Abstract
Objectives
Genetic factors may play a role in fibrosis progression in patients with chronic hepatitis C (CHC). A cirrhosis risk score (CRS7) with 7 SNPs was previously shown to correlate with cirrhosis in patients with CHC. This study aimed to assess the validity of CRS7 as a marker of fibrosis progression and cirrhosis and as a predictor of clinical outcomes in patients with CHC.
Methods
A total of 938 patients (677 Caucasians, 165 African Americans, and 96 Hispanic/Other) in the HALT-C Trial were studied. CRS7 was categorized a priori as high risk (n=440), medium risk (n=310) or low risk (n=188). Patients were assessed for four possible outcomes: fibrosis progression, cirrhosis, clinical outcomes (decompensation or hepatocellular carcinoma [HCC]), or HCC alone.
Results
29% (142/493) developed an increase in fibrosis score by ≥ 2 points on follow-up biopsies, 58% had cirrhosis on one or more biopsies, 35% developed at least one clinical outcome, and 13% developed HCC. CRS7 (trend test) was associated with risk for fibrosis progression (p=0.04) with adjusted hazard ratio (HR) of 1.27 (95%CI: 1.01–1.58) and with cirrhosis (p=0.05) with adjusted odds ratio (OR) of 1.19 (1.00–1.41). Rates of HCC and clinical outcomes were increased in patients with higher CRS7 scores, but were not statistically significant (p=0.12 clinical outcomes, and p=0.07 HCC). A SNP in AZIN1 was significantly associated with fibrosis progression.
Conclusions
CRS7 was validated as a predictor of fibrosis progression and cirrhosis among HALT-C patients, who all had advanced fibrosis. CRS7 was not predictive of clinical outcome.
Keywords: cirrhosis risk score, CRS7, Hepatitis C Antiviral Long-term Treatment against Cirrhosis (HALT-C), hepatitis C, hepatic decompensation, hepatocellular carcinoma (HCC), liver fibrosis
INTRODUCTION
Progression of chronic liver disease to cirrhosis, clinical decompensation, and hepatocellular carcinoma (HCC) is variable, even for liver disease caused by the same etiological agent. For example, while chronic hepatitis C is the most important cause of liver failure and HCC in western countries, most patients with chronic hepatitis C either do not progress to cirrhosis or do so only slowly [1]. The reasons for variability in disease progression and the ability to predict who will develop severe progressive disease are not well defined. Host factors such as older age at the time of infection, male gender, obesity, and heavy alcohol consumption have been shown to be associated with accelerated fibrosis progression [2, 3] but have limited utility in predicting which patients will progress to cirrhosis. Laboratory indicators such as high serum aspartate/alanine aminotransferase (AST/ALT) ratio, AST-platelet ratio index (APRI), bilirubin, and international normalized ratio of prothrombin time (INR), and low platelet count can distinguish between patients with cirrhosis and those with early stage fibrosis with reasonable accuracy because they reflect disease severity. These tests, however, are of limited value in predicting which patients with early stage liver disease will subsequently progress to cirrhosis.
Evidence is accumulating to indicate that genetic factors influence the natural history of chronic liver disease [4]. Genetic polymorphisms may play a role in susceptibility to the etiologic agent and subsequent liver injury. Genetic variability may also contribute to differences in immune response as well as regeneration and repair. Predisposition to disease progression or treatment response based on genetic risk is intensely pursued as a means to personalized medicine, but has met with limited success to date from the perspective of assisting compelling and actionable patient management decisions. Recent studies showed that a single nucleotide polymorphism (SNP) in the region of the interleukin 28B (IL-28B) gene is associated with spontaneous recovery from acute hepatitis C virus (HCV) infection and response to interferon and ribavirin treatment of chronic hepatitis C [5, 6]. Other investigators have focused on the search for genetic markers that are associated with the risk of cirrhosis. To date, most of these studies have been cross-sectional and included small numbers of patients, and the findings have not been validated in subsequent studies [7].
A recent study in the United States of 574 Caucasian patients with chronic hepatitis C (420 in the training set and 154 in the validation set) who had either no fibrosis or bridging fibrosis/cirrhosis, identified 7 SNPs that were significantly associated with bridging fibrosis/cirrhosis [8]. A cirrhosis risk score (CRS7), which was calculated based on the genotypes of these 7 SNPs in each patient, yielded an area under the receiver operating characteristic curve (AUROC) of 0.75 (95% CI: 0.70–0.80) in detecting patients with bridging fibrosis/cirrhosis in the training set and an AUROC of 0.73 (95% CI: 0.56–0.89) in the validation set. These data were derived from a cross-sectional study, so the ability of CRS7 to predict the timing of future risks of cirrhosis could not be assessed. In another study that included 271 Italian patients whose initial biopsies showed Metavir fibrosis stages F0, 1 or 2 on the initial biopsy and who had follow-up biopsies after an interval of at least 60 months (mean 109 months), the mean CRS7 was found to be significantly higher in patients with fibrosis progression (defined as increase in Metavir fibrosis score by ≥ 1) than in those who did not experience fibrosis progression [9]. A subsequent smaller study involving 56 patients in Belgium and Germany, who had paired biopsies with an interval of ≥ 5 years, found that CRS7 was the only variable associated with fibrosis progression [10].
The Hepatitis C Antiviral Long-term Treatment against Cirrhosis (HALT-C) Trial is a prospective study that enrolled more than 1000 patients with chronic hepatitis C of all race and ethnicity followed for up to 8.7 years for histological and clinical outcomes. The availability of genetic samples from a large cohort of these well-characterized patients with serial liver biopsies provided a unique opportunity to evaluate CRS7 and the individual SNPs as predictors of cirrhosis and clinical outcomes.
PATIENTS AND METHODS
Design and Patients
The design of the HALT-C trial has been described previously [11, 12]. Briefly, patients enrolled were those with chronic hepatitis C who had failed to achieve a sustained virologic response (SVR) during previous treatment with interferon with or without ribavirin and who had advanced fibrosis on liver biopsy (Ishak fibrosis score ≥3) and no history of hepatic decompensation or HCC. Patients were re-treated with a combination of pegylated interferon alfa-2a 180 mcg weekly and ribavirin 1–1.2 g/day (lead-in), and those who had undetectable HCV RNA at week 20 continued combination therapy until week 48. Lead-in non-responders and responders who subsequently experienced breakthrough or relapse, as well as those patients who had not responded to pegylated interferon and ribavirin treatment outside the HALT-C Trial (Express patients), were randomized to maintenance therapy (pegylated interferon alfa-2a 90 mcg weekly) or to no further treatment for 3.5 years. Patients were evaluated quarterly for study outcomes of decompensated liver disease and HCC. Liver biopsies were repeated 1.5 and 3.5 years after randomization. Upon completion of the randomized trial, all patients were invited to continue 6-monthly follow-up without treatment. At each visit, patients were assessed clinically and blood was drawn for complete blood counts, hepatic panel (albumin, AST, ALT, bilirubin, and alkaline phosphatase), creatinine, INR, and alpha fetoprotein (AFP). Hepatic ultrasound was performed at randomization, 6 months after randomization and then every 12 months during the randomized trial and every 6 months thereafter. Patients with suspicious lesions on ultrasound or elevated AFP were further evaluated by computed tomography (CT) or magnetic resonance imaging (MRI) of the liver. All patients underwent endoscopy to evaluate for esophageal varices at randomization. All liver biopsies were reviewed and scored by a panel of 12 hepatic pathologists. Fibrosis was staged using the Ishak score (range of 0–6) [13]. The mean length of the biopsies was 1.8 cm, 63% of the biopsies were longer than 1.5 cm and 75% had at least 10 portal triads.
