Abstract
Histone deacetylases (HDACs) are part of a vast family of enzymes that have crucial roles in numerous biological processes, largely through their repressive influence on transcription. The expression of many HDAC isoforms in eukaryotic cells raises questions about their possible specificity or redundancy, and whether they control global or specific programmes of gene expression. Recent analyses of HDAC knockout mice have revealed highly specific functions of individual HDACs in development and disease. Mutant mice lacking individual HDACs are a powerful tool for defining the functions of HDACs in vivo and the molecular targets of HDAC inhibitors in disease.
The distinctive patterns of gene expression that are associated with specialized embryonic and adult cell types, as well as the modulation of specific gene programmes in response to physiological and pathological signalling, require multiple levels of transcriptional control. This control occurs through transcriptional regulators that bind specific DNA sequences, leading to the modification of chromatin structure, which in turn controls the accessibility of DNA to regulatory factors. The main factor influencing chromatin structure is the state of amino-acid residues within histone tails, which serve as targets for a variety of reversible post-translational modifications that modulate nucleosome structure and gene transcription, both positively and negatively1,2. Acetylation, one of the most widespread modifications of histones, serves as a key modulator of chromatin structure and gene transcription, and provides a mechanism for coupling extracellular signals with the genome by regulated acetylation and deacetylation3.
Histone acetylation modulates transcription in multiple ways. Acetylation of ε-amino groups of lysine residues within histone tails neutralizes their positive charge, thereby relaxing chromatin structure. This interferes with the generation of higher-order chromatin structures, and increasing the accessibility of transcription factors to their target genes4. Acetylated histones also serve as binding sites for bromodomain proteins, which often act as transcriptional activators. Conversely, histone deacetylation favours transcriptional repression by allowing for chromatin compaction5. Direct acetylation and deacetylation of transcription factors has also been shown to have positive and negative consequences on gene transcription, respectively6.
Histone acetylation is a dynamic process controlled by the antagonistic actions of two large families of enzymes — the histone acetyltransferases (HATs) and the histone deacetylases (HDACs). The balance between the actions of these enzymes serves as a key regulatory mechanism for gene expression and governs numerous developmental processes and disease states.
The vast majority of studies of HDAC functions have involved biochemical analyses in vitro, studies in cultured cells with HDAC inhibitors, HDAC knockdown by small interfering RNA (siRNA), or overexpression of HDACs. Although these experiments demonstrate the biochemical functions of HDACs as transcriptional repressors, they are non-predictive of their role in vivo. An important question regarding the functions and mechanisms of action of HDACs in vivo is whether HDACs act primarily to control global changes in the state of chromatin or whether they also have more specific functions in the regulation of key downstream genes and transcriptional programmes. The existence of many HDAC isoforms in eukaryotic cells also raises questions about possible specificity or redundancy of functions. Ongoing human clinical trials that are investigating the use of HDAC inhibitors as a treatment for a variety of disorders mean that it is vital these questions are answered. The recent creation of knockout mice lacking HDAC genes has revealed highly specific functions for individual HDAC isoforms during development and adulthood. These mutant mice are a powerful tool for defining the functions of HDACs in vivo and for identifying the molecular targets of HDAC inhibitors in disease. In this Review, we discuss the developmental and physiological functions of HDACs that are revealed by gene deletions in mice, and how these studies can inform future efforts to exploit HDACs in the settings of human disease.
Control of gene expression by HDACs
HDACs lack intrinsic DNA-binding activity and are recruited to target genes via their direct association with transcriptional activators and repressors, as well as their incorporation into large multiprotein transcriptional complexes2,4. Thus, the specificity of HDACs for regulation of distinct gene programmes depends on cell identity and the spectrum of available partner proteins in a cell, in addition to the signalling milieu of the cell. Although diminished histone acetylation at promoter regions generally correlates with gene silencing, consistent with the well-established functions of HDACs as transcriptional repressors, there is also evidence that HDACs can activate some genes. In yeast, for example, the HDAC Hos2 is required for gene activation, and deletion of the HDAC1 and 2 homologue, Rpd3, leads to repression of transcription at telomeric loci7–9. HDACs have also been linked to transcriptional activation of a subset of genes in higher eukaryotes10 but, in settings in which HDAC inhibition leads to downregulation of specific genes, it is difficult to rule out possible secondary effects that result in transcriptional repression. It should be noted, however, that deletion or inhibition of HDACs often results in the upregulation or down-regulation of approximately equivalent percentages of genes10–12.
It has also become clear in recent years that HDACs can act on numerous cellular substrates in addition to histones, and that acetylation might rival phosphorylation in its importance6. In this regard, class IIa deacetylases possess only minimal HDAC activity against acetylated histones, despite extensive evolutionary conservation of their deacetylase domain, pointing to the possible importance of other types of cellular substrates for their actions13. How these many facets of acetylation and deacetylation are controlled and integrated, and how they influence the expression of specific genes as well as global changes in gene expression, are important issues for the field.
The HDAC superfamily
The HDAC superfamily is vast and ancient, dating back to prokaryotes. Here we focus on the mammalian HDACs and the lessons learned from genetic deletion models.
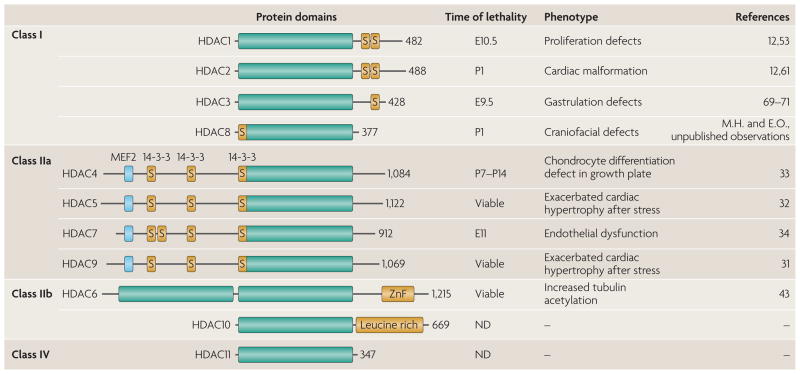
Mammalian genomes encode 11 proteins with a highly conserved deacetylase domain (FIG. 1). These proteins can be classified into four families (class I, IIa, IIb and IV), which differ in structure, enzymatic function, subcellular localization and expression patterns. In addition to these classical HDACs, mammalian genomes encode another group of deacetylases, the sirtuins, which are sometimes referred to as class III HDACs. There have been several recent reviews on the sirtuins14–17, so they will not be covered here.
Figure 1. The histone deacetylase (HDAC) superfamily, showing protein domains, loss-of-function phenotypes in mice and time point of lethality of the knockouts.
Green rectangles indicate the conserved HDAC domain; numbers following the HDAC domain indicate the number of amino acids. Myocyte enhancer factor 2 (MEF2)-binding sites are marked by a blue square, and binding sites for the 14-3-3 chaperone protein are also shown. E, embryonic day; ND, not determined; P, days postnatal; S, serine phosphorylation sites; ZnF, zinc finger.
Class I HDACs
The class I HDAC family consists of HDAC1, 2, 3 and 8, which share homology with Rpd3 — a founding member from budding yeast18–20. These HDACs are expressed ubiquitously, localized predominantly to the nucleus and display high enzymatic activity toward histone substrates. They possess relatively simple structures, consisting of the conserved deacetylase domain with short amino- and carboxy-terminal extensions.
HDAC1 and HDAC2 are nearly identical and are generally found together in repressive complexes such as the sin3, NuRD, CoREST and PRC2 complexes21. HDAC3 is found in distinct complexes such as the N-CoR–SMRT complex, whereas no complex has been described for HDAC8 (REF. 19).
Class IIa HDACs
HDAC4, 5, 7 and 9 belong to the class IIa HDAC family. These HDACs have large N-terminal extensions with conserved binding sites for the transcription factor myocyte enhancer factor 2 (MEF2) and the chaperone protein 14-3-3, which render HDACs signal responsive. Following phosphorylation by kinases, such as calcium/calmodulin-dependent protein kinase (CaMK) and protein kinase D (PKD), these HDACs bind 14-3-3 and shuttle from the nucleus to the cytoplasm22–25. The dissociation of class II HDACs from MEF2 allows the HAT p300 to associate with MEF2 via the HDAC docking site, thereby converting MEF2 from a transcriptional repressor to a transcriptional activator26–30. The regulated phosphorylation of class IIa HDACs provides a mechanism for linking extracellular signals with transcription and has key roles in numerous tissues during development and disease.
In contrast to other HDACs, class IIa HDACs show relatively restricted expression patterns. HDAC5 and HDAC9 are highly enriched in muscles, the heart and brain31,32. HDAC4 is highly expressed in the brain and growth plates of the skeleton33, and HDAC7 is enriched in endothelial cells and thymocytes34 (T-cell precursors derived from the thymus).
The precise mechanism whereby class IIa HDACs repress transcription has not been fully elucidated. Highly purified recombinant class IIa HDACs possess only minimal catalytic activity, and the activity of class IIa HDACs purified from mammalian cells has been shown to be due to contaminating class I HDACs13,35,36. Moreover, MEF2-interacting transcription repressor (MITR), which is a splice variant of HDAC9 that lacks the HDAC domain, is as effective in repression of MEF2-target genes as the full-length HDAC9 protein, indicating that the intrinsic catalytic activity of class IIa HDACs is not required for repression37–39.
