Abstract
There is considerable evidence for exposure of humans to an unknown ethylating agent, and some studies indicate that cigarette smoking may be one source of this exposure. Therefore, we have developed a liquid chromatography-nanoelectrospray-high resolution tandem mass spectrometry-selected reaction monitoring (LC-NSI-HRMS/MS-SRM) method for the analysis of 7-ethyl-Gua in human leukocyte DNA, a readily available source of DNA. [15N5]7-Ethyl-Gua was used as the internal standard. Leukocyte DNA was isolated and treated by thermal neutral hydrolysis. The hydrolysate was partially purified by solid-phase extraction. The fraction containing 7-ethyl-Gua was analyzed by LC-NSI-HRMS/MS-SRM using the transition m/z 180 [M + H]+ → m/z 152.05669 [Gua + H]+ for 7-ethyl-Gua and m/z 185 → m/z 157.04187 for the internal standard. The detection limit was approximately 10 amol on column, while the limit of quantitation was about 8 fmol/μmol Gua starting with 180 μg DNA (corresponding to 36 μg DNA on-column). Leukocyte DNA samples from 30 smokers and 30 nonsmokers were analyzed. Clear peaks for 7-ethyl-Gua and the internal standard were observed in most of the samples. The mean (± S.D.) level of 7-ethyl-Gua measured in leukocyte DNA from smokers was 49.6 ± 43.3 (range 14.6 – 181) fmol/μmol Gua while that from nonsmokers was 41.3 ± 34.9 (range 9.64 – 157) fmol/μmol Gua. Although a significant difference between smokers and nonsmokers was not observed, the method described here is unique in the use of high resolution mass spectrometry and establishes for the first time the presence of 7-ethyl-Gua in human leukocyte DNA.
Introduction
Tobacco smoke causes various adverse health outcomes including cancer, cardiovascular and pulmonary diseases. When individuals inhale cigarette smoke, they are arguably exposed to more than 5,000 chemicals. Many of these compounds are rapidly absorbed by cells in the body producing disease-causing cellular changes via receptor interactions, inflammation, oxidative stress, DNA damage, and other mechanisms. The complex mixture of chemicals present in tobacco smoke includes more than 70 carcinogens, many of which can react with DNA resulting in the formation of DNA adducts. DNA adducts play a critical role in carcinogenesis because they cause gene mutations and loss of normal cellular growth control mechanisms.1,2
Most of the known carcinogens in tobacco smoke such as polycyclic aromatic hydrocarbons, tobacco-specific nitrosamines and aromatic amines undergo P450-dependent metabolism to form electrophilic species that covalently react with DNA nucleotides. Only a few compounds such as ethylene oxide and acetaldehyde do not require metabolic activation to react with DNA. In addition to these known compounds, evidence for the presence in cigarette smoke of a direct acting DNA ethylating agent of unknown structure has been recently reported.3 Thus, Singh et al. measured levels of 7-ethyl-Gua in calf thymus DNA exposed to cigarette smoke and showed a correlation between levels of 7-ethyl-Gua and the number of cigarettes used to generate the smoke.3 Consistent with these findings, several studies have shown an increase of ethylated DNA bases in smokers. Levels of 3-ethyl-Ade and 7-ethyl-Gua in urine, O4-ethyl-dThd in lung, and ethylvaline in hemoglobin were higher in smokers than in nonsmokers in some studies.4–9
Alkylation of the 7 position of guanine is frequently the main base modification which occurs in the reactions of alkylating agents with nucleic acids.10,11 Quantitation of ethylated DNA adducts in human tissues could provide an approach to investigate the possible role of direct ethylating agents in human cancer. With this goal in mind, we previously developed a liquid chromatography-electrospray ionization-tandem mass spectrometry-selected reaction monitoring method for quantitation of 7-ethyl-Gua in human hepatic DNA.12 This adduct was detected in all human liver DNA samples analyzed, with an average level of about 42 fmol/μmol Gua. In the present study, we have extended this research by developing a highly sensitive and specific liquid chromatography-nanoelectrospray-high resolution tandem mass spectrometry-selected reaction monitoring (LC-NSI-HRMS/MS-SRM) method for the analysis of 7-ethyl-Gua in human leukocyte DNA, a readily available source of DNA for epidemiologic studies. The method was applied to investigate levels of 7-ethyl-Gua in leukocyte DNA from 30 smokers and 30 nonsmokers.
Experimental Procedures
HPLC-UV analysis
Quantitation of Gua was carried out with an Agilent 1100 capillary flow HPLC with a diode array UV detector operated at 254 nm (Agilent Technologies, Palo Alto, CA). A 0.5 × 25 cm Luna 5 μm C18 column (Phenomenex, Torrance, CA) was used with a 35 min linear gradient from 5 to 40% CH3OH in H2O at a flow rate of 10 μL/min.
Chemicals
7-Ethyl-Gua and [15N5]7-ethyl-Gua were prepared as described.12 Ethanol was obtained from AAPER Alcohol and Chemical Co. (Shelbyville, KY). Puregene DNA purification solutions were obtained from Qiagen (Valencia, CA). Calf thymus DNA was purchased from Worthington Biochemical Corporation (Lakewood, NJ). All other chemicals were purchased from Sigma-Aldrich (St. Louis, MO).
