Abstract
Many receptors that activate cells of the immune system are multisubunit membrane protein complexes in which ligand recognition and signaling functions are contributed by separate protein modules. Receptors and signaling subunits assemble through contacts among basic and acidic residues in their transmembrane domains to form the functional complexes. Here we report the nuclear magnetic resonance (NMR) structure of the membrane-embedded, heterotrimeric assembly formed by association of the DAP12 signaling module with the natural killer (NK) cell–activating receptor NKG2C. The main intramembrane contact site is formed by a complex electrostatic network involving five hydrophilic transmembrane residues. Functional mutagenesis demonstrated that similar polar intramembrane motifs are also important for assembly of the NK cell–activating NKG2D-DAP10 complex and the T cell antigen receptor (TCR)–invariant signaling protein CD3 complex. This structural motif therefore lies at the core of the molecular organization of many activating immunoreceptors.
Most activating immunoreceptors are composed of multiple single-pass transmembrane proteins that assemble in the endoplasmic reticulum to form functional complexes that are then exported to the cell surface. Ligand-binding and signal-transducing functions are contributed by separate protein modules that couple a broad array of ligand specificities to common intracellular signaling pathways1, most of which use the well-studied cytoplasmic immunoreceptor tyrosine-based activation motifs (ITAMs)2. The mechanisms used to link extracellular ligand binding to the phosphorylation of intracellular signaling domains for this class of modular immunoreceptors remain largely unknown. The lack of structural information about the membrane-embedded portions, which are the main points of physical contact between receptors and signaling modules, represents a considerable challenge to the development of comprehensive mechanistic models. The ITAM-bearing CD3δε, CD3γε and ζζ signaling modules assemble with the T cell antigen receptor (TCR) in a process that requires a pair of acidic transmembrane residues in each signaling dimer but only a single basic transmembrane residue in the receptor3–5. Other activating immunoreceptors have similar requirements for assembly1, including the Fc receptor for immunoglobulin A6,7, the natural killer cell (NK cell)–activating receptor complex of DAP10 (A000749) and NKG2D (A001666)8,9, and more than a dozen different receptors expressed in lymphoid and myeloid cells that associate with the ITAM-bearing signaling dimer DAP12 (A000750; also called KARAP)10,11. Published mutagenesis studies have suggested that the key intramembrane contacts are mediated not by simple one-to-one, charge-paired salt bridges but by a considerably more complex electrostatic network3,6,8,12. Those studies demonstrated that two acidic residues in the dimeric signaling modules are absolutely required for effective assembly with receptors through a single basic transmembrane residue. Those stoichiometric data therefore highlighted the need for detailed structural and biochemical studies to show how these unusual polar interactions direct the intramembrane assembly of activating immunoreceptor complexes and how this fits with the present models of intact receptor complexes.
To gain structural insight into the assembled state of a representative immunoreceptor complex, we determined the solution nuclear magnetic resonance (NMR) structure of the heterotrimeric transmembrane complex formed by assembly of the DAP12 signaling module with the NK cell–activating receptor NKG2C (A001665)13. NKG2C forms a heterodimer with the C-type lectin CD94 and recognizes the human nonclassical major histocompatibility complex class I molecule HLA-E14 (Qa-lb in mice), delivering activating signals through the ITAM motifs in the DAP12 cytoplasmic tails2. Along with its inhibitory counterpart NKG2A, the DAP12-NKG2C-CD94 complex is thought to have an important role in maintaining the balance of positive and negative signals that govern NK cell tolerance and responsiveness15. NKG2C is also expressed by γδ T cells and a subset of CD8+ T cells and may therefore have a further role in non-NK cell–mediated immunity. We found that the heterotrimeric structure of the transmembrane portion of DAP12 (DAP12TM) assembled with the transmembrane portion of NKG2C (NKG2CTM) had an unexpectedly complex electrostatic network at the core of the assembly. This network was composed of a pair of aspartic acid residues in the DAP12 dimer interface and two adjacent threonine residues that functioned together to guide association with a basic lysine residue in the receptor transmembrane domain. Analysis of sequence alignments and functional mutagenesis in an in vitro assembly assay demonstrate that this particular electrostatic network also lies at the core of the NK cell-activating NKG2D-DAP10 complex8,9 and of the critically important TCR-CD3 complex. This arrangement may therefore constitute a shared structural element that guides the assembly of many immunoreceptor complexes. The DAP12-NKG2C trimeric structure reported here provides an illustration of how intramembrane protein interactions can determine the molecular architecture of a multisubunit transmembrane receptor.
Results
Covalently stabilized transmembrane peptide complexes for NMR
Establishing a system that would allow us to obtain a structure of an assembled DAP12-receptor complex presented considerable technical challenges. Receptors that associate with DAP12 are both type I membrane proteins (amino terminus extracellular) and type II membrane proteins (amino terminus cytosolic)1,16 that adopt either parallel or antiparallel orientations relative to DAP12, and both orientations are probably possible when free peptides are combined in detergent micelles. Furthermore, titration of free peptide onto DAP12TM dimer would produce a mixture of assembled and unassembled peptides, introducing an additional level of complexity. We focused our efforts on the transmembrane sequence of the type II NKG2C protein13 because its antiparallel orientation relative to DAP12 afforded us the opportunity to engineer a covalently stabilized, stoichiometric three-transmembrane peptide complex (Fig. 1a). CD94 does not participate in intramembrane assembly, and transmembrane-derived peptides are sufficient for stable association of NKG2C with DAP12 (ref. 6). We therefore constructed a covalent trimeric complex by fusion of NKG2CTM to the carboxy-terminal end of one DAP12TM strand through an optimized flexible linker sequence (Supplementary Fig. 1) and covalent capture of the second DAP12 strand through a native disulfide bond in the amino-terminal extracellular stalk region. In the pure disulfide-linked DAP12TM-DAP12TM-NKG2CTM trimeric peptide product, the transmembrane domain of NKG2C would be expected to assemble with DAP12 by folding back along the helix dimer in its natural antiparallel orientation (Fig. 1a, right).
