Abstract
Auxin is an essential hormone, but its biosynthetic routes in plants have not been fully defined. In this paper, we show that the TRYPTOPHAN AMINOTRANSFERASE OF ARABIDOPSIS (TAA) family of amino transferases converts tryptophan to indole-3-pyruvate (IPA) and that the YUCCA (YUC) family of flavin monooxygenases participates in converting IPA to indole-3-acetic acid, the main auxin in plants. Both the YUCs and the TAAs have been shown to play essential roles in auxin biosynthesis, but it has been suggested that they participate in two independent pathways. Here, we show that all of the taa mutant phenotypes, including defects in shade avoidance, root resistance to ethylene and N-1-naphthylphthalamic acid (NPA), are phenocopied by inactivating YUC genes. On the other hand, we show that the taa mutants in several known auxin mutant backgrounds, including pid and npy1, mimic all of the well-characterized developmental defects caused by combining yuc mutants with the auxin mutants. Furthermore, we show that overexpression of YUC1 partially suppresses the shade avoidance defects of taa1 and the sterile phenotypes of the weak but not the strong taa mutants. In addition, we discovered that the auxin overproduction phenotypes of YUC overexpression lines are dependent on active TAA genes. Our genetic data show that YUC and TAA work in the same pathway and that YUC is downstream of TAA. The yuc mutants accumulate IPA, and the taa mutants are partially IPA-deficient, indicating that TAAs are responsible for converting tryptophan to IPA, whereas YUCs play an important role in converting IPA to indole-3-acetic acid.
Auxin is an essential regulator for various plant developmental processes. Indole-3-acetic acid (IAA), the main auxin in plants, can be synthesized from tryptophan (Trp) -dependent and -independent pathways (1). Several auxin biosynthesis routes have been proposed (Fig. 1A), but none of the proposed pathways in plants have been fully determined (1). In some plant pathogenic bacteria, IAA is synthesized from Trp by the Trp monooxygenase iaaM and the hydrolase iaaH (Fig. 1A). The iaaM converts Trp to indole-3-acetamide (IAM) that is subsequently hydrolyzed to IAA by iaaH (2). Plants also make IAM, but the biosynthesis routes of IAM in plants are not defined (3). IAM can be converted to IAA by Arabidopsis AMIDASE1 (4). Trp can also be converted into indole-3-acetaldoxime (IAOx) by the P450 enzymes CYP79B2 and -B3 (Fig. 1A) (5). The routes from IAOx to IAA are not understood, although both IAM and indole-3-acetonitrile have been suggested as intermediates (Fig. 1A) (3). IAOx is probably not a main auxin biosynthesis intermediate, because (i) a complete elimination of IAOx production in Arabidopsis only leads to subtle growth defects (5), (ii) the enzymes CYP79B2 and -B3 seemed only to exist in a small group of plants, and (iii) there is no detectable IAOx in rice and maize (3).
Fig. 1.
Trp-dependent auxin biosynthesis routes. (A) The proposed pathways for converting Trp to IAA. (B) A complete two-step auxin biosynthesis pathway. IPA, indole-3-pyruvate; TAM, tryptamine; IAOx, indole-3-acetaldoxime; IAM, indole-3-acetamide; IAN, indole-3-acetonitrile; IAAld, indole-3-acetaldehyde; IAA, indole-3-acteic acid. A solid line indicates that a gene has been suggested for the step, whereas no genes have been suggested for the step indicated by a dotted line.