The class IIa HDACs have been shown to recruit class I HDACs through their C-terminal HDAC domain, which probably accounts for a portion of their repressive activity35. In addition, the regulatory domains of class IIa HDACs interact with other transcriptional repressors, such as heterochromatin protein 1 (HP1) and C-terminal-binding protein (CTBP)37,38,40. Thus, they function as adaptors to nucleate multiple types of transcriptional regulators and to confer signal-responsiveness to downstream target genes.
Recently, the biochemical basis for the different activities of class I and class IIa HDACs has been elucidated. In the catalytic pocket of most HDACs, an ultra-conserved tyrosine acts as a transition-state stabilizer in the deacetylation reaction41. This tyrosine is changed to a histidine in vertebrate class IIa HDACs, and this conservative amino-acid change reduces the catalytic activity of the vertebrate enzymes more than 1,000-fold36,42. Although this activity is still measurable in vitro, it is unclear if it is of any biological relevance in vivo, especially given the observation that class IIa HDACs do not need their catalytic domain in order to be potent repressors.
Class IIb HDACs
HDAC6 and HDAC10 form the class IIb family. HDAC6 is the main cytoplasmic deacetylase in mammalian cells43, whereas little is known about the functions of HDAC10 (REFS 44,45). Among the targets directly deacetylated by HDAC6 are cytoskeletal proteins such as α-tubulin and cortactin, transmembrane proteins such as the interferon receptor IFNαR, and chaperones46–50. HDAC6 is distinct from all other HDACs, as it harbours two deacetylase domains and a C-terminal zinc finger.
Class IV HDAC
HDAC11 is the sole class IV HDAC. expression of HDAC11 is enriched in the brain, heart, muscle, kidney and testis, but little is known about its function51,52. It is composed of a deacetylase domain that shows homology to class I and II HDAC domains, with small N- and C-terminal extensions.
Roles of class I HDACs in development
The ubiquitous expression, high deacetylase activity towards common substrates and high homology between class I HDACs suggests functional redundancy among these HDACs in vivo. However, deletion of each member of the class I HDAC family in mice leads to lethality in all cases, demonstrating the unique roles of each HDAC in the control of specific gene expression programmes.
HDAC1
HDAC1-null mice die before embryonic day 10.5 (E10.5) and display severe proliferation defects and general growth retardation12,53. Proliferation defects can also be observed in HDAC1-null embryonic stem (ES) cells and are associated with increased expression of the cyclin-dependent kinase inhibitors p21 and p27 (REF. 54). These cells show a significant reduction in total HDAC activity and modest hyperacetylation of histones H3 and H4, indicating that HDAC1 is a major deacetylase in ES cells. surprisingly, 3% of genes are downregulated and ~5% of genes are upregulated in HDAC1-null ES cells54, suggesting that HDAC1 does not function simply as a global repressor of transcription, but instead regulates specific gene programmes by repressing or activating certain promoters.
Deletion of hdac1 in zebrafish causes a variety of lethal defects in skeletal and neuronal elements. The specific target genes responsible for these phenotypes have not been defined, but they seem to be downstream of canonical and non-canonical Wnt signalling55–59. None of the other HDACs has been genetically analysed in detail in zebrafish, although a recent report described disrupted liver development following morpholino- mediated knockdown of hdac3 mRNA (REF. 60). surprisingly, conditional deletion of HDAC1 in tissues such as the heart, brain, skeletal muscle and smooth muscle is well tolerated in mice12, although this is probably due to redundancy with HDAC2 in later development and postnatal life (see below)12.
HDAC2
There is disagreement regarding the function of HDAC2 in vivo. One study found that HDAC2-null mice die within the first 24 hours after birth with severe cardiac malformations, including obliteration of the lumen of the right ventricle owing to excessive proliferation of cardiomyocytes, as well as bradycardia12 (FIG. 2). By contrast, other studies have reported that mice harbouring a lacZ insertion in Hdac2, which is purported to create a null mutation, are viable61. The basis for these conflicting results is unclear. It is possible that different genetic backgrounds account for this difference. Alternatively, the lacZ insertion allele might be ‘leaky’ and allow adequate expression of HDAC2 for viability, a phenomenon that has previously been described for gene trap approaches62.
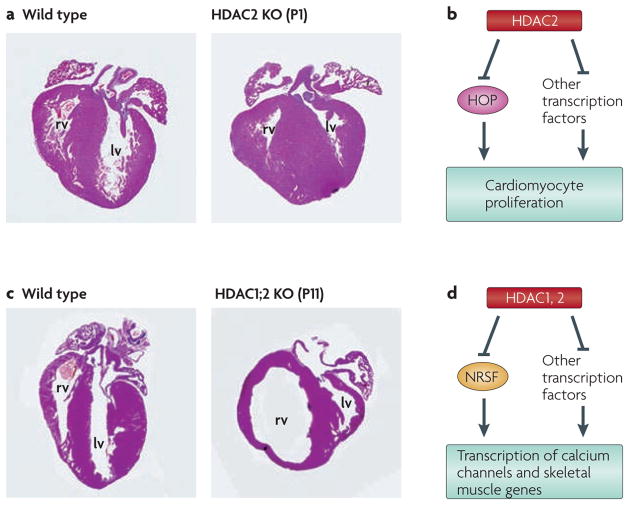
Figure 2. Control of heart development by histone deacetylase 1 (HDAC1) and HDAC2.
a | Histological sections of hearts from wild type and HDAC2 knockout (KO) mice at postnatal day 1 (P1). Note the excessive number of cardiomyocytes in the mutant heart, which fill the chambers of the left ventricle (lv) and right ventricle (rv). b | Schematic of the role of HDAC2 in the repression of cardiomyocyte proliferation through inhibition of homeodomain-only protein (HOP). c | Histological sections of hearts from wild-type mice and mice with a cardiac deletion of HDAC1 and 2 at P11. Note the dilatation of the right ventricle in the mutant, which is indicative of heart failure. d | Schematic of the redundant roles of HDAC1 and 2 in regulation of calcium channel and skeletal muscle genes in cardiomyocytes via repression of neuron-restrictive silencer factor (NRSF) and other transcription factors. Parts a and c are reproduced, with permission, from REF. 12 © (2007) Cold Spring Harbor Laboratory Press.
The transcriptional targets of HDAC2 in the heart remain to be fully defined. However, the homeodomain-only protein (HOP), which functions as a positive and negative regulator of cardiomyocyte proliferation, has been shown to interact with HDAC2 (FIG. 2). Deletion of HOP also results in hyperproliferation of developing cardiomyocytes, suggesting that HDAC2 and HOP reside in a transcriptionally repressive complex to regulate cardiac proliferation and differentiation during development63–65.
Redundant roles of HDAC1 and 2 in cardiac growth and development
Conditional null alleles for class I HDACs have permitted an analysis of their functions in specific tissues, bypassing the early lethality associated with global gene deletion. Given the lethal phenotypes resulting from global deletion of HDAC1 and HDAC2, it was surprising to find that deletion of either HDAC1 or HDAC2 in a variety of tissues, including the heart, brain, endothelial cells, smooth muscle, and neural crest cells did not yield obvious phenotypes. By contrast, deletion of both genes together results in severe phenotypes in all tissues examined, pointing to redundant functions of these HDACs during later development and adulthood12.
Conditional deletion of HDAC1 and 2 together in the cardiac lineage has shown that a single wild-type allele of either gene is sufficient to support normal development, whereas deletion of all HDAC1 and 2 alleles results in neonatal lethality, accompanied by cardiac arrhythmias, dilated cardiomyopathy, and upregulation of genes encoding skeletal muscle-specific contractile proteins and calcium channels in the heart12 (FIG. 2). The earlier that HDAC1 and HDAC2 are deleted, the more dramatic the phenotype. In the heart, deletion of HDAC1 and HDAC2 at E8.5 causes lethality 2 days later, whereas mice with a cardiac deletion at E10.5 survived for more than 3 weeks. Transcriptional analysis in these animals revealed that only 1.6% of transcripts were upregulated, and that deletion of HDAC1 and HDAC2 in the heart derepressed specific gene programmes involved in Ca2+ ion handling and in contractility. Cardiac expression of multiple fetal calcium channels is transcriptionally regulated by neuron-restrictive silencer factor (NRSF) through the recruitment of both class I and class IIa HDACs66,67. A dominant negative mutant of NRSF that is unable to bind repressors results in activation of the fetal gene programme, arrhythmogenesis and sudden death68. Thus, loss of HDAC1 and HDAC2 allows for the loss of repression by NRSF and other transcription factors, resulting in aberrant transcriptional activity of genes involved in calcium flux and contractility, leading to cardiac arrhythmia and sudden death.
HDAC3
HDAC3 mutant mice die before E9.5 owing to defects in gastrulation69–71. The target genes responsible for this early phenotype are unknown, although loss of Hdac3 seems to be associated with defective DNA double-stranded break repair70. Conditional deletions of HDAC3 have so far been described for the liver and heart. Loss of HDAC3 in the liver disrupts lipid and cholesterol homeostasis, leading to an accumulation of lipids and a decrease in glycogen storage71. These changes are caused by derepression of a gene programme that usually is under the control of nuclear hormone receptors such as the thyroid hormone receptor and peroxisome proliferator-activated receptor gamma (PPARγ), which control key steps in lipid and cholesterol biosynthesis early in the postnatal liver. Only minor increases could be observed in bulk histone acetylation and on the promoters of dysregulated genes, indicating that other class I HDACs are also likely to play a part in liver homeostasis.