Figure 1.
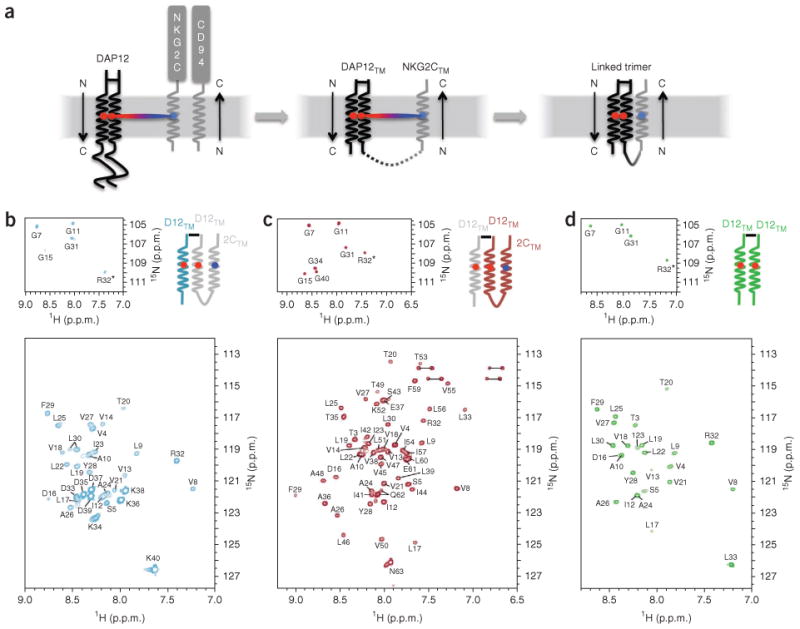
Construct design and labeling strategy. (a) Assembly of DAP12 with the NKG2C-CD94 heterodimer in the membrane (left); assembly of DAP12TM with NKG2CTM (middle; NKG2C ectodomain and CD94 omitted as they do not participate in the transmembrane assembly); and covalent peptide construct representing the membrane-embedded portion of the trimolecular complex (right), (b-d) The tr-HSQC spectra of trimer samples segmentally labeled with 15N-2H on the DAP12TM (D12TM)-only strand (b) or on the DAP12TM-NKG2CTM (2CTM) strand (c), and of the DAP12TM homodimer alone (d), recorded for samples prepared in 250 mM tetradecylphosphocholine with 25 mM SDS in 20 mM phosphate buffer, pH 6.8, and recorded at 600 MHz and 303 K. Top row, separate view of glycine region. Amino acid positions are noted with single-letter designations and position number. Data are representative of at least three experiments each.
A key feature of our design was that the two DAP12TM segments in the trimer, which were identical in sequence and thus indistinguishable by NMR in a uniformly labeled sample, were produced from separate fusion proteins and could therefore be independently labeled with different stable-isotope mixtures (Fig. 1b,c). We collected transverse relaxation-optimized17 1H-15N-labeled heteronuclear single-quantum coherence (tr-HSQC) spectra for two trimer samples that were identical in amino acid sequence but labeled with 15N exclusively on the DAP12TM-only strand (Fig. 1b) or the DAP12TM-NKG2CTM strand (Fig. 1c). The decrease in spectroscopic complexity achieved with this segmental labeling strategy was critical for the unambiguous assignment of backbone and side-chain proton resonances. We also produced a DAP12TM peptide homodimer alone (tr-HSQC; Fig. 1d) using methods similar to those described before for structural studies of the transmembrane portion of the TCR-associated ζζ transmembrane dimer18 to allow separate NMR analysis and structural comparisons with the trimer. For structure determination, we constructed a panel of segmentally labeled trimer samples to assign backbone and side-chain proton resonances and collect nuclear Overhauser enhancement (NOE)-based distance restraints on the two segments separately and we then combined the restraints for determination of the intact structures.
Structure of the DAP12TM-DAP12TM-NKG2CTM complex
As predicted, the NKG2CTM sequence adopted a helical conformation and packed in an antiparallel orientation along the lateral surface of the DAP12TM-DAP12TM dimer (Fig. 2a and Supplementary Fig. 2a; structure statistics, Table 1). The DAP12TM-DAP12TM interface was approximately symmetrical and placed two polar residues, Asp16 and Thr20, near the interface (Fig. 2a). Little surface complementarity existed between NKG2CTM and the DAP12TM-DAP12TM dimer, although the interface extended along the entire length of the receptor transmembrane helix and constituted a total contact surface area of 537±21 Å2. This is consistent with the ability of DAP12 to assemble with many different receptor transmembrane domain sequences. A broad and shallow groove formed by the dimer interface accommodated the receptor transmembrane helix, and the side chains of Tyr28 and Phe29 from DAP12TM created an aromatic base that further restrained the position of the cytosolic end of the NKG2CTM helix. It has been shown that substitution of the transmembrane aspartic acid residue in DAP12 with alanine causes a profound defect in assembly with NKG2C and other receptors6,12,13. Unexpectedly, we found that substitution of the threonine residue at position 20 with alanine caused a similar defect in assembly of the full-length proteins in an in vitro translation-based assembly assay3 (Fig. 2b, T20A) even though the critical aspartic acid residues remained unchanged. In contrast, the simultaneous substitution of three glycine residues that form two tandem glycophorin A-like GXXXG motifs (where ‘X’ is any amino acid)19–21 with bulkier residues in DAP12 caused no measurable defects in assembly (Fig. 2b, 3GL and 3GF), which effectively ruled out the possibility of a role for this common helix association motif in the assembly of DAP12-NKG2C. The receptor transmembrane helix packs loosely into the space created by the small Thr20 and Ala24 side chains, and substitution of the alanine at position 24 with phenylalanine indeed caused appreciably less receptor assembly due to steric incompatibility (Fig. 2b, A24F). Notably, these features represent the general requirements for the assembly of DAP12 with many different receptors, as the same substitutions produced similar effects on assembly with KIR2DS3, a type I receptor of the immunoglobulin superfamily22 (Fig. 2c) and with versions of both of these receptors in which all transmembrane residues except the critical lysine residue were replaced with leucine12 (data not shown).