DNA isolation from Human Leukocytes
DNA was isolated using the DNA purification from buffy coat protocol (Qiagen) with several modifications. Briefly, 3 mL of RBC cell lysis solution were added to 1 mL of buffy coat prepared from 10 mL of whole blood. The white blood cell pellet was collected by centrifugation (2000 g × 10 min), treated with 3 mL of cell lysis solution and incubated at room temperature overnight. A solution of RNase A (4 mg/mL) was added (15 μL) and the sample was incubated at room temperature for 2 h. One mL of protein precipitation solution was added to the cell lysate and the mixture was centrifuged (2000 g × 15 min) to remove proteins. DNA was precipitated from the supernatant by addition of 4 mL of isopropanol. The DNA pellet was washed with 1 mL of 70% ethanol in H2O, and then 1 mL of 100 % ethanol. DNA was dried in a stream of N2 and stored at −20 ºC until use. Buffy coat (1 mL) from 10 mL of blood typically yielded approximately 0.1 mg DNA. The purity of the DNA was determined by measuring its UV absorption at 230, 260 and 280 nm. The ratios of A260:A230 and A260:A280 were greater than 2.0 and 1.7 respectively. RNA contamination was assessed by HPLC analysis for uridine in the hydrolysates when performing enzymatic hydrolysis on portions of the DNA samples. No uridine was detected.
Sample enrichment
DNA hydrolysis and sample enrichment and purification were carried out as previously reported.12 Briefly, DNA (188 ± 114 ug) was dissolved in 1 mL of 10 mM sodium cacodylate buffer containing 25 fmol of [15N5]7-ethyl-Gua and heated at 100 °C for 1 h. A 50 μL aliquot was removed for Gua analysis. The remaining material was loaded on a centrifree MPS device (MW cutoff of 30000 amu; Amicon, Beverly, MA) to remove high molecular weight material. The filtrate was applied to a solid-phase extraction cartridge [Strata-X 33 μm, 30 mg/1 mL (Phenomenex)]. The sample was loaded on the cartridge and washed with 1 mL H2O and 1 mL CH3OH 5%, and eluted with 70% CH3OH. The 70% CH3OH fraction was collected and evaporated to dryness. The residue was dissolved in H2O and analyzed by LC-NSI-HRMS/MS-SRM.
LC-NSI-HRMS/MS-SRM
Samples were resuspended in 10 μL of H2O (average DNA concentration about 19 μg/μL). Separation was performed on a Nano2D-LC HPLC (Eksigent, Dublin, CA) system equipped with a 1μL injection loop. One μL of sample was injected onto a capillary column (75 μm ID, 10 cm length, 15 μm orifice) created by hand packing a commercially available fused-silica emitter (New Objective, Woburn MA) with Luna C18 bonded separation media (Phenomenex, Torrance, CA). The flow rate was 300 nL/min with a 12 min linear gradient of 2 to 55% CH3OH, followed by a 5:95 H2O:CH3OH hold for 2 min and a 14 min re-equilibration at 98:2 H2O:CH3OH. Samples were analyzed by nanoelectrospray using an LTQ-Orbitrap Velos instrument (Thermo Scientific, Waltham, MA). The nanoelectrospray source voltage was set at 1.6 kV. The capillary temperature was 350 °C and the S-Lens RF Level was set at 40%. Adducts were quantified by HRMS/MS-SRM of 7-ethyl-Gua at m/z 180 [M + H]+ → m/z 152.05669 [Gua + H]+ and [15N5]7-ethyl-Gua at m/z 185 → m/z 157.04187 with accurate mass monitoring of the fragment ions at 5 ppm (152.05669 ± 0.0008 and 157.04187 ± 0.0008 respectively) utilizing the Orbitrap detector. These two SRM events were performed using the HCD collision cell with a 1 amu isolation width, collision energy of 60% and the resolution set at 30,000 (at 400 amu) with an actual resolution of 55,000 (at 180 and 185 amu). A full scan event was also performed over 100–500 amu mass range at a resolution setting of 30,000 to monitor for the accurate mass at 5 ppm of the molecular ion of the analyte m/z 180.08799 and of the internal standard m/z 185.07317, to confirm analyte identity.
A calibration curve was constructed before each analysis using a standard solution of 7-ethyl-Gua and [15N5]7-ethyl-Gua. A constant amount of [15N5]7-ethyl-Gua (5 fmol) was mixed with differing amounts of 7-ethyl-Gua (0.05, 0.1, 1, 5, 25, and 100 fmol) and analyzed.