The HALT-C Trial protocol was approved by the local Institutional Review Boards (IRB) of all the HALT-C Trial sites. Patients provided written informed consent for participation in the HALT-C Trial and only those who provided consent for genetic testing were included in this study. This ancillary study was approved by the Data Coordinating Center IRB on February 1, 2009.
Outcome definitions
Patients were assessed for four possible outcomes: fibrosis progression, cirrhosis, clinical outcome and HCC. Fibrosis progression was defined as an increase in Ishak fibrosis score by ≥ 2 points on at least one of the follow-up biopsies 1.5 or 3.5 years after randomization. Cirrhosis was defined as the presence of an Ishak fibrosis score of 5 or 6 on any biopsy, whether baseline or follow-up, or an explanted liver. Clinical outcomes included an increase in Child-Turcotte-Pugh (CTP) score to ≥ 7 on two consecutive occasions 3 months apart, variceal bleeding, ascites, spontaneous bacterial peritonitis, hepatic encephalopathy, HCC, or liver-related death during an observation period of up to 8.7 years (median 6.1 years). Diagnostic criteria were established for each of the clinical outcomes, and an Outcomes Review Panel adjudicated all reported outcomes. Only first clinical outcomes that met the predefined criteria were included in this analysis. For HCC, both definite and presumed HCC were included [14]. Definite HCC required histologic confirmation of the tumor or a new mass lesion on imaging with AFP levels increasing to >1,000 ng/mL. Presumed HCC was defined as a new mass lesion on ultrasound in the absence of histology and an AFP of <1,000 ng/mL in conjunction with one of the following characteristics: a) 2 liver imaging studies showing a mass lesion with characteristics of HCC (vascular enhancement with or without wash out), or b) a progressively enlarging lesion on ultrasound leading to death of the patient, or c) 1 additional imaging study showing a mass lesion with characteristics of HCC that either increased in size over time or was accompanied by increasing AFP levels.
Genetic testing
DNA was extracted at SeraCare (Gaithersburg, MD) from frozen whole blood using the Gentra Systems Puregene kit or from either Epstein-Barr transformed B-lymphocytes or frozen peripheral blood mononuclear cells using Qiagen DNA purification columns (Qiagen Inc, Valencia, CA). Blind coded DNA samples were sent from the HALT-C sample repository at SeraCare to Celera Diagnostics (Alameda, CA) for genetic testing. Genotyping of SNPs was carried out by allele-specific real-time PCR at a high throughput facility. For each allele-specific PCR reaction, 0.3 ng of DNA was amplified. Genotypes were automatically determined by an in-house software program followed by manual curation without knowledge of the phenotype. Genotyping accuracy in prior studies was ~99%. Primer sequences are available upon request.
CRS7 scores were derived as described previously [8]. The scores range from 0 to 1 with higher scores indicating higher risk of cirrhosis. Patients were classified as having high risk (CRS7 >0.7), intermediate risk (CRS7 0.5–0.7), or low risk (CRS7 <0.5) of cirrhosis using cutoffs based on a receiver operating characteristic (ROC) curve in the study in which CRS7 was developed.
Statistical Methods
Baseline patient characteristics for binary variables were assessed for linear trend across the low, intermediate, and high CRS7 risk groups by the Cochran-Armitage trend test [15]. Continuous and ordinal baseline variables were evaluated for correlation with CRS7 risk group by Kendall’s Tau-b correlation test. Differences in distribution of CRS7 risk group among the various sources of patients were assessed by the Cochran-Mantel-Haenszel Mean Score Test [16]. Differences in the distribution of CRS7 in HALT-C and non HALT-C subjects were assessed by the Kruskal-Wallis Test. Deviation from Hardy-Weinberg equilibrium was assessed by exact test of the allele frequencies [17]. Multiple statistical models were generated to assess the association of CRS7 risk group and the various liver disease outcomes. For fibrosis progression, cirrhosis, and clinical outcome analyses that included both African American and Caucasian subjects, two models were fit: one where CRS7 risk group was adjusted for race alone and another where CRS7 risk group was adjusted for race, treatment assignment, patient source of randomization (lead-in, breakthrough, relapse or express), and study site. Two models with and without adjustment were also fit for analyses including only Caucasian subjects. For the HCC outcome models that included both African American and Caucasian subjects, CRS7 risk group was adjusted for race alone and for race, treatment assignment, patient source of randomization, study site, and baseline cirrhosis status. Two HCC models with and without adjustment were fit for analyses including only Caucasian subjects.
The power to detect associations between CRS7 risk group and various endpoints was approximated by calculating the power to detect an association using the Cochran-Armitage trend test.
Fibrosis Progression and Cirrhosis
Exact times for ≥ 2-point fibrosis progression are unknown, but rather are only known to have occurred during the intervals of time between the scheduled biopsies (interval censored) at 1.5 and 3.5 years of follow-up.
Fibrosis progression hazard ratios for the intermediate and high CRS7 risk groups were estimated by proportional hazard models where the probabilities of progression were complementary log-log transformed to accommodate interval censoring of subjects for biopsies taken from 0–1.5 years and 1.5–3.5 years. Hazard ratios for these models were interpreted as the increase (or decrease) in risk of fibrosis progression that a patient in the high or intermediate CRS7 risk group assumes (at any time) relative to patients in the low CRS7 risk group. A test for linear trend of the proportion of subjects having fibrosis progression was conducted across the CRS7 low, intermediate, and high risk groups (coded in the model as 0, 1, and 2, respectively).
For cirrhosis detected on any biopsy, the association with the CRS7 risk group was evaluated by logistic regression. Odds ratios were reported for high and intermediate CRS7 risk groups relative to the low CRS7 risk group. Logistic regression models were also used to test for a linear trend of the proportion of subjects having cirrhosis across the CRS7 low, intermediate, and high risk groups.
