Abstract
A defect in germ-cell (sperm and oocyte) development is the leading cause of male and female infertility. Control of translation through the binding of deleted in azoospermia (DAZ)-like (DAZL) to the 3′-UTRs of mRNAs, via a highly conserved RNA recognition motif (RRM), has been shown to be essential in germ-cell development. Crystal structures of the RRM from murine DAZL (Dazl), both alone and in complex with RNA sequences from the 3′-UTRs of mRNAs regulated by Dazl, reveal high-affinity sequence-specific recognition of a GUU triplet involving an extended, kinked, pair of β-strands. Recognition of the GUU triplet is maintained, whereas the identity and position of bases flanking this triplet varies. The Dazl RRM is thus able to recognize GUU triplets in different sequence contexts. Mutation of bases within the GUU triplet reduces the affinity of binding. Together with the demonstration that multiple Dazl RRMs can bind to a single RNA containing multiple GUU triplets, these structures suggest that the number of DAZL molecules bound to GUU triplets in the 3′-UTR provides a method for modulating the translation of a target RNA. The conservation of RNA binding and structurally important residues between members of the DAZ family, together with the demonstration that mutation of these residues severely impairs RNA binding, indicate that the mode of RNA binding revealed by these structures is conserved in proteins essential for gamete development from flies to humans.
Two percent of men are infertile because of severe abnormalities in sperm production (1). The characterization of deletions in the long arm of the Y chromosome in infertile men led to the identification of a region termed the azoospermia factor locus (AZFc), implicated in azoospermia and oligospermia defects in 5–10% of cases of male infertility (2–4). In humans, this region contains the four deleted in azoospermia (DAZ) genes that encode proteins containing one or more RNA recognition motifs and up to 15 copies of the DAZ repeat—a 24-residue sequence rich in glutamate, tyrosine, and proline residues (4).
DAZ is the youngest member in evolutionary terms of the DAZ gene family and is found only in humans, apes, and Old World monkeys (4). BOULE, the ancestral member of the family, is conserved from flies to humans, and a gene duplication event has led to the presence of the final family member, DAZ-like (DAZL), in vertebrates (5).
DAZL and BOULE are both single-copy, autosomal genes that encode proteins containing a single, highly conserved RRM and a single DAZ repeat. Disruption of murine Boule results in azoospermia because of a global arrest of spermatogenesis (6), whereas knockout of the murine Dazl gene results in azoospermia and absence of oocytes because of loss of germ cells (7), indicating that BOULE is required for spermatogenesis and DAZL for gametogenesis in both sexes. Furthermore, overexpression of DAZL promotes primordial germ-cell formation from human embryonic stem cells (8).
Xenopus DAZL (xDAZL) has also been shown to be critically involved in primordial germ-cell development (9). xDAZL has been demonstrated to bind RNA in vitro with a preference for poly-U and poly-G with the interaction with poly-U maintained in high salt conditions (2 M NaCl), indicating that binding involved hydrophobic, probably base-stacking, interactions (10). Human DAZL (hDAZL) and DAZ were also shown to preferentially bind poly-U (again with reduced sensitivity to ionic strength) and poly-G (11). Deletion of the hDAZL RRM abolished binding, demonstrating that the RRM is necessary for RNA binding by DAZL (11). Point mutations of conserved aromatic residues within the murine Dazl RRM also severely impaired RNA binding (12). Further studies using the dual approach of in vitro selection and three-hybrid screening with murine Dazl defined a Dazl consensus sequence of U3[G/C]U3 although the preference for G or C was markedly different in the sequences returned by each experiment with the in vitro selection data indicating a strong preference for (GUn)n rich sequences (13). The target sequence of zebrafish DAZL (zDAZL) was identified by in vitro selection as GUUC, and UV–cross-linking experiments showed that zDAZL specifically bound this sequence in vitro with mutation to CAUC or GUAG abolishing binding (14). A single point mutation, F91A (equivalent to F84 in Dazl and hDAZL), was sufficient to abolish RNA binding, confirming the RRM is essential for RNA recognition by DAZL (14). A recent study suggests that zDAZL binds to multiple repeats of the sequence GUUU in the 3′-UTR of HuB mRNA to enhance translation (15).
A number of in vivo targets of Dazl have been identified (16–18). Coimmunoprecipitation with Dazl from UV-crosslinked mouse testes extracts identified the mRNAs encoding mouse vasa homologue (Mvh) and a synaptonemal complex component (Sycp3) (16, 17). The male phenotypes of Mvh and Sycp3 null mice are similar to those of Dazl null mice (19, 20). Dazl was shown to bind specifically to sequences from the 3′-UTR of these mRNAs, and translation reporter assays in Xenopus oocytes indicated that Dazl could stimulate the translation of RNAs containing the Mvh or Sycp3 3′-UTRs. Importantly, the surviving germ cells of Dazl null testes contain significantly reduced levels of Sycp3 and Mvh proteins (16, 17). These results demonstrate that, in vivo, enhanced translation of these mRNAs via Dazl binding is essential for germ-cell development.