The most important plant auxin biosynthetic enzymes are the YUCCA (YUC) family of flavin-containing monooxygenases (6, 7) and the TRYPTOPHAN AMINOTRANSFERASE OF ARABIDOPSIS (TAA) (8, 9) family of aminotransferases, because inactivating members of either family causes dramatic developmental defects (Fig. 1A). Both families are widely distributed among all sequenced plant genomes. The YUC genes were first identified as auxin biosynthesis enzymes, because overexpression of YUCs leads to auxin overproduction in Arabidopsis (7). YUC genes are essential for embryogenesis, seedling development, vascular patterning, and flower development (6, 10). Furthermore, the yuc phenotypes can be rescued by expressing the iaaM gene under the control of a YUC promoter (6). The TAA genes were independently isolated from three genetic screens. Inactivation of TAA1 leads to altered responses to shade (9), ethylene (8), and the auxin transport inhibitor NPA (11). Furthermore, simultaneously inactivating TAA1 and its close homologs (TAR genes) lead to defects in embryogenesis, vascular patterning, and flower development (8). YUCs were proposed to catalyze the conversion of tryptamine to N-hydroxyl tryptamine, which may be converted to IAOx (7, 12). However, our recent biochemical analysis indicates that YUCs do not play a major role in IAOx production (3). Recent work has also questioned whether YUCs are involved in the tryptamine pathway (13). Flavin monooxygeanses are known to use a broad range of substrates in vitro, because they use the stable C4a-hydroperoxyl flavin, an activated intermediate, for catalysis (14). Therefore, additional genetic and biochemical analyses are needed to define the in vivo roles of YUCs in auxin biosynthesis. The TAA family proteins catalyze the conversion of Trp to indole-3-pyruvate (IPA), but the in vitro data suggest that the reaction from IPA to Trp is actually more favorable (9). Furthermore, the mechanism by which IPA is converted to IAA is still not understood. It has been widely speculated that IPA is converted to indole-3-acetaldehyde (IAAld) by IPA decarboxylase. IAAld then is believed to be converted to IAA by aldehyde oxidases or dehydrogenases (Fig. 1A) (1).
Interestingly, some of the yuc mutant phenotypes are very similar to those phenotypes of taa mutants. For example, yuc1 yuc4 yuc10 yuc11 quadruple mutants fail to make the basal parts of Arabidopsis embryos (10), a phenotype that is also observed in wei8 tar1 tar2-1 (8). Mutant alleles of taa1 are also called wei8 (8), sav3 (9), and tir2 (11), which reflect the three genetic screens that identified the mutant alleles. We keep using the wei8, sav3, and tir2 allele names in this paper so that readers can easily track the alleles of taa1 that are used. A list of mutants used in this work is shown in Table S1. Vascular defects in yuc1 yuc2 yuc4 yuc6 are also very similar to those defects in wei8 tar2-1 (6, 8). Both yuc and wei8 tar2 mutants also have defects in flower development (6, 8). The observed phenotypic similarities between yuc and taa mutants have prompted speculations that YUC and TAA may participate in the same pathway to produce auxin (1, 15). The observation that inactivation of the SPI1/YUC gene in the vt2/taa1 mutant background in maize did not further enhance the phenotypes of vt2 also suggests that the YUC and TAA genes might belong to the same pathway (16).
Although there are many similarities between yuc and taa mutants, there are several key differences. First, the yuc mutants analyzed so far have not been shown to affect shade avoidance responses, whereas taa1/sav3 alone showed dramatic defects in shade avoidance (9). Second, the yuc mutants have not been reported to affect ethylene responses in roots, but taa mutants are insensitive to the inhibitory effects of ethylene in roots (8). Third, the yuc mutants have not been shown to alter responses to NPA in roots, whereas taa is resistant to NPA (11). Fourth, the wei8/sav3 alone dramatically reduced IAA levels in seedlings, whereas quadruple yuc mutants did not show a significant decrease in auxin levels (9), although the yuc mutants have phenotypes much more severe than those phenotypes of wei8 or sav3.
Some of the phenotypic defects observed in yuc mutants have not been observed in taa mutants or have not been analyzed. For example, the floral defects of yuc1 yuc4 seem to be different from those defects in wei8 tar2 mutants (6). The yuc1 yuc4 double mutants have fewer floral organs, whereas wei8 tar2 flowers are defective; however, no floral organs are missing. We have shown that yuc mutants have synergistic interactions with known auxin mutants, including pid, pin1, and npy1 (10, 17, 18). It will be informative to analyze the genetic interactions between the known auxin and taa mutants.
