evidence that NifL may inhibit the enhancer binding function of NifA (8, 71).
NifL may have inhibitory functions analogous to the functions of other adaptors of the EBP family, including PspA, which controls the activity of PspF in E. coli (30), and HrpV, which inactivates HrpR-HrpS in Pseudomonas syringae (80). However, PspF and HrpR-HrpS do not contain an N-terminal regulatory domain, and NifL, PspA, and HrpV do not exhibit detectable sequence homology. Like NifL, however, PspA appears to inhibit two functional states of the PspF AAA+ domain, productive interactions with σ54 and nucleotide interactions (30). However, PspA interacts directly with the AAA+ domain to inhibit the ATPase activity of PspF, whereas in the NifL-NifA interaction this function requires the GAF domain.
CONCLUDING REMARKS
The NifL and NifA proteins constitute a remarkably intricate multidomain regulatory complex in which complementary interactions between the partners are finely tuned to integrate signals of redox oxygen, carbon, and the fixed-nitrogen status. Although the complexities of the interactions are only just beginning to emerge, it is evident that at least some of the mechanisms for signal communication are different in K. pneumoniae and A. vinelandii. The NifL and NifA proteins from these organisms are 30 and 57% identical, respectively, and yet the two systems have evidently adapted to the physiologies of their hosts. Nitrogen regulation provides an obvious example. The two PII-like proteins in K. pneumoniae regulate the nitrogen response at both levels of the cascade. The uridylylation status of GlnB controls transcription of the nifLA operon via the level of NtrC-P. Under conditions of nitrogen deprivation this also allows activation of GlnK expression, which is absolutely required to prevent formation of the inhibitory complex between Kp NifL and Kp NifA. However the precise role of GlnK in the nitrogen response of the K. pneumoniae NifL-NifA system remains unclear since the uridylylation state of GlnK does not apparently influence its function in preventing NifL-mediated inhibition. In contrast, A. vinelandii contains only a single PII-like protein, which regulates the activity of Av NifL in response to uridylylation. The nonmodified form of GlnK activates NifL, thus favoring formation of the NifL-NifA complex under conditions of nitrogen excess. These contrasting mechanisms nevertheless qualitatively provide similar nitrogen regulatory switches. However, the K. pneumoniae system is likely to be more stringent, only permitting nitrogen fixation under conditions of extreme nitrogen deprivation, perhaps reflecting the high energetic penalty for nitrogen fixation under anaerobic growth conditions.
Unlike many other members of the EBP family of transcriptional activators, Kp NifA and Av NifA constitutively activate transcription in the absence of their partner NifL proteins. In contrast, the activities of many transcriptional activators of the EBP family are regulated directly by the amino-terminal regulatory domain, and a partner protein is not required. Indeed, in many diazotrophic representatives of the α and β subgroups of the Proteobacteria, NifA directly integrates signals for fixed nitrogen and oxygen in the absence of a NifL-like protein. What, therefore, are the advantages of regulating nitrogen fixation by this unusual two-component regulatory system? Clearly, a two-component system is likely to be more sophisticated as it provides more opportunities for interdomain communication and signaling interactions and it permits regulation via spatial separation of the partners. The similarity between NifL and the HPKs suggests that the NifL-NifA system may have evolved from a conventional two-component system in which the original equivalent of NifA may have been a response regulator. Loss of the kinase activity of the ancient precursor of NifL and acquisition of a GAF domain at the N terminus of NifA, via a domain swap, may have facilitated evolution of this fascinating regulatory system.
Acknowledgments
Work in our laboratory is supported by the UK Biotechnology and Biological Sciences Research Council.
We thank Gary Sawers for his helpful comments on the manuscript.