Clinical Outcomes and HCC
Cox proportional hazards models were used to estimate the relative hazard of CRS7 risk group on time to hepatic decompensation and HCC through 8.7 years of observation. Hazard ratios for high and intermediate CRS7 risk groups were reported relative to the low CRS7 risk group. Kaplan-Meier estimates of the survivor functions for clinical outcome and HCC at 8 years of follow up were taken as estimates of the cumulative incidences of outcomes. Proportional hazards models were also used to test for a linear trend of the proportion of subjects having clinical outcome and HCC in the CRS7 low, intermediate, and high risk groups. Survival times were right censored and tied survival times were handled by the method of Efron [18].
All analyses except deviation from Hardy-Weinberg equilibrium (see above) were performed in SAS 9.1.3 software. Reported confidence intervals were generated by large sample normal approximation methods. All p-values were derived from two-sided tests.
RESULTS
Among the 1050 randomized patients, 166/191 (86.9%) African-American, 686/752 (91.2%) Caucasian patients and 99/107 (92.5%) Hispanic patients and patients of other race or ethnicity consented to genetic testing. Of these 951 patients, 13 did not have adequate DNA available for testing. The characteristics of the 938 patients included in this analysis are shown in Table 1. The mean age of the patients was 50.1 years and 70.7% were men. There were 677 Caucasian (72.2%) and 165 (17.6%) African-American patients. Five-hundred and fifty-three (59.0%) patients had bridging fibrosis at baseline (Ishak 2, 3 or 4); of these, 493 (89.2 %) were evaluated for fibrosis progression. Sixty patients with missing follow-up biopsy data were not evaluated for fibrosis progression. Reasons for missing follow-up biopsy data were that the patient reached a clinical outcome, withdrew, missed the visit, had an adverse event, had low platelets, refused the biopsy or had an inadequate biopsy. All 938 randomized patients were evaluated for cirrhosis, clinical outcome and HCC.
Table 1.
Baseline Characteristics of Patients
| All Patients (N = 938)
|
Caucasian Only (N = 677)
|
Fibrosis Progression Evaluated Pts (N = 493)
|
||||
|---|---|---|---|---|---|---|
| Mean/% | SD | Mean/% | SD | Mean/% | SD | |
| Demographics | ||||||
| Age, y | 50.11 | 7.1 | 49.9 | 6.9 | 50.2 | 7.1 |
| Gender, Male | 70.7% | - | 73.6% | - | 70.0% | -- |
| Race/ethnicity | ||||||
| African-American | 17.6% | - | 0% | - | 19.3% | - |
| Caucasian | 72.2% | 100% | 73.4% | |||
| Hispanic and Other | 10.2% | 0 | 7.3% | |||
| Source of randomization | - | |||||
| Lead-in | 63.2% | - | 61.2% | - | 61.0% | - |
| Express | 22.4% | - | 22.6% | - | 21.1% | - |
| Breakthrough | 3.3% | - | 3.1% | - | 3.7% | - |
| Relapser | 11.1% | - | 13.1% | - | 14.2% | - |
| Treatment assignment | - | - | ||||
| Peginterferon | 49.4% | - | 49.8% | - | 49.9% | - |
| Control | 50.6% | - | 50.2% | - | 50.1% | - |
| Body Mass Index (BMI) | 30.0 | 5.5 | 29.7 | 5.2 | 29.7 | 5.5 |
| Viral Factors | ||||||
| Log10 HCV RNA, IU/mL | 6.42 | 0.53 | 6.47 | 0.51 | 6.52 | 0.47 |
| Duration of infection, y | 28.1 | 8.0 | 28.1 | 7.7 | 27.7 | 7.9 |
| Laboratory Values | ||||||
| Hemoglobin, g/dL | 15.0 | 1.4 | 15.1 | 1.4 | 15.1 | 1.4 |
| White blood cells, x1000/mm3 | 5.78 | 1.88 | 5.84 | 1.88 | 5.91 | 1.81 |
| Platelets, x1000/mm3 | 164.3 | 65.1 | 163.9 | 65.3 | 187.8 | 60.3 |
| Albumin, g/dL | 3.86 | 0.39 | 3.90 | 0.39 | 3.95 | 0.34 |
| AST, U/L | 87.2 | 57.0 | 85.5 | 58.3 | 76.6 | 53.4 |
| ALT, U/L | 106.4 | 76.7 | 105.8 | 77.3 | 101.3 | 80.8 |
| Alkaline Phosphatase, U/L | 100.0 | 45.4 | 93.0 | 38.4 | 91.4 | 42.2 |
| Total bilirubin, mg/dL | 0.79 | 0.41 | 0.80 | 0.41 | 0.72 | 0.36 |
| Prothrombin time, INR | 1.04 | 0.11 | 1.04 | 0.10 | 1.01 | 0.10 |
| AFP, ng/mL | 17.3 | 28.4 | 15.5 | 27.9 | 13.4 | 23.8 |
| Liver histology | ||||||
| Ishak fibrosis score | - | |||||
| 5 or 6 | 41.0% | - | 40.5% | - | 0% | - |
| <5 | 59.0% | - | 59.5% | - | 100% | - |
| Esophageal varices on EGD | 25.9% | - | 28.2% | - | 15.8% | - |
| CRS7 | 0.64 | 0.18 | 0.63 | 0.19 | 0.64 | 0.18 |
Table 2 shows baseline characteristics of the patients by CRS7 risk group. Four-hundred and forty (47%) patients were in the high CRS7 risk group, 310 (33%) were in the intermediate CRS7 risk group and 188 (20%) were in the low CRS7 risk group. High and intermediate CRS7 risk groups had a significantly higher proportion of African-American patients, Hispanic patients and patients of other races. Caucasians had lower proportions of patients in the high and intermediate CRS7 risk groups. Differences were also observed in the distributions of age and serum alkaline phosphatase across the CRS7 risk groups. The CRS7 risk groups were otherwise similar in demographic, laboratory and other baseline factors including Ishak fibrosis score.
Table 2.