In this paper, we show that all of the phenotypes observed in taa mutants, including root resistance to ethylene treatment, altered shade avoidance responses, and root resistance to NPA treatments, are phenocopied by inactivating certain combinations of YUC genes. We also show that all of the synergistic effects between yuc and known auxin mutants are observed when TAA genes are inactivated in the known auxin mutant backgrounds. Furthermore, we show that overexpression of YUC1 in taa1/sav3 can partially rescue the shade avoidance defects. Overexpression of YUC1 can also rescue the sterile phenotypes of wei8 tar2-2, a weak taa double mutant. However, the characteristic long hypocotyl phenotypes associated with YUC overexpression are not observed in the strong wei8 tar2-1 mutant background. Our genetic analysis put the YUC and TAA genes in the same auxin biosynthesis pathway, with YUCs downstream of TAAs. We discovered that IPA levels in wei tar2 are decreased, whereas in yuc1 yuc2 yuc6, they are increased, further supporting our hypothesis that YUCs function downstream of TAAs. We propose that auxin is synthesized by a two-step pathway in which TAAs convert Trp into IPA and YUCs are responsible for converting IPA into IAA (Fig. 1B).
Results
yuc Mutants Phenocopied the taa Mutants.
Alleles of taa1 were isolated from genetic screens for mutants with altered responses to ethylene (wei8 mutants) (8), shade (sav3 mutants) (9), and NPA (tir2 mutants) (11). Interestingly, no yuc mutants were identified from the three genetic screens. It is likely that genetic redundancy among the YUC genes might have prevented the identification of yuc mutants in the screens.
Our previous work on yuc mutants mainly focused on the development of aerial parts of Arabidopsis (6, 10). We hypothesized that a different set of YUC genes might be responsible for root development. We investigated whether the yuc3, yuc5, yuc7, yuc8, and yuc9 quintuple mutants (yuc Q) displayed resistance to ethylene in roots. The five YUC genes form two distinct clades in the YUC phylogenetic tree (6). As shown in Fig. 2 A and B, the yuc Q mutants were strongly resistant to the treatment of 1-aminocyclopropane-1-carboxylic-acid (ACC), an ethylene biosynthesis precursor. In the presence of 10 μM ACC, root elongation in WT Arabidopsis seedlings was almost completely inhibited (Fig. 2A). In contrast, the yuc Q mutants displayed healthy and elongated roots (Fig. 2A). The ethylene resistance of the yuc Q roots was much stronger than the resistance of wei8 tar2-1 double mutants, which are known to confer ethylene resistance in roots (Fig. 2 A and B). Our data showed that YUC genes also play an important role in root responses to ethylene.
Fig. 2.
Inactivation of YUC genes phenocopied the taa mutants. (A) The yuc3 yuc5 yuc7 yuc8 yuc9 quintuple mutants (yuc Q) displayed strong resistance to ethylene in roots. (B) Measurements of root length. Both wei8 tar2-1 and yuc Q had longer roots in the presence of 10 μM ACC. (C) The yuc1-163 yuc4 double mutants displayed defects in shade avoidance similar to the defects observed in sav3-1/taa1. (D) The yuc Q displayed resistance to the auxin transport inhibitor NPA. Roots of WT plants became swollen, but both wei8 tar2-1 and yuc quintuple mutants had normal root tips. Error bars refer to SD. (Scale bar: 1 mm.)
We previously analyzed shade avoidance responses of the yuc Q mutants, because YUC8 seemed to be induced by shade conditions (9). However, the yuc Q mutants did not show obvious defects in shade avoidance responses under our conditions (9). Our previous experiments had clearly shown that YUC1, YUC2, YUC4, and YUC6 are the predominant YUCs functioning in aerial parts of Arabidopsis (6). Unfortunately, the yuc1 yuc4 double null mutants are completely sterile, making analysis of yuc1 yuc4 in shade avoidance responses unfeasible. We used a weak allele of yuc1 (yuc1-163), which showed vascular and floral defects when combined with yuc4 mutant. The yuc1-163 yuc4 could generate a small amount of seeds that allowed us to analyze the shade avoidance response. As shown in Fig. 2C, under shade conditions, hypocotyls of WT plants were much elongated. In contrast, hypocotyls of sav3-1 were only one-half as long as those hypocotyls of WT plants. The yuc1-163 yuc4 was clearly shade-resistant (Fig. 2C), indicating that YUC genes also participate in shade avoidance responses.
Inactivation of TAA1/TIR2 caused root resistance to the auxin transport inhibitor NPA (11) (Fig. 2D). NPA leads to auxin accumulation in root tips and causes root tip swelling in WT plants (Fig. 2D). We also investigated whether YUC genes play a role in mediating root responses to NPA treatments. Like the taa mutants wei8 tar2-1, the yuc Q mutants did not have a swollen root tip (Fig. 2D), indicating that yuc Q mutants were also resistant to NPA treatments.