Deletion of HDAC3 in cardiomyocytes also led to a dramatic upregulation of ligand-induced lipid storage in the heart69. These mice survive until 3–4 months of age, at which point they show massive cardiac hypertrophy and derepression of genes that control fatty-acid uptake and metabolism. In the heart, these gene programmes are under the control of the nuclear receptor PPARα, and derepression by loss of HDAC3 leads to abnormalities that mimic the metabolic derangements observed in diabetic cardiomyopathies. Furthermore, loss of HDAC3 in the heart results in robust interstitial fibrosis, which is phenotypically independent of rampant PPARα activity. However, it is currently unknown whether the transcription factors that regulate the fibrotic gene programme are directly repressed by HDAC3. Overexpresion of HDAC3 in the heart leads to increased thickness of the myocardium, which is due to increased cardiomyocyte hyperplasia without hypertrophy72.
Class IIa HDACs in development and physiology
Each of the four class IIa HDACs have been deleted in mice and, although each gene seems to be dedicated to specific programmes of tissue-specific gene expression, commonalities between the different loss-of-function phenotypes point to similar mechanisms of action. Many of these modes of action reflect the repressive influence of these HDACs on the expression and function of the MEF2 transcription factor, as well as their signal responsiveness. Importantly, there is a high degree of redundancy between the class IIa HDACs. It is thus possible that each tissue has a hard-wired threshold for class IIa HDAC repression, and that the observed phenotypes reflect the cell types or gene programmes that are most sensitive to MEF2 and HDAC activity.
Regulation of skeletogenesis by HDAC4
HDAC4 has a central role in the formation of the skeleton33. Most of the bones in the vertebrate skeleton are formed from a cartilaginous template in which chondrocytes undergo hypertrophy, which is followed by apoptosis. Thereafter, osteoblasts, blood vessels and other cell types invade and produce the mature bone matrix73. HDAC4 is expressed in prehypertrophic chondrocytes in vivo, and mice with a global deletion of HDAC4 die during the first week of life owing to ectopic ossification of endochondral cartilage, which prevents expansion of the rib cage and leads to an inability to breathe (FIG. 3). This lethal phenotype is accompanied by precocious and ectopic hypertrophy of chondrocytes, resulting in the conversion of cartilaginous skeletal elements to ossified bone. Runt related transcription factor 2 (RUNX2) and the MEF2C transcription factor, which interact with HDAC4, have vital roles in the control of chondrocyte hypertrophy and bone formation74. In the absence of HDAC4, transcriptional activation of these factors is unrestrained, leading to excessive bone formation33. Consistent with this mechanism, forced expression of RUNX2 or a constitutively active form of MEF2 in developing chondrocytes mimics the HDAC4 loss-of-function phenotype75. Conversely, forced expression of a signal-resistant mutant form of HDAC4 in chondrocytes in vivo inhibits chondrocyte hypertrophy and differentiation (FIG. 3).
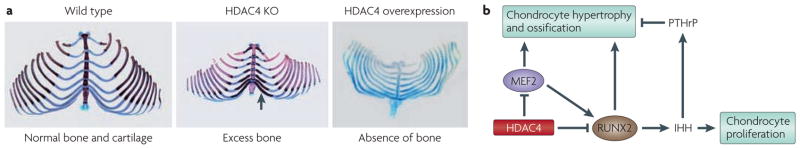
Figure 3. Control of chondrocyte hypertrophy by histone deacetylase 4 (HDAC4).
a | Ribs from neonatal mice stained for bone (red) and cartilage (blue). Deletion of HDAC4 results in ossification of cartilage by the arrowhead), whereas overexpression of HDAC4 in the cartilage of transgenic mice prevents ossification. b | Schematic of the repressive influence of HDAC4 on myocyte enhancer factor 2 (MEF2) and runt related transcription factor 2 (RUNX2) in the pathway for chondrocyte proliferation and hypertrophy. IHH, Indian hedgehog; KO, knockout; PTHrP, parathyroid hormone-related peptide. Part a is reproduced, with permission, from REF. 33 © (2007) Cell Press.
Thus, by repressing the activity of MEF2C and RUNX2 in developing chondrocytes, HDAC4 is able to delay chondrocyte hypertrophy and thereby control the timing and extent of ossification of endochondral bones (FIG. 3). MEF2 directly regulates the expression of extracellular matrix protein genes, such as collagen type X alpha 1, and of vascular endothelial growth factor (VEGF), which is required for angiogenesis in the late stages of chondrocyte development75. In addition, RUNX2 activates the expression of the secreted growth factor Indian hedgehog (IHH), which has a number of functions in endochondral bone development. These functions are mediated by enhancing chondrocyte proliferation and stimulating the synthesis of parathyroid hormone-related peptide (PTHrP), which in turn inhibits differentiation of prehypertrophic to hypertrophic chondrocytes76. RUNX2 expression is also controlled by MEF2, and RUNX2 is a target for regulation by HDAC4 (REF. 75).
Control of cardiovascular growth and function by HDAC5 and 9
Mice lacking either HDAC5 or HDAC9 are viable, whereas compound mutant mice lacking both HDAC5 and 9 show a propensity for lethal ventricular septal defects and thin-walled myocardium, which typically arise from abnormalities in growth and maturation of cardiomyocytes32. Given the interaction between class IIa HDACs and MEF2, and the central role of MEF2 in the control of cardiomyocyte differentiation, the developmental cardiac defects in these double mutant mice probably result from super-activation of MEF2. This would be expected to lead to precocious differentiation and cell-cycle withdrawal of cardiomyocytes, causing hypocellularity of the myocardium. In addition, class IIa HDACs participate in multiprotein complexes and modulate the activities of numerous other transcription factors involved in myocardial growth, such as the serum response factor, myocardin and calmodulin binding transcription activator 2 (CAMTA2)77. Thus, the absence of HDAC5 and 9 probably perturbs the precisely coordinated gene expression programmes required for myocyte differentiation, proliferation and morphogenesis that underlie heart formation.
The adult heart typically responds to stress by a pathological growth response that ultimately leads to loss of cardiac function78–80. MEF2 is sufficient and necessary to drive the pathological cardiac hypertrophy and heart failure that takes place in response to injury81. HDAC5 and 9 have redundant roles in the suppression of cardiac growth in response to stress signalling. Mice lacking either HDAC5 or 9 are hypersensitive to cardiac stress resulting from excess workload or neurohumoral signalling (FIG. 4). These stimuli typically activate the calcineurin and CaMK–PKD pathways, which in turn lead to phosphorylation of class IIa HDACs, promoting their nuclear export82. Deletion of class IIa HDACs eliminates the counter-regulatory mechanism that restrains cardiac growth and sensitizes MEF2 and perhaps other transcription factors so that they become activated by stress-dependent intracellular signals.
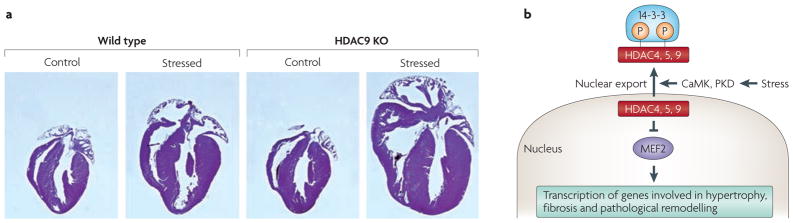
Figure 4. Control of pathological cardiac hypertrophy by class IIa histone deacetylases (HDACs).
a | Histological sections of hearts from wild-type and HDAC9 knockout (KO) adult mice. Mice were subjected to cardiac stress by expression of a cardiac-specific transgene encoding activated calcineurin, which drives pathological hypertrophy. Note that HDAC9 knockout mice have normal hearts in the absence of stress, but display cardiomegaly in response to stress, owing to loss of the growth-inhibitory function of HDAC9. b | Schematic of the repressive influence of class IIa HDACs on myocyte enhancer factor 2 (MEF2) and pathological cardiac remodelling. Stress-inducible kinases, such as calcium/calmodulin-dependent protein kinase (CaMK) and protein kinase D (PKD), induce the phosphorylation of class IIa HDACs, which creates docking sites for the 14-3-3 chaperone protein, resulting in nuclear export with consequent activation of MEF2 and its downstream target genes, which are involved in cardiac remodelling. Part a is reproduced, with permission, from REF. 31 (2007) © Cell Press.
Functions of class IIa HDACs in skeletal muscle
Numerous functions for class IIa HDACs have been described in skeletal muscle. Skeletal muscle fibres differ in their contractile and metabolic properties, which reflect different patterns of gene expression83. Slow-twitch, or type I, myofibres exhibit an oxidative metabolism, are rich in mitochondria, are heavily vascularized and are resistant to fatigue. By contrast, fast-twitch, or type II, myofibres exhibit glycolytic metabolism, are involved in rapid bursts of contraction and fatigue rapidly. The calcium-dependent protein kinases CaMK and PKD have been implicated in the transduction of calcium signals that upregulate the expression of oxidative, slow fibre-specific genes in skeletal muscle84. MEF2 is a target for calcium signalling in skeletal muscle and is a key regulator of the slow myofibre phenotype. This function of MEF2 is mediated through its regulation by class IIa HDACs: in slow myofibres, class IIa HDACs are selectively degraded by the proteasome and MEF2 exerts a transcriptional activation function. Among the target genes of MEF2 in slow myofibres are type IIx myosin heavy chain, myosin light chain 2, slow troponin I and myoglobin. In support of this mode of function of class IIa HDACs in skeletal muscle, genetic deletion of class IIa HDACs in this tissue derepresses MEF2 and results in conversion of fast fibres to slow fibres85 (FIG. 5).