Baseline Characteristics of Patients in the High, Intermediate and Low CRS7 Risk Groups
| CRS7 Risk Group(N = 938) | p-value | ||||||
|---|---|---|---|---|---|---|---|
| High, >0.7 (n= 440)
|
Intermediate, 0.5–0.7 (n= 310)
|
Low, <0.5 (n= 188)
|
|||||
| Mean/% | SD | Mean/% | SD | Mean/% | SD | ||
| Demographics | |||||||
| Age, y | 49.6 | 7.0 | 50.3 | 7.4 | 50.9 | 6.8 | 0.02* |
| Gender, Male | 72.3% | - | 68.7% | - | 70.2% | - | 0.46§ |
| Race/ethnicity | - | - | |||||
| African-American | 19.6% | - | 20.0% | - | 9.0% | ||
| Caucasian | 68.6 | - | 68.1% | - | 87.2% | <0.00 | |
| Hispanic and Other | 11.8 | - | 11.9% | - | 3.7% | - | 1‡ |
| Source of randomization | - | - | 0.53‡ | ||||
| Lead-in | 62.7% | - | 65.2% | - | 61.2% | - | |
| Express | 22.3% | - | 21.9% | - | 23.4% | - | |
| Breakthrough | 3.6% | - | 3.2% | - | 2.7% | - | |
| Relapser | 11.4% | - | 9.7% | - | 12.8% | - | |
| Treatment assignment | - | ||||||
| Peginterferon | 47.7% | - | 49.4% | - | 53.2% | - | 0.22§ |
| Control | 52.3% | - | 50.6% | - | 46.8% | - | |
| Body Mass Index (BMI) | 29.9 | 5.6 | 30.1 | 5.2 | 30.1 | 5.5 | 0.46* |
| Viral Factors | |||||||
| Log10 HCV RNA, IU/mL | 6.45 | 0.50 | 6.39 | 0.55 | 6.40 | 0.55 | 0.19* |
| Duration of infection, y | 28.3 | 8.1 | 27.9 | 8.1 | 28.2 | 7.7 | 0.81* |
| Laboratory Values | |||||||
| Hemoglobin, g/dL | 15.0 | 1.4 | 14.9 | 1.5 | 15.0 | 1.3 | 0.50* |
| White blood cells, x1000/mm3 | 5.82 | 1.98 | 5.69 | 1.80 | 5.85 | 1.80 | 0.99* |
| Platelets, x1000/mm3 | 165.5 | 67.0 | 163.7 | 65.6 | 162.3 | 60.1 | 0.70* |
| Albumin, g/dL | 3.85 | 0.39 | 3.83 | 0.39 | 3.93 | 0.41 | 0.09* |
| AST, U/L | 83.4 | 55.4 | 94.9 | 61.9 | 83.4 | 51.3 | 0.28* |
| ALT, U/L | 102.2 | 77.9 | 115.1 | 81.5 | 101.7 | 63.9 | 0.35* |
| Alkaline Phosphatase, U/L | 100.4 | 42.3 | 104.0 | 48.4 | 92.7 | 46.5 | 0.03* |
| Total bilirubin, mg/dL | 0.79 | 0.40 | 0.81 | 0.41 | 0.78 | 0.42 | 0.82* |
| Prothrombin time, INR | 1.04 | 0.10 | 1.04 | 0.11 | 1.04 | 0.12 | 0.98* |
| AFP, ng/mL | 15.9 | 22.0 | 18.1 | 27.9 | 19.2 | 40.0 | 0.39* |
| Liver histology | |||||||
| Ishak fibrosis score | - | - | - | ||||
| 5 or 6 | 42.5% | - | 40.6% | - | 38.3% | - | 0.32§ |
| <5 | 57.5% | - | 59.4% | - | 61.7% | - | |
| Esophageal varices on EGD | 26.2% | - | 25.9% | - | 25.1% | - | 0.80§ |
| CRS7 | 0.80 | 0.06 | 0.60 | 0.04 | 0.35 | 0.10 | - |
Kendall’s Tau-b Correlation Test,
Cochran-Armitage Trend Test,
Cochran-Mantel-Haenszel Mean Score Test. All p-values are derived from two-sided tests.
Table 3 shows individual SNP carrier frequencies by race for the 7 SNPs included in the CRS7 score, as well as the CRS7 score as a continuous variable. The HALT-C population is shown for all patients and by race. Comparison groups are from HapMap and two cohorts of patients with chronic hepatitis C that were included in previous studies on genetic markers of fibrosis conducted at Celera Diagnostics [8, 19]. No significant differences in CRS7 score were observed between HALT-C and non-HALT-C sample sets (p=0.72 for African American subjects, p=0.21 for Caucasian subjects).
Table 3.
Individual SNP Carrier Frequencies and CRS7 Mean (SD) by Race
| SNP | Gene | Risk | HALT-C | Cau (HapMap) (N=57) | AA (Internal Celera)* (N=216) | YRI (HapM ap) | |||
|---|---|---|---|---|---|---|---|---|---|
| All (N=938) | Cau (N=677) | AA (N=165) | Hisp & Others (N=96) | ||||||
| rs62522600 | AZIN1 | GG | 0.91 | 0.88 | 0.98 | 0.95 | 0.87 | 0.84 | n/a |
| rs4986791 | TLR4 | CC | 0.92 | 0.90 | 0.96 | 0.97 | 0.87 | 0.95 | 0.98 |
| rs886277 | TRPM5 | CC/CT | 0.66 | 0.63 | 0.79 | 0.67 | 0.64 | 0.65 | 0.86 |
| rs2290351 | AP3S2 | AA/AG | 0.44 | 0.39 | 0.59 | 0.53 | 0.42 | 0.56 | 0.61 |
| rs4290029 | BC008027 | GG | 0.67 | 0.70 | 0.54 | 0.68 | 0.72 | 0.72 | 0.52 |
| rs17740066 | STXBP5L | AA/AG | 0.18 | 0.21 | 0.02 | 0.21 | 0.19 | 0.04 | 0.00 |
| rs2878771 | AQP2 | GG | 0.67 | 0.64 | 0.77 | 0.71 | 0.67 | 0.68 | 0.76 |
| CRS7 | - | - | 0.64 (0.18) | 0.63 (0.19) | 0.67 (0.16) | 0.69 (0.15) | 0.61 (0.18) | 0.67 (0.16) | n/a |
Fibrosis Progression
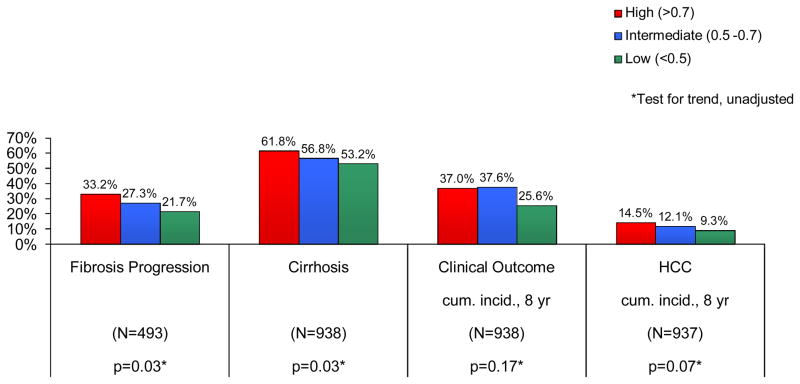
Among the patients with bridging fibrosis on baseline liver biopsy, 29% (142/493) had fibrosis progression defined as ≥ 2-point increase in Ishak fibrosis score: 33.2%, 27.3% and 21.7% in the high, intermediate and low CRS7 risk groups, respectively (p=0.03) (Fig. 1). In multivariate models predicting fibrosis progression and adjusted for race, patient type, clinical site and treatment assignment, the Hazard Ratio (HR) for CRS7 (trend test) was 1.27 (95%CI: 1.01–1.58, p=0.04). The HR per 0.1-unit change in continuous CRS7 was 1.13 (95% CI 1.04–1.22, p=0.003) (Table 4). A model fit with CRS7 adjusted for race, gender, and the interaction effects of CRS7 and gender, did not indicate any differential risk of fibrosis progression between women and men across the CRS7 risk groups (p for interaction with intermediate CRS7=0.49, with high CRS7=0.72, data not shown). Results for models with Caucasians only were similar.