Method Characterization and Sample Analysis
Accuracy and precision were determined by adding 7-ethyl-Gua (0.05, 0.5, 5, 25 and 50 fmol) and 25 fmol of internal standard to 0.3 mg calf thymus DNA and analyzing samples in triplicate.18 7-Ethyl-Gua already present in calf thymus DNA (about 70 fmol/μmol Gua) was subtracted from each value. Sensitivity was determined by estimating the limit of detection for standard solutions of 7-ethyl-Gua while the limit of quantitation was ascertained in human buffy coat DNA samples. The limit of quantitation was determined by adding 7-ethyl-Gua (0.5, 0.7, 1, 2 and 5 fmol) and internal standard (50 fmol) to human buffy coat DNA samples (180 μg DNA) and analyzing samples in triplicate. 7-Ethyl-Gua already present in human buffy coat DNA (about 30 fmol/μmol Gua)) was subtracted from each value. The limit of quantitation was defined by identification of the measurement with a coefficient of variation lower than 5 %.19 Recovery was determined by adding [15N5]7-ethyl-Gua (50 fmol) to 180 μg human leukocyte DNA and processing the samples as described above. These samples were then compared to the same amount of leukocyte DNA processed without [15N5]7-ethyl-Gua, which was added just before LC-NSI-HRMS/MS-SRM analysis.18
Buffer blanks containing internal standard were processed as described above and analyzed to check the LC-NSI-HRMS/MS-SRM baseline and possible contamination. Calf thymus DNA (0.2 mg) with internal standard added as above was used as a positive control to determine inter-day precision and accuracy. Each set of samples was run together with buffer blanks and positive controls.
Study Subjects
The study was approved by the University of Minnesota Human Research Protection Programs Institutional Review Board. Blood samples from 30 smokers and 30 nonsmokers were obtained from the University of Minnesota Tobacco Research Programs repository. All subjects were age 18 years or older, not pregnant or breastfeeding, consumed less than 21 alcoholic drinks per week, and were in good physical and mental health. Additional criteria for smokers included smoking at least 10 cigarettes per day (CPD), having been a smoker for at least 5 years with no change greater than 50% in CPD or brand in the last year, and not using any other tobacco products in the last 6 months. Nonsmokers were required to have smoked less than 100 cigarettes in their lifetime and were not using any tobacco products regularly. Smoking status was confirmed by expired carbon monoxide (CO) levels. CO levels were assessed by having participants take a deep breath and hold it for 20 s before exhaling in to a carbon monoxide monitor (Bedfont Scientific, Upchurch, UK). Leukocytes were separated from 10 – 20 mL of freshly collected blood as described above, and DNA was isolated.
Results