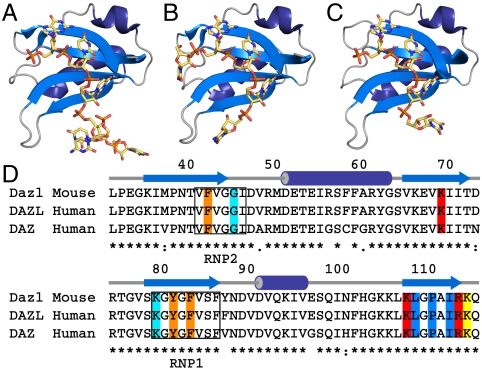
The RRMs of Dazl and hDAZL differ in only one position: Y/F88. In the RRMs of the human DAZ proteins, this residue is valine, and there are several other substitutions leading to an overall 90% identity of DAZ vs. Dazl in the RRM (Fig. 1D). The characteristic RNP1 and RNP2 motifs containing aromatic residues that are frequently involved in recognition of nucleic acid by RRMs (21) are completely conserved between Dazl/hDAZL and the human DAZ proteins (Fig. 1D). These motifs, together with the N- and C-terminal residues of the RRM, are the most highly conserved regions of DAZL between species suggesting that the structural basis for the specificity of RNA recognition by DAZ-family proteins is also conserved. The mechanism of translational regulation by DAZ-family members appears to be conserved across evolution. Ectopic expression of Xenopus DAZL or human BOULE in Drosophila can rescue the boule null phenotype, and human DAZ expression in mice rescues the Dazl null phenotype (10, 22, 23). However, this redundancy cannot be complete because disruption of either mouse Dazl or Boule results in male infertility despite partially overlapping expression patterns during spermatogenesis (2). Therefore, insight from structures of the Dazl RRM in complex with known mRNA targets are directly applicable to recognition of RNA by human DAZL and DAZ proteins.
Fig. 1.
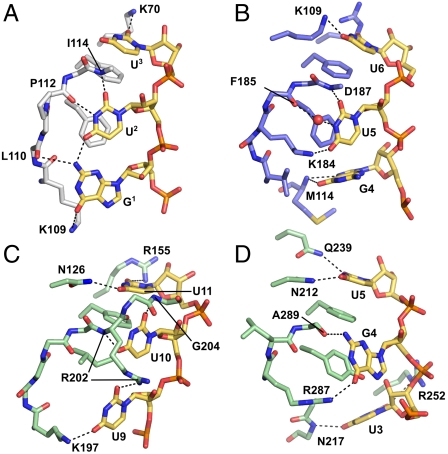
Dazl:RNA complexes, and the residues involved in RNA recognition. (A) Structure of Dazl32–132 with RNA sequence UUGUUCUU. (B) Structure of Dazl32–117 with Mvh 3′-UTR sequence UGUUC. (C) Structure of Dazl32–117 with Sycp3 3′-UTR sequence UUGUUU. Residues 35–116 only are shown for clarity. (D) Sequence alignment of RRMs from M. musculus Dazl (residues 32–117) and Homo sapiens DAZL and DAZ. Secondary structure elements and the conserved RNP1 and RNP2 motifs are indicated. Interactions between residues and the GUU motif are highlighted: orange, stacking interactions with base/ribose; red, side-chain hydrogen bonds to RNA; blue, backbone hydrogen bonds to RNA; yellow, both side-chain and backbone hydrogen bonds to RNA; cyan, hydrogen bonds to RNA via a bridging water molecule.
Previously, no structural information for any member of the DAZ family was available. To address this, we have solved high-resolution crystal structures of both the apo Dazl RRM, and three complexes with different RNA sequences (Fig. S1 and Fig. 1 A–C).
Results
Structure of the RNA Recognition Motif of Dazl.
The crystal structure of residues 35–118 of Dazl was determined at a resolution of 1.7 Å (Fig. S1 and Table S1) by molecular replacement using human TIA-1 RRM2 [Protein Data Bank (PDB) ID code 3BS9] as the search model. The Dazl RRM adopts the canonical βαββαββ RRM fold but with a shorter α2 helix than often observed in RRM structures. This enables the adjacent β1 and β4 strands to be extended and kinked around the conserved P39 and P112 residues twisting the surface of the β-sheet. Residues in the kinked β1 and β4 strands are almost completely conserved between DAZL proteins from different species, suggesting this conformation is important for RNA recognition by DAZL. This correlation between recognition of RNA and the kinked β1 and β4 strands is confirmed in the crystal structures of Dazl in complex with RNA described below.
Structure of the Dazl RRM in Complex with RNA Targets.
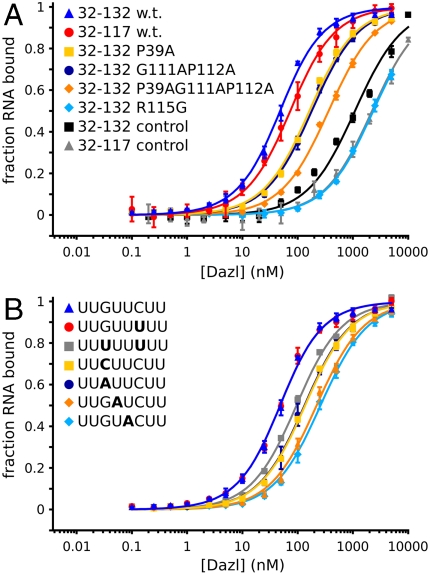
In order to define the structural basis of RNA recognition by the DAZL RRM, a construct encompassing residues 32–132 of Dazl (with a stabilizing C120S mutation) was produced. This construct, referred to as Dazl32–132, contains the RRM and also the region previously reported as necessary and sufficient for homodimerization by yeast two-hybrid—residues 80–132 (12). Dazl32–132 specifically binds with high affinity (Kd = 38 nM) an 8-nt RNA UUGUUCUU representing a combination of sequences from the previously reported Dazl binding sites in the Mvh and Sycp3 3′-UTRs (16, 17) (Fig. 2A and Table 1). To our surprise, Dazl32–132 appears monomeric by size exclusion chromatography (Fig. S2).
Fig. 2.
Mutations of residues critical for RNA binding and bases within the GUU motif impair RNA binding. (A) Binding isotherms for Dazl32–117 and wild-type and mutant Dazl32–132 with 8-nt RNA target, UUGUUCUU, and a negative control sequence, AAUUGUACAUAA. (B) Binding of Dazl32–132 to RNA sequences UUGUUCUU, UUGUUUUU, UUAUUCUU, UUCUUCUU, UUGAUCUU, UUGUACUU, and UUUUUUUU. All RNAs used were 3′-fluorescein labeled. Data represent mean and standard deviation from three experiments. The curves are fit by nonlinear least-squares regression; Kd for each interaction is listed in Table 1.