Fig. 1.
Outcomes of all patients according to CRS7 Risk Group
Table 4.
CRS7 Risk Group and Risks of Outcomes
| All patients – Multivariate§§ | Caucasians only - Multivariate§§ | ||||||
|---|---|---|---|---|---|---|---|
| CRS7 Risk Group | N | OR/HR (95% CI) | p-value | N | OR/HR (95% CI) | p-value | |
| Fibrosis Progression* | High | 226 | 1.61 (1.00–2.59) | 0.05 | 158 | 1.60 (0.95–2.70) | 0.08 |
| Int | 161 | 1.28 (0.76–2.15) | 0.35 | 114 | 1.30 (0.74–2.30) | 0.36 | |
| Low | 106 | - | - | 90 | - | - | |
| Trend | 493 | 1.27 (1.01–1.58) | 0.04 | 362 | 1.26 (0.98–1.62) | 0.07 | |
| CRS7a Continuous | 493 | 1.13 (1.04–1.22) | 0.003 | 362 | 1.14 (1.04–1.25) | 0.004 | |
|
| |||||||
| Cirrhosis§ | High | 440 | 1.39 (0.98–1.99) | 0.07 | 302 | 1.39 (0.93–2.07) | 0.11 |
| Int | 310 | 1.13 (0.77–1.64) | 0.53 | 211 | 1.00 (0.66–1.53) | 1.00 | |
| Low | 188 | - | - | 164 | - | - | |
| Trend | 938 | 1.19 (1.00–1.41) | 0.05 | 677 | 1.19 (0.98–1.45) | 0.08 | |
| CRS7a Continuous | 938 | 1.07 (0.99–1.15) | 0.07 | 677 | 1.08 (0.99–1.17) | 0.08 | |
|
| |||||||
| Clinical outcome‡ | High | 440 | 1.36 (0.95–1.95) | 0.09 | 302 | 1.34 (0.89–2.02) | 0.16 |
| Int | 310 | 1.30 (0.89–1.90) | 0.17 | 211 | 1.47 (0.97–2.25) | 0.07 | |
| Low | 188 | - | - | 164 | - | - | |
| Trend | 938 | 1.14 (0.97–1.35) | 0.12 | 677 | 1.12 (0.93–1.36) | 0.23 | |
| CRS7a Continuous | 938 | 1.05 (0.98–1.13) | 0.15 | 677 | 1.05 (0.97–1.14) | 0.21 | |
|
| |||||||
| HCC‡ | High | 440 | 1.65 (0.84–3.24) | 0.15 | 302 | 1.88 (0.88–4.01) | 0.10 |
| Int | 309 | 1.09 (0.52–2.28) | 0.83 | 210 | 0.89 (0.36–2.17) | 0.79 | |
| Low | 188 | - | - | 164 | - | - | |
| Trend | 937 | 1.34 (0.97–1.85) | 0.07 | 676 | 1.48 (1.00–2.18) | 0.05 | |
| CRS7a Continuous | 937 | 1.09 (0.96–1.25) | 0.19 | 676 | 1.14 (0.97–1.33) | 0.11 | |
Complementary Log-Log Model (HR).
Logistic Regression Model (OR),
Cox Proportional Hazards Model (HR).
Fibrosis Progression, cirrhosis and clinical outcome multivariate models were adjusted for race, patient type, clinical site and treatment assignment.
OR/HR for 0.1-unit change in continuous CRS7
Multivariate HCC models were adjusted for race, treatment assignment, patient type, clinical site and cirrhosis at baseline.
All p-values are derived from two-sided tests.
Cirrhosis
Cirrhosis was present on one or more biopsies in 58% (548/938) patients, 61.8%, 56.8% and 53.2% in the high, intermediate and low CRS7 risk groups, respectively (p=0.03) (Fig. 1). In multivariate models predicting cirrhosis and adjusted for race, treatment assignment, patient type and clinical site, the Odds Ratio (OR) for CRS7 (trend test) was 1.19 (95%CI: 1.00–1.41, p=0.05). The OR per 0.1-unit change in continuous CRS7 was 1.07 (95% CI 0.99–1.15, p=0.07) (Table 4). A model fit with CRS7 risk group adjusted for race, gender, and the interaction effects of CRS7 and gender, did not indicate any differential risk of cirrhosis between women and men across the CRS7 risk groups (p for interaction with intermediate CRS7 = 0.42, with high CRS7=0.63, data not shown). Results for models with Caucasians only were similar.
Clinical Outcome
Two hundred and forty-eight patients developed at least one clinical outcome (cumulative incidence = 34.8% at 8 years of follow up), 37.0%, 37.6% and 25.6% in the high, intermediate and low CRS7 risk groups, respectively (p=0.17) (Fig. 1). In multivariate models predicting clinical outcome and adjusted for race, patient type, clinical site and treatment assignment, the HR for CRS7 (trend test) was 1.14 (95%CI: 0.97–1.35, p=0.12). The HR per 0.1-unit change in continuous CRS7 was 1.05 (95% CI 0.98–1.13, p=0.15) (Table 4). Results for models with Caucasians only were similar.
HCC
Seventy-six patients met HALT-C criteria for HCC (cumulative incidence = 12.6% at 8 years of follow up), 14.5%, 12.1% and 9.3% in the high, intermediate and low CRS7 risk groups respectively, p=0.07 (Fig. 1). In multivariate models predicting HCC and adjusted for race, patient type, clinical site, treatment assignment and presence of cirrhosis at baseline, the HR for CRS7 (trend test) was 1.34 (95%CI: 0.97–1.85, p=0.07). The HR per 0.1-unit change in continuous CRS7 was 1.09 (95% CI 0.96–1.25, p=0.19) (Table 4). Results for models with Caucasians only were similar.