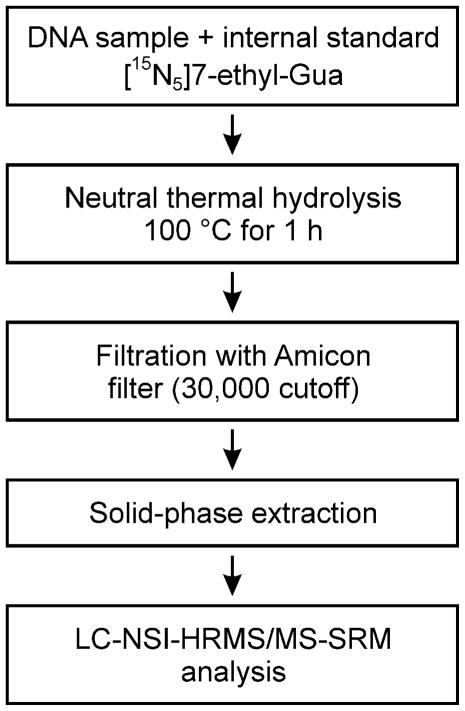
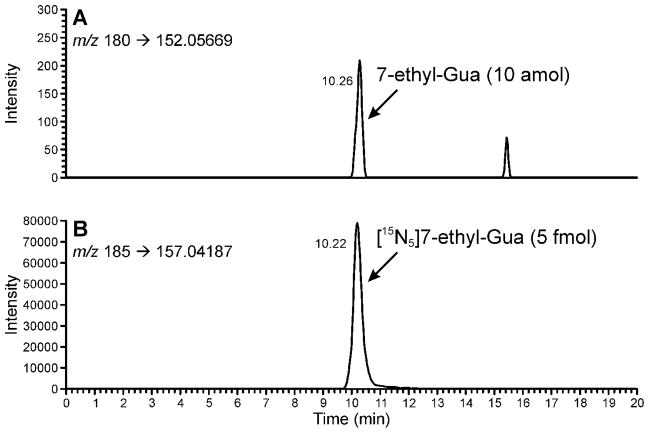
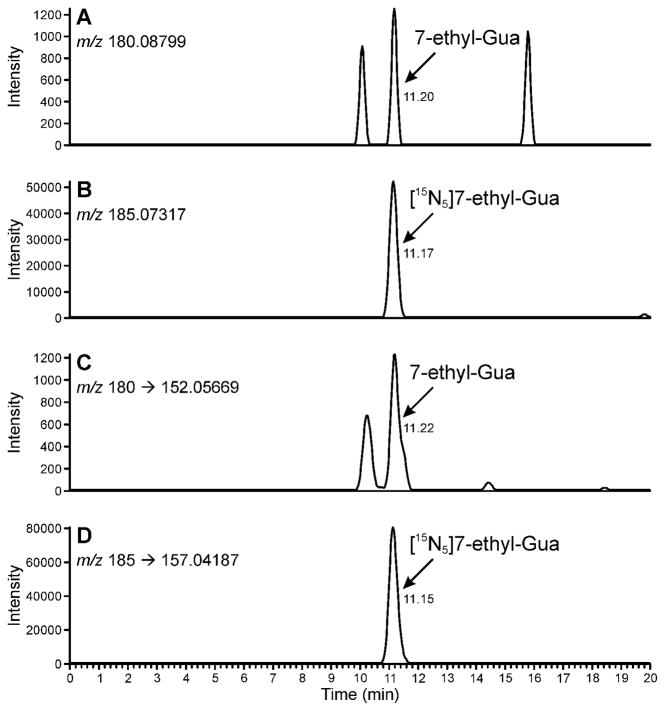
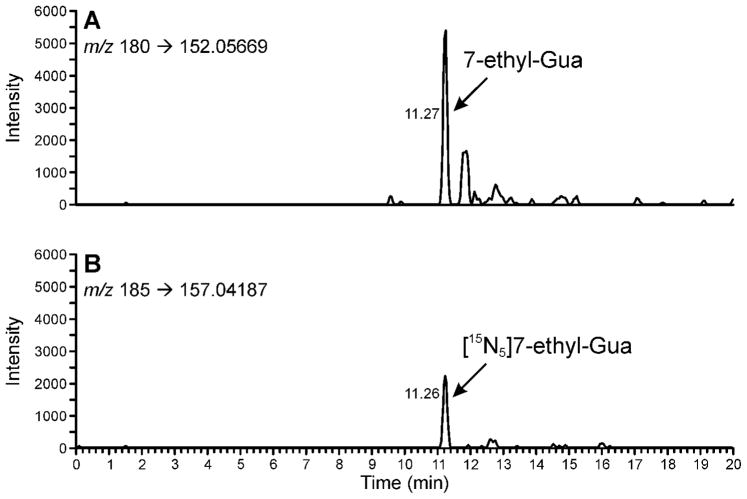
DNA was analyzed following the method outlined in Scheme 1. It was dissolved in 10mM sodium cacodylate buffer and [15N5]7-ethyl-Gua was added as internal standard. Neutral thermal hydrolysis was performed to release 7-ethyl-Gua, the samples were purified by solid-phase extraction and analyzed by LC-NSI-HRMS/MS-SRM, monitoring the transition at m/z 180 [M + H]+ → m/z 152.05669 [Gua + H]+ for 7-ethyl-Gua and the corresponding transition m/z 185 [M + H]+ → m/z 157.04187 [Gua + H]+ for the internal standard. Monitoring of these transitions was the most sensitive method for 7-ethyl-Gua quantitation. A chromatogram obtained upon analysis of a standard mixture of 10 amol of 7-ethyl-Gua and 5 fmol of the internal standard is illustrated in Figure 1. Additional monitoring for the accurate mass of the molecular ion of the analyte and the internal standard was used for confirmation of analyte identity when sufficient sample was present as shown in the upper channels of Figure 2. The limit of detection was about 10 amol on column, based on a coefficient of variation (CV) of 40% when this amount was injected. The limit of quantitation was about 100 amol on column based on a CV of 3.8 % when this amount was injected, corresponding to about 8 fmol/μmol Gua or about 2 adducts per 109 nucleotides starting from 180 μg DNA.
Scheme 1.
Analytical scheme for determination of 7-ethyl-Gua in human leukocyte DNA
Figure 1.
Chromatograms obtained upon analysis of a standard mixture of 10 amol of 7-ethyl-Gua and 5 fmol [15N5]7-ethyl-Gua using LC-NSI-HRMS/MS-SRM. Panel (A) shows the results from the transition at m/z 180 [M + H]+ → m/z 152.05669 [Gua + H]+ for 7-ethyl-Gua. Panel (B) shows the corresponding transition m/z 185 [M + H]+ → m/z 157.04187 [Gua + H]+ for the internal standard. Results are shown with a 5 ppm mass tolerance.
Figure 2.
Chromatograms obtained upon LC-NSI-HRMS/MS-SRM analysis of human leukocyte DNA (129 μg, 12.9 μg on column) containing 59.4 fmol/μmol Gua. The relatively higher amount of analyte in this sample allowed confirmation of its identity by additional monitoring of the accurate mass of the molecular ion of 7-ethyl-Gua and the internal standard. Panel (A) shows the result from monitoring of the accurate mass of 7-ethyl-Gua (m/z 180.08799). Panel (B) shows the result from the monitoring of the accurate mass of [15N5]7-ethyl-Gua (m/z 185.07317). Panel (C) shows the results from the transition at m/z 180 [M + H]+ → m/z 152.05669 [Gua + H]+ for 7-ethyl-Gua and panel (D) shows the corresponding transition m/z 185 [M + H]+ → m/z 157.04187 [Gua + H]+ for the internal standard. Results are shown with a 5 ppm mass tolerance.