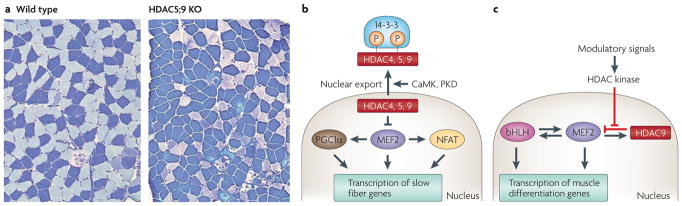
Figure 5. Control of slow myofibre gene expression by class IIa histone deacetylases (HDACs).
a | Histological sections of soleus muscle from wild-type and HDAC5;9 double mutant knockout (KO) mice stained for type I myosin heavy chain, a marker of type I slow myofibres. Note the increase in slow myofibres after deletion of class IIa HDACs. b | Schematic of the repressive influence of class IIa HDACs on myocyte enhancer factor 2 (MEF2), which acts together with PGC-1α (peroxisome proliferator-activated receptor gamma, coactivator 1 alpha) and NFAT (nuclear factor of activated T-cells) to promote the formation of slow myofibres. Signalling by calcium/calmodulin-dependent protein kinase (CaMK) and protein kinase D (PKD) induces the phosphorylation of class IIa HDACs, which creates docking sites for the 14-3-3 chaperone protein, resulting in nuclear export with consequent activation of slow myofibre genes. c | A MEF2-dependent negative feedback loop for the control of HDAC9 expression during muscle differentiation. Myogenic basic helix-loop-helix (bHLH) transcription factors activate the expression of MEF2, which then amplifies and sustains the expression of myogenic bHLH genes. Myogenic bHLH factors and MEF2 also cooperate to activate skeletal muscle differentiation genes. In addition, MEF2 activates the expression of HDAC9, which in turn represses MEF2 activity. Signals that influence myogenesis activate HDAC kinases and thereby repress HDAC9 activity, providing a ‘rheostat’ mechanism for the control of myogenesis. Part a is reproduced from REF. 85.
HDAC9 has also been shown to modulate the response of skeletal muscle to motor innervation. Electrical activity from motor neurons represses the expression of many muscle genes, including those encoding acetylcholine receptor subunits. In response to denervation, these genes are derepressed, resulting in hypersensitivity of the muscle fibre to acetylcholine. Mice lacking HDAC9 are extremely sensitive to denervation-induced changes in gene expression, whereas mice that overexpress HDAC9 in skeletal muscle are rendered insensitive to the effects of denervation86.
In addition to being a signal-responsive modulator of gene transcription, the expression of HDAC9 is tightly modulated. A highly conserved MEF2-binding site in the proximal promoter of the HDAC9 gene drives the expression of HDAC9, thereby establishing a negative feedback loop in which MEF2 drives the expression of its own repressor87 (FIG. 5). This feedback loop is thought to provide robustness and fine-tuning to the gene programmes controlled by HDAC9 and MEF2, and to provide a myogenic ‘rheostat’ that modulates muscle differentiation in response to extracellular cues.
Control of endothelial function by HDAC7
During embryogenesis, HDAC7 is specifically expressed in the endothelial cells that form the inner lining of the cardiovascular system34. Genetic deletion of HDAC7 in mice results in embryonic lethality, owing to a loss of integrity of endothelial-cell interactions and consequent rupture of blood vessels and haemorrhaging (FIG. 6). Vascular disruption in HDAC7-null mice is accompanied by upregulation of matrix metalloproteinase 10 (MMP10), an endoprotease that is secreted by endothelial cells and that degrades the extracellular matrix, thereby perturbing endothelial-cell and smooth muscle-cell interactions. The inappropriate expression of MMP10 can be traced to its regulation by MEF2; in the absence of HDAC7, MEF2 activity is elevated, leading to pathological levels of MMP10 (FIG. 6). Concurrently, tissue inhibitor of metalloproteinase 1 (TIMP1) is downregulated in endothelial cells, presumably as a secondary consequence of vascular demise. The downregulation of TIMP1 in the face of enhanced expression of MMP10 would be expected to further exacerbate vascular destruction34.
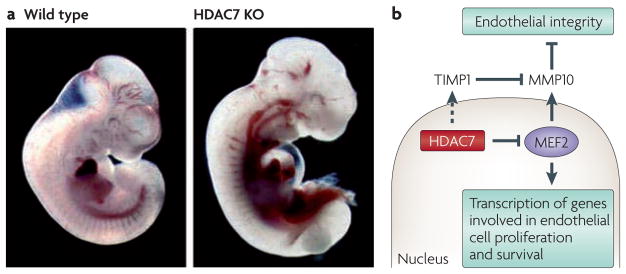
Figure 6. Control of endothelial integrity by histone deacetylase 7 (HDAC7).
a | Wild-type and HDAC7 knockout (KO) embryos at embryonic day 10.5 (E10.5). The absence of HDAC7 results in vascular rupture, pericardial oedema and haemorrhaging throughout the mutant embryos. b | Schematic of the role of HDAC7 in maintenance of vascular integrity. HDAC7 is expressed specifically in endothelial cells, where it represses the activity of myocyte enhancer factor 2 (MEF2). In the absence of HDAC7, MEF2 activity is elevated, resulting in upregulation of matrix metalloproteinase 10 (MMP10) and degradation of cell–cell interactions required for vascular integrity. Deletion of HDAC7 also leads to downregulation of tissue inhibitor of metalloproteinase 1 (TIMP1), presumably through indirect mechanisms, which further enhances MMP10 activity.
Part a is reproduced, with permission, from REF. 34 © (2007) Cell Press.
The involvement of HDAC7 in the control of MMP10 expression and vascular integrity has potentially important implications for a variety of human disorders. Vascular leakage causes circulatory collapse and contributes to the pathogenesis of numerous usually life-threatening diseases, such as atherosclerosis and aneurysm. Moreover, the imbalance between MMP and TIMP activity has been shown to profoundly influence vascular integrity following myocardial infarction and during tumour angiogenesis88,89. Hence, strategies to maintain the repressive influence of HDAC7 on MEF2 — for example, through inhibition of the kinase cascades that lead to the phosphorylation of HDAC7 and its dissociation from MEF2 — would be expected to be beneficial with respect to maintaining vascular integrity.
Control of cytoskeletal dynamics by HDAC6
Mice with a deletion of HDAC6 are the only HDAC mutant animals published so far that do not have an obvious phenotype. However, they do display a dramatic increase in acetylated tubulin, in line with the notion that HDAC6 is the main tubulin deacetylase43. Given the large body of experimental in vitro evidence that clearly shows that HDAC6 has important functions in modulating the misfolded protein response and cytoskeletal dynamics, the lack of phenotype in the HDAC6 mutants is somewhat surprising. It is possible that redundancy with HDAC10 can explain the lack of in vivo phenotype.
Therapeutic actions of HDAC inhibitors
The involvement of histone acetylation and deacetylation in so many aspects of development and tissue homeostasis might suggest that systemic inhibition of HDACs with pharmacologic inhibitors would result in nonspecific and catastrophic effects as a consequence of global derepression of gene expression. Thus, it is striking that systemic HDAC inhibition with compounds that broadly inhibit most or all HDACs is well tolerated in vivo and blocks numerous disease-associated gene expression programmes in a seemingly specific manner.
Given the dramatic phenotypes that result from HDAC gene deletions, why are HDAC inhibitors so well tolerated in vivo? we propose three explanations, which are not mutually exclusive. First, a genetic deletion of an HDAC results in the complete absence of the enzyme, whereas inhibitors do not result in complete inhibition of activity. Second, a genetic deletion of an HDAC eliminates the gene product permanently, whereas the actions of an inhibitor are transient. Third, and perhaps most importantly, HDACs participate in multiprotein transcriptional complexes. Genetic deletion of an HDAC perturbs the complexes in which it would normally be associated, whereas inhibitors are believed to block enzymatic activity without necessarily disrupting the repressive complex.
Classical HDAC inhibitors such as trichostatin A (TSA) or suberoylanilide hydroxamic acid (SAHA) are mostly ‘pan–HDAC’ inhibitors; that is, they block, with similar affinities, the activity of all isoforms except class IIa HDACs. For example, the IC50 values for SAHA are: HDAC1 = 37.1 nM; HDAC3 = 44.6 nM; and HDAC6 = 40.9 nM90. Given that different HDAC isoforms govern dramatically different gene expression programmes in development and disease, it seems plausible that isoform-selective inhibitors should lead to improved efficacy and drug safety. The recent mechanistic insights into the biochemistry of class IIa HDACs should also be taken into account36,41,91. Many screening studies used class IIa HDACs purified from mammalian cells for the development of class IIa isoform-specific inhibitors and, surprisingly, compounds were identified that blocked class IIa but not class I activity. These compounds probably function as small-molecule inhibitors of protein–protein interactions and not as bona fide HDAC inhibitors92. As these molecules are entering clinical trials, it is important to realize that they might show biological properties distinct from classical HDAC inhibitors.