REFERENCES
- 1.Aravind, L., and C. P. Ponting. 1997. The GAF domain: an evolutionary link between diverse phototransducing proteins. Trends Biochem. Sci. 22:458-459. [DOI] [PubMed] [Google Scholar]
- 2.Arcondéguy, T., R. Jack, and M. Merrick. 2001. P(II) signal transduction proteins, pivotal players in microbial nitrogen control. Microbiol. Mol. Biol. Rev. 65:80-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arcondéguy, T., D. Lawson, and M. Merrick. 2000. Two residues in the T-loop of Klebsiella pneumoniae GlnK determine NifL-dependent nitrogen control of nif gene expression. J. Biol. Chem. 275:38452-38456. [DOI] [PubMed] [Google Scholar]
- 4.Arcondéguy, T., W. C. van Heeswijk, and M. Merrick. 1999. Studies on the roles of GlnK and GlnB in regulating Klebsiella pneumoniae NifL-dependent nitrogen control. FEMS Microbiol. Lett. 180:263-270. [DOI] [PubMed] [Google Scholar]
- 5.Atkinson, M. R., T. A. Blauwkamp, V. Bondarenko, V. Studitsky, and A. J. Ninfa. 2002. Activation of the glnA, glnK, and nac promoters as Escherichia coli undergoes the transition from nitrogen excess growth to nitrogen starvation. J. Bacteriol. 184:5358-5363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Austin, S., M. Buck, W. Cannon, T. Eydmann, and R. Dixon. 1994. Purification and in vitro activities of the native nitrogen fixation control proteins NIFA and NIFL. J. Bacteriol. 176:3460-3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Austin, S., N. Henderson, and R. Dixon. 1990. Characterisation of the Klebsiella pneumoniae nitrogen-fixation regulatory proteins NIFA and NIFL in vitro. Eur. J. Biochem. 187:353-360. [DOI] [PubMed] [Google Scholar]
- 8.Barrett, J., P. Ray, A. Sobczyk, R. Little, and R. Dixon. 2001. Concerted inhibition of the transcriptional activation functions of the enhancer-binding protein NIFA by the anti-activator NIFL. Mol. Microbiol. 39:480-494. [DOI] [PubMed] [Google Scholar]
- 9.Barrios, H., B. Valderrama, and E. Morett. 1999. Compilation and analysis of sigma(54)-dependent promoter sequences. Nucleic Acids Res. 27:4305-4313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berger, D. K., F. Narberhaus, and S. Kustu. 1994. The isolated catalytic domain of NIFA, a bacterial enhancer-binding protein, activates transcription in vitro: activation is inhibited by NIFL. Proc. Natl. Acad. Sci. 91:103-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bibikov, S. I., R. Biran, K. Rudd, and J. Parkinson. 1997. A signal transducer for aerotaxis in Escherichia coli. J. Bacteriol. 179:4075-4079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blanco, G., M. Drummond, P. Woodley, and C. Kennedy. 1993. Sequence and molecular analysis of the nifL gene of Azotobacter vinelandii. Mol. Microbiol. 9:869-880. [DOI] [PubMed] [Google Scholar]
- 13.Blauwkamp, T. A., and A. J. Ninfa. 2002. Physiological role of the GlnK signal transduction protein of Escherichia coli: survival of nitrogen starvation. Mol. Microbiol. 46:203-214. [DOI] [PubMed] [Google Scholar]
- 14.Bongaerts, J., S. Zoske, U. Weidner, and G. Unden. 1995. Transcriptional regulation of the proton translocating NADH dehydrogenase genes (nuoA-N) of Escherichia coli by electron acceptors, electron donors and gene regulators. Mol. Microbiol. 16:521-534. [DOI] [PubMed] [Google Scholar]
- 15.Bordes, P., S. R. Wigneshweraraj, J. Schumacher, X. Zhang, M. Chaney, and M. Buck. 2003. The ATP hydrolyzing transcription activator phage shock protein F of Escherichia coli: identifying a surface that binds sigma 54. Proc. Natl. Acad. Sci. 100:2278-2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Buck, M., M. T. Gallegos, D. J. Studholme, Y. Guo, and J. D. Gralla. 2000. The bacterial enhancer-dependent σ54 (σN) transcription factor. J. Bacteriol. 182:4129-4136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cannon, W. V., M. T. Gallegos, and M. Buck. 2000. Isomerization of a binary sigma-promoter DNA complex by transcription activators. Nat. Struct. Biol. 7:594-601. [DOI] [PubMed] [Google Scholar]