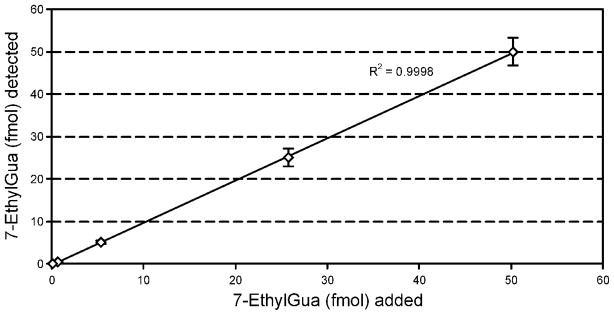
A calibration curve for 7-ethyl-Gua and the internal standard was linear in the range of concentrations found in the samples analyzed (0.5 – 100 fmol) (R2= 0.99). Accuracy was determined by adding 7-ethyl-Gua (0.05, 0.5, 5, 25 and 50 fmol) and 25 fmol of internal standard to 0.3 mg of calf thymus DNA and analyzing samples in triplicate. The amount of 7-ethyl-Gua present in calf thymus DNA was subtracted from each value. The results are summarized in Figure 3. The added and measured amounts of 7-ethyl-Gua correlated (R2 = 0.99).
Figure 3.
Relationship of added to detected 7-ethyl-Gua. Various amounts of the DNA adduct were added to calf thymus DNA (0.3 mg, 30 μg on column) and analyzed by the method described in the text; R2 = 0.99. 7-Ethyl-Gua present in the calf thymus DNA was subtracted from each value
The recovery of the assay was about 70%. The precision of the method was investigated by analyzing a leukocyte DNA sample with an amount of 7-ethyl-Gua (10 fmol/μmol Gua) relatively lower than the amounts found in the samples analyzed. The measurement was performed in triplicate on three separate days. As shown in Table 1, the interday CV was 3.7%.
Table 1.
A leukocyte DNA sample containing 7-ethyl-Gua (~10 fmol/μmol Gua) was used to assess the precision of the method. Three aliquots of the leukocyte DNA were analyzed on three separate days and the coefficient of variation (% CV) of the measurement was calculated.
| 7-Ethyl-Gua (fmol/μmol Gua)
| ||||
|---|---|---|---|---|
| day 1 | day 2 | day 3 | interday | |
| mean ± SD | mean ± SD | mean ± SD | average | % CV |
| 11.1 ± 0.45 | 10.5 ± 0.30 | 11.4 ± 0.24 | 11.0 ± 0.41 | 3.7 |
The method was applied to investigate the effect of cigarette smoking on levels of 7-ethyl-Gua in human leukocyte DNA. Samples from 30 smokers and 30 nonsmokers were analyzed. CO levels were measured for all study participants to confirm their smoking status. Smokers had CO levels ≥ 10 ppm (range 10–38 ppm) while non smokers had CO levels ≤ 3 ppm (range 0–3 ppm). The mean age of the smokers was 41 ± 10 (range 20 – 54) and the group included 15 women. The mean age of the nonsmokers was 33 ±11 years (range 20 – 54) and the group included 14 women. A clear peak for 7-ethyl-Gua was observed in 28 samples from smokers and 28 samples from nonsmokers and it co-eluted with the corresponding [15N5] labeled analogue.
The results are summarized in Table 2. The mean (± S.D.) level of 7-ethyl-Gua measured in leukocyte DNA from the nonsmokers was 41.3 ± 34.9 fmol/μmol Gua (range 9.64 – 157) while that from smokers was 49.6 ± 43.3 (range 14.6 – 181) fmol/μmol Gua. No significant difference between the two groups was observed (p = 0.13, based on a two-sample t-test of the log-normal means). The effects of age and gender were investigated but did not show any significant effect on the difference in DNA adduct levels by smoking status (p = 0.32).
Table 2.
Effect of smoking on levels of 7-ethyl-Gua in leukocyte DNA
| Nonsmokers | Smokers | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| sample | gender | age | amount of DNA analyzed (μg) | 7-ethyl-Gua (fmol/μmol Gua) | sample | gender | age | CPDb | amount of DNA analyzed (μg) | 7-ethyl-Gua (fmol/μmol Gua) |
| 1 | F | 30 | 197 | 16.3 | 31 | M | 50 | 34 | 612 | 18.6 |
| 2 | M | 31 | 129 | 65.6 | 32 | F | 54 | 20 | 165 | 75.7 |
| 3 | M | 44 | 105 | 157 | 33 | M | 20 | 10 | 259 | 61.3 |
| 4 | M | 52 | 100 | 98.5 | 34 | F | 56 | 20 | 38 | ND |
| 5 | M | 39 | 12 | NDa | 35 | F | 53 | 38 | 15 | 146 |
| 6 | F | 25 | 414 | 46.8 | 36 | M | 51 | 30 | 14 | 70.2 |
| 7 | M | 24 | 35 | 103 | 37 | F | 38 | 15 | 86 | 120 |
| 8 | F | 25 | 132 | 18.8 | 38 | M | 30 | 15 | 31 | 118 |
| 9 | M | 24 | 127 | 10.2 | 39 | M | 46 | 30 | 357 | 73.9 |
| 10 | F | 25 | 174 | 15.2 | 40 | F | 23 | 10 | 129 | 59.4 |
| 11 | M | 23 | 132 | 64.3 | 41 | M | 51 | 40 | 92 | 23.1 |
| 12 | F | 23 | 252 | 20.1 | 42 | M | 40 | 19 | 225 | 18.4 |
| 13 | M | 30 | 145 | 12.8 | 43 | M | 40 | 20 | 252 | 181 |
| 14 | F | 30 | 199 | 14.9 | 44 | F | 31 | 20 | 173 | 25.1 |
| 15 | F | 27 | 180 | 51.1 | 45 | F | 37 | 10 | 223 | 14.6 |