Table 1.
RNA-binding analyses of Dazl
| Dazl |
RNA * |
Kd, nM |
Krel† |
| 32–117 | UUGUUCUU | 63.6 ± 2.4 | |
| 32–132 | UUGUUCUU | 38.2 ± 1.9 | 1 |
| 32–132 | UUGUUUUU | 38.0 ± 1.9 | 1.0 |
| 32–132 | UUCUUCUU | 126.4 ± 3.8 | 3.3 |
| 32–132 | UUAUUCUU | 122.9 ± 3.8 | 3.2 |
| 32–132 | UUGAUCUU | 210.2 ± 8.0 | 5.5 |
| 32–132 | UUGUACUU | 249.5 ± 7.9 | 6.5 |
| 32–132 | UUUUUUUU | 85.2 ± 4.2 | 2.2 |
| P39A | UUGUUCUU | 153.0 ± 6.8 | 4.0 |
| G111AP112A | UUGUUCUU | 170.6 ± 4.6 | 4.5 |
| P39AG111AP112A | UUGUUCUU | 353.4 ± 8.2 | 9.3 |
| R115G | UUGUUCUU | > 2,000‡ | |
| 32–117 | AAUUGUACAUAA | > 2,000‡ | |
| 32–132 | AAUUGUACAUAA | > 2,000‡ |
*Bases that differ from UUGUUCUU are indicated in boldface.
†Krel reports the affinity of Dazl32–132 for the RNA relative to that of Dazl32–132 for the 8-nt RNA UUGUUCUU.
‡Saturable binding could not be achieved at the highest protein concentration; therefore, a lower limit for the Kd is reported.
The structure of Dazl32–132 in complex with UUGUUCUU was determined by single wavelength anomalous dispersion (SAD) at a resolution of 1.35 Å (Table S1 and Figs. 1A and 3B). In this structure (referred to as 32–132:8-nt), we observe electron density for residues 32–117 of Dazl and for 6-nt of RNA (G3-U8). A zinc ion necessary for crystallization (and used for phasing) coordinates N3 of the C6 base in one asymmetric unit and the side chains of E55 and H104 in the adjacent asymmetric unit. The fourth atom coordinated by the zinc ion is a side-chain nitrogen of H123. This histidine is the only residue from the region C-terminal to the RRM fold for which we observe electron density, demonstrating that although this region is present in the crystals, it is disordered. None of the zinc-mediated interactions are at the canonical RNA-binding surface and represent a fortuitous crystal contact. Analysis of interfaces in the crystal with PISA (24) indicates that the largest interface (646.2 Å2) is between the protein and RNA, indicating that Dazl32–132 (which contains the region previously described as sufficient for homodimerization) binds RNA as a monomer.
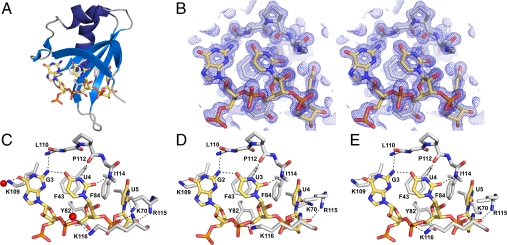
Fig. 3.
GUU triplet recognition by the Dazl RRM. (A) Overview showing orientation of Fig. 4 C–E; for clarity, Fig. 4B is rotated slightly. (B) Stereo view of initial zinc–SAD phased electron density map contoured at 2σ. (C) Recognition of the GUU triplet in the 32–132:8-nt structure (D) Recognition of the GUU triplet in the 32–117:Mvh structure. (E) Recognition of the GUU triplet in the 32–117:Sycp3 structure. Hydrogen bonds are shown as dashed lines.
The RRM of Dazl Alone Mediates Specific RNA Recognition.
As no dimerization interface, and no direct interaction with RNA, was observed for residues C-terminal to K116 in the 32–132:8-nt structure, a construct encompassing residues 32–117 of Dazl (referred to as Dazl32–117) was produced. The affinity of Dazl32–117 for the 8-nt RNA is essentially identical to that of Dazl32–132 (Kd = 64 vs. 38 nM; Fig. 2A and Table 1) demonstrating that residues C-terminal to the RRM are not involved in sequence-specific RNA recognition by Dazl.
Dazl Recognizes a GUU Triplet.
The structures of Dazl32–117 with a 5-nt sequence from the 3′-UTR of Mvh, UGUUC (32–117:Mvh) and with a 6-nt sequence from the 3′-UTR of Sycp3, UUGUUU (32–117:Sycp3) were solved at resolutions of 1.6 and 1.45 Å, respectively, by molecular replacement using the RRM from the 32–132:8-nt structure as the search model (Fig. 1 B and C). In the 32–117:Mvh structure, there are two protein:RNA complexes in the asymmetric unit, and we observe electron density for all 5-nt of RNA in the first complex and 4-nt of RNA (G2-C5) in the second. The protein:RNA contacts (including those involving bridging water molecules) are identical in both complexes. In the 32–117:Sycp3 structure, we observe electron density for 4-nt of RNA (G3-U6). Again, in both of these structures the largest interface (704.4/609.8 Å2 32–117:Mvh chain A/B, 555.6 Å2 32–117:Sycp3) is between protein and RNA, demonstrating that protein–protein interactions are not required for RNA binding.
Comparison of the three complex structures (Figs. 1 A–C and 3 C–E) reveals that the Dazl RRM specifically recognizes a GUU triplet. These bases are in identical positions in all three structures, whereas the bases 5′ or 3′ to this triplet are in different orientations in each structure. A detailed comparison of the protein:RNA hydrogen bonds, including those via bridging water molecules (Fig. S3), reveals that the interaction between the Dazl RRM and the GUU triplet is essentially identical in all three structures.