Individual SNPs and outcomes
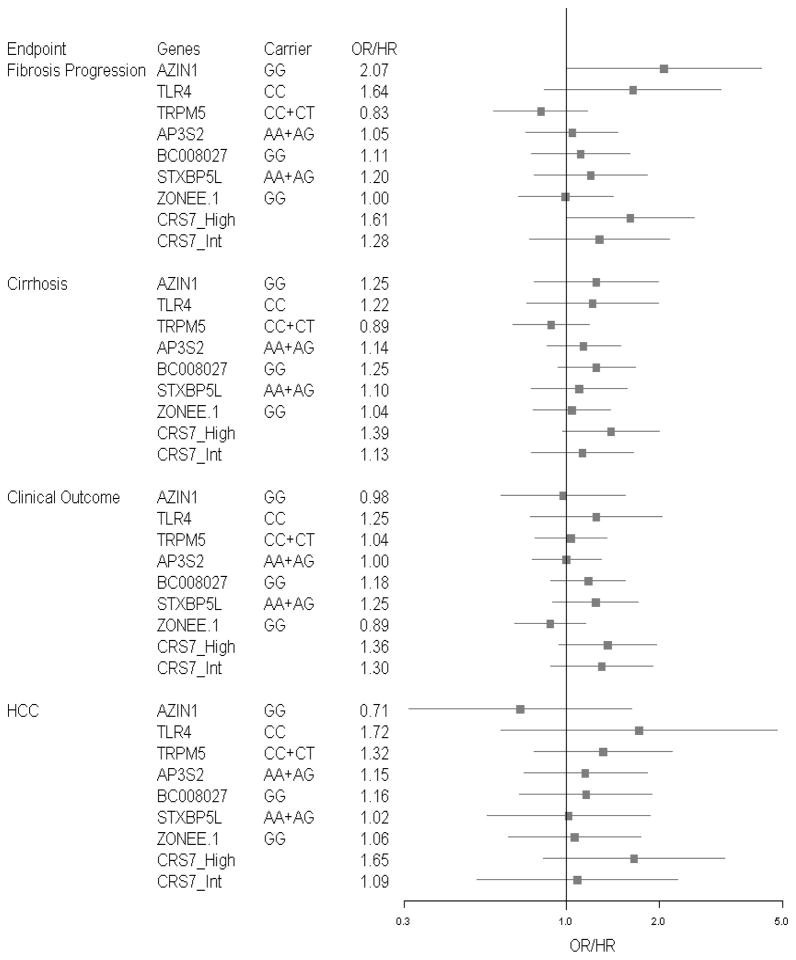
Figure 2 shows ORs and HRs for the individual SNPs and CRS7 risk groups for all outcomes. Of the 7 SNPs, AZIN1 was significantly associated with fibrosis progression. The other 6 individual SNPs studied were not associated with any of the outcomes analyzed.
Fig. 2.
Forest plots of individual SNPs and CRS7 risk groups for HALT-C patients – all outcomes
DISCUSSION
This analysis demonstrated that among patients with chronic hepatitis C, a higher rate of progression of fibrosis is associated with higher CRS7 scores, thus extending and validating the results of the original studies in which CRS7 was first developed and evaluated. However, the association is modest and is unlikely to be used to alter patient management. Because all patients in the HALT-C Trial had advanced fibrosis before enrollment, this cohort posed a stringent test for the predictive signature and suggests that the cirrhosis risk score is involved along the disease continuum. Indeed, the study by Marcolongo et al. found that the association between CRS7 and fibrosis progression was most evident in patients who had Metavir F0 on the initial biopsy [9].
Although CRS7 was derived originally from a cohort of Caucasian patients, it is noteworthy that our results were similar when the analysis included Caucasians, African-Americans, Hispanic and other ethnicities or Caucasians alone. It is possible that race is a potential confounder since it is associated with both allele frequencies and the outcomes of interest. We have attempted to address this in two ways. First, the odds ratios and hazard ratios among all randomized subjects were estimated from multivariate regression models which included race as a covariate, thus controlling for differences in allele frequencies. Second, we compared results among Caucasians only and all subjects and found that the results were similar. We acknowledge that appropriately powered studies including other ethnic groups should be analyzed separately to validate the utility of CRS7 as a predictor of fibrosis progression and cirrhosis in non-Caucasians.
CRS7 was less accurate in predicting clinical outcomes and HCC than in predicting severity of fibrotic liver disease. Statistical power to observe an association of CRS7 with the outcomes studied was constrained by the total number of subjects and the number of events observed for each outcome. The power to observe a significant trend test reached 90% when the odds/hazard ratio (trend test) was at least 1.3 for clinical outcome or at least 1.7 for HCC. Thus, this study would not have reliably found associations between CRS7 and clinical outcome or HCC of a lesser magnitude. Patients with high and intermediate risk scores in this study had a greater risk of clinical outcomes than patients with low risk scores, but the trend across scores was not statistically significant (p=0.12). Patients with high risk scores were more likely to develop HCC, statistically significant in unadjusted analysis, and a trend of borderline statistical significance in multivariate analysis (p=0.07). This association of HCC with high CRS7 was unexpected because of the relatively small number of patients (only 76) who developed HCC. Additional studies that include a larger number of well-characterized patients with HCC and controls should be performed to further evaluate the possible association between CRS7 and HCC and to determine if this is observed in patients with other etiologies of liver disease.
There are several limitations of this study that merit consideration. First, HALT-C patients entered the trial with advanced disease relative to the cross-sectional study from which CRS7 was derived. If some of the genetic variants in CRS7 score were associated with early steps in fibrosis progression, they would not have been observed in this cohort. Second, even though treatment in the HALT-C trial was reported not to alter outcomes, this study differed from earlier studies in that half of the patients were randomized to receive low dose pegylated interferon for up to 3.5 years. Third, the lower hazard for the score categories may be due to regression to the mean commonly observed with replication studies. Candidate genes are chosen for verification precisely because of their strong association in an initial study. In a replication study, the association may not be as strong.
There are many strengths in this study. Reported genetic associations have a long history of false positive results [20], i.e., not validated in external cohorts. The previous validations of CRS7 were performed in relatively small sample sets (N=154 subjects in the Huang et al. validation set, N=271 in Marcolongo’s study and N=56 in Trepo’s study). The large number of patients in this study (n=938) provides a needed additional level of evidence for this reported association [8–10]. In addition, previous longitudinal studies of CRS7 assessed subjects with varying intervals between biopsies while HALT-C provides a well-defined interval between biopsies (biopsies were performed at baseline, 1.5 and 3.5 years of follow-up), allowing estimates of the absolute risk of fibrosis progression across CRS7 risk groups during this time period. Further, the rigor of histological assessment was higher in HALT-C than in previous studies. In those studies, fibrosis staging was assessed by individual site pathologists while in the HALT-C trial, all biopsies were staged centrally by a panel of expert liver pathologists and fibrosis stage was assigned based on consensus of this panel.