| 16 | F | 20 | 175 | 15.3 | 46 | M | 52 | 15 | 221 | 19.2 |
| 17 | M | 40 | 233 | 86.3 | 47 | F | 32 | 15 | 527 | 18.0 |
| 18 | M | 44 | 142 | 14.0 | 48 | F | 39 | 17 | 312 | 17.5 |
| 19 | F | 23 | 138 | ND | 49 | M | 52 | 10 | 186 | 31.7 |
| 20 | M | 49 | 312 | 48.8 | 50 | F | 46 | 20 | 330 | 46.9 |
| 21 | M | 22 | 331 | 9.64 | 51 | F | 44 | 18 | 112 | 26.7 |
| 22 | M | 22 | 193 | 15.1 | 52 | F | 22 | 15 | 255 | 34.4 |
| 23 | M | 31 | 121 | 46.8 | 53 | F | 41 | 15 | 133 | 34.5 |
| 24 | M | 30 | 176 | 33.3 | 54 | F | 26 | 15 | 93 | 36.6 |
| 25 | M | 27 | 298 | 45.5 | 55 | F | 38 | 20 | 137 | 24.2 |
| 26 | F | 54 | 236 | 24.5 | 56 | M | 52 | 10 | 225 | 16.7 |
| 27 | F | 46 | 171 | 26.6 | 57 | M | 52 | 15 | 177 | 20.6 |
| 28 | F | 51 | 129 | 50.5 | 58 | M | 47 | 20 | 122 | 36.6 |
| 29 | F | 21 | 164 | 22.5 | 59 | M | 41 | 15 | 98 | ND |
| 30 | F | 47 | 317 | 24.2 | 60 | M | 40 | 20 | 200 | 21.1 |
|
| ||||||||||
| mean ± SD: | 33 ± 11 | 182 ± 87.5 | 41.3 ± 34.9 | mean ± SD: | 41 ± 10 | 19 ± 8 | 193 ± 136 | 49.6 ± 43.3 | ||
ND: not detected
CPD: cigarettes per day
Although in this study the average amount of DNA used per analysis was 188 ± 114 μg, the analysis was feasible with amounts of DNA smaller than 20 μg as starting material. Figure 4 shows a chromatogram obtained upon analysis of a 14 μg DNA sample. This sample contained 7-ethyl-Gua (70.2 fmol/μmol Gua) corresponding to 130 amol injected on column.
Figure 4.
Chromatograms obtained upon LC-NSI-HRMS/MS-SRM analysis of human leukocyte DNA (14 μg, 1,4 μg on column) for 7-ethyl-Gua by the method described in the text. This sample contained 70.2 fmol/μmol Gua of 7-ethyl-Gua corresponding to 130 amol injected on column (1 μl injected out of a 10 μl sample). Panel (A), analyte; panel (B), internal standard. Results are shown with a 5 ppm mass tolerance.
Discussion
We have developed a sensitive high resolution mass spectrometry method for quantitation of 7-ethyl-Gua in human leukocyte DNA. The accuracy and precision of the method, which uses a 15N-labelled internal standard, were established. To our knowledge this is the first method developed for the analysis of 7-ethyl-Gua in leukocyte DNA. Our approach featured a unique application of high resolution mass spectrometry to achieve the requisite sensitivity and specificity, allowing quantitation of this adduct in samples containing low amounts of DNA.
In our previous study of 7-ethyl-Gua in human hepatic DNA, we used a triple quadrupole-based method.12 We were unsuccessful in using that method to quantify 7-ethyl-Gua in human leukocyte DNA because its levels were below the limit of detection, mainly due to matrix effects in this relatively low resolution approach. The level of background noise and matrix effects often increase as the mass of the analyte decreases. Alkylated DNA adducts such as 7-ethyl-Gua bearing a short chain alkyl group and no deoxyribose moiety have lower m/z values compared to many other DNA adducts. This results in parent ion values that can be difficult to distinguish from intense background noise and matrix effect signals that increase in intensity at low m/z values. Also, the relatively high polarity of 7-ethyl-Gua makes it more difficult to improve significantly the purity of the sample through either solid-phase extraction or by optimizing liquid chromatography parameters, which are the usual approaches used to decrease background noise.13 These limitations were addressed by using a nanoelectrospray ionization (NSI) source and the LTQ Orbitrap Velos instrument. The efficiency of the conversion of analytes to ionized species increases with decreasing flow rate.14 The NSI source allowed optimal operation at lower flow rates. Furthermore, the mass spectrometer couples a dual pressure linear ion trap for rapid MSn scanning and an Orbitrap mass analyzer to achieve high resolution capability, thus reducing background noise and ensuring analyte identity based on high resolution mass accuracy. Thus, we achieved a limit of detection of 10 amol on column and a limit of quantitation of 100 amol on column corresponding to approximately 8 fmol/μmol Gua, suitable for quantitation of 7-ethyl-Gua in human leukocyte DNA.