Our data showed that the three main characteristic phenotypes associated with taa mutants were also observed in yuc mutants. Interestingly, Arabidopsis plants use the same TAA genes in both the shade avoidance and root responses to ethylene and NPA. By contrast, Arabidopsis uses two different sets of YUC genes for shade avoidance and root responses to ethylene and NPA. YUC1 and YUC4 were the main YUCs used in shade avoidance responses, and Arabidopsis relied primarily on YUC3, YUC5, YUC7, YUC8, and YUC9 for responses to ethylene and NPA in roots.
Characteristic yuc Phenotypes Were Mimicked by Inactivating TAA Genes.
We previously showed that yuc mutants displayed dramatic genetic interactions with known auxin mutants pin1, pid, and npy1 (10, 17, 18). We investigated whether taa mutants could mimic the synergistic genetic interactions observed between yuc and known auxin mutants.
When yuc1 yuc4 was combined with pin1, the development of true leaves in the triple mutants was completely abolished (10). The no-leaf phenotypes could also be observed when yuc1 yuc4 were grown on media containing NPA (10) (Fig. 3A). Clearly, the development of true leaves in both wei8 tar2-1 and wei8 tar2-2 was completely abolished by NPA, whereas under the same conditions, WT seedlings developed true leaves (Fig. 3A). Interestingly, roots of both yuc1 yuc4 and WT plants were swollen (Fig. 3A), whereas wei8 tar2 mutants were resistant to NPA in roots (Figs. 2D and 3A). Our data showed that taa mutants could phenocopy yuc1 yuc4 in terms of the development of true leaves in the presence of NPA (Fig. 3A). The NPA experiments also showed that YUC1 and YUC4 were the main YUCs in regulating the development of aerial parts, but YUC1 and YUC4 were not the main YUCs in root responses to NPA.
Fig. 3.
Mutations in TAA genes could mimic yuc mutants. (A) Inactivation of WEI8 and TAR2 caused the same phenotypes as those of yuc1 yuc4 in aerial parts in the presence of NPA. NPA completely abolished the initiation of true leaves in both taa and yuc mutants (white arrows). Note that yuc1 yuc4 was not resistant to NPA in the root, whereas wei8 tar2 was resistant to NPA in roots (red arrows). (B) The triple mutants of wei8 tar2-1 pid completely deleted cotyledons. (C) Mutations in npy1 enhanced wei8 tar2 mutants. The wei8 tar2 npy1 developed pin-like inflorescences and failed to make flowers. (Scale bar: A and B, 1 mm; C, 1 cm.)
We previously showed that both YUC1 and YUC4 were important for cotyledon development (17, 18). Inactivation of both YUC1 and YUC4 in the pid mutant background leads to a complete deletion of cotyledons (17, 18). When we combined wei8 tar2-1 with pid, the resulting triple mutants also failed to develop any cotyledons (Fig. 3B), showing that both the YUCs and TAAs play similar roles in cotyledon development.
Another well-characterized genetic interaction between yuc and known auxin mutants is the enhancement of yuc1 yuc4 double mutants by npy1 (17). The yuc1 yuc4 npy1 triple mutants completely eliminated the formation of flowers but still developed pin-like inflorescences (17). Simultaneous inactivation of TAA1/WEI8, TAR2, and NPY1 also abolished the formation of flowers (Fig. 3C). The wei8 tar2-2 npy1 developed many pin-like inflorescences (Fig. 3C). Combining the strong mutants wei8 tar2-1 with npy1 led to even stronger phenotypes (Fig. 3C). Both the yuc1 yuc4 npy1 and wei8 tar2 npy1 mutants developed similar pin-like inflorescences, but the wei8 tar2 npy1 had more severe phenotypes. The observed differences could be caused by the fact that additional YUC genes are involved in inflorescences development (6). In fact, the juvenile plants of wei8 tar2-1 were similar to those plants of yuc1 yuc2 yuc4 yuc6 quadruple mutants (6, 8). Our data showed that the taa mutants could mimic all of the yuc phenotypes.
Genetic Interactions Between yuc and taa Mutants.