HDAC inhibitors from multiple chemical classes have entered clinical trials, and SAHA (marketed as Vorinostat, brand name Zolinza) has been approved for treatment of cutaneous manifestations of advanced, refractory T-cell lymphoma in a select group of patients93. The exact mechanism for the effect of HDAC inhibitors on tumour cells is currently unknown, and numerous explanations, such as changes in gene transcription, direct induction of apoptosis, production of reactive oxygen species and induction of cell-cycle arrest, have been proposed92,94–96. The specific HDAC isoforms that mediate this antiproliferative effect also remain to be clearly identified. Genetic deletion of HDAC3 leads to cell-cycle dependent DNA damage coupled with defective double-stranded break repair70. HDAC3-null cells are thus sensitized to ionizing radiation, a phenomenon that has also been observed with HDAC inhibitors97. Therefore, some of the effects observed with HDAC inhibition might be mediated via HDAC3, although the involvement of other isoforms can not be ruled out98. The existence of conditional alleles for all class I HDACs might allow the creation of transformed cancer cell lines with conditional alleles for all the different class I HDAC isoforms (and their combinations), which would make a systematic analysis of HDAC requirement in cancer cells possible. These studies could then be extended by crossing conditional HDAC-null alleles into tumour-prone genetic backgrounds.
One of the most perplexing aspects of HDAC biology is that pharmacological inhibition of HDAC activity provides a therapeutic benefit in such a wide variety of disease states. TABLE 1 gives an overview of the disease states in which HDAC inhibition has been shown to be beneficial as well as the proposed mechanisms involved. These range from infectious and immunological diseases to traumatic shock, and from cardiac hypertrophy to neurodegenerative disease99–105, and in certain circumstances HDAC inhibitors are even able to ‘cure’ genetic disease in humans106. Although a unifying theory explaining how reduced deacetylase activity is beneficial in such diverse pathophysiological states is currently unknown, it is tempting to speculate that most of these diseases have an epigenetic component (that is, aberrant histone acetylation), and that treatment with HDAC inhibitors resets the epigenetic memory of the cell to a pre-disease state.
Table 1.
Clinical and experimental use of histone deacetlyase (HDAC) inhibitors in diverse disease states
| Disease | Proposed mechanism | Refs |
|---|---|---|
| Humans | ||
| HIV infection | De-silencing of latent virus | 99 |
| Cutaneous T-cell lymphoma | Upregulation of tumor-suppressor genes, induction of apoptosis | 108, 109 |
| GPI deficiency | Increased PIGM expression by hyper-acetylation of histones on promoter | 106 |
| Ulcerative colitis | Inhibition of NF-κB activation | 110 |
| Sickle cell disease | Increase in fetal haemoglobin expression | 111 |
| Mice | ||
| Arthritis | Inhibition of TNFα expression and of inflammation | 112 |
| Asthma | Inhibition of cytokine expression and T-cell infiltration | 113 |
| Autoimmune encephalitis | Upregulation of antioxidant, antiexcitotoxicity and proneuronal factors | 114 |
| Colitis | Suppression of pro-inflammatory cytokines | 115 |
| Cardiac hypertrophy | Unknown | 102, 116 |
| Dementia | Dendrite sprouting, increased synapse number | 100 |
| Graft versus host disease | Reduction of pro-inflammatory cytokines | 101, 117 |
| Hepatitis | Inhibition of TNFα and INFγ | 118 |
| Muscular dystrophy | Induction of follistatin | 119 |
| Systemic lupus erythematosus | Downregulation of pro-inflammatory cytokines | 120 |
| Spinal muscular atrophy | Activation of survival motor neuron 2 gene | 121 |
| Rats | ||
| Haemorrhagic shock | Reduction of TNFα expression | 122 |
| Brain trauma | Inhibiting neuroinflammation | 104 |
GPI, glycosylphosphatidylinositol; INFγ, interferon gamma, NF-κB, nuclear factor kappa B; PIGM, phosphatidylinositol glycan anchor biosynthesis, class M; TNFα, tumour-necrosis factor alpha.
Issues for the future
Although the interest in HDAC biology has intensified with the successful introduction of HDAC inhibitors in the clinical setting, we are still far from understanding the intricacies of protein acetylation. The number of identified acetylated non-histone proteins is rapidly increasing, raising questions regarding whether phenotypes resulting from HDAC gene deletion or from pharmacological inhibition reflect changes in chromatin structure and transcription, or if they reflect hyperacetylation of non-histone proteins. In the cases of non-histone substrates, it will be important to identify the proteins and understand how acetylation influences their actions. It also remains to be determined whether different HDAC isoforms have specific targets in vivo, as suggested by the specific phenotypes resulting from genetic deletion of individual HDACs. It is expected that unbiased profiling of hyperacetylated proteins in knockout or inhibitor-treated cells using mass spectrometry approaches will be a valuable tool in answering these questions. A recent study in human cancer cells showed the feasibility of this approach107.
Given the variety of pre-clinical studies in which HDAC inhibitors have shown a therapeutic benefit, another major challenge will be to decipher the role of individual HDACs in specific disease processes and to develop isoform-specific HDAC inhibitors. Because HDACs can affect multiple targets, it will be important to develop inhibitors that selectively block the pathological actions of HDACs. Solving these challenges will most likely broaden the therapeutic window and possibly lead to the clinical application of HDAC inhibitors in a variety of non-oncological disease states.
Acknowledgments
We apologize to the many authors in the field whose work we were not able to cite because of space constraints. Research in the Olson laboratory has been supported by grants from the National Institutes of Health, the D.W. Reynolds Clinical Cardiovascular Research Center, the Robert A. Welch Foundation and the Sandler Foundation for Asthma Research. M.H was supported by a grant from the Deutsche Forschungsgemeinschaft (DFG, HA 3335/2-1).
Glossary
- Transition state
A particular state corresponding to the highest energy in a chemical reaction
- Morpholino
A stable antisense oligonucleotide that is commonly used in zebrafish and Xenopus laevis to inhibit either the translation or splicing of mRNAs
- Gene trap
A mutation strategy that uses insertion vectors to trap or isolate transcripts from flanking genes
- Gastrulation
The process in animal embryos in which the endoderm and mesoderm move from the outer surface of the embryo to the inside, where they give rise to the internal organs
- Interstitial fibrosis
Proliferation of fibroblasts resulting in increased collagen production and consequent organ dysfunction
- Chondrocyte
The type of cell that produces and maintains the cartilaginous matrix
- Osteoblast
The type of cell that is responsible for bone formation by secreting and mineralizing the bone matrix
- Ossification
The process of bone formation in which soft connective tissue is converted into mineralized tissue
- Endochondral bone
The parts of the skeleton which form by endochondral ossification, a process in which cartilage is replaced by bone.
- Rheostat
A module for fine tuning a molecular circuit
- IC50
Inhibition concentration 50%. The concentration of inhibitor that is required to inhibit 50% of the activity of an enzyme compared with an unhibited control.
Footnotes
Competing interests statement
The authors declare competing financial interests; see web version for details.