Figure 2.
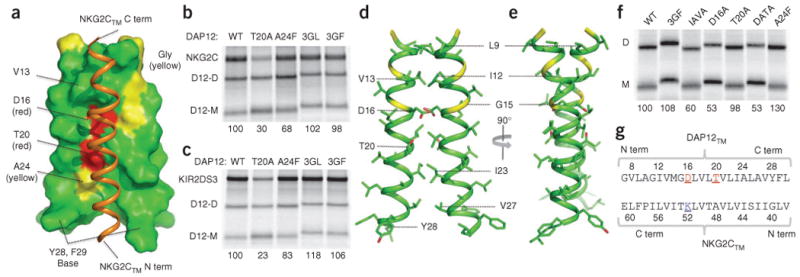
Structure of the DAP12TM-DAP12TM-NKG2CTM complex, (a) One structure from the ensemble of the fifteen trimer structures of lowest energy (helical portions only), showing surface features of the DAP12TM dimer (all NKG2CTM (orange ribbon) side chains omitted for clarity); amino acid and position included for regions of interest (bundle views, sample NOE strips and assigned methyl spectra, Supplementary Figs. 2 and 3). C term, carboxyl terminus; N term, amino terminus. (b,c) In vitro translation–based assay of the effects of DAP12 transmembrane substitutions on its assembly with NKG2C (b) or KIR2DS3 (c), assessed with 35S-labeled proteins extracted in 0.5% digitonin and immunoprecipitated with monoclonal antibody to the hemagglutinin tag (DAP12). Numbers below lanes indicate assembly efficiency, calculated as the ratio of receptor to DAP12 dimer and presented relative to assembly efficiency with wild-type DAP12 (set as 100%). WT, wild-type; T20A, substitution of alanine for threonine at position 20; A24F, substitution of phenylalanine for alanine at position 24; 3GL and 3GF, triple substitution of leucine (3GL) or phenylalanine (3GF) for Gly7, Gly11 and Gly15; D12-D, DAP12 dimer; D12-M, DAP12 monomer. Data are from at least two experiments with similar results (mean). (d,e) Two views of one structure from the ensemble of fifteen DAP12TM dimer structures (helical portions only, from Gly7 to Leu30): red, side-chain oxygen atoms; yellow, glycine residues at positions 7, 11 and 15, as in a (bundle views, sample NOE strips and assigned methyl spectra, Supplementary Figs. 2 and 3). (f) In vitro translation analysis of DAP12 transmembrane substitutions in homodimer formation as in b,c. IAVA, double substitution of alanine for isoleucine at position 12 and alanine for valine at position 13; D16A, substitution of alanine for aspartic acid at position 16; DATA, double substitution of alanine for aspartic acid at position 16 and alanine for threonine at position 20. Numbers below lanes indicate assembly efficiency, calculated as the ratio of DAP12 dimer to DAP12 monomer and presented relative to assembly efficiency with wild-type DAP12 (set as 100%). Data are from at least two experiments with similar results (mean). (g) Alignment of human DAP12 and NKG2C transmembrane sequences; numbers indicate amino acid positions in the engineered NMR constructs. Letter color and underlining: red, electronegative; blue, electropositive.
Table 1. NMR and refinement statistics for protein structures.
| DAP12TM-DAP12TM | DAP12TM-DAP12TM-NKG2CTM | |
|---|---|---|
| NMR-derived constraints | ||
| NOE | 172 | 238 |
| Intrahelical | 146 | 211 |
| Interhelical | 26 | 27 |
| Hydrogen bondsa | 80 | 108 |
| Dihedral anglesa | 92 | 134 |
| ϕ | 46 | 67 |
| ψ | 46 | 67 |
| Structure statisticsb | ||
| Violations (mean ± s.d.) | ||
| Distance constraints (Å) | 0.048 ± 0.004 | 0.057 ± 0.002 |
| Dihedral angle constraints (°) | 1.747 ± 0.400 | 1.569 ± 0.299 |
| Deviations from idealized geometry | ||
| Bond length (Å) | 0.004 ± 0.000 | 0.004 ± 0.000 |
| Bond angle (°) | 0.575 ± 0.025 | 0.564 ± 0.006 |
| Improper (°) | 0.377 ± 0.022 | 0.393 ± 0.016 |
| Coordinate precision (Å)c | ||
| Heavy | 0.754 | 0.890 |
| Backbone | 0.595 | 0.615 |
| Ramachandran plot statistics (%)d | ||
| Most favored | 97.6 | 97.4 |
| Additionally allowed | 2.4 | 2.0 |
| Generally allowed | 0.0 | 0.6 |
| Disallowed | 0.0 | 0.0 |
Backbone hydrogen bond and dihedral restraints of the α-helix are applied for regions confirmed to be α-helical according to local NOE pattern and characteristic chemical shifts (assessed with the TALOS software package for predicting polypeptide secondary structure30).