The Kinked and Extended β1 and β4 Strands Govern the Sequence Specificity.
The GUU triplet binds across the β-sheet of the Dazl RRM with each base positioned over a separate strand (Figs. 1 A–C and 3A): the guanine (referred to as G1) on β4, the first uracil (U2) on β1, and the second uracil (U3) on β3. U2 and U3 stack on the side chains of F43 and F84, respectively (Fig. 3 C–E). Y82 makes van der Waals interactions with the ribose sugars of G1 and U2. The phosphates in the GUU triplet are recognized by hydrogen bonds to K116; the phosphate between G1 and U2 is bound by the side chain (via a bridging water molecule in the 32–132:8-nt structure), and the phosphate between U2 and U3 by the amide nitrogen. The bases of G1 and U2 bind into a trough between the side chain of K109 and the backbone of residues in the β4 strand, and the base of U3 is inserted into a pocket formed by the side chains of R115 and K70 (Fig. S4). R115 is hydrogen bonded to the 2′ OH of U3, thus discriminating between RNA and DNA. The side chain of K70 is hydrogen bonded to O2 of U3 in two of the three structures (Fig. 3 C–E), and N3 and O2 of this base make hydrogen bonds to an ordered network of water molecules that form hydrogen bonds with the side chains of T41, E68, S86, and R115. G1 and U2 are recognized via sequence-specific hydrogen bonds: O6 of G1 with the side chain of K109 (via a bridging water molecule in the 32–132:8-nt structure), N2 of G1 with the carbonyl oxygen of L110 and N3 and O2 of U2 with the carbonyl oxygen of P112 and the amide nitrogen of I114, respectively. A hydrogen bond between N2 of G1 and O4 of U2 further discriminates for a GU pair. These structures, therefore, enable us to define the structural basis for preferential binding of GUU by Dazl.
These sequence-specifying interactions are made possible by the kink induced in β4 by P112, which is stabilized by the equivalent kink in the adjacent β1 strand induced by P39. Mutation of either P39 or the glycine–proline pair G111P112 to alanine reduces the affinity of Dazl for the 8-nt RNA UUGUUCUU around fourfold (Fig. 2A and Table 1), and mutating both together decreases binding by nearly 10-fold. The sequence of the β4 strand—KLGPAIRK—is completely conserved in mammalian DAZL proteins, and all residues are crucial for governing the sequence specificity. The hydrophobic side chain of L110 is buried into the core of the RRM fold, and the side chain of I114 makes a hydrophobic interaction with the side chain of I37 in β1, allowing the backbone atoms of these residues to make hydrogen bonds with the bases of G1 and U2 because of the kink in β4 induced by the glycine–proline pair. The side chains of the charged K109, R115, and K116 all interact directly with the GUU triplet. The role of A113 in selecting U3 is discussed later.
Substitutions in the GUU Triplet Decrease Affinity.
In order to verify the specificity of interactions revealed by the crystal structures, we tested the effect of mutating bases in the 8-nt sequence UUGUUCUU on the affinity of RNA binding by Dazl32–132. (Fig. 2B and Table 1). The relative affinity (Krel) when C6 was exchanged with U was 1.0, indicating the identity of this base has no affect on the affinity of interaction. This is as expected from comparison of the 32–117:Mvh and 32–117:Sycp3 structures, where this nucleotide differs (C in the former, U in the latter) and is in different locations in the two structures (Fig. 1 B and C and Fig. S4) with no interaction between the base and Dazl32–117 (Fig. S3). In contrast, mutations within the GUU triplet markedly reduce the affinity: Mutation of G1 to C or A reduces the affinity by more than threefold (Krel = 3.3/3.2), and when either U2 or U3 is replaced by A the affinity is further reduced (Krel = 5.5/6.5). Mutation to poly-U appears to have an intermediate affect on the affinity (Krel = 2.2), although the presence of multiple equivalent binding sites in this RNA complicates analysis.
The R115G SNP Severely Impairs RNA Binding.
A missense mutation in hDAZL identified in a woman with spontaneous premature ovarian failure is R115G (25). This individual was homozygous for this mutation and underwent premature ovarian failure aged 34, having had no children. The side chain of R115 directly contacts RNA stacking on U3 in the GUU triplet and forming an RNA-specifying hydrogen bond with the 2′-OH. Mutation of this residue to glycine would destroy these interactions. In our assay, the RNA binding of this mutant is at least 50-fold weaker than wild type (Fig. 2A). Therefore, these structures and in vitro binding data strongly indicate that the pathology associated with the R115G mutation is due to disruption of RNA binding by the DAZL RRM.
Multiple Dazl RRMs Can Bind to Repeated GUU Triplets in a Single RNA.