- 18.Carlton, J. M., S. V. Angiuoli, B. B. Suh, T. W. Kooij, M. Pertea, J. C. Silva, M. D. Ermolaeva, J. E. Allen, J. D. Selengut, H. L. Koo, J. D. Peterson, M. Pop, D. S. Kosack, M. F. Shumway, S. L. Bidwell, S. J. Shallom, S. E. van Aken, S. B. Riedmuller, T. V. Feldblyum, J. K. Cho, J. Quackenbush, M. Sedegah, A. Shoaibi, L. M. Cummings, L. Florens, J. R. Yates, J. D. Raine, R. E. Sinden, M. A. Harris, D. A. Cunningham, P. R. Preiser, L. W. Bergman, A. B. Vaidya, L. H. van Lin, C. J. Janse, A. P. Waters, H. O. Smith, O. R. White, S. L. Salzberg, J. C. Venter, C. M. Fraser, S. L. Hoffman, M. J. Gardner, and D. J. Carucci. 2002. Genome sequence and comparative analysis of the model rodent malaria parasite Plasmodium yoelii yoelii. Nature 419:512-519. [DOI] [PubMed] [Google Scholar]
- 19.Chaney, M., R. Grande, S. R. Wigneshweraraj, W. Cannon, P. Casaz, M. T. Gallegos, J. Schumacher, S. Jones, S. Elderkin, A. E. Dago, E. Morett, and M. Buck. 2001. Binding of transcriptional activators to sigma 54 in the presence of the transition state analog ADP-aluminum fluoride: insights into activator mechanochemical action. Genes Dev. 15:2282-2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Colnaghi, R., P. Rudnick, L. He, A. Green, D. Yan, E. Larson, and C. Kennedy. 2001. Lethality of glnD null mutations in Azotobacter vinelandii is suppressible by prevention of glutamine synthetase adenylylation. Microbiology 147:1267-1276. [DOI] [PubMed] [Google Scholar]
- 21.Contreras, A., M. Drummond, A. Bali, G. Blanco, E. Garcia, G. Bush, C. Kennedy, and M. Merrick. 1991. The product of the nitrogen fixation regulatory gene nfrX of Azotobacter vinelandii is functionally and structurally homologous to the uridylyltransferase encoded by glnD in enteric bacteria. J. Bacteriol. 173:7741-7749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Coutts, G., G. Thomas, D. Blakey, and M. Merrick. 2002. Membrane sequestration of the signal transduction protein GlnK by the ammonium transporter AmtB. EMBO J. 21:536-545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Desnoues, N., M. Lin, X. Guo, L. Ma, R. Carreno-Lopez, and C. Elmerich. 2003. Nitrogen fixation genetics and regulation in a Pseudomonas stutzeri strain associated with rice. Microbiology 149:2251-2262. [DOI] [PubMed] [Google Scholar]
- 24.Dixon, R. 1998. The oxygen-responsive NIFL-NIFA complex: a novel two-component regulatory system controlling nitrogenase synthesis in γ-proteobacteria. Arch. Microbiol. 169:371-380. [DOI] [PubMed] [Google Scholar]
- 25.Drummond, M. H., A. Contreras, and L. A. Mitchenall. 1990. The function of isolated domains and chimaeric proteins constructed from the transcriptional activators NifA and NtrC of Klebsiella pneumoniae. Mol. Microbiol. 4:29-37. [DOI] [PubMed] [Google Scholar]
- 26.Drummond, M. H., and J. C. Wootton. 1987. Sequence of nifL from Klebsiella pneumoniae: mode of action and relationship to two families of regulatory proteins. Mol. Microbiol. 1:37-44. [DOI] [PubMed] [Google Scholar]
- 27.Dutta, R., and M. Inouye. 2000. GHKL, an emergent ATPase/kinase superfamily. Trends Biochem. Sci. 25:24-28. [DOI] [PubMed] [Google Scholar]
- 28.Edwards, R., and M. Merrick. 1995. The role of uridylyltransferase in the control of Klebsiella pneumoniae nif gene regulation. Mol. Gen. Genet. 247:189-198. [DOI] [PubMed] [Google Scholar]