Statistics are calculated and averaged over an ensemble of the 15 structures with lowest energy.
The precision of the atomic coordinates is defined as the average r.m.s. difference between the 15 final structures and their mean coordinates. Unstructured residues of the DAP12TM dimer (1–6 and 31–33) and the DAP12TM-DAP12TM-NKG2CTM trimer (1–6, 31–37 and 61–63) were excluded.
The plot statistics are as evaluated with the PROCHECK program for protein structure validation, with exclusion of unstructured regions as in the coordinate precision values above (footnote c).
Structural comparison of DAP12TM and ζζTM homodimers
We separately determined the structure of a DAP12TM-DAP12TM homodimer without a receptor for comparison with the trimeric complex and the published ζζTM structure18. This structure had a packing arrangement similar to that in the trimer (main features, Fig. 2d,e). Favorable van der Waals contacts at positions Leu9, Ile12-Val13 and Leu19-Thr20 accounted for most of the buried surface area in the amino-terminal portion of the dimer interface, and the helices diverged slightly below Leu19-Thr20. The total contact area at the dimer interface was 440±9 Å2. Notably, substitution of the threonine at position 20 with alanine had no adverse effect on the formation of the DAP12 dimer (Fig. 2f, T20A); together with the disruptive effects of this substitution on assembly with receptor transmembrane domains, this suggests that Thr20 has a direct role in the polar interaction with lysine in the trimeric DAP12-receptor assembly. We aligned the native DAP12TM and NKG2CTM sequences (in their natural antiparallel orientation) for clarification (Fig. 2g). A methionine at position 14 in the DAP12TM sequence that was replaced with valine to avoid secondary cleavage during cyanogen bromide treatment is located distant from the dimer and trimer interfaces, and this substitution had no measurable effect on assembly in vitro (data not shown).
DAP12 and ζζ show little amino acid sequence homology and associate with nonoverlapping sets of activating receptors1, yet they activate the same ITAM-dependent signaling pathways2 and follow similar rules for association with their respective receptor partners. Despite the apparent biochemical similarities in assembly, the di-aspartate site of DAP12TM was unique in several ways (Fig. 3a,b). First, it was situated in a position in the membrane distinct from its position in ζζTM. Second, the aspartic acid side chains adopted different conformations in the two structures. Third, a hydroxyl-bearing side chain, in addition to the aspartic acid, seemed to be involved in the contact of DAP12TM with the receptor. A notable feature of the ζζTM-ζζTM homodimer structure is that one or more water molecules are near the aspartic acid side chains18, and we have proposed that water contributes to the hydrogen-bonding network that stabilizes the acidic groups in the interface18. The Thr20 hydroxyl in DAP12TM was oriented toward the aspartic acid side chain one turn above (Fig. 3c) so that it was able to form a hydrogen bond with one of the carboxyl oxygens (and possibly backbone carbonyl oxygen) of Asp16 and may therefore have a role similar to that of water in the ζζTM structure. We cannot, however, rule out the possibility of the presence of water in the DAP12TM interface, because the Thr20 hydroxyl proton has a chemical shift very similar to that of water.
Figure 3.
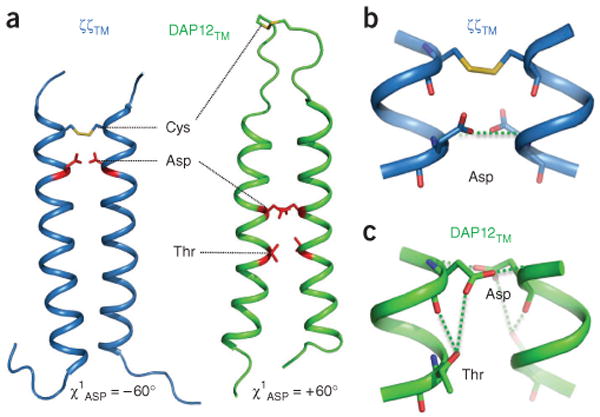
Structural comparison of ζζTM and DAP12TM receptor-binding sites, (a) En face view of ribbon structures showing the relative locations of disulfide bonds and side chains that participate in the receptor-binding sites in the ζζTM-ζζTM dimer (blue ribbons; Protein Data Bank, 2HAC) and DAP12TM-DAP12TM dimer (green ribbons); all other side chains omitted for clarity. Numbers below indicate χ1 values for aspartic acid side chains. (b,c) Enlargement of the receptor-binding sites of homodimers of ζζTM (b) and DAP12TM (c); green dotted lines indicate putative hydrogen bonds.