DATABASES
UniProtKB: http://www.uniprot.org
HDAC1 | HDAC2 | HDAC3 | HDAC4 | HDAC5 | HDAC6 | HDAC7 | HDAC8 | HDAC9 | HDAC10 | HDAC11 | MEF2
FURTHER INFORMATION
Olson laboratory homepage:
http://www4.utsouthwestern.edu/olsonlab
ALL LINKS ARE ACTIVE IN THE ONLINE PDf
References
- 1.Cheung WL, Briggs SD, Allis CD. Acetylation and chromosomal functions. Curr Opin Cell Biol. 2000;12:326–333. doi: 10.1016/s0955-0674(00)00096-x. [DOI] [PubMed] [Google Scholar]
- 2.Grunstein M. Histone acetylation in chromatin structure and transcription. Nature. 1997;389:349–352. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- 3.Clayton AL, Hazzalin CA, Mahadevan LC. Enhanced histone acetylation and transcription: a dynamic perspective. Mol Cell. 2006;23:289–296. doi: 10.1016/j.molcel.2006.06.017. [DOI] [PubMed] [Google Scholar]
- 4.Shahbazian MD, Grunstein M. Functions of site-specific histone acetylation and deacetylation. Annu Rev Biochem. 2007;76:75–100. doi: 10.1146/annurev.biochem.76.052705.162114. [DOI] [PubMed] [Google Scholar]
- 5.Ruthenburg AJ, Li H, Patel DJ, Allis CD. Multivalent engagement of chromatin modifications by linked binding modules. Nature Rev Mol Cell Biol. 2007;8:983–994. doi: 10.1038/nrm2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kouzarides T. Acetylation: a regulatory modification to rival phosphorylation? EMBO J. 2000;19:1176–1179. doi: 10.1093/emboj/19.6.1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rundlett SE, et al. HDA1 and RPD3 are members of distinct yeast histone deacetylase complexes that regulate silencing and transcription. Proc Natl Acad Sci USA. 1996;93:14503–14508. doi: 10.1073/pnas.93.25.14503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang A, Kurdistani SK, Grunstein M. Requirement of Hos2 histone deacetylase for gene activity in yeast. Science. 2002;298:1412–1414. doi: 10.1126/science.1077790. [DOI] [PubMed] [Google Scholar]
- 9.De Nadal E, et al. The MAPK Hog1 recruits Rpd3 histone deacetylase to activate osmoresponsive genes. Nature. 2004;427:370–374. doi: 10.1038/nature02258. [DOI] [PubMed] [Google Scholar]
- 10.Nusinzon I, Horvath CM. Histone deacetylases as transcriptional activators? Role reversal in inducible gene regulation. Sci STKE. 2005;296:re11. doi: 10.1126/stke.2962005re11. [DOI] [PubMed] [Google Scholar]
- 11.Glaser KB, et al. Gene expression profiling of multiple histone deacetylase (HDAC) inhibitors: defining a common gene set produced by HDAC inhibition in T24 and MDA carcinoma cell lines. Mol Cancer Ther. 2003;2:151–163. [PubMed] [Google Scholar]
- 12.Montgomery RL, et al. Histone deacetylases 1 and 2 redundantly regulate cardiac morphogenesis, growth, and contractility. Genes Dev. 2007;21:1790–1802. doi: 10.1101/gad.1563807. Reports the first conditional knockout of a class I HDAC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lahm A, et al. Unraveling the hidden catalytic activity of vertebrate class IIa histone deacetylases. Proc Natl Acad Sci USA. 2007;104:17335–17340. doi: 10.1073/pnas.0706487104. Describes the mechanism for the loss of enzymatic activity in class IIa HDACs. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schwer B, Verdin E. Conserved metabolic regulatory functions of sirtuins. Cell Metab. 2008;7:104–112. doi: 10.1016/j.cmet.2007.11.006. [DOI] [PubMed] [Google Scholar]
- 15.Longo VD, Kennedy BK. Sirtuins in aging and age-related disease. Cell. 2006;126:257–268. doi: 10.1016/j.cell.2006.07.002. [DOI] [PubMed] [Google Scholar]
- 16.Guarente L. Sirtuins as potential targets for metabolic syndrome. Nature. 2006;444:868–874. doi: 10.1038/nature05486. [DOI] [PubMed] [Google Scholar]
- 17.Bordone L, Guarente L. Calorie restriction, SIRT1 and metabolism: understanding longevity. Nature Rev Mol Cell Biol. 2005;6:298–305. doi: 10.1038/nrm1616. [DOI] [PubMed] [Google Scholar]
- 18.Taunton J, Hassig CA, Schreiber SL. A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p. Science. 1996;272:408–411. doi: 10.1126/science.272.5260.408. A seminal paper describing the cloning of the first mammalian HDAC. [DOI] [PubMed] [Google Scholar]
- 19.Yang XJ, Seto E. The Rpd3/Hda1 family of lysine deacetylases: from bacteria and yeast to mice and men. Nature Rev Mol Cell Biol. 2008;9:206–218. doi: 10.1038/nrm2346. An elegant review about the classical HDAC family. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kurdistani SK, Grunstein M. Histone acetylation and deacetylation in yeast. Nature Rev Mol Cell Biol. 2003;4:276–284. doi: 10.1038/nrm1075. [DOI] [PubMed] [Google Scholar]
- 21.Yang XJ, Seto E. Collaborative spirit of histone deacetylases in regulating chromatin structure and gene expression. Curr Opin Genet Dev. 2003;13:143–153. doi: 10.1016/s0959-437x(03)00015-7. [DOI] [PubMed] [Google Scholar]
- 22.Vega RB, et al. Protein kinases C and D mediate agonist-dependent cardiac hypertrophy through nuclear export of histone deacetylase 5. Mol Cell Biol. 2004;24:8374–8385. doi: 10.1128/MCB.24.19.8374-8385.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McKinsey TA, Zhang CL, Lu J, Olson EN. Signal-dependent nuclear export of a histone deacetylase regulates muscle differentiation. Nature. 2000;408:106–111. doi: 10.1038/35040593. The first study to show that class IIa HDACs are signal-responsive modulators of gene expression. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lu J, McKinsey TA, Nicol RL, Olson EN. Signal-dependent activation of the MEF2 transcription factor by dissociation from histone deacetylases. Proc Natl Acad Sci USA. 2000;97:4070–4075. doi: 10.1073/pnas.080064097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Passier R, et al. CaM kinase signaling induces cardiac hypertrophy and activates the MEF2 transcription factor in vivo. J Clin Invest. 2000;105:1395–1406. doi: 10.1172/JCI8551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu J, McKinsey TA, Zhang CL, Olson EN. Regulation of skeletal myogenesis by association of the MEF2 transcription factor with class II histone deacetylases. Mol Cell. 2000;6:233–244. doi: 10.1016/s1097-2765(00)00025-3. [DOI] [PubMed] [Google Scholar]
- 27.Youn HD, Grozinger CM, Liu JO. Calcium regulates transcriptional repression of myocyte enhancer factor 2 by histone deacetylase 4. J Biol Chem. 2000;275:22563–22567. doi: 10.1074/jbc.C000304200. [DOI] [PubMed] [Google Scholar]
- 28.Wang AH, et al. HDAC4, a human histone deacetylase related to yeast HDA1, is a transcriptional corepressor. Mol Cell Biol. 1999;19:7816–7827. doi: 10.1128/mcb.19.11.7816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miska EA, et al. HDAC4 deacetylase associates with and represses the MEF2 transcription factor. EMBO J. 1999;18:5099–5107. doi: 10.1093/emboj/18.18.5099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sparrow DB, et al. MEF-2 function is modified by a novel co-repressor, MITR. EMBO J. 1999;18:5085–5098. doi: 10.1093/emboj/18.18.5085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang CL, et al. Class II histone deacetylases act as signal-responsive repressors of cardiac hypertrophy. Cell. 2002;110:479–488. doi: 10.1016/s0092-8674(02)00861-9. Describes the first knockout of a class IIa HDAC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chang S, et al. Histone deacetylases 5 and 9 govern responsiveness of the heart to a subset of stress signals and play redundant roles in heart development. Mol Cell Biol. 2004;24:8467–8476. doi: 10.1128/MCB.24.19.8467-8476.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vega RB, et al. Histone deacetylase 4 controls chondrocyte hypertrophy during skeletogenesis. Cell. 2004;119:555–566. doi: 10.1016/j.cell.2004.10.024. [DOI] [PubMed] [Google Scholar]
- 34.Chang S, et al. Histone deacetylase 7 maintains vascular integrity by repressing matrix metalloproteinase 10. Cell. 2006;126:321–334. doi: 10.1016/j.cell.2006.05.040. Reports the first conditional class IIa HDAC knockout. [DOI] [PubMed] [Google Scholar]
- 35.Fischle W, et al. Enzymatic activity associated with class II HDACs is dependent on a multiprotein complex containing HDAC3 and SMRT/N-CoR. Mol Cell. 2002;9:45–57. doi: 10.1016/s1097-2765(01)00429-4. The first definitive report showing that class IIa HDAC enzymatic activity is due to the copurification of class I HDACs. [DOI] [PubMed] [Google Scholar]
- 36.Jones P, et al. Probing the elusive catalytic activity of vertebrate class IIa histone deacetylases. Bioorg Med Chem Lett. 2008;18:1814–1819. doi: 10.1016/j.bmcl.2008.02.025. [DOI] [PubMed] [Google Scholar]
- 37.Zhang CL, McKinsey TA, Lu JR, Olson EN. Association of COOH-terminal-binding protein (CtBP) and MEF2-interacting transcription repressor (MITR) contributes to transcriptional repression of the MEF2 transcription factor. J Biol Chem. 2001;276:35–39. doi: 10.1074/jbc.M007364200. [DOI] [PubMed] [Google Scholar]
- 38.Zhang CL, McKinsey TA, Olson EN. Association of class II histone deacetylases with heterochromatin protein 1: potential role for histone methylation in control of muscle differentiation. Mol Cell Biol. 2002;22:7302–7312. doi: 10.1128/MCB.22.20.7302-7312.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang CL, McKinsey TA, Olson EN. The transcriptional corepressor MITR is a signal-responsive inhibitor of myogenesis. Proc Natl Acad Sci USA. 2001;98:7354–7359. doi: 10.1073/pnas.131198498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dressel U, et al. A dynamic role for HDAC7 in MEF2-mediated muscle differentiation. J Biol Chem. 2001;276:17007–17013. doi: 10.1074/jbc.M101508200. [DOI] [PubMed] [Google Scholar]