- 29.Egener, T., A. Sarkar, D. E. Martin, and B. Reinhold-Hurek. 2002. Identification of a NifL-like protein in a diazotroph of the beta-subgroup of the Proteobacteria, Azoarcus sp. strain BH72. Microbiology 148:3203-3212. [DOI] [PubMed] [Google Scholar]
- 30.Elderkin, S., S. Jones, J. Schumacher, D. Studholme, and M. Buck. 2002. Mechanism of action of the Escherichia coli phage shock protein PspA in repression of the AAA family transcription factor PspF. J. Mol. Biol. 320:23-37. [DOI] [PubMed] [Google Scholar]
- 31.Eydmann, T., E. Söderbäck, T. Jones, S. Hill, S. Austin, and R. Dixon. 1995. Transcriptional activation of the nitrogenase promoter in vitro: adenosine nucleosides are required for inhibition of NIFA activity by NIFL. J. Bacteriol. 177:1186-1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fischer, H. M. 1994. Genetic regulation of nitrogen fixation in rhizobia. Microbiol. Rev. 58:352-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fischer, H. M., and H. Hennecke. 1987. Direct response of Bradyrhizobium japonicum nifA-mediated nif gene regulation to cellular oxygen status. Mol. Gen. Genet. 209:621-626. [DOI] [PubMed] [Google Scholar]
- 34.Govantes, F., E. Andujar, and E. Santero. 1998. Mechanism of translational coupling in the nifLA operon of Klebsiella pneumoniae. EMBO J. 17:2368-2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Govantes, F., J. A. Molina-Lopez, and E. Santero. 1996. Mechanism of coordinated synthesis of the antogonistic regulatory proteins NifL and NifA of Klebsiella pneumoniae. J. Bacteriol. 178:6817-6823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Grabbe, R., K. Klopprogge, and R. A. Schmitz. 2001. Fnr is required for NifL-dependent oxygen control of nif gene expression in Klebsiella pneumoniae. J. Bacteriol. 183:1385-1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grabbe, R., and R. A. Schmitz. 2003. Oxygen control of nif gene expression in Klebsiella pneumoniae depends on NifL reduction at the cytoplasmic membrane by electrons derived from the reduced quinone pool. Eur. J. Biochem. 270:1555-1566. [DOI] [PubMed] [Google Scholar]
- 38.He, L., E. Soupene, A. Ninfa, and S. Kustu. 1998. Physiological role for the GlnK protein of enteric bacteria: relief of NifL inhibition under nitrogen-limiting conditions. J. Bacteriol. 180:6661-6667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.He, L. H., E. Soupene, and S. Kustu. 1997. NtrC is required for control of Klebsiella pneumoniae NifL activity. J. Bacteriol. 179:7446-7455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hefti, M., J. Hendle, C. Enroth, J. Vervoort, and P. A. Tucker. 2001. Crystallization and preliminary crystallographic data of the PAS domain of the NifL protein from Azotobacter vinelandii. Acta Crystallogr. D Biol. Crystallogr. 57:1895-1896. [DOI] [PubMed] [Google Scholar]
- 41.Hefti, M. H., F. J. Milder, S. Boeren, J. Vervoort, and W. J. van Berkel. 2003. A His-tag based immobilization method for the preparation and reconstitution of apoflavoproteins. Biochim. Biophys. Acta 1619:139-143. [DOI] [PubMed] [Google Scholar]
- 42.Hefti, M. H., C. J. Van Vugt-Van der Toorn, R. Dixon, and J. Vervoort. 2001. A novel purification method for histidine-tagged proteins containing a thrombin cleavage site. Anal. Biochem. 295:180-185. [DOI] [PubMed] [Google Scholar]
- 43.Henderson, N., S. A. Austin, and R. A. Dixon. 1989. Role of metal ions in negative regulation of nitrogen fixation by the nifL gene product from Klebsiella pneumoniae. Mol. Gen. Genet. 216:484-491. [Google Scholar]
- 44.Hill, S. 1992. Physiology of nitrogen fixation in free-living heterotrophs, p. 87-134. In G. Stacey, R. H. Burris, and H. J. Evans (ed.), Biological nitrogen fixation. Chapman and Hall, New York, N.Y.