Structure and conservation at the assembly site
The structure of the critical contact site in the trimer (Fig. 4) suggested the mechanism by which Thr20 participates in receptor assembly. This side-chain hydroxyl is ideally positioned to stabilize Asp16 in the +60° rotamer through intrahelical hydrogen bonds (Fig. 4), and in some conformations the lysine side chain may directly contact Thr20 through an additional interhelical hydrogen bond. In addition to being highly conserved in vertebrate DAP12 sequences (Fig. 5a), a conserved hydroxyl-bearing side chain is also present at position +4 (relative to the aspartic acid) in the NK cell signaling module DAP10 (Fig. 5b) and in the CD3δ and CD3ε chains associated with the TCR (Fig. 5c). Substitution of the serine residue at this position in DAP10 resulted in more than 60% less assembly with the NK cell–activating receptor NKG2D (Fig. 5d), which suggested an arrangement similar to that observed in the assembly of DAP12 with NKG2C. Furthermore, replacement of threonine with alanine in transmembrane domain of either CD3δ or CD3ε caused considerable defects in the assembly of TCRα-CD3δ-CD3ε, and the double-mutant CD3δ-CD3ε dimer had almost no detectable propensity to assemble with TCRα (Fig. 5e). Notably, CD3γ has a glutamic acid in place of aspartic acid and does not have a threonine at position +4 but instead has a serine at position +3 (Fig. 5c). As noted in published studies3, the association of TCRβ with CD3γ-CD3ε was also weaker than the association of TCRα with CD3δ-CD3ε (Fig. 5e,f). Despite those differences, the assembly of TCRβ-CD3γ-CD3ε was similarly sensitive to substitutions of the hydroxyl-bearing residues in the transmembrane domain of either CD3ε or CD3γ (Fig. 5f), which confirmed a role for these side chains in both TCR-CD3 trimeric complexes. These data indicate that many important immunoreceptors assemble around this polar five-amino acid transmembrane motif, which suggests the possibility that electrostatic networks similar to the one reported here for the DAP12-NKG2C trimer are relevant for many immunoreceptor complexes (Fig. 6).
Figure 4.
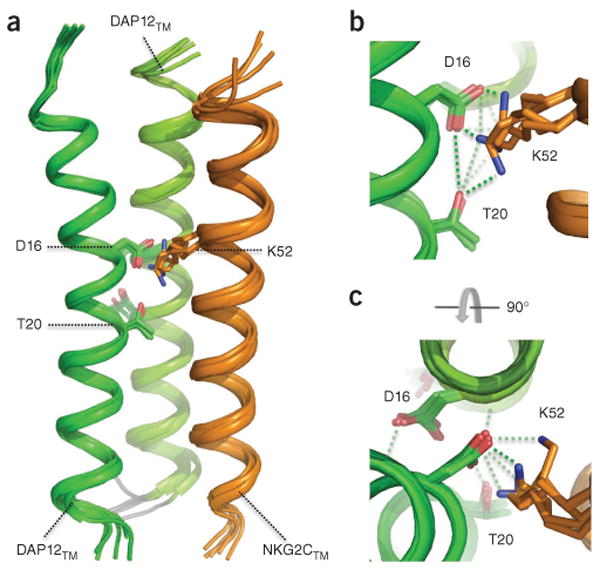
Structure of the electrostatic network at the DAP12TM-DAP12TM-NKG2CTM binding site, (a) Bundle of five selected ribbon structures of the DAP12TM-DAP12TM-NKG2CTM covalent trimer showing possible configurations of the critical binding site (all other side chains omitted for clarity). (b,c) Enlarged views of the electrostatic network at Asp16-Thr20-Lys52, presented in the same orientation as in a (b) and presented in an axial view from above (c); green dotted lines indicate putative hydrogen bonds.
Figure 5.
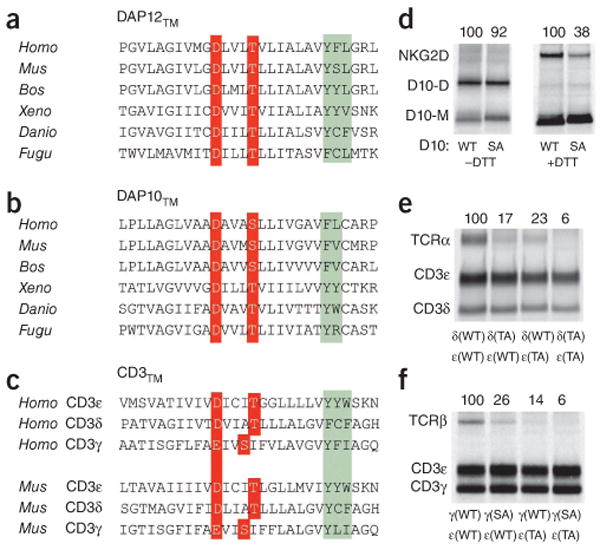
A similar electrostatic network governs the assembly of NKG2C-DAP12, NKG2D-DAP10 and TCR-CD3 complexes, (a-c) Sequence alignment of vertebrate DAP12TM (a) and DAP10TM (b) and human and mouse CD3TM (c); red, acidic and hydroxyl-bearing residues involved in receptor binding; green, conserved and semiconserved aromatic residues that suggest further similarities in helix packing. Homo, Homo sapiens; Mus, Mus musculus; Bos, Bos taurus; Xeno, Xenopus laevis; Danio, Danio rerio; Fugu, Fugu rubripes. (d-f) In vitro analysis of the effect of substitution of alanine for serine or threonine in the transmembrane domains on the assembly of DAP10 with NKG2D (d), TCRα with CD3δ-CD3ε (e) or TCRβ with CD3γ-CD3ε (f), assessed as described above (Fig. 2), with monoclonal antibody to hemagglutinin (DAP10; d) or to CD3 (UCH-T1; e,f) used for immunoprecipitation. (d) Formation of disulfide-linked dimers of wild-type DAP10 (D10: WT) and mutant DAP10 with serine-to-alanine substitution (D10: SA), assessed in nonreducing conditions (without dithiothreitol (–DTT); left) or reducing conditions (because oxidized NKG2D runs as a diffuse band that is difficult to quantify; +DTT; right). (e,f) δ(WT), ε(WT) or γ(WT), wild-type CD3δ, CD3ε or CD3γ; δ(TA) or ε(TA), mutant CD3δ or CD3ε with threonine-to-alanine substitution; γ(SA), mutant CDγ with serine-to-alanine substitution. Numbers above lanes indicate assembly efficiency, calculated as the ratio of DAP10 dimer to DAP10 monomer (b, left), NKG2D to total DAP10 (b, right) or TCR to CD3ε (c) and presented relative to assembly efficiency with wild-type DAP12 (set as 100%). Data are from at least three experiments with similar results (mean).