- 41.Bottomley MJ, et al. Structural and functional analysis of the human HDAC4 catalytic domain reveals a regulatory structural zinc-binding domain. J Biol Chem. 2008;283:26694–26704. doi: 10.1074/jbc.M803514200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schuetz A, et al. Human HDAC7 harbors a class IIa histone deacetylase-specific zinc binding motif and cryptic deacetylase activity. J Biol Chem. 2008;283:11355–11363. doi: 10.1074/jbc.M707362200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang Y, et al. Mice lacking histone deacetylase 6 have hyperacetylated tubulin but are viable and develop normally. Mol Cell Biol. 2008;28:1688–1701. doi: 10.1128/MCB.01154-06. Describes the first knockout of a class IIb HDAC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fischer DD, et al. Isolation and characterization of a novel class II histone deacetylase, HDAC10. J Biol Chem. 2002;277:6656–6666. doi: 10.1074/jbc.M108055200. [DOI] [PubMed] [Google Scholar]
- 45.Guardiola AR, Yao TP. Molecular cloning and characterization of a novel histone deacetylase HDAC10. J Biol Chem. 2002;277:3350–3356. doi: 10.1074/jbc.M109861200. [DOI] [PubMed] [Google Scholar]
- 46.Tang X, et al. Acetylation-dependent signal transduction for type I interferon receptor. Cell. 2007;131:93–105. doi: 10.1016/j.cell.2007.07.034. [DOI] [PubMed] [Google Scholar]
- 47.Zhang X, et al. HDAC6 modulates cell motility by altering the acetylation level of cortactin. Mol Cell. 2007;27:197–213. doi: 10.1016/j.molcel.2007.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kovacs JJ, et al. HDAC6 regulates Hsp90 acetylation and chaperone-dependent activation of glucocorticoid receptor. Mol Cell. 2005;18:601–607. doi: 10.1016/j.molcel.2005.04.021. [DOI] [PubMed] [Google Scholar]
- 49.Matsuyama A, et al. In vivo destabilization of dynamic microtubules by HDAC6-mediated deacetylation. EMBO J. 2002;21:6820–6831. doi: 10.1093/emboj/cdf682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hubbert C, et al. HDAC6 is a microtubule-associated deacetylase. Nature. 2002;417:455–458. doi: 10.1038/417455a. [DOI] [PubMed] [Google Scholar]
- 51.Gao L, Cueto MA, Asselbergs F, Atadja P. Cloning and functional characterization of HDAC11, a novel member of the human histone deacetylase family. J Biol Chem. 2002;277:25748–25755. doi: 10.1074/jbc.M111871200. [DOI] [PubMed] [Google Scholar]
- 52.Liu H, Hu Q, Kaufman A, D’Ercole AJ, Ye P. Developmental expression of histone deacetylase 11 in the murine brain. J Neurosci Res. 2008;86:537–543. doi: 10.1002/jnr.21521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lagger G, et al. Essential function of histone deacetylase 1 in proliferation control and CDK inhibitor repression. EMBO J. 2002;21:2672–2681. doi: 10.1093/emboj/21.11.2672. Reports the first knockout of a class I HDAC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zupkovitz G, et al. Negative and positive regulation of gene expression by mouse histone deacetylase 1. Mol Cell Biol. 2006;26:7913–7928. doi: 10.1128/MCB.01220-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yamaguchi M, et al. Histone deacetylase 1 regulates retinal neurogenesis in zebrafish by suppressing Wnt and Notch signaling pathways. Development. 2005;132:3027–3043. doi: 10.1242/dev.01881. [DOI] [PubMed] [Google Scholar]
- 56.Cunliffe VT. Histone deacetylase 1 is required to repress Notch target gene expression during zebrafish neurogenesis and to maintain the production of motoneurones in response to hedgehog signalling. Development. 2004;131:2983–2995. doi: 10.1242/dev.01166. [DOI] [PubMed] [Google Scholar]
- 57.Cunliffe VT, Casaccia-Bonnefil P. Histone deacetylase 1 is essential for oligodendrocyte specification in the zebrafish CNS. Mech Dev. 2006;123:24–30. doi: 10.1016/j.mod.2005.10.005. [DOI] [PubMed] [Google Scholar]
- 58.Pillai R, Coverdale LE, Dubey G, Martin CC. Histone deacetylase 1 (HDAC-1) required for the normal formation of craniofacial cartilage and pectoral fins of the zebrafish. Dev Dyn. 2004;231:647–654. doi: 10.1002/dvdy.20168. [DOI] [PubMed] [Google Scholar]
- 59.Nambiar RM, Ignatius MS, Henion PD. Zebrafish colgate/hdac1 functions in the non-canonical Wnt pathway during axial extension and in Wnt-independent branchiomotor neuron migration. Mech Dev. 2007;124:682–698. doi: 10.1016/j.mod.2007.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Farooq M, et al. Histone deacetylase 3 (hdac3) is specifically required for liver development in zebrafish. Dev Biol. 2008;317:336–353. doi: 10.1016/j.ydbio.2008.02.034. [DOI] [PubMed] [Google Scholar]
- 61.Trivedi CM, et al. Hdac2 regulates the cardiac hypertrophic response by modulating Gsk3 beta activity. Nature Med. 2007;13:324–331. doi: 10.1038/nm1552. [DOI] [PubMed] [Google Scholar]
- 62.Voss AK, Thomas T, Gruss P. Efficiency assessment of the gene trap approach. Dev Dyn. 1998;212:171–180. doi: 10.1002/(SICI)1097-0177(199806)212:2<171::AID-AJA3>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 63.Chen F, et al. Hop is an unusual homeobox gene that modulates cardiac development. Cell. 2002;110:713–723. doi: 10.1016/s0092-8674(02)00932-7. [DOI] [PubMed] [Google Scholar]
- 64.Kook H, et al. Cardiac hypertrophy and histone deacetylase-dependent transcriptional repression mediated by the atypical homeodomain protein Hop. J Clin Invest. 2003;112:863–871. doi: 10.1172/JCI19137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shin CH, et al. Modulation of cardiac growth and development by HOP, an unusual homeodomain protein. Cell. 2002;110:725–735. doi: 10.1016/s0092-8674(02)00933-9. [DOI] [PubMed] [Google Scholar]
- 66.Kuwahara K, et al. The neuron-restrictive silencer element-neuron-restrictive silencer factor system regulates basal and endothelin 1-inducible atrial natriuretic peptide gene expression in ventricular myocytes. Mol Cell Biol. 2001;21:2085–2097. doi: 10.1128/MCB.21.6.2085-2097.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nakagawa Y, et al. Class II HDACs mediate CaMK-dependent signaling to NRSF in ventricular myocytes. J Mol Cell Cardiol. 2006;41:1010–1022. doi: 10.1016/j.yjmcc.2006.08.010. [DOI] [PubMed] [Google Scholar]
- 68.Kuwahara K, et al. NRSF regulates the fetal cardiac gene program and maintains normal cardiac structure and function. EMBO J. 2003;22:6310–6321. doi: 10.1093/emboj/cdg601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Montgomery RL, et al. Maintenance of cardiac energy metabolism by histone deacetylase 3 in mice. J Clin Invest. 2008;118:3588–3597. doi: 10.1172/JCI35847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bhaskara S, et al. Deletion of histone deacetylase 3 reveals critical roles in S phase progression and DNA damage control. Mol Cell. 2008;30:61–72. doi: 10.1016/j.molcel.2008.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Knutson SK, et al. Liver-specific deletion of histone deacetylase 3 disrupts metabolic transcriptional networks. EMBO J. 2008;27:1017–1028. doi: 10.1038/emboj.2008.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Trivedi CM, Lu MM, Wang Q, Epstein JA. Transgenic over-expression of Hdac3 in the heart produces increased postnatal cardiac myocyte proliferation but does not induce hypertrophy. J Biol Chem. 2008;283:26484–26489. doi: 10.1074/jbc.M803686200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Karsenty G. The complexities of skeletal biology. Nature. 2003;423:316–318. doi: 10.1038/nature01654. [DOI] [PubMed] [Google Scholar]
- 74.Cohen MM., Jr The new bone biology: pathologic, molecular, and clinical correlates. Am J Med Genet A. 2006;140:2646–2706. doi: 10.1002/ajmg.a.31368. [DOI] [PubMed] [Google Scholar]
- 75.Arnold MA, et al. MEF2C transcription factor controls chondrocyte hypertrophy and bone development. Dev Cell. 2007;12:377–89. doi: 10.1016/j.devcel.2007.02.004. [DOI] [PubMed] [Google Scholar]
- 76.Kronenberg HM. PTHrP and skeletal development. Ann NY Acad Sci. 2006;1068:1–13. doi: 10.1196/annals.1346.002. [DOI] [PubMed] [Google Scholar]
- 77.Song K, et al. The transcriptional coactivator CAMTA2 stimulates cardiac growth by opposing class II histone deacetylases. Cell. 2006;125:453–466. doi: 10.1016/j.cell.2006.02.048. [DOI] [PubMed] [Google Scholar]
- 78.McKinsey TA, Olson EN. Toward transcriptional therapies for the failing heart: chemical screens to modulate genes. J Clin Invest. 2005;115:538–546. doi: 10.1172/JCI24144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Backs J, Olson EN. Control of cardiac growth by histone acetylation/deacetylation. Circ Res. 2006;98:15–24. doi: 10.1161/01.RES.0000197782.21444.8f. [DOI] [PubMed] [Google Scholar]
- 80.Hill JA, Olson EN. Cardiac plasticity. N Engl J Med. 2008;358:1370–1380. doi: 10.1056/NEJMra072139. [DOI] [PubMed] [Google Scholar]
- 81.Kim Y, et al. The MEF2D transcription factor mediates stress-dependent cardiac remodeling in mice. J Clin Invest. 2008;118:124–32. doi: 10.1172/JCI33255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Backs J, Song K, Bezprozvannaya S, Chang S, Olson EN. CaM kinase II selectively signals to histone deacetylase 4 during cardiomyocyte hypertrophy. J Clin Invest. 2006;116:1853–1864. doi: 10.1172/JCI27438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bassel-Duby R, Olson EN. Signaling pathways in skeletal muscle remodeling. Annu Rev Biochem. 2006;75:19–37. doi: 10.1146/annurev.biochem.75.103004.142622. [DOI] [PubMed] [Google Scholar]