- 45.Hill, S., S. Austin, T. Eydmann, T. Jones, and R. Dixon. 1996. Azotobacter vinelandii NIFL is a flavoprotein that modulates transcriptional activation of nitrogen-fixation genes via a redox-sensitive switch. Proc. Natl. Acad. Sci. 93:2143-2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hill, S., C. Kennedy, E. Kavanagh, R. Goldberg, and R. Hanau. 1981. Nitrogen fixation gene (nifL) involved in oxygen regulation of nitrogenase synthesis in K. pneumoniae. Nature 290:424-426. [DOI] [PubMed] [Google Scholar]
- 47.Ho, Y. S., L. M. Burden, and J. H. Hurley. 2000. Structure of the GAF domain, a ubiquitous signaling motif and a new class of cyclic GMP receptor. EMBO J. 19:5288-5299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hopper, S., and A. Böck. 1995. Effector-mediated stimulation of ATPase activity by the σ54-dependent transcriptional activator FHLA from Escherichia coli. J. Bacteriol. 177:2798-2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jack, R., M. De Zamaroczy, and M. Merrick. 1999. The signal transduction protein GlnK is required for NifL-dependent nitrogen control of nif gene expression in Klebsiella pneumoniae. J. Bacteriol. 181:1156-1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kanacher, T., A. Schultz, J. U. Linder, and J. E. Schultz. 2002. A GAF-domain-regulated adenylyl cyclase from Anabaena is a self-activating cAMP switch. EMBO J. 21:3672-3680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kennedy, C. 1977. Linkage map of the nitrogen fixation (nif) genes in Klebsiella pneumoniae. Mol. Gen. Genet. 157:199-204. [DOI] [PubMed] [Google Scholar]
- 52.Kiley, P. J., and H. Beinert. 2003. The role of Fe-S proteins in sensing and regulation in bacteria. Curr. Opin. Microbiol. 6:181-185. [DOI] [PubMed] [Google Scholar]
- 53.Kim, Y. M., K. J. Ahn, T. Beppu, and T. Uozomi. 1986. Nucleotide sequence of the nifLA operon of Klebsiella oxytoca NG13 and characterisation of the gene products. Mol. Gen. Genet. 205:253-259. [DOI] [PubMed] [Google Scholar]
- 54.Klopprogge, K., R. Grabbe, M. Hoppert, and R. A. Schmitz. 2002. Membrane association of Klebsiella pneumoniae NifL is affected by molecular oxygen and combined nitrogen. Arch. Microbiol. 177:223-234. [DOI] [PubMed] [Google Scholar]
- 55.Klopprogge, K., and R. A. Schmitz. 1999. NifL of Klebsiella pneumoniae: redox characterization in relation to the nitrogen source. Biochim. Biophys. Acta 1431:462-470. [DOI] [PubMed] [Google Scholar]
- 56.Klopprogge, K., J. Stips, and R. A. Schmitz. 2002. The inhibitory form of NifL from Klebsiella pneumoniae exhibits ATP hydrolyzing activity only when synthesized under nitrogen sufficiency. Biochim. Biophys. Acta 1594:243-254. [DOI] [PubMed] [Google Scholar]
- 57.Lee, H.-S., F. Narberhaus, and S. Kustu. 1993. In vitro activity of NifL,a signal transduction protein for biological nitrogen fixation. J. Bacteriol. 175:7683-7688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lei, S., L. Pulakat, and N. Gavini. 1999. Genetic analysis of nif regulatory genes by utilizing the yeast two-hybrid system detected formation of a NifL-NifA complex that is implicated in regulated expression of nif genes. J. Bacteriol. 181:6535-6539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li, J., and V. Stewart. 1992. Localization of upstream sequence elements required for nitrate and anaerobic induction of fdn (formate dehydrogenase-N) operon expression in Escherichia coli K-12. J. Bacteriol. 174:4935-4942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Little, R., V. Colombo, A. Leech, and R. Dixon. 2002. Direct interaction of the NifL regulatory protein with the GlnK signal transducer enables the Azotobacter vinelandii NifL-NifA regulatory system to respond to conditions replete for nitrogen. J. Biol. Chem. 277:15472-15481. [DOI] [PubMed] [Google Scholar]
- 61.Little, R., and R. Dixon. 2003. The amino-terminal GAF domain of Azotobacter vinelandii NifA binds 2-oxoglutarate to resist inhibition by NifL under nitrogen-limiting conditions. J. Biol. Chem. 278:28711-28718. [DOI] [PubMed] [Google Scholar]
- 62.Little, R., S. Hill, S. Perry, S. Austin, F. Reyes-Ramirez, R. Dixon, and P. Macheroux. 1999. Properties of NifL, a regulatory flavoprotein containing a PAS domain, p. 737-740. In S. Ghisla, P. Kroneck, P. Macheroux, and H. Sund (ed.), Flavins and flavoproteins 1999. Rudolf Weber, Berlin, Germany.