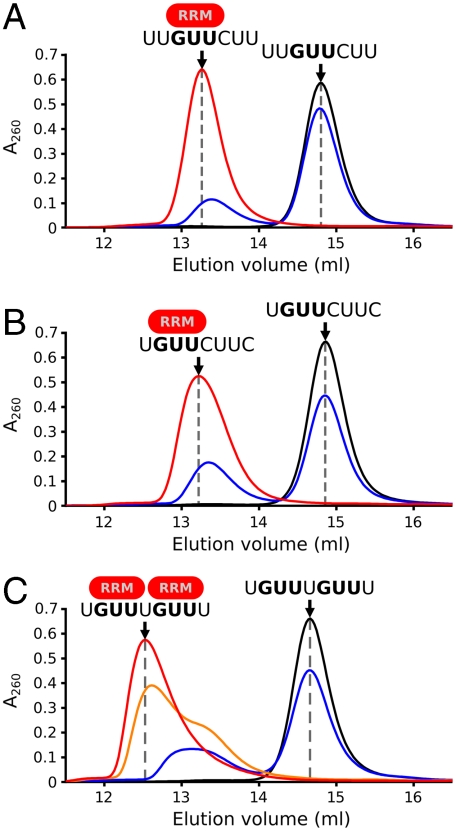
The 3′-UTRs of Mvh and Sycp3 contain multiple GUU triplets with different flanking sequences. We investigated whether such mRNA sequences could recruit multiple copies of the Dazl RRM by analytical size exclusion chromatography. The complex of Dazl32–117 with an 8-nt sequence from the Mvh 3′-UTR (UGUUCUUC) (Fig. 4B) elutes at the same volume as the monomeric Dazl32–117 : UUGUUCUU complex (Fig. 4A), indicating that this is also a 1∶1 complex and that a second Dazl RRM does not bind to the CUU triplet [as would be suggested by the previously defined Dazl consensus sequence of Un[G/C]Un (13)]. In contrast, the complex formed with a 9-nt sequence from the Sycp3 3′-UTR containing two GUU triplets (UGUUUGUUU) elutes earlier with a shoulder at the elution volume of the 1∶1 complexes (Fig. 4C, orange curve), indicating an equilibrium between a 1∶1 complex and a larger complex. When the Dazl32–117 : RNA ratio is increased, the shoulder disappears and the complex elutes in a single peak (Fig. 4C, red curve), suggesting that two RRMs are bound to a single RNA.
Fig. 4.
Multiple copies of the Dazl RRM can bind a single RNA containing more than one GUU triplet. Analytical size exclusion chromatograms of (A) monomeric complex formed with UUGUUCUU RNA. Dazl:RNA ratios: black, 0∶1; blue, 1∶2; red, 5∶1. (B) Monomeric complex formed with 8-nt sequence from Mvh 3′-UTR, UGUUCUUC. Dazl:RNA ratios: black, 0∶1; blue, 1∶2; red, 2∶1. (C) Dimeric complex formed with 9-nt from Sycp3 3′-UTR, UGUUUGUU. Dazl:RNA ratios: black, 0∶1; blue, 1∶2; orange, 2∶1; red 3∶1.
Discussion
High-Affinity Recognition of a GU Pair Through Kinked β-Strands.
The Dazl RRM differs from a canonical RRM fold because of the unusual conformation of the extended and kinked β1 and β4 strands. A search of the Protein Data Bank with the protein structure comparison service (26) failed to find similarly extended and kinked β1 and β4 strands in the over 300 structures of RRM folds. The secondary structure of the antiparallel β1 and β4 strands is maintained because of the alignment of the absolutely conserved residues P39 and P112. Also, in all four Dazl structures a structurally conserved water molecule satisfies the hydrogen bond donors and acceptors that would normally form interstrand hydrogen bonds in a β-sheet (Fig. S5). This arrangement is critical because prior to RNA binding it positions the backbone amides and carbonyls of residues P110 to I114 into the correct orientation to form sequence-specifying hydrogen bonds with the Watson–Crick edges of the G1U2 pair of the GUU triplet. Mutation of these prolines severely impairs RNA binding by Dazl (Fig. 2A and Table 1).
Structural comparison of the complexes of the Dazl RRM with GUU, RRM2 of CUG-BP1 with GUU (27), and RRMs 1 and 2 of Sex-lethal with UUU and UGU, respectively (28) (Fig. 5 and Table S2) reveals that while recognition of the second base in the triplet by the backbone of residues at positions i and i + 2 in the β4 strand is commonly observed, the kink in β4 of Dazl additionally allows recognition of the first base of the triplet by the backbone of the residue at position i - 2.
Fig. 5.
Sequence-specific RNA recognition by (A) Dazl, (B) CUG-BP1 RRM2 (PDB ID code 3NMR), (C) Sex-lethal RRM1, and (D) Sex-lethal RRM2 (PDB ID code 1B7F). Residues making sequence-specifying hydrogen bonds are labeled.
It has recently been shown that RNA binding by RRMs is driven by large favorable enthalpy changes—predominantly due to stacking of bases onto the conserved aromatic residues in the RNP1 and RNP2 motifs—that overcome the unfavorable entropy of binding (29). This is also the case for Dazl because substitution of the two stacking bases has the greatest affect on affinity (Fig. 2B). The preordering of the backbone of residues in the RNA interacting β4 strand would act to reduce the unfavorable entropic cost of the increased ordering of residues that are solvent exposed in the absence of RNA but bound to RNA in the complex. This arrangement may, therefore, represent another way in which the RRM has evolved toward specific high-affinity recognition for the target RNA sequence.
The Dazl RRM specifically recognizes a GUU triplet.
The Dazl consensus sequence was previously defined as Un[G/C]Un (13). In our structures, a C in position one of the GUU triplet would not be able to satisfy the sequence-specifying hydrogen bonds with K109, L110, and U2 without substantial distortion of the RNA backbone. Hydrogen bonds to L110 or U2 would also not be formed by A in this position. When the G in this position is mutated to C or A, the affinity is reduced by more than threefold (Fig. 2B). The U in the central position of the triplet is clearly specified by hydrogen bonds between the backbone of P112 and I114 in the β4 strand with the Watson–Crick edge of this base, and mutation to A reduces binding over fivefold. In contrast, the specificity at position three is less clearly defined. Although sterically a purine would not be accommodated in the pocket occupied by U3, and A in this position reduces binding over sixfold, there are no hydrogen bonds that would discriminate between U and C. An explanation is provided by the environment of position 4 on the pyrimidine ring—this is in close proximity to the methyl group of A113, and in all the structures O4 is not hydrogen bonded to any water molecules. Calculation of the solvation energy gain of the interface (ΔiG) with PISA (24) shows that substitution of U3 with C makes ΔiG more unfavorable by 3.15 kcal/mol. This indicates that Dazl will bind GUU with higher affinity than GUC for steric reasons rather than because of differences in hydrogen bonding.