Figure 6.
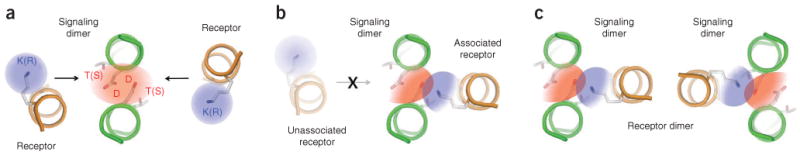
Proposed model for intramembrane immunoreceptor complex assembly. (a) Dimeric signaling modules (CD3, DAP10 or DAP12; green coils) exist as metastable intermediates in the endoplasmic reticulum membrane, whereas newly synthesized receptor subunits (orange coils) are either incorporated into complexes or rapidly degraded4,27. The electrostatic network in the signaling dimer (red shading) shows a symmetrical electron distribution and may have multiple opportunities to assemble with basic residues (blue shading) from receptor subunits. K(R), lysine or arginine; T(S), threonine or serine, (b) Once a receptor subunit has associated with an available binding site, an asymmetric redistribution of electronegativity in the network (color gradients) may render the opposite side of the signaling module unable to bind a second receptor. This trimeric assembly therefore represents the fundamental structural unit that organizes immunoreceptor complexes, (c) In a step that is either subsequent to or simultaneous and cooperative with that in b, receptors that form dimers (such as TCR, NKG2D and the mouse Ly49 family) combine two or more trimeric units to form the final complex. The relative orientation of the individual trimeric units cannot be determined from structural or biochemical data available at present.
Discussion
The genetically dissociated ligand-binding and signal-transducing modules of most activating immunoreceptors assemble through polar contacts among their transmembrane α-helices1,4. We sought to understand how the particular arrangement of one basic residue and two acidic residues creates a stable and specific contact site deep in the lipid bilayer that directs the assembly of many important receptor complexes. DAP12 is a particularly interesting example because the structure it forms must support stable association with many different transmembrane sequences that have only a single lysine residue in common10,12. These receptors represent both type I and type II membrane proteins, which associate in parallel or antiparallel orientations relative to the transmembrane helices of DAP12. The structures presented here have shown how the DAP12 transmembrane sequence creates both a specific homodimer interface that supports the proper arrangement of polar residues for receptor binding and a composite surface that enables stable association of receptor transmembrane helices in a manner that is largely indifferent to amino acid sequence context. The finding that conserved glycophorin A–like GXXXG motifs in the transmembrane domains of DAP12 did not participate in intracomplex helix packing was unexpected. This sequence forms the core of a highly stable transmembrane helix interface in the native, noncovalent glycophorin A dimer19,20, and similar configurations have been observed in many helical membrane proteins23,24 and model peptides generated semi-randomly21. However, our structures and in vitro assembly data demonstrated that all glycine residues in the DAP12 dimer interface were outside the surfaces that mediated helix-helix contact in the trimeric complex. Given the frequency with which this motif is observed at helix-helix interfaces21,24, it is possible that these sequences participate in other transmembrane interactions that contribute to receptor function. Our structures therefore provide a guide for future mutagenesis studies testing the hypothesis that immunoreceptor activation involves structural rearrangements transmitted through the transmembrane domains.
Aside from substitution of the critical aspartic acid residues, the most striking effect of DAP12TM substitution on receptor assembly was that assembly was diminished by 70% or more when the threonine at position 20 was replaced with alanine. Thr20 does not contribute hydrogen bonds across the DAP12TM dimer interface, and published studies have established that the properly placed receptor transmembrane lysine is necessary and sufficient for assembly with DAP12 (ref. 12). Thus, Thr20 does not require contacts with NKG2CTM sequences other than the lysine side chain, and other receptors show the same sensitivity to substitutions at this site. These results indicate a role for Thr20 in the lysine–aspartic acid interaction. On the basis of the structure of this region in both the dimer and trimer, we propose that the role of Thr20 could be to stabilize the Asp16 side chain in a position that favors contact with the lysine amino group. In the trimer, the degree of conformational variability observed in the bundle of low-energy structures also leaves open the possibility that the lysine head group accesses Thr20 directly, and these possibilities are not mutually exclusive. Inspection of other signaling module transmembrane sequences showed that the DXXXT/S motif (where ‘T/S’ indicates threonine or serine) in DAP12 was also conserved in DAP10, CD3δ and CD3ε, which suggested that this extended polar network is an important element in all of these assemblies. Indeed, a serine-to-alanine substitution in DAP10 resulted in a defect in its assembly with NKG2D similar to that observed for DAP12-NKG2C. Analysis of TCR-CD3 interactions demonstrated that removal of even a single hydroxyl was sufficient to disrupt the interactions on both sides of the CD3δ-CD3ε-TCRα-TCRβ-CD3γ-CD3ε hexameric complex. On the basis of the DAP12TM-DAP12TM-NKG2CTM structure as a model of TCR-CD3 assembly, we propose that placement of the serine residue in the CD3γ transmembrane domain at position +3 instead of position +4 could position this residue within the trimeric interface, where it may participate more directly in the Asp-Thr-Lys interaction. However, experimental confirmation of this model will require further structural analysis. Consistent with the similarity of the requirements for intramembrane assembly of all of these transmembrane domains, both NKG2C and NKG2D transmembrane peptides can assemble with CD3 hetero-dimers in vitro. However, this is prevented in the full-length receptor proteins because of steric incompatibility of the ectodomains12. All human DAP12-associating receptors have lysine residues in their transmembrane domains, whereas the DAP10-associated NKG2D has arginine at the analogous position, and exchanging these basic residues is not well tolerated12. Together these features provide a high degree of selectivity for the assembly of NKG2C with DAP12 rather than with DAP10 or other signaling modules.