We directly tested whether there were genetic interactions between yuc and wei8 tar2 mutants. We chose yuc1 yuc4 and wei8 tar2-1, because both double mutants displayed strong developmental phenotypes (6, 8). The yuc1 yuc4 wei8 tar2 quadruple mutants did not make any hypocotyls and roots, a phenotype that was not observed in either yuc1 yuc4 or wei8 tar2-1 (Fig. 4A). Interestingly, the yuc1 yuc4 wei8 tar2 quadruple phenotypes were very similar to those phenotypes of yuc1 yuc4 yuc10 yuc11 and wei8 tar1 tar2-1 (8, 10).
Fig. 4.
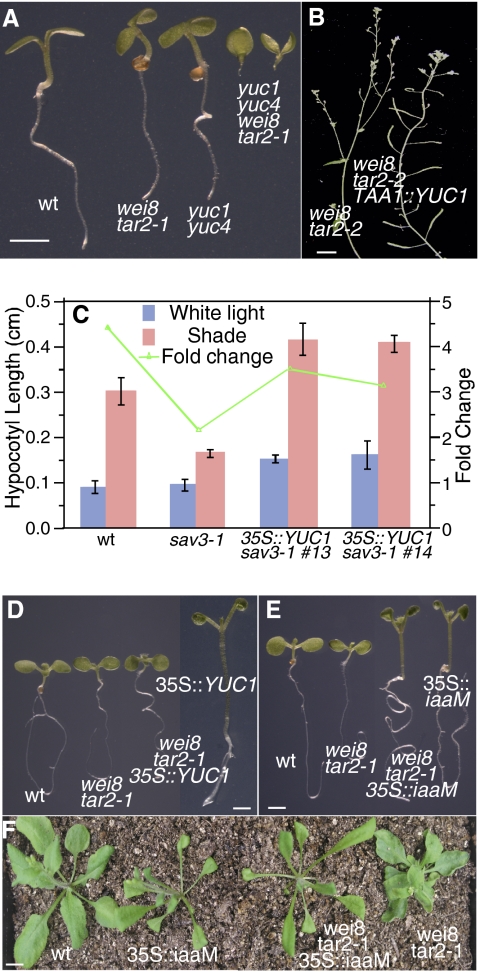
Genetic interactions between YUCs and TAAs in Arabidopsis. (A) Synergistic interactions between wei8 tar2-1 and yuc1 yuc4. The yuc1 yuc4 wei8 tar2-1 failed to make hypocotyls and roots. (Scale bar: 2 mm.) (B) Expression of YUC1 cDNA under the control of TAA1 promoter partially rescued the sterile phenotypes of wei8 tar2-2. (Scale bar: 1 cm.) (C) Overexpression of YUC1 cDNA using the 35S promoter partially rescued the shade avoidance defects in sav3-1. Error bars refer to SD. (D) Auxin overproduction phenotypes of 35S::YUC1 are suppressed in wei8 tar2-1 background. (Scale bar: 1 mm.) (E) Overexpression of iaaM in wei8 tar2-1 caused auxin overproduction in both WT and wei8 tar2-1 backgrounds. (Scale bar: 1 mm.) (F) The adult phenotypes of wei8 tar2-1 were also partially suppressed by 35S::iaaM. (Scale bar: 1 cm.)
We expressed YUC1 cDNA under the control of the TAA1 promoter (9) in wei8 tar2-2 background. The wei8 tar2-2 double mutants were weaker than wei8 tar2-1 but still sterile (Fig. 4B). Expression of YUC1 partially rescued the sterile phenotype of wei8 tar2-2 (Fig. 4B).
We also overexpressed YUC1 cDNA using the Cauliflower mosaic virus (CaMV) 35S promoter in the taa1/sav3-1 background. Overexpression of YUC1 in sav3-1 leads to longer hypocotyl, a characteristic phenotype associated with auxin overproduction (6, 7) (Fig. 4C). Overexpression of YUC1 also partially suppressed the shade avoidance phenotypes of sav3-1 (Fig. 4C).
When we introduced 35S::YUC1 construct into wei8 tar2-1 by transforming wei8 tar2-1+/− plants, we did not observe the typical auxin overproduction phenotypes in wei8 tar2-1 plants (Fig. 4D). Because the tar2-1 is a T-DNA line, we investigated whether 35S::YUC1 was silenced in the wei8 tar2-1 background. We found that the expression levels of YUC1 in 35S::YUC1 wei8 tar2-1 were higher than in WT (Fig. S1), suggesting that overproduction of auxin by 35S::YUC1 is dependent on active TAA genes.