GUU Triplet Recognition Explains Effect of Mutations in the Mvh 3′-UTR.
The differing effect of identical mutations at different sites within the Mvh 3′-UTR on Dazl binding were unaccounted for by the previously defined Dazl consensus sequence of Un[G/C]Un but are easily explained in the light of the structures described above. Mutation (by exchange of U and C) of sites with sequence AUUCUUA and AUUCUUC or deletion of a further site with sequence AUUCUUA had no effect on Dazl binding. In contrast, identical mutation of two sites with sequence GUUCUUC abolished Dazl binding (17). Our structures allow a reinterpretation of this conflicting effect of mutation of UUCUU in differing sequence contexts: The removal of GUU triplets prevents Dazl binding, whereas removal of CUU has no effect on Dazl binding. In addition, Dazl also bound a region of Mvh 3-UTR encompassing only the noninteracting sites but with an additional 3′ U4GU3 site (17). This also confirms binding via GUU triplets.
A Redefined DAZL Consensus Sequence.
The structures presented here, supported by measurement of the affinity of Dazl binding to different RNA sequences, demonstrate that Dazl recognizes GUU triplets. The location of the nucleotide at position four (either U or C) varies between complexes, and GUUC and GUUU bind with equal affinity. The structures and binding data, together with the conservation of RNA interacting residues and inspection of the nucleotides surrounding GUU triplets in the 3′-UTR of known DAZL targets, suggest that an optimal DAZL binding site can be defined as GUU[U/C]. However, the lack of sequence-specifying contacts for the bases flanking the GUU triplet (Fig. S3) may enable binding of GUU triplets in the context of any flanking nucleotide. The short length of this target sequence (consistent with the number of nucleotides bound by a single RRM) means that searching mRNA databases for potential targets of DAZL-mediated regulation is likely to return many false positives. In vivo, the location of a GUU triplet with respect to any secondary structural elements present in the 3′-UTR or overlap with the target sequence of another RNA-binding protein is likely to be as important for identifying a DAZL binding site as the identity of the bases flanking the triplet. The identical specificity of in vitro RNA-binding by residues 1–137 of Dazl to that of the full length protein and the abolition of this binding by point mutations within the RRM (12–14, 16, 17) suggests that any additional mRNA target specificity in vivo is unlikely to derive from regions of the protein outside the RRM. More likely, if required, additional specificity comes from interactions with RNA-binding protein partners such as DAZAP1 or PUM2 (30, 31).
Dazl Binds RNA as a Monomer.
We found no evidence that Dazl was able to form specific homotypic interactions, but did observe that the addition of residues 118–132 to the RRM lead to a propensity for nonspecific aggregation. In all the structures, Dazl is monomeric with no protein–protein interfaces larger than are typically observed as crystal contacts. We therefore suggest that the previously reported homodimerization (12) is an artifact caused by nonspecific aggregation of Dazl in the yeast two-hybrid assay.
Cooccupancy of Dazl RRMs on a Single RNA.
We have shown that it is possible to accommodate two Dazl RRMs on a 9-nt RNA containing two GUU motifs separated by a single U. Thus, it is likely that multiple molecules of DAZL bind to a 3′-UTR containing multiple GUU triplets. Binding does not appear to be cooperative, which is consistent with the differing spacing of GUU triplets both between known in vivo RNA targets and between the 3′-UTRs of the same mRNA in different species (Table S3) (16, 17). If protein–protein interaction between RRMs were important for modulating binding, the spacing of the individual DAZL binding sites would likely be conserved, especially between mice and rats in which the proteins differ in only a single position (I225L), but this is not the case. A rationale for the presence of multiple DAZL binding sites in the 3′-UTRs of target mRNAs is indicated by the increase in translation of a target RNA bound by multiple Dazl molecules over that due to binding of a single Dazl molecule in tethered translation stimulation assays in Xenopus oocytes (32). A similar effect was seen for zDAZL binding directly to RNA containing multiple copies of the zDAZL target sequence in the 3′-UTR in both CV-1 and mouse fibroblast cells (14). The proposed method for DAZL-mediated stimulation of translation involves direct recruitment of PABP to the 3′-UTRs of target mRNAs via the protein–protein interaction between DAZL and PABP (32). This is predicted to increase end-to-end complex formation leading to enhanced ribosomal subunit recruitment in a manner analogous to cytoplasmic polyadenylation (2). This method implies that recruitment of differing numbers of DAZL (and therefore PABP) molecules to the 3′-UTRs of target mRNAs—i.e., a “dose” effect—could provide a method for modulation of translation during gametogenesis.
Implications for RNA Recognition by DAZ/BOULE.
All of the residues that interact with RNA are conserved between Dazl and DAZ (Fig. 1D), therefore, we propose that the RRMs in the human DAZ proteins also recognize GUU triplets. Although the RRM is the most highly conserved region between Dazl, Drosophila boule and mammalian BOULE, the K109N substitution in boule and BOULE means that we cannot be certain that these proteins recognize GUU triplets in the same way as Dazl. However, the almost complete conservation of the RNP1 and RNP2 motifs and the conservation of the proline residues in β1 and β4 that stabilize the kinked, RNA-binding, β4 strand suggest that the mode of RNA recognition is conserved from boule in flies to the DAZ proteins in humans. Cross family member rescue data (as described above) does suggest that at least some critical targets can be recognized in a sufficiently similar manner to retain functional regulation.