Biochemical studies of intramembrane receptor assembly have consistently shown that only one receptor transmembrane domain associates with a dimeric signaling module, yet there is a strict requirement for two transmembrane aspartic acid residues in dimeric signaling modules for the formation of the assembly-competent state3,6,8,12,18. Although our structures showed that only one of the two aspartic acid residues in the DAP12TM dimer interface was in a position to directly contact the receptor lysine, we conclude that all five polar residues must be considered members of a continuous electrostatic network that forms the core of the trimolecular transmembrane helix assembly. As shown by our mutagenesis analyses, this network is profoundly altered by substitution of even one carboxylate or one hydroxyl group. The addition of the basic group after assembly with a receptor transmembrane domain may cause an asymmetric redistribution of electronegativity on the two faces of the signaling dimer, which could explain why receptor assembly on one side renders the opposing face unable to bind a second receptor transmembrane domain. An example of such an effect can be found in the catalytic dyad of the aspartyl protease of human immunodeficiency virus type 1, in which two chemically identical active-site aspartic acid residues (one from each monomer at the homodimer interface) have different apparent acid dissociation constant values of approximately 3 and 6 (ref. 25). In this example, the distribution of electronegativity in one acidic group is altered by a change in the ionization state of a nearby group, and such effects could be even more important in the absence of bulk water and competing ions in the lipid bilayer interior26. DAP12 associates with more than a dozen different receptors in cells of the human lymphoid and myeloid lineages and almost two dozen receptors in mice16, and our data suggest that most if not all DAP12-associated receptors use similar structural features for assembly. Given the biochemical evidence demonstrating that TCR-CD3 and NKG2D-DAP10 also assemble with similar transmembrane sequence motifs, the structure of the membrane-embedded DAP12-NKG2C trimolecular complex reported here may provide an instructive model for many intramembrane-assembling receptors.
Methods
Methods and any associated references are available in the online version of the paper at http://www.nature.com/natureimmunology/.
Supplementary Material
Acknowledgments
We thank S.C. Blacklow (Harvard Medical School) for pMM-LR6 vector; M.J. Call and members of the Chou lab for reading the manuscript and discussions; and J. Pyrdol for assistance with initial construct design and expression trials. Supported by the Helen Hay Whitney Foundation (M.E.C.), the Charles A. King Trust (M.E.C.) and the US National Institutes of Health (R01AI054520 to K.W.W., R01HL084329 to J.J.C. and EB002026 to the Center for Magnetic Resonance at the Massachusetts Institute of Technology where NMR data were collected).
Footnotes
Accession codes. UCSD-Nature Signaling Gateway (http://www.signaling-gateway.org): A000749, A001666, A000750 and A001665; Protein Data Bank: DAP12-DAP12 homodimer, 2L34; DAP12-DAP12-NKG2C heterotrimer, 2L35.
Note: Supplementary information is available on the Nature Immunology website.
Author Contributions: M.E.C., K.W.W. and J.J.C. conceived of the study; M.E.C. designed and did all biochemical experiments, produced transmembrane peptide constructs and prepared NMR samples; M.E.C. and J.J.C. collected and analyzed NMR data and calculated structures; M.E.C. wrote the paper; and all authors contributed to editing of the manuscript.
Competing Financial Interests: The authors declare no competing financial interests.
Reprints and permissions information is available online at http://npg.nature.com/reprintsandpermissions/.