The method was used to investigate levels of 7-ethyl-Gua in leukocyte DNA samples from smokers and nonsmokers. 7-Ethyl-Gua was detected in 56 DNA samples out of 60 analyzed with a mass tolerance of 5 ppm, providing convincing evidence for its presence in human leukocyte DNA. Average levels of this DNA adduct in the subjects analyzed were about 45 fmol/μmol Gua or 1.1 adducts per 108 nucleotides, which is comparable to the levels which we found in human hepatic DNA,12 and similar to those observed in our recent study of acrolein-DNA adducts in human leukocyte DNA.15 These levels are in the lower range of reported “endogenous” DNA adducts, similar to reports of “etheno” DNA adduct levels which have been attributed to lipid peroxidation.16
We observed no significant differences in levels of 7-ethyl-Gua between smokers and nonsmokers, indicating that this adduct in leukocyte DNA has sources other than direct ethylation by cigarette smoke. The origin of 7-ethyl-Gua in human DNA is not known. One possible source could be ethanol metabolism, possibly through reaction with DNA of ethyl glucuronide or related conjugates. Another possible source is from degradation of DNA adducts with more complex structures. For example, we have recently detected 7-carboxyethyl-Gua in human hepatic DNA (24 of 24 samples analyzed, about 75 adducts per 109 nucleotides), although analyses of leukocyte DNA have not been performed.17 It is possible that this adduct could be converted in part to 7-ethyl-Gua, but further studies are required.
While our data definitively establish the presence of 7-ethyl-Gua in human leukocyte DNA, they do not provide much insight on the possible role of smoking in its formation. Cigarette smoke contains a DNA ethylating agent3 but it is possible that this agent does not reach leukocyte DNA. In a previous study, we have reported approximately 50% higher levels of N-terminal N-ethylvaline in globin from smokers compared to nonsmokers.7 The ethylating agent may preferably bind to more readily available reactive sites such as the N-terminal valine in globin, thus preventing the detection of smoking related differences in leukocyte DNA. Godschalk et al reported about 2-fold higher levels of O4-ethyl-dThd in lung DNA of smokers (n = 13) compared to non-smokers (n = 11), based on a 32P-postlabelling method,6 and this finding has recently been confirmed in a larger study.20 Contradictory results have been obtained in studies of 7-ethyl-Gua and 3-ethyl-Ade in the urine of smokers and nonsmokers. Levels of 7-ethyl-Gua were reported to be higher in smokers’ urine.5 However, levels of 3-ethyl-Ade have yet to be clearly related to smoking and the results were not conclusive.8,9 Urinary levels of these ethylated bases could also be influenced by diet.21
In summary, we describe a LC-NSI-HRMS/MS-SRM method for the quantitation of 7-ethyl-Gua in human leukocyte DNA. The method is very sensitive and allows the measurement of 7-ethyl-Gua in relatively small amounts of DNA. The method potentially can be applied to other readily available sources of DNA. Further studies are required to determine the effects of smoking on DNA ethylation in humans, perhaps using DNA from cell types more directly exposed to smoke. This is an important question because it may reflect the immediate deleterious consequences of smoking, as discussed in the recent Surgeon General’s report.1
Acknowledgments
Funding Support:
This study was supported in part by NIH contract N01-PC-64402, Laboratory Assessment of Tobacco Use Behavior and Exposure to Toxins. The shared resources of the Masonic Cancer Center are supported in part by Cancer Center Support Grant CA-77598. Funds for the purchase of the Orbitrap Velos mass spectrometer were provided by NIH Shared Instrumentation Grant S10-RR-024618.
Mass spectrometry was carried out in the Analytical Biochemistry Shared Resource of the Masonic Cancer Center and statistical analysis was performed by Robin Bliss of the Biostatistics and Informatics Shared Resource of the Masonic Cancer Center. We thank Bob Carlson for graphic and editorial assistance. We also thank Elizabeth Thompson for help with sample collection.