In conclusion, the structures we report reveal that the Dazl RRM specifically recognizes a GUU triplet by means of a unique kinked β4 strand stabilized by two absolutely conserved proline residues. This mode of binding allows high-affinity recognition of this triplet in the context of varying flanking sequences and also for a pair of RRMs to cooccupy an RNA molecule with two GUU triplets separated by a single U. We suggest this allows subtle modulation of the level of translation of target RNAs through recruitment of differing numbers of DAZL molecules due to the presence of multiple GUU triplets in their 3′-UTRs, which is presumably also sensitive to cellular DAZL protein concentrations.
Methods
Detailed methods for all procedures are available in SI Methods.
Protein Expression and Purification.
Constructs were expressed either as a GST fusion (35–118) and purified by glutathione affinity, HRV 3C protease cleavage and gel filtration, or as His6–SUMO (small ubiquitin-like modifier) fusions (32–132 and 32–117) and purified by Ni2+ affinity, Ulp1 protease cleavage, and gel filtration. P39A, G111AP112A, P39AG111AP112A, and R115G mutants of Dazl32–132 were expressed and purified as wild type.
Crystallization and Structure Determination.
Crystals of apo Dazl were grown by hanging drop vapor diffusion [2.1 M Li2SO4, 50 mM HEPES pH 7.5, 5% (vol/vol) glycerol, 291 K], cryoprotected with 20% (vol/vol) glycerol and data collected at 100 K at Synchrotron Radiation Source Daresbury beamline PX14.2. Crystals of Dazl:RNA complexes were grown by sitting drop vapor diffusion at 277 K [10 mM magnesium formate, 20% (wt/vol) PEG 3350, 0.1 mM zinc acetate (32–132:8-nt); 0.1 M magnesium acetate, 18% (wt/vol) PEG 3350 (32–117:Mvh) or 10 mM magnesium formate, 25% (wt/vol) PEG 3350, 10 mM zinc acetate (32–117:Sycp3)], cryoprotected with 20% (vol/vol) PEG 400, and data collected at 100 K at Diamond Light Source beamlines I03 and I02.
The apo Dazl structure was solved by molecular replacement with PHASER (33) using 3BS9 as the search model. The 32–132:8-nt complex was solved by SAD using SHELX (34). The 32–117:Mvh and 32–117:Sycp3 complexes were solved by molecular replacement using the RRM from the 32–132:8-nt structure as the search model. Structures were validated with MOLPROBITY (35) and have 100% of residues in the favored region of the Ramachandran plot with all RNA geometry correct. Data collection and refinement statistics are shown in Table S1.
Fluorescence Polarization.
Assays were carried out as described previously (36) with the following modifications: Buffer used was 20 mM HEPES pH 7.5, 100 mM NaCl, 0.01% (vol/vol) Triton X-100, and the top protein concentration was 5 μM (10 μM for AAUUGUACAUA controls).
Analytical Size Exclusion Chromatograpy.
RNA and Dazl:RNA complexes were resolved at 277 K in 10 mM HEPES pH 7.5, 100 mM NaCl on a Superdex 75 10/300 column. Complexes were incubated on ice for 30 min before loading on the column.
Supplementary Material
Acknowledgments.
We thank Helen Walden for the kind gift of reagents, the beamline scientists at both Daresbury and Diamond synchrotrons for continuing help and support, and Nicola Gray and Howard Cooke for plasmids and comments on the manuscript. This work was supported by the Biotechnology and Biological Sciences Research Council Grant BB/E020070/1.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1105211108/-/DCSupplemental.
Data deposition footnote: The coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 2XS2, 2XS5, 2XS7, and 2XSF).
References
- 1.Hull MG, et al. Population study of causes, treatment, and outcome of infertility. Br Med J Clin Res Ed. 1985;291:1693–1697. doi: 10.1136/bmj.291.6510.1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brook M, Smith J, Gray N. The DAZL and PABP families: RNA-binding proteins with interrelated roles in translational control in oocytes. Reproduction. 2009;137:595–617. doi: 10.1530/REP-08-0524. [DOI] [PubMed] [Google Scholar]
- 3.Reynolds N, Cooke H. Role of the DAZ genes in male fertility. Reprod Biomed Online. 2005;10:72–80. doi: 10.1016/s1472-6483(10)60806-1. [DOI] [PubMed] [Google Scholar]
- 4.Reijo R, et al. Diverse spermatogenic defects in humans caused by Y chromosome deletions encompassing a novel RNA-binding protein gene. Nat Genet. 1995;10:383–393. doi: 10.1038/ng0895-383. [DOI] [PubMed] [Google Scholar]
- 5.Xu EY, Moore FL, Pera RA. A gene family required for human germ cell development evolved from an ancient meiotic gene conserved in metazoans. Proc Natl Acad Sci USA. 2001;98:7414–7419. doi: 10.1073/pnas.131090498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.VanGompel MJ, Xu EY. A novel requirement in mammalian spermatid differentiation for the DAZ-family protein Boule. Hum Mol Genet. 2010;19:2360–2369. doi: 10.1093/hmg/ddq109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ruggiu M, et al. The mouse Dazla gene encodes a cytoplasmic protein essential for gametogenesis. Nature. 1997;389:73–77. doi: 10.1038/37987. [DOI] [PubMed] [Google Scholar]
- 8.Kee K, Angeles V, Flores M, Nguyen H, Reijo Pera RA. Human DAZL, DAZ and BOULE genes modulate primordial germ-cell and haploid gamete formation. Nature. 2009;462:222–225. doi: 10.1038/nature08562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Houston DW, King ML. A critical role for Xdazl, a germ plasm-localized RNA, in the differentiation of primordial germ cells in Xenopus. Development. 2000;127:447–456. doi: 10.1242/dev.127.3.447. [DOI] [PubMed] [Google Scholar]