- 84.Kim MS, et al. Protein kinase D1 stimulates MEF2 activity in skeletal muscle and enhances muscle performance. Mol Cell Biol. 2008;28:3600–3609. doi: 10.1128/MCB.00189-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Potthoff MJ, et al. Histone deacetylase degradation and MEF2 activation promote the formation of slow-twitch myofibers. J Clin Invest. 2007;117:2459–2467. doi: 10.1172/JCI31960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mejat A, et al. Histone deacetylase 9 couples neuronal activity to muscle chromatin acetylation and gene expression. Nature Neurosci. 2005;8:313–321. doi: 10.1038/nn1408. [DOI] [PubMed] [Google Scholar]
- 87.Haberland M, et al. Regulation of HDAC9 gene expression by MEF2 establishes a negative-feedback loop in the transcriptional circuitry of muscle differentiation. Mol Cell Biol. 2007;27:518–525. doi: 10.1128/MCB.01415-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lindsey ML, Mann DL, Entman ML, Spinale FG. Extracellular matrix remodeling following myocardial injury. Ann Med. 2003;35:316–326. doi: 10.1080/07853890310001285. [DOI] [PubMed] [Google Scholar]
- 89.Jiang Y, Goldberg ID, Shi YE. Complex roles of tissue inhibitors of metalloproteinases in cancer. Oncogene. 2002;21:2245–2252. doi: 10.1038/sj.onc.1205291. [DOI] [PubMed] [Google Scholar]
- 90.Lee AY, et al. Quantitative analysis of histone deacetylase-1 selective histone modifications by differential mass spectrometry. J Proteome Res. 2008 Oct 31; doi: 10.1021/pr800510p. [DOI] [PubMed] [Google Scholar]
- 91.Jones P, et al. 2-Trifluoroacetylthiophenes, a novel series of potent and selective class II histone deacetylase inhibitors. Bioorg Med Chem Lett. 2008;18:3456–3461. doi: 10.1016/j.bmcl.2008.02.026. [DOI] [PubMed] [Google Scholar]
- 92.Bolden JE, Peart MJ, Johnstone RW. Anticancer activities of histone deacetylase inhibitors. Nature Rev Drug Discov. 2006;5:769–784. doi: 10.1038/nrd2133. [DOI] [PubMed] [Google Scholar]
- 93.Duvic M, et al. Phase 2 trial of oral vorinostat (suberolanilide hydroxamic acid, SAHA) for refractory cutaneous T-cell lymphoma (CTCL) Blood. 2007;109:31–39. doi: 10.1182/blood-2006-06-025999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Xu WS, Parmigiani RB, Marks PA. Histone deacetylase inhibitors: molecular mechanisms of action. Oncogene. 2007;26:5541–5552. doi: 10.1038/sj.onc.1210620. [DOI] [PubMed] [Google Scholar]
- 95.Marks PA. Discovery and development of SAHA as an anticancer agent. Oncogene. 2007;26:1351–1356. doi: 10.1038/sj.onc.1210204. [DOI] [PubMed] [Google Scholar]
- 96.Marks PA, Breslow R. Dimethyl sulfoxide to vorinostat: development of this histone deacetylase inhibitor as an anticancer drug. Nature Biotechnol. 2007;25:84–90. doi: 10.1038/nbt1272. [DOI] [PubMed] [Google Scholar]
- 97.Karagiannis TC, El-Osta A. Modulation of cellular radiation responses by histone deacetylase inhibitors. Oncogene. 2006;25:3885–3893. doi: 10.1038/sj.onc.1209417. [DOI] [PubMed] [Google Scholar]
- 98.Zimmermann S, et al. Reduced body size and decreased intestinal tumor rates in HDAC2-mutant mice. Cancer Res. 2007;67:9047–9054. doi: 10.1158/0008-5472.CAN-07-0312. [DOI] [PubMed] [Google Scholar]
- 99.Lehrman G, et al. Depletion of latent HIV-1 infection in vivo: a proof-of-concept study. Lancet. 2005;366:549–555. doi: 10.1016/S0140-6736(05)67098-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Fischer A, Sananbenesi F, Wang X, Dobbin M, Tsai LH. Recovery of learning and memory is associated with chromatin remodelling. Nature. 2007;447:178–182. doi: 10.1038/nature05772. [DOI] [PubMed] [Google Scholar]
- 101.Reddy P, et al. Histone deacetylase inhibition modulates indoleamine 2,3-dioxygenase-dependent DC functions and regulates experimental graft-versus-host disease in mice. J Clin Invest. 2008;118:2562–2573. doi: 10.1172/JCI34712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kong Y, et al. Suppression of class I and II histone deacetylases blunts pressure-overload cardiac hypertrophy. Circulation. 2006;113:2579–2588. doi: 10.1161/CIRCULATIONAHA.106.625467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Granger A, et al. Histone deacetylase inhibition reduces myocardial ischemia-reperfusion injury in mice. FASEB J. 2008;22:3549–3560. doi: 10.1096/fj.08-108548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zhang B, et al. HDAC inhibitor increases histone H3 acetylation and reduces microglia inflammatory response following traumatic brain injury in rats. Brain Res. 2008;1226:181–191. doi: 10.1016/j.brainres.2008.05.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Williams JA, et al. Valproic acid prevents brain injury in a canine model of hypothermic circulatory arrest: a promising new approach to neuroprotection during cardiac surgery. Ann Thorac Surg. 2006;81:2235–2241. doi: 10.1016/j.athoracsur.2005.12.060. discussion 2241–2242. [DOI] [PubMed] [Google Scholar]
- 106.Almeida AM, et al. Targeted therapy for inherited GPI deficiency. N Engl J Med. 2007;356:1641–1647. doi: 10.1056/NEJMoa063369. Describes the ‘cure’ of an inherited genetic disorder using an HDAC inhibitor. [DOI] [PubMed] [Google Scholar]
- 107.Kim SC, et al. Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol Cell. 2006;23:607–618. doi: 10.1016/j.molcel.2006.06.026. An elegant screen for the ‘acetylome’. [DOI] [PubMed] [Google Scholar]
- 108.Mann BS, Johnson JR, Cohen MH, Justice R, Pazdur R. FDA approval summary: vorinostat for treatment of advanced primary cutaneous T-cell lymphoma. Oncologist. 2007;12:1247–1252. doi: 10.1634/theoncologist.12-10-1247. This study highlights the first US Food and Drug Administration (FDA) approval of an HDAC inhibitor. [DOI] [PubMed] [Google Scholar]
- 109.Olsen EA, et al. Phase IIb multicenter trial of vorinostat in patients with persistent, progressive, or treatment refractory cutaneous T-cell lymphoma. J Clin Oncol. 2007;25:3109–3115. doi: 10.1200/JCO.2006.10.2434. [DOI] [PubMed] [Google Scholar]
- 110.Luhrs H, et al. Butyrate inhibits NF-kappaB activation in lamina propria macrophages of patients with ulcerative colitis. Scand J Gastroenterol. 2002;37:458–466. doi: 10.1080/003655202317316105. [DOI] [PubMed] [Google Scholar]
- 111.Atweh GF, et al. Sustained induction of fetal hemoglobin by pulse butyrate therapy in sickle cell disease. Blood. 1999;93:1790–1797. [PMC free article] [PubMed] [Google Scholar]
- 112.Chung YL, Lee MY, Wang AJ, Yao LF. A therapeutic strategy uses histone deacetylase inhibitors to modulate the expression of genes involved in the pathogenesis of rheumatoid arthritis. Mol Ther. 2003;8:707–717. doi: 10.1016/s1525-0016(03)00235-1. [DOI] [PubMed] [Google Scholar]
- 113.Choi JH, et al. Trichostatin A attenuates airway inflammation in mouse asthma model. Clin Exp Allergy. 2005;35:89–96. doi: 10.1111/j.1365-2222.2004.02006.x. [DOI] [PubMed] [Google Scholar]
- 114.Camelo S, et al. Transcriptional therapy with the histone deacetylase inhibitor trichostatin A ameliorates experimental autoimmune encephalomyelitis. J Neuroimmunol. 2005;164:10–21. doi: 10.1016/j.jneuroim.2005.02.022. [DOI] [PubMed] [Google Scholar]
- 115.Glauben R, et al. Histone hyperacetylation is associated with amelioration of experimental colitis in mice. J Immunol. 2006;176:5015–5022. doi: 10.4049/jimmunol.176.8.5015. [DOI] [PubMed] [Google Scholar]
- 116.Antos CL, et al. Dose-dependent blockade to cardiomyocyte hypertrophy by histone deacetylase inhibitors. J Biol Chem. 2003;278:28930–28937. doi: 10.1074/jbc.M303113200. [DOI] [PubMed] [Google Scholar]
- 117.Reddy P, et al. Histone deacetylase inhibitor suberoylanilide hydroxamic acid reduces acute graft-versus-host disease and preserves graft-versus-leukemia effect. Proc Natl Acad Sci USA. 2004;101:3921–3926. doi: 10.1073/pnas.0400380101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Leoni F, et al. The histone deacetylase inhibitor ITF2357 reduces production of pro-inflammatory cytokines in vitro and systemic inflammation in vivo. Mol Med. 2005;11:1–15. doi: 10.2119/2006-00005.Dinarello. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Minetti GC, et al. Functional and morphological recovery of dystrophic muscles in mice treated with deacetylase inhibitors. Nature Med. 2006;12:1147–1150. doi: 10.1038/nm1479. [DOI] [PubMed] [Google Scholar]
- 120.Mishra N, Reilly CM, Brown DR, Ruiz P, Gilkeson GS. Histone deacetylase inhibitors modulate renal disease in the MRL-lpr/lpr mouse. J Clin Invest. 2003;111:539–552. doi: 10.1172/JCI16153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Avila AM, et al. Trichostatin A increases SMN expression and survival in a mouse model of spinal muscular atrophy. J Clin Invest. 2007;117:659–671. doi: 10.1172/JCI29562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Sailhamer EA, et al. Acetylation: a novel method for modulation of the immune response following trauma/hemorrhage and inflammatory second hit in animals and humans. Surgery. 2008;144:204–216. doi: 10.1016/j.surg.2008.03.034. [DOI] [PubMed] [Google Scholar]