- 63.Little, R., F. Reyes-Ramirez, Y. Zhang, W. C. van Heeswijk, and R. Dixon. 2000. Signal transduction to the Azotobacter vinelandii NIFL-NIFA regulatory system is influenced directly by interaction with 2-oxoglutarate and the PII regulatory protein. EMBO J. 19:6041-6050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Macheroux, P., S. Hill, S. Austin, T. Eydmann, T. Jones, S.-O. Kim, R. Poole, and R. Dixon. 1998. Electron donation to the flavoprotein NifL, a redox-sensing transcriptional regulator. Biochem. J. 332:413-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Meletzus, D., P. Rudnick, N. Doetsch, A. Green, and C. Kennedy. 1998. Characterization of the glnK-amtB operon of Azotobacter vinelandii. J. Bacteriol. 180:3260-3264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Merrick, M., S. Hill, H. Hennecke, M. Hahn, R. Dixon, and C. Kennedy. 1982. Repressor properties of the nifL gene product of Klebsiella pneumoniae. Mol. Gen. Genet. 185:75-81. [Google Scholar]
- 67.Minchin, S. D., S. Austin, and R. A. Dixon. 1988. The role of activator binding sites in transcriptional control of the divergently transcribed nifF and nifLA promoters from Klebsiella pneumoniae. Mol. Microbiol. 2:433-442. [DOI] [PubMed] [Google Scholar]
- 68.Money, T., J. Barrett, R. Dixon, and S. Austin. 2001. Protein-protein interactions in the complex between the enhancer binding protein NIFA and the sensor NIFL from Azotobacter vinelandii. J. Bacteriol. 183:1359-1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Money, T., T. Jones, R. Dixon, and S. Austin. 1999. Isolation and properties of the complex between the enhancer binding protein NIFA and the sensor NIFL. J. Bacteriol. 181:4461-4468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Morett, E., W. Cannon, and M. Buck. 1988. The DNA-binding domain of the transcriptional activator protein NifA resides in its carboxy terminus, recognises the upstream activator sequences of nif promoters and can be separated from the positive control function of NifA. Nucleic Acids Res. 16:11469-11488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Morett, E., R. Kreutzer, W. Cannon, and M. Buck. 1990. The influence of the Klebsiella pneumoniae regulatory gene nifL upon the transcriptional activator protein NifA. Mol. Microbiol. 4:1253-1258. [DOI] [PubMed] [Google Scholar]
- 72.Morett, E., and L. Segovia. 1993. The sigma 54 bacterial enhancer-binding protein family: mechanism of action and phylogenetic relationship of their functional domains. J. Bacteriol. 175:6067-6074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Moshiri, F., J. Kim, C. Fu, and R. Maier. 1994. The FeSII protein of Azotobacter vinelandii is not essential for aerobic nitrogen fixation but confers significant protection to oxygen-mediated inactivation of nitrogenase in vitro and in vivo. Mol. Microbiol. 14:101-114. [DOI] [PubMed] [Google Scholar]
- 74.Narberhaus, F., H.-S. Lee, R. A. Schmitz, L. He, and S. Kustu. 1995. The C-terminal domain of NIFL is sufficient to inhibit NIFA activity. J. Bacteriol. 177:5078-5087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Neuwald, A. F., L. Aravind, J. L. Spouge, and E. V. Koonin. 1999. AAA+: a class of chaperone-like ATPases associated with the assembly, operation, and disassembly of protein complexes. Genome Res. 9:27-43. [PubMed] [Google Scholar]