- 10.Houston DW, Zhang J, Maines JZ, Wasserman SA, King ML. A Xenopus DAZ-like gene encodes an RNA component of germ plasm and is a functional homologue of Drosophila boule. Development. 1998;125:171–180. doi: 10.1242/dev.125.2.171. [DOI] [PubMed] [Google Scholar]
- 11.Tsui S, Dai T, Warren ST, Salido EC, Yen PH. Association of the mouse infertility factor DAZL1 with actively translating polyribosomes. Biol Reprod. 2000;62:1655–1660. doi: 10.1095/biolreprod62.6.1655. [DOI] [PubMed] [Google Scholar]
- 12.Ruggiu M, Cooke HJ. In vivo and in vitro analysis of homodimerisation activity of the mouse Dazl1 protein. Gene. 2000;252:119–126. doi: 10.1016/s0378-1119(00)00219-5. [DOI] [PubMed] [Google Scholar]
- 13.Venables JP, Ruggiu M, Cooke HJ. The RNA-binding specificity of the mouse Dazl protein. Nucleic Acids Res. 2001;29:2479–2483. doi: 10.1093/nar/29.12.2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maegawa S, Yamashita M, Yasuda K, Inoue K. Zebrafish DAZ-like protein controls translation via the sequence ‘GUUC’. Genes Cells. 2002;7:971–984. doi: 10.1046/j.1365-2443.2002.00576.x. [DOI] [PubMed] [Google Scholar]
- 15.Wiszniak SE, Dredge BK, Jensen KB. HuB (elavl2) mRNA is restricted to the germ cells by post-transcriptional mechanisms including stabilisation of the message by DAZL. PLoS One. 2011;6:e20773. doi: 10.1371/journal.pone.0020773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reynolds N, Collier B, Bingham V, Gray N, Cooke H. Translation of the synaptonemal complex component Sycp3 is enhanced in vivo by the germ cell specific regulator Dazl. RNA. 2007;13:974–981. doi: 10.1261/rna.465507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reynolds N, et al. Dazl binds in vivo to specific transcripts and can regulate the pre-meiotic translation of Mvh in germ cells. Hum Mol Genet. 2005;14:3899–3909. doi: 10.1093/hmg/ddi414. [DOI] [PubMed] [Google Scholar]
- 18.Jiao X, Trifillis P, Kiledjian M. Identification of target messenger RNA substrates for the murine deleted in azoospermia-like RNA-binding protein. Biol Reprod. 2002;66:475–485. doi: 10.1095/biolreprod66.2.475. [DOI] [PubMed] [Google Scholar]
- 19.Yuan L, et al. The murine SCP3 gene is required for synaptonemal complex assembly, chromosome synapsis, and male fertility. Mol Cell. 2000;5:73–83. doi: 10.1016/s1097-2765(00)80404-9. [DOI] [PubMed] [Google Scholar]
- 20.Tanaka SS, et al. The mouse homolog of Drosophila Vasa is required for the development of male germ cells. Genes Dev. 2000;14:841–853. [PMC free article] [PubMed] [Google Scholar]
- 21.Auweter SD, Oberstrass FC, Allain FH. Sequence-specific binding of single-stranded RNA: Is there a code for recognition? Nucleic Acids Res. 2006;34:4943–4959. doi: 10.1093/nar/gkl620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Slee R, et al. A human DAZ transgene confers partial rescue of the mouse Dazl null phenotype. Proc Natl Acad Sci USA. 1999;96:8040–8045. doi: 10.1073/pnas.96.14.8040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu EY, et al. Human BOULE gene rescues meiotic defects in infertile flies. Hum Mol Genet. 2003;12:169–175. doi: 10.1093/hmg/ddg017. [DOI] [PubMed] [Google Scholar]
- 24.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 25.Tung JY, et al. Novel missense mutations of the Deleted-in-AZoospermia-Like (DAZL) gene in infertile women and men. Reprod Biol Endocrinol. 2006;4:40. doi: 10.1186/1477-7827-4-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krissinel E, Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr D Biol Crystallogr. 2004;60:2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- 27.Teplova M, Song J, Gaw HY, Teplov A, Patel DJ. Structural insights into RNA recognition by the alternate-splicing regulator CUG-binding protein 1. Structure. 2010;18:1364–1377. doi: 10.1016/j.str.2010.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Handa N, et al. Structural basis for recognition of the tra mRNA precursor by the Sex-lethal protein. Nature. 1999;398:579–585. doi: 10.1038/19242. [DOI] [PubMed] [Google Scholar]
- 29.McLaughlin KJ, Jenkins JL, Kielkopf CL. Large favorable enthalpy changes drive specific RNA recognition by RNA recognition motif proteins. Biochemistry. 2011;50:1429–1431. doi: 10.1021/bi102057m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moore FL, et al. Human Pumilio-2 is expressed in embryonic stem cells and germ cells and interacts with DAZ (Deleted in AZoospermia) and DAZ-like proteins. Proc Natl Acad Sci USA. 2003;100:538–543. doi: 10.1073/pnas.0234478100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tsui S, et al. Identification of two novel proteins that interact with germ-cell-specific RNA-binding proteins DAZ and DAZL1. Genomics. 2000;65:266–273. doi: 10.1006/geno.2000.6169. [DOI] [PubMed] [Google Scholar]
- 32.Collier B, Gorgoni B, Loveridge C, Cooke H, Gray N. The DAZL family proteins are PABP-binding proteins that regulate translation in germ cells. EMBO J. 2005;24:2656–2666. doi: 10.1038/sj.emboj.7600738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McCoy AJ, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sheldrick GM. A short history of SHELX. Acta Crystallogr A. 2008;64:112–122. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- 35.Chen VB, et al. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jenkins H, Baker-Wilding R, Edwards TA. Structure and RNA binding of the mouse Pumilio-2 Puf domain. J Struct Biol. 2009;167:271–276. doi: 10.1016/j.jsb.2009.06.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.