Finally, this study included Caucasians, African-Americans as well as Hispanics and patients of other races while the other studies included Caucasians only. The results showed that CRS7 was predictive of fibrosis progression and of cirrhosis not only among Caucasians but also in the combined population of Caucasians, African-Americans, and to a much lesser extent other ethnic groups.
In conclusion, in a carefully characterized cohort of patients with chronic hepatitis C and advanced fibrosis, CRS7 was associated with fibrosis progression and with cirrhosis, the basis on which it was originally constructed. The level of characterization of CRS7 prior to this study, the prior adjustment for demographic and clinical risk factors for individual SNPs, and use of a “machine learning” algorithm to optimize included SNPs rather than simple assembly of associated individual SNPs to generate the score may have been key to the observed confirmation. Associations with clinical outcomes, including hepatic decompensation and HCC were also observed, but the associations were not as strong and the validity of these associations needs to be confirmed. Among the individual SNPs, AZIN1 was most strongly associated with fibrosis progression and cirrhosis. The association of a genetic risk score (CRS7) with disease progression should encourage mechanistic studies of the included genes to gain insight into the potential underlying biological mechanisms involved. The successful confirmation of this risk score should encourage further investigation of these gene variants and the combined score in other randomized controlled trials involving the endpoints studied.
Acknowledgments
This study was supported by the National Institute of Diabetes & Digestive & Kidney Diseases (contract numbers are listed below). Additional support was provided by the National Institute of Allergy and Infectious Diseases (NIAID), the National Cancer Institute, the National Center for Minority Health and Health Disparities and by General Clinical Research Center and Clinical and Translational Science Center grants from the National Center for Research Resources, National Institutes of Health (grant numbers are listed below). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Center for Research Resources or the National Institutes of Health. Additional funding to conduct this study was supplied by Hoffmann-La Roche, Inc., and Celera Corporation through a Cooperative Research and Development Agreements (CRADAs) with the National Institutes of Health.
The authors express their gratitude to the HALT-C patients and to Joe Catanese and the Celera High Throughput group for generating genotyping data, to Dr. David Ross and Computational Biology group for bioinformatics support, and to Yonghong Li for continued discussions of results. In addition to the authors of this manuscript, the following individuals were instrumental in the planning, conduct and/or care of patients enrolled in this study at each of the participating institutions as follows:
University of Massachusetts Medical Center, Worcester, MA: (Contract N01-DK-9-2326) Gyongyi Szabo, MD, Barbara F. Banner, MD, Maureen Cormier, RN, Donna Giansiracusa, RN
University of Connecticut Health Center, Farmington, CT: (Grant M01RR-06192) Herbert L. Bonkovsky, MD, Gloria Borders, RN, Michelle Kelley, RN, ANP
Saint Louis University School of Medicine, St Louis, MO: (Contract N01-DK-9-2324) Adrian M. Di Bisceglie, MD, Bruce Bacon, MD, Brent Neuschwander-Tetri, MD, Elizabeth M. Brunt, MD, Debra King, RN
Massachusetts General Hospital, Boston, MA: (Contract N01-DK-9-2319, Grant M01RR-01066; Grant 1 UL1 RR025758-01, Harvard Clinical and Translational Science Center) Jules L. Dienstag, MD, Raymond T. Chung, MD, Andrea E. Reid, MD, Atul K. Bhan, MD, Wallis A. Molchen, David P. Lundmark
University of Colorado Denver, School of Medicine, Aurora, CO: (Contract N01-DK-9-2327, Grant M01RR-00051, Grant 1 UL1 RR 025780-01), Gregory T. Everson, MD, Thomas Trouillot, MD, Marcelo Kugelmas, MD, S. Russell Nash, MD, Jennifer DeSanto, RN, Carol McKinley, RN
University of California - Irvine, Irvine, CA: (Contract N01-DK-9-2320, Grant M01RR-00827) Timothy R. Morgan, MD, John C. Hoefs, MD, John R. Craig, MD, M. Mazen Jamal, MD, MPH, Muhammad Sheikh, MD, Choon Park, RN
University of Texas Southwestern Medical Center, Dallas, TX: (Contract N01-DK-9-2321, Grant M01RR-00633, Grant 1 UL1 RR024982-01, North and Central Texas Clinical and Translational Science Initiative) William M. Lee, MD, Thomas E. Rogers, MD, Peter F. Malet, MD, Janel Shelton, Nicole Crowder, LVN, Rivka Elbein, RN, BSN, Nancy Liston, MPH
University of Southern California, Los Angeles, CA: (Contract N01-DK-9-2325, Grant M01RR-00043) Karen L. Lindsay, MD, MMM, Sugantha Govindarajan, MD, Carol B. Jones, RN, Susan L. Milstein, RN
University of Michigan Medical Center, Ann Arbor, MI: (Contract N01-DK-9-2323, Grant M01RR-00042, Grant 1 UL1 RR024986, Michigan Center for Clinical and Health Research) Robert J. Fontana, MD, Joel K. Greenson, MD, Pamela A. Richtmyer, LPN, CCRC, R. Tess Bonham, BS
Virginia Commonwealth University Health System, Richmond, VA: (Contract N01-DK-9-2322, Grant M01RR-00065) Mitchell L. Shiffman, MD, Richard K. Sterling, MD, MSc, Melissa J. Contos, MD, A. Scott Mills, MD, Charlotte Hofmann, RN, Paula Smith, RN
Liver Diseases Branch, National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, Bethesda, MD: Marc G. Ghany, MD, T. Jake Liang, MD, David Kleiner, MD, PhD, Yoon Park, RN, Elenita Rivera, RN, Vanessa Haynes-Williams, RN
National Institute of Diabetes and Digestive and Kidney Diseases, Division of Digestive Diseases and Nutrition, Bethesda, MD: Leonard B. Seeff, MD, Patricia R. Robuck, PhD, Jay H. Hoofnagle, MD, Elizabeth C. Wright, PhD
University of Washington, Seattle, WA: (Contract N01-DK-9-2318) Chihiro Morishima, MD, David R. Gretch, MD, PhD, Minjun Chung Apodaca, BS, ASCP, Rohit Shankar, BC, ASCP, Natalia Antonov, M. Ed.