YUCs and the iaaM Gene Behaved Differently in wei8 tar2 Mutants.
We previously showed that expression of the bacterial auxin biosynthesis gene iaaM rescued yuc mutant phenotypes (6, 7). Expression of iaaM also rescued the shade avoidance phenotypes of sav3-1 (9). We investigated whether iaaM could rescue the developmental defects of wei8 tar2 mutants. As shown in Fig. 4E, overexpression of iaaM led to the typical auxin overproduction phenotypes in both WT and wei8 tar2 mutants. The iaaM gene also partially rescued the defects of wei8 tar2 phenotypes at juvenile and adult stages (Fig. 4F). Our work shows that YUCs and iaaM genes probably use different mechanisms for auxin biosynthesis.
Both YUCs and TAA Genes Affected the Homeostasis of IPA.
We directly measured the IPA contents in WT plants, yuc mutants, and taa mutants. Because the strong yuc and taa mutants have dramatic developmental defects, we chose yuc1 yuc2 yuc6 triple mutants and wei8 tar2-2 double mutants to analyze the IPA contents. The yuc1 yuc2 yuc6 mutants are similar to wei8 tar2-2. Both the yuc1 yuc2 yuc6 and wei8 tar2-2 are sterile and have similar sizes. As shown in Fig. 5, the IPA levels in wei8 tar2-2 were reduced dramatically. In contrast, the yuc1 yuc2 yuc6 had elevated levels of IPA (Fig. 5). Our data indicate that TAA genes are involved in the production of IPA, whereas YUC genes are involved in metabolism of IPA (Fig. 1B).
Fig. 5.
IPA levels in auxin biosynthesis mutants. Inactivation of yuc1 yuc2 yuc6 caused IPA accumulation, whereas the wei8 tar2-2 mutants had less IPA than WT plants. Error bar refers to SD.
Discussion
Both YUCs and TAAs were reported to participate in Trp-dependent auxin biosynthesis. Here, we have shown that the YUCs and TAAs participate in the same auxin biosynthesis pathway and that YUCs work downstream of TAAs. Our data indicate that TAAs are responsible for making IPA from Trp and that YUCs are required for converting IPA to IAA.
We conclude that YUCs and TAAs participate in the same pathway to convert Trp into IAA. First, inactivation of YUC genes caused the same phenotypes as those phenotypes observed in taa mutants (Fig. 2). Second, all of the yuc phenotypes were mimicked by taa mutants or taa mutant combinations (Fig. 3). The yuc1 yuc4 pid and wei8 tar2-1 pid displayed the exact same no cotyledon phenotypes (Fig. 3). The fact that the yuc and taa mutants had similar phenotypes in every aspect of growth and developmental processes that we have analyzed is indicative that both gene families participate in the same pathway. Alternatively, YUCs and TAAs may participate in parallel auxin biosynthesis pathways. The similar phenotypes observed in yuc and taa mutants could simply be a reflection of decreased auxin levels in the mutants. The latter interpretation is consistent with the observation that yuc1 yuc4 and wei8 tar2 enhanced each other (Fig. 4). However, yuc1 yuc4 mutants are not null for YUC functions because of the existence of other YUCs. The wei8 tar2 mutants are not null for TAA activity either. Therefore, the synergistic genetic interactions between yuc1 yuc4 and wei8 tar2 are also compatible with the interpretation that YUCs and TAAs participate in the same pathway. The hypothesis that YUCs and TAAs are in the same pathway is also supported by the studies on YUC overexpression lines. Overexpression of YUC1 in WT background leads to dramatic auxin overproduction phenotypes (Fig. 4). However, the YUC1 overexpression phenotypes were weakened in the taa1/sav3 background (Fig. 4). We did not observe the long hypocotyl phenotypes associated with YUC1 overexpression in wei8 tar2-1, suggesting that YUC1 overexpression-mediated auxin overproduction is dependent on TAA functions. We were concerned that perhaps hypocotyls of wei8 tar2-1 simply could not elongate. However, when we overexpressed iaaM in wei8 tar2-1, the auxin overproduction phenotypes were clearly observed (Fig. 4).