References
- 1.Call ME, Wucherpfennig KW. Common themes in the assembly and architecture of activating immune receptors. Nat Rev Immunol. 2007;7:841–850. doi: 10.1038/nri2186. [DOI] [PubMed] [Google Scholar]
- 2.Humphrey MB, Lanier LL, Nakamura MC. Role of ITAM-containing adapter proteins and their receptors in the immune system and bone. Immunol Rev. 2005;208:50–65. doi: 10.1111/j.0105-2896.2005.00325.x. [DOI] [PubMed] [Google Scholar]
- 3.Call ME, Pyrdol J, Wiedmann M, Wucherpfennig KW. The organizing principle in the formation of the T cell receptor-CD3 complex. Cell. 2002;111:967–979. doi: 10.1016/s0092-8674(02)01194-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Call ME, Wucherpfennig KW. The T cell receptor: critical role of the membrane environment in receptor assembly and function. Annu Rev Immunol. 2005;23:101–125. doi: 10.1146/annurev.immunol.23.021704.115625. [DOI] [PubMed] [Google Scholar]
- 5.Exley M, Terhorst C, Wileman T. Structure, assembly and intracellular transport of the T cell receptor for antigen. Semin Immunol. 1991;3:283–297. [PubMed] [Google Scholar]
- 6.Feng J, Garrity D, Call ME, Moffett H, Wucherpfennig KW. Convergence on a distinctive assembly mechanism by unrelated families of activating immune receptors. Immunity. 2005;22:427–438. doi: 10.1016/j.immuni.2005.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kuster H, Thompson H, Kinet JP. Characterization and expression of the gene for the human Fc receptor γ subunit. Definition of a new gene family. J Biol Chem. 1990;265:6448–6452. [PubMed] [Google Scholar]
- 8.Garrity D, Call ME, Feng J, Wucherpfennig KW. The activating NKG2D receptor assembles in the membrane with two signaling dimers into a hexameric structure. Proc Natl Acad Sci USA. 2005;102:7641–7646. doi: 10.1073/pnas.0502439102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu J, et al. An activating immunoreceptor complex formed by NKG2D and DAP10. Science. 1999;285:730–732. doi: 10.1126/science.285.5428.730. [DOI] [PubMed] [Google Scholar]
- 10.Lanier LL, Corliss BC, Wu J, Leong C, Phillips JH. Immunoreceptor DAP12 bearing a tyrosine-based activation motif is involved in activating NK cells. Nature. 1998;391:703–707. doi: 10.1038/35642. [DOI] [PubMed] [Google Scholar]
- 11.Tomasello E, et al. Gene structure, expression pattern, and biological activity of mouse killer cell activating receptor-associated protein (KARAP)/DAP-12. J Biol Chem. 1998;273:34115–34119. doi: 10.1074/jbc.273.51.34115. [DOI] [PubMed] [Google Scholar]
- 12.Feng J, Call ME, Wucherpfennig KW. The assembly of diverse immune receptors is focused on a polar membrane-embedded interaction site. PLoS Biol. 2006;4:e142. doi: 10.1371/journal.pbio.0040142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lanier LL, Corliss B, Wu J, Phillips JH. Association of DAP12 with activating CD94/NKG2C NK cell receptors. Immunity. 1998b;8:693–701. doi: 10.1016/s1074-7613(00)80574-9. [DOI] [PubMed] [Google Scholar]
- 14.Braud VM, et al. HLA-E binds to natural killer cell receptors CD94/NKG2A, B and C. Nature. 1998;391:795–799. doi: 10.1038/35869. [DOI] [PubMed] [Google Scholar]
- 15.Lanier LL. Up on the tightrope: natural killer cell activation and inhibition. Nat Immunol. 2008;9:495–502. doi: 10.1038/ni1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lanier LL. DAP10- and DAP12-associated receptors in innate immunity. Immunol Rev. 2009;227:150–160. doi: 10.1111/j.1600-065X.2008.00720.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pervushin K, Riek R, Wider G, Wuthrich K. Attenuated T2 relaxation by mutual cancellation of dipole-dipole coupling and chemical shift anisotropy indicates an avenue to NMR structures of very large biological macromolecules in solution. Proc Natl Acad Sci USA. 1997;94:12366–12371. doi: 10.1073/pnas.94.23.12366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Call ME, et al. The structure of the ζζ transmembrane dimer reveals features essential for its assembly with the T cell receptor. Cell. 2006;127:355–368. doi: 10.1016/j.cell.2006.08.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lemmon MA, Treutlein HR, Adams PD, Brunger AT, Engelman DM. A dimerization motif for transmembrane α-helices. Nat Struct Biol. 1994;1:157–163. doi: 10.1038/nsb0394-157. [DOI] [PubMed] [Google Scholar]
- 20.MacKenzie KR, Prestegard JH, Engelman DM. A transmembrane helix dimer: structure and implications. Science. 1997;276:131–133. doi: 10.1126/science.276.5309.131. [DOI] [PubMed] [Google Scholar]
- 21.Russ WP, Engelman DM. The GxxxG motif: a framework for transmembrane helix-helix association. J Mol Biol. 2000;296:911–919. doi: 10.1006/jmbi.1999.3489. [DOI] [PubMed] [Google Scholar]
- 22.Vilches C, Parham P. KIR: diverse, rapidly evolving receptors of innate and adaptive immunity. Annu Rev Immunol. 2002;20:217–251. doi: 10.1146/annurev.immunol.20.092501.134942. [DOI] [PubMed] [Google Scholar]
- 23.Senes A, Engel DE, DeGrado WF. Folding of helical membrane proteins: the role of polar, GxxxG-like and proline motifs. Curr Opin Struct Biol. 2004;14:465–479. doi: 10.1016/j.sbi.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 24.Senes A, Gerstein M, Engelman DM. Statistical analysis of amino acid patterns in transmembrane helices: the GxxxG motif occurs frequently and in association with beta-branched residues at neighboring positions. J Mol Biol. 2000;296:921–936. doi: 10.1006/jmbi.1999.3488. [DOI] [PubMed] [Google Scholar]
- 25.Trylska J, et al. Thermodynamic linkage between the binding of protons and inhibitors to HIV-1 protease. Protein Sci. 1999;8:180–195. doi: 10.1110/ps.8.1.180. 10.1110/ps.8.1.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smith R, Brereton IM, Chai RY, Kent SB. Ionization states of the catalytic residues in HIV-1 protease. Nat Struct Biol. 1996;3:946–950. doi: 10.1038/nsb1196-946. [DOI] [PubMed] [Google Scholar]
- 27.Bonifacino JS, Cosson P, Klausner RD. Colocalized transmembrane determinants for ER degradation and subunit assembly explain the intracellular fate of TCR chains. Cell. 1990;63:503–513. doi: 10.1016/0092-8674(90)90447-m. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