- 76.Ninfa, A., and M. Atkinson. 2000. PII signal transduction proteins. Trends Microbiol. 8:172-179. [DOI] [PubMed] [Google Scholar]
- 77.Pioszak, A. A., P. Jiang, and A. J. Ninfa. 2000. The Escherichia coli PII signal transduction protein regulates the activities of the two-component system transmitter protein NRII by direct interaction with the kinase domain of the transmitter module. Biochemistry 39:13450-13461. [DOI] [PubMed] [Google Scholar]
- 78.Ponting, C. P., and L. Aravind. 1997. PAS: a multifunctional domain family comes to light. Curr. Biol. 7:R674-R677. [DOI] [PubMed] [Google Scholar]
- 79.Poole, R. K., and S. Hill. 1997. Respiratory protection of nitrogenase activity in Azotobacter vinelandii—roles of the terminal oxidases. Biosci. Rep. 17:303-317. [DOI] [PubMed] [Google Scholar]
- 80.Preston, G., W. L. Deng, H. C. Huang, and A. Collmer. 1998. Negative regulation of hrp genes in Pseudomonas syringae by HrpV. J. Bacteriol. 180:4532-4537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Raina, R., U. K. Bageshwar, and H. K. Das. 1993. The Azotobacter vinelandii nifL-like gene: nucleotide sequence analysis and regulation of expression. Mol. Gen. Genet. 237:400-406. [DOI] [PubMed] [Google Scholar]
- 82.Ray, P., K. J. Smith, R. A. Parslow, R. Dixon, and E. I. Hyde. 2002. Secondary structure and DNA binding by the C-terminal domain of the transcriptional activator NifA from Klebsiella pneumoniae. Nucleic Acids Res. 30:3972-3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Rebbapragada, A., M. S. Johnson, G. P. Harding, A. J. Zuccarelli, H. M. Fletcher, I. B. Zhulin, and B. L. Taylor. 1997. The Aer protein and the serine chemoreceptor sense intracellular energy levels and transduce oxygen, redox, and energy signals for Escherichia coli behaviour. Proc. Natl. Acad. Sci. 94:10541-10546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Reyes-Ramirez, F., R. Little, and R. Dixon. 2002. Mutant forms of the Azotobacter vinelandii transcriptional activator NifA resistant to inhibition by the NifL regulatory protein. J. Bacteriol. 184:6777-6785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Reyes-Ramirez, F., R. Little, and R. Dixon. 2001. Role of Escherichia coli nitrogen regulatory genes in the nitrogen response of the Azotobacter vinelandii NifL-NifA complex. J. Bacteriol. 183:3076-3082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Rudnick, P., C. Kunz, M. K. Gunatilaka, E. R. Hines, and C. Kennedy. 2002. Role of GlnK in NifL-mediated regulation of NifA activity in Azotobacter vinelandii. J. Bacteriol. 184:812-820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Schmitz, R., L. He, and S. Kustu. 1996. Iron is required to relieve inhibitory effects of NifL on transcriptional activation by NifA in Klebsiella pneumoniae. J. Bacteriol. 178:4679-4687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Schmitz, R. A. 1997. NifL of Klebsiella pneumoniae carries an N-terminally bound FAD cofactor, which is not directly required for the inhibitory function of NifL. FEMS Microbiol. Lett. 157:313-318. [DOI] [PubMed] [Google Scholar]
- 89.Schmitz, R. A., K. Klopprogge, and R. Grabbe. 2002. Regulation of nitrogen fixation in Klebsiella pneumoniae and Azotobacter vinelandii: NifL, transducing two environmental signals to the nif transcriptional activator NifA. J. Mol. Microbiol. Biotechnol. 4:235-242. [PubMed] [Google Scholar]