We place YUCs downstream of TAAs in auxin biosynthesis. We discovered that the taa mutants made less IPA than WT, but yuc mutants accumulated much more IPA (Fig. 5). A logic interpretation is that TAAs make IPA from Trp and that YUCs participate in the conversion of IPA to IAA. This interpretation is also consistent with our findings that overexpression of YUC1 partially rescued the shade avoidance phenotypes of sav3-1 and that expression of YUC1 partially rescued wei8 tar2-2 (Fig. 4). The conversion of the residual IPA in the weak taa mutants was accelerated in YUC overexpression lines, thus making more IAA to partially rescue the weak taa mutant phenotypes.
Although all of the genetic data indicate that YUCs and TAAs participate in the same auxin biosynthetic pathway, we have been puzzled by the fact that yuc mutants did not have lower levels of IAA than WT plants, whereas taa1 alone had a dramatic reduction in IAA levels (8, 9). It is puzzling, because yuc mutants such as yuc1 yuc2 yuc4 yuc6 quadruple mutants had much more severe phenotypes than the phenotypes of taa1/sav3 (6, 8, 10). We hypothesize that the paradoxical results may be partially caused by the nonenzymatic conversion of IPA to IAA during the process of IAA measurement given that yuc mutants accumulate much more IPA (Fig. 5). It is well-known that IPA is very labile in aqueous solutions and that IPA is easily converted to IAA in vitro nonenzymatically (19). It will be necessary to redo the IAA analysis in the yuc mutants by removing IPA first.
It is evident that TAAs convert Trp to IPA by removing the amino group from Trp. However, it is not obvious how YUCs participate in IPA metabolism. We propose that YUCs convert IPA to IAA using a mechanism analogous to the mechanism of lactate monooxygenases, which convert lactate to acetic acid and CO2 (20, 21). Lactate is first converted to pyruvate by transferring the two electrons to the flavin cofactor in lactate monooxygenase. The reduced flavin binds oxygen and subsequently reacts with pyruvate to release CO2 and acetic acid. YUCs probably use NADPH to reduce the flavin cofactor. After the flavin is reduced, the resulting FADH2 can bind oxygen and convert IPA to IAA.
Materials and Methods
The wei8, tar2-1, and tar2-2 mutants were described in ref. 8, and sav3-1 was reported in ref. 9. The npy1 mutant used in this study was the npy1-2 T-DNA allele (SALK-108406) (17). Genotyping of the npy1 mutant has been described (17). The yuc1 and yuc4 mutants were described in ref. 6. The yuc3, yuc5, yuc7, yuc8, and yuc9 quadruple mutants were described in ref. 9. The pid allele was the SALK-049736 line as previously reported (17). Methods for genotyping the various mutants used in this work were described previously (6, 8, 9, 17).
Shade avoidance assay was conducted according to the procedures described previously (9). Ethylene responses were measured using 4-d-old dark-grown seedlings. Root length was measured using the National Institutes of Health image software.
Methods for IPA analysis are shown in SI Methods.
Supplementary Material
Acknowledgments
We thank members of the Y.Z. laboratory for comments. This work is supported by the National Institutes of Health Grant R01GM68631 (to Y.Z.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1108436108/-/DCSupplemental.