Nonstandard Abbreviations
- NSI
nanoelectrospray ionization
References
- 1.U.S. Department of Health and Human Services. How Tobacco Smoke Causes Disease: The Biology and Behavioral Basis for Smoking-Attributable Disease: A Report of the Surgeon General. Atlanta, GA: U.S. Department of Health and Human Services, Centers for Disease Control and Prevention, National Center for Chronic Disease Prevention and Health Promotion, Office on Smoking and Health; 2010. [Google Scholar]
- 2.International Agency for Research on Cancer. IARC Monographs on the Evaluation of Carcinogenic Risks to Humans. Vol. 83. IARC; Lyon, France: 2004. Tobacco smoke and involuntary smoking; pp. 53–119. [PMC free article] [PubMed] [Google Scholar]
- 3.Singh R, Kaur B, Farmer PB. Detection of DNA damage derived from a direct acting ethylating agent present in cigarette smoke by use of liquid chromatography-tandem mass spectrometry. Chem Res Toxicol. 2005;18:249–256. doi: 10.1021/tx049793j. [DOI] [PubMed] [Google Scholar]
- 4.Kopplin A, Eberle-Adamkiewicz G, Glusenkamp KH, Nehls P, Kirstein U. Urinary excretion of 3-methyladenine and 3-ethyladenine after controlled exposure to tobacco smoke. Carcinogenesis. 1995;16:2637–2641. doi: 10.1093/carcin/16.11.2637. [DOI] [PubMed] [Google Scholar]
- 5.Chao MR, Wang CJ, Chang LW, Hu CW. Quantitative determination of urinary N7-ethylguanine in smokers and non-smokers using an isotope dilution liquid chromatography/tandem mass spectrometry with on-line analyte enrichment. Carcinogenesis. 2006;27:146–151. doi: 10.1093/carcin/bgi177. [DOI] [PubMed] [Google Scholar]
- 6.Godschalk R, Nair J, van Schooten FJ, Risch A, Drings P, Kayser K, Dienemann H, Bartsch H. Comparison of multiple DNA adduct types in tumor adjacent human lung tissue: effect of cigarette smoking. Carcinogenesis. 2002;23:2081–2086. doi: 10.1093/carcin/23.12.2081. [DOI] [PubMed] [Google Scholar]
- 7.Carmella SG, Chen M, Villalta PW, Gurney JG, Hatsukami DK, Hecht SS. Ethylation and methylation of hemoglobin in smokers and non-smokers. Carcinogenesis. 2002;23:1903–1910. doi: 10.1093/carcin/23.11.1903. [DOI] [PubMed] [Google Scholar]
- 8.Feng S, Roethig HJ, Liang Q, Kinser R, Jin Y, Scherer G, Urban M, Engl J, Riedel K. Evaluation of urinary 1-hydroxypyrene, S-phenylmercapturic acid, trans,trans-muconic acid, 3-methyladenine, 3-ethyladenine, 8-hydroxy-2′-deoxyguanosine and thioethers as biomarkers of exposure to cigarette smoke. Biomarkers. 2006;11:28–52. doi: 10.1080/13547500500399730. [DOI] [PubMed] [Google Scholar]
- 9.Prevost V, Shuker DE. Cigarette smoking and urinary 3-alkyladenine excretion in man. Chem Res Toxicol. 1996;9:439–444. doi: 10.1021/tx9501041. [DOI] [PubMed] [Google Scholar]
- 10.Singer B, Grunberger D. Molecular Biology of Mutagens and Carcinogens. Plenum Press; New York: 1983. pp. 45–96. [Google Scholar]
- 11.Singer B, Essigmann JM. Site-specific mutagenesis: Retrospective and prospective. Carcinogenesis. 1991;12:949–955. doi: 10.1093/carcin/12.6.949. [DOI] [PubMed] [Google Scholar]
- 12.Chen L, Wang M, Villalta PW, Hecht SS. Liquid chromatography–electrospray ionization tandem mass spectrometry analysis of 7-ethylguanine in human liver DNA. Chem Res Toxicol. 2007;20:1498–1502. doi: 10.1021/tx700147f. [DOI] [PubMed] [Google Scholar]
- 13.Krauss M, Hollender J. Analysis of nitrosamines in wastewater: exploring the trace level quantification capabilities of a hybrid linear ion trap/orbitrap mass spectrometer. Anal Chem. 2008;80:834–842. doi: 10.1021/ac701804y. [DOI] [PubMed] [Google Scholar]
- 14.Smith RD, Shen Y, Tang K. Ultrasensitive and quantitative analysis from combined separation-mass spectrometry for the characterization of proteomes. AccChem Res. 2004;37:269–278. doi: 10.1021/ar0301330. [DOI] [PubMed] [Google Scholar]
- 15.Zhang S, Balbo S, Wang M, Hecht SS. Analysis of acrolein-derived 1,N2- propanodeoxyguanosine adducts in human leukocyte DNA from smokers and nonsmokers. Chem Res Toxicol. 2011;24:119–124. doi: 10.1021/tx100321y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.De Bont R, van Larebeke N. Endogenous DNA damage in humans: a review of quantitative data. Mutagenesis. 2004;19:169–185. doi: 10.1093/mutage/geh025. [DOI] [PubMed] [Google Scholar]
- 17.Cheng G, Wang M, Villalta PW, Hecht SS. Detection of 7-(2′-carboxyethyl)guanine but not 7-carboxymethylguanine in human liver DNA. Chem Res Toxicol. 2010;23:1089–1096. doi: 10.1021/tx100062v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.U.S FDA. Guidance for Industry: Bioanalytical Method Validation. 2001 www.fda.gov/cedr/guidance/index.htm.
- 19.Dolan JW. Calibration Curves, Part II: What are the Limits? LCGC North America. 2009;27:306–312. [Google Scholar]
- 20.Anna L, Kovács K, Gyorffy E, Schoket B, Nair J. Smoking-related O4-ethylthymidine formation in human lung tissue and comparisons with bulky DNA adducts. Mutagenesis. 2011 Jul;26(4):523–527. doi: 10.1093/mutage/ger011. [DOI] [PubMed] [Google Scholar]
- 21.Tricker AR, Preussmann R. Carcinogenic N-nitrosamines in the diet: occurrence, formation, mechanisms and carcinogenic potential. Mutation Research. 1991;259:277–289. doi: 10.1016/0165-1218(91)90123-4. [DOI] [PubMed] [Google Scholar]