New England Research Institutes, Watertown, MA: (Contract N01-DK-9-2328) Kristin K. Snow, MSc, ScD, Anne M. Stoddard, ScD, Deepa Naishadham, MA, MS
Inova Fairfax Hospital, Falls Church, VA: Zachary D. Goodman, MD, PhD, Fanny Monge, Michelle Parks
Data and Safety Monitoring Board Members: (Chair) Gary L. Davis, MD, Guadalupe Garcia-Tsao, MD, Michael Kutner, PhD, Stanley M. Lemon, MD, Robert P. Perrillo, MD
Abbreviations
- AFP
alpha fetoprotein
- ALT
alanine aminotransferase
- APRI
AST-platelet ratio index
- AST
aspartate
- AUROC curve
area under the receiver operating characteristic curve
- AZIN
antizyme inhibitor
- CHC
chronic hepatitis C
- CI
confidence interval
- CRS
cirrhosis risk score
- CT
computed tomography
- HALT-C
Hepatitis C Antiviral Long-term Treatment against Cirrhosis
- HCC
hepatocellular carcinoma
- HCV
hepatitis C virus
- HR
hazard ratio
- INR
international normalized ratio of prothrombin time
- MRI
magnetic resonance imaging
- OR
odds ratio
- SNP
single nucleotide polymorphism
- SVR
sustained virologic response
- TLR
toll like receptor
Footnotes
The HALT-C Trial was registered with clinicaltrials.gov (#NCT00006164).
Conflicts of Interest and Source of Funding: A. S. Lok is a consultant and receives research support from Hoffmann-La Roche, Inc. R. J. Lagier, C. M. Rowland, and J. J. Sninsky are employees and have equity interest with Celera Corporation. T. M. Curto and J. E. Everhart have no financial relationship related to this project.
References
- 1.Ghany MG, Strader DB, Thomas DL, Seeff LB. Diagnosis, management, and treatment of hepatitis C: an update. Hepatology. 2009;49:1335–1374. doi: 10.1002/hep.22759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Poynard T, Bedossa P, Opolon P. Natural history of liver fibrosis progression in patients with chronic hepatitis C. The OBSVIRC, METAVIR, CLINIVIR, and DOSVIRC groups. Lancet. 1997;349:825–832. doi: 10.1016/s0140-6736(96)07642-8. [DOI] [PubMed] [Google Scholar]
- 3.Seeff LB. The history of the “natural history” of hepatitis C (1968–2009) Liver Int. 2009;29 (Suppl 1):89–99. doi: 10.1111/j.1478-3231.2008.01927.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Karlsen TH, Melum E, Franke A. The utility of genome-wide association studies in hepatology. Hepatology. 2010;51:1833–1842. doi: 10.1002/hep.23564. [DOI] [PubMed] [Google Scholar]
- 5.Ge D, Fellay J, Thompson AJ, Simon JS, Shianna KV, Urban TJ, Heinzen EL, et al. Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature. 2009;461:399–401. doi: 10.1038/nature08309. [DOI] [PubMed] [Google Scholar]
- 6.Thomas DL, Thio CL, Martin MP, Qi Y, Ge D, O’Huigin C, Kidd J, et al. Genetic variation in IL28B and spontaneous clearance of hepatitis C virus. Nature. 2009;461:798–801. doi: 10.1038/nature08463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bataller R, North KE, Brenner DA. Genetic polymorphisms and the progression of liver fibrosis: a critical appraisal. Hepatology. 2003;37:493–503. doi: 10.1053/jhep.2003.50127. [DOI] [PubMed] [Google Scholar]
- 8.Huang H, Shiffman ML, Friedman S, Venkatesh R, Bzowej N, Abar OT, Rowland CM, et al. A 7 gene signature identifies the risk of developing cirrhosis in patients with chronic hepatitis C. Hepatology. 2007;46:297–306. doi: 10.1002/hep.21695. [DOI] [PubMed] [Google Scholar]
- 9.Marcolongo M, Young B, Dal Pero F, Fattovich G, Peraro L, Guido M, Sebastiani G, et al. A seven-gene signature (cirrhosis risk score) predicts liver fibrosis progression in patients with initially mild chronic hepatitis C. Hepatology. 2009;50:1038–1044. doi: 10.1002/hep.23111. [DOI] [PubMed] [Google Scholar]
- 10.Trepo E, Potthoff A, Pradat P, Bakshi R, Young B, Lagier R, Moreno C, et al. Role of a cirrhosis risk score for the early prediction of fibrosis progression in hepatitis C patients with minimal liver disease. J Hepatol. 2011;55:38–44. doi: 10.1016/j.jhep.2010.10.018. [DOI] [PubMed] [Google Scholar]
- 11.Lee WM, Dienstag JL, Lindsay KL, Lok AS, Bonkovsky HL, Shiffman ML, Everson GT, et al. Evolution of the HALT-C Trial: pegylated interferon as maintenance therapy for chronic hepatitis C in previous interferon nonresponders. Control Clin Trials. 2004;25:472–492. doi: 10.1016/j.cct.2004.08.003. [DOI] [PubMed] [Google Scholar]
- 12.Di Bisceglie AM, Shiffman ML, Everson GT, Lindsay KL, Everhart JE, Wright EC, Lee WM, et al. Prolonged therapy of advanced chronic hepatitis C with low-dose peginterferon. N Engl J Med. 2008;359:2429–2441. doi: 10.1056/NEJMoa0707615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ishak K, Baptista A, Bianchi L, Callea F, De Groote J, Gudat F, Denk H, et al. Histological grading and staging of chronic hepatitis. J Hepatol. 1995;22:696–699. doi: 10.1016/0168-8278(95)80226-6. [DOI] [PubMed] [Google Scholar]
- 14.Lok AS, Seeff LB, Morgan TR, di Bisceglie AM, Sterling RK, Curto TM, Everson GT, et al. Incidence of hepatocellular carcinoma and associated risk factors in hepatitis C-related advanced liver disease. Gastroenterology. 2009;136:138–148. doi: 10.1053/j.gastro.2008.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Slager SL, Schaid DJ. Case-control studies of genetic markers: power and sample size approximations for Armitage’s test for trend. Hum Hered. 2001;52:149–153. doi: 10.1159/000053370. [DOI] [PubMed] [Google Scholar]
- 16.Landis JR, Heyman ER, Koch GG. Average partial association in three-way contingency tables: a review and discussion of alternative tests. Intl Statistical Rev. 1978;46:237–254. [Google Scholar]
- 17.Weir BS. Genetic Data Analysis II. Sunderland: Sinauer Associates Inc; 1996. [Google Scholar]
- 18.Efron B. The efficience of Cox’s likelihood function for censored data. Am J Stat Assoc. 1977;72:557–565. [Google Scholar]
- 19.Huang H, Shiffman ML, Cheung RC, Layden TJ, Friedman S, Abar OT, Yee L, et al. Identification of two gene variants associated with risk of advanced fibrosis in patients with chronic hepatitis C. Gastroenterology. 2006;130:1679–1687. doi: 10.1053/j.gastro.2006.02.032. [DOI] [PubMed] [Google Scholar]
- 20.Ioannidis JP, Tarone R, McLaughlin JK. The False-positive to False-negative Ratio in Epidemiologic Studies. Epidemiology. 2011;22:450–456. doi: 10.1097/EDE.0b013e31821b506e. [DOI] [PubMed] [Google Scholar]