References
- 1.Zhao Y. Auxin biosynthesis and its role in plant development. Annu Rev Plant Biol. 2010;61:49–64. doi: 10.1146/annurev-arplant-042809-112308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Comai L, Kosuge T. Cloning characterization of iaaM, a virulence determinant of Pseudomonas savastanoi. J Bacteriol. 1982;149:40–46. doi: 10.1128/jb.149.1.40-46.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sugawara S, et al. Biochemical analyses of indole-3-acetaldoxime-dependent auxin biosynthesis in Arabidopsis. Proc Natl Acad Sci USA. 2009;106:5430–5435. doi: 10.1073/pnas.0811226106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lehmann T, Hoffmann M, Hentrich M, Pollmann S. Indole-3-acetamide-dependent auxin biosynthesis: A widely distributed way of indole-3-acetic acid production? Eur J Cell Biol. 2010;89:895–905. doi: 10.1016/j.ejcb.2010.06.021. [DOI] [PubMed] [Google Scholar]
- 5.Zhao Y, et al. Trp-dependent auxin biosynthesis in Arabidopsis: Involvement of cytochrome P450s CYP79B2 and CYP79B3. Genes Dev. 2002;16:3100–3112. doi: 10.1101/gad.1035402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheng Y, Dai X, Zhao Y. Auxin biosynthesis by the YUCCA flavin monooxygenases controls the formation of floral organs and vascular tissues in Arabidopsis. Genes Dev. 2006;20:1790–1799. doi: 10.1101/gad.1415106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhao Y, et al. A role for flavin monooxygenase-like enzymes in auxin biosynthesis. Science. 2001;291:306–309. doi: 10.1126/science.291.5502.306. [DOI] [PubMed] [Google Scholar]
- 8.Stepanova AN, et al. TAA1-mediated auxin biosynthesis is essential for hormone crosstalk and plant development. Cell. 2008;133:177–191. doi: 10.1016/j.cell.2008.01.047. [DOI] [PubMed] [Google Scholar]
- 9.Tao Y, et al. Rapid synthesis of auxin via a new tryptophan-dependent pathway is required for shade avoidance in plants. Cell. 2008;133:164–176. doi: 10.1016/j.cell.2008.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheng Y, Dai X, Zhao Y. Auxin synthesized by the YUCCA flavin monooxygenases is essential for embryogenesis and leaf formation in Arabidopsis. Plant Cell. 2007;19:2430–2439. doi: 10.1105/tpc.107.053009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yamada M, Greenham K, Prigge MJ, Jensen PJ, Estelle M. The TRANSPORT INHIBITOR RESPONSE2 gene is required for auxin synthesis and diverse aspects of plant development. Plant Physiol. 2009;151:168–179. doi: 10.1104/pp.109.138859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Expósito-Rodríguez M, et al. Cloning and biochemical characterization of ToFZY, a tomato gene encoding a flavin monooxygenase involved in a tryptophan-dependent auxin biosynthesis pathway. J Plant Growth Regul. 2007;26:329–340. [Google Scholar]
- 13.Tivendale ND, et al. Reassessing the role of N-hydroxytryptamine in auxin biosynthesis. Plant Physiol. 2010;154:1957–1965. doi: 10.1104/pp.110.165803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ziegler DM. Flavin-containing monooxygenases: Enzymes adapted for multisubstrate specificity. Trends Pharmacol Sci. 1990;11:321–324. doi: 10.1016/0165-6147(90)90235-z. [DOI] [PubMed] [Google Scholar]
- 15.Strader LC, Bartel B. A new path to auxin. Nat Chem Biol. 2008;4:337–339. doi: 10.1038/nchembio0608-337. [DOI] [PubMed] [Google Scholar]
- 16.Phillips KA, et al. vanishing tassel2 encodes a grass-specific tryptophan aminotransferase required for vegetative and reproductive development in maize. Plant Cell. 2011;23:550–566. doi: 10.1105/tpc.110.075267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cheng Y, Qin G, Dai X, Zhao Y. NPY1, a BTB-NPH3-like protein, plays a critical role in auxin-regulated organogenesis in Arabidopsis. Proc Natl Acad Sci USA. 2007;104:18825–18829. doi: 10.1073/pnas.0708506104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cheng Y, Qin G, Dai X, Zhao Y. NPY genes and AGC kinases define two key steps in auxin-mediated organogenesis in Arabidopsis. Proc Natl Acad Sci USA. 2008;105:21017–21022. doi: 10.1073/pnas.0809761106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bentley JA, Farrar KR, Housley S, Smith GF, Taylor WC. Some chemical and physiological properties of 3-indolylpyruvic acid. Biochem J. 1956;64:44–49. doi: 10.1042/bj0640044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Müh U, Williams CH, Jr, Massey V. Lactate monooxygenase. II. Site-directed mutagenesis of the postulated active site base histidine 290. J Biol Chem. 1994;269:7989–7993. [PubMed] [Google Scholar]
- 21.Müh U, Massey V, Williams CH., Jr Lactate monooxygenase. I. Expression of the mycobacterial gene in Escherichia coli and site-directed mutagenesis of lysine 266. J Biol Chem. 1994;269:7982–7988. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.