- 90.Senior, P. J. 1975. Regulation of nitrogen metabolism in Escherichia coli and Klebsiella aerogenes: studies with the continuous culture technique. J. Bacteriol. 123:407-418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Shingler, V. 1996. Signal sensing by sigma 54-dependent regulators: derepression as a control mechanism. Mol. Microbiol. 19:409-416. [DOI] [PubMed] [Google Scholar]
- 92.Siddavattam, D., H. D. Steibl, R. Kreutzer, and W. Klingmuller. 1995. Regulation of nif gene expression in Enterobacter agglomerans: nucleotide sequence of the nifLA operon and influence of temperature and ammonium on its transcription. Mol. Gen. Genet. 249:629-636. [DOI] [PubMed] [Google Scholar]
- 93.Söderbäck, E., F. Reyes-Ramirez, T. Eydmann, S. Austin, S. Hill, and R. Dixon. 1998. The redox-and fixed nitrogen-responsive regulatory protein NIFL from Azotobacter vinelandii comprises discrete flavin and nucleotide-binding domains. Mol. Microbiol. 28:179-192. [DOI] [PubMed] [Google Scholar]
- 94.Stock, A. M., V. L. Robinson, and P. N. Goudreau. 2000. Two-component signal transduction. Annu. Rev. Biochem. 69:183-215. [DOI] [PubMed] [Google Scholar]
- 95.Studholme, D. J., and R. Dixon. 2003. Domain architectures of σ54-dependent transcriptional activators. J. Bacteriol. 185:1757-1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Taylor, B. L., and I. B. Zhulin. 1999. PAS domains: internal sensors of oxygen, redox potential, and light. Microbiol. Mol. Biol. Rev. 63:479-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Thorneley, R. N., and D. J. Lowe. 1983. Nitrogenase of Klebsiella pneumoniae. Kinetics of the dissociation of oxidized iron protein from molybdenum-iron protein: identification of the rate-limiting step for substrate reduction. Biochem. J. 215:393-403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Toukdarian, A., and C. Kennedy. 1986. Regulation of nitrogen metabolism in Azotobacter vinelandii: isolation of ntr and glnA genes and construction of ntr mutants. EMBO J. 5:399-407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Vale, R. D. 2000. AAA proteins. Lords of the ring. J. Cell Biol. 150:F13-F19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wang, Y.-K., J. Lee, J. Brewer, and T. Hoover. 1997. A conserved region in the σ54-dependent activator DctD is involved in both binding to RNA polymerase and coupling ATP hydrolysis to activation. Mol. Microbiol. 26:373-386. [DOI] [PubMed] [Google Scholar]
- 101.Woodley, P., and M. Drummond. 1994. Redundancy of the conserved His residue in Azotobacter vinelandii NifL, a histidine protein kinase homologue which regulates transcription of nitrogen fixation genes. Mol. Microbiol. 13:619-626. [DOI] [PubMed] [Google Scholar]
- 102.Wootton, J. C., and M. Drummond. 1989. The Q-linker: a class of interdomain sequences found in bacterial multidomain regulatory proteins. Protein Eng. 2:535-543. [DOI] [PubMed] [Google Scholar]
- 103.Zhang, X., M. Chaney, S. R. Wigneshweraraj, J. Schumacher, P. Bordes, W. Cannon, and M. Buck. 2002. Mechanochemical ATPases and transcriptional activation. Mol. Microbiol. 45:895-903. [DOI] [PubMed] [Google Scholar]
- 104.Zhulin, I. B., B. L. Taylor, and R. Dixon. 1997. PAS domain S-boxes in Archea, Bacteria and sensors for oxygen and redox. Trends Biochem. Sci. 22:331-333. [DOI] [PubMed] [Google Scholar]