Abstract
The ybxI gene of Bacillus subtilis 168 encodes a preprotein of 267 amino acid residues, including a putative signal peptide of 23 residues. The YbxI primary structure exhibits high similarity scores with two members of the superfamily of the serine penicillin-recognizing enzymes: the class D β-lactamases and the hydrophilic carboxy-terminal domains of the BlaR and MecR penicillin receptors. To determine the function and the activity of this putative penicillin-recognizing enzyme, we have subcloned the ybxI gene in the pET-26b expression vector. Transformation of Escherichia coli BL21(DE3) by the recombinant plasmid pCIP51 resulted in the export of the mature YbxI in the periplasm as a water-soluble protein. The recombinant protein was purified to 95% homogeneity. YbxI interacts with several β-lactam antibiotics and can hydrolyze some of them. YbxI is not inactivated by clavulanic acid. The YbxI function and its enzymatic activity in B. subtilis remain unknown. The acyl-enzyme obtained after incubation of YbxI with a fluorescent derivative of ampicillin can be detected by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, confirming that YbxI can be acylated by β-lactam antibiotics. YbxI does not hydrolyze some of the standard substrates of d-alanyl-d-alanine peptidases, the targets of penicillin. YbxI belongs to the penicillin-recognizing enzyme family but has an activity intermediate between those of a penicillin-binding protein and a β-lactamase.
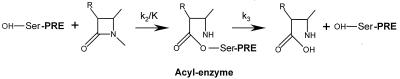
Two families of bacterial serine enzymes recognize penicillin and other β-lactam antibiotics and form a superfamily of evolutionarily related proteins named serine penicillin-recognizing enzymes (PREs) (21). The first family contains the d-alanyl-d-alanine peptidases (dd-transpeptidases) involved in the biosynthesis or in the control of the cross-linking of the peptidoglycan, the main constituent of the bacterial cell wall. dd-peptidases are inactivated by β-lactam antibiotics, as a result of the acylation of their active serine by the antibiotic, which yields a very stable covalent adduct (Fig. 1). Because of these properties, these enzymes are also referred to as penicillin-binding proteins (PBPs) (10). The members of the second family, the serine β-lactamases are subdivided into three classes (A, C, and D) and efficiently hydrolyze the amide bond of the β-lactam nucleus, to yield products devoid of antibiotic activity (12, 31). For serine β-lactamases, the catalytic pathway involves the formation of an acyl-enzyme similar to that obtained in the interaction of the β-lactam antibiotic with the dd-peptidases, but in the case of β-lactamases the acyl-enzyme is very unstable. Although one might think that they would be very different, the two families of enzymes share many biochemical characteristics and very similar three-dimensional (3D) structures (17, 22, 33, 36). Along their primary structures, three conserved peptide sequences, important for recognition of the substrate or catalysis, have been identified by comparison of their 3D structures (Table 1). Sequence identities between the class D and the class A and C enzymes are on average 16% (29). Nevertheless, these proteins are thought to have a common evolutionary history (15, 21, 24).
FIG. 1.
Interaction between the serine PREs with β-lactam antibiotics. k2/K and k3 are the acylation and the deacylation rate constants, respectively. For β-lactamases, k3 is large (≥1,000 s−1 for good substrates), and for dd-peptidases and PBPs k3 is low (≤10−3 s−1).
TABLE 1.
Conserved active-site elements of the serine penicillin-recognizing enzymes
| Enzyme | Element
|
||
|---|---|---|---|
| 1 | 2 | 3 | |
| β-Lactamases | |||
| Class A | SXXKa | SDN | (K/R)(S/T)G |
| Class C | SXXK | YAN | KTG |
| Class D | SXXK | SXV | K(T/S)G |
| dd-Transpeptidases | S(T/S)FK | (Y/S)XN | (H/K)(T/S)G |
| YbxI | STFK | SAI | KTG |
| BlaR/MecR-CTD | STYK | S(T/V)(T/N) | KTG |
Note that several class A enzymes also exhibit an STFK sequence in the first element.
By database comparison, three open reading frames, penP, ybbE, and ybxI, have been found in the B. subtilis 168 genome which encode putative β-lactamases. PenP has been overproduced and characterized as a class A β-lactamase (J. M. van Dijl, unpublished data), whereas YbbE remains uncharacterized, and YbxI has been postulated to be a class D β-lactamase because its primary structure exhibits a high similarity with the members of this group of serine β-lactamases. A phylogenetic tree was constructed by Barlow and Hall (3), in which the ybxI nucleotide sequence was referred to as B. subtilis z99105. No promoter sequence was identified upstream of the coding sequences for these three genes, in agreement with the fact that no β-lactamase activity was detected in growing B. subtilis 168 cells or in the supernatant. To determine if YbxI is a class D β-lactamase, it was overproduced in the periplasm of Escherichia coli, purified to homogeneity, and analyzed for its capacity to hydrolyze β-lactam antibiotics and dd-peptidase substrates. YbxI was shown to be a β-lactamase of very low activity which is not inactivated by clavulanic acid. Indeed, the recombinant protein interacts with several β-lactam antibiotics and can hydrolyze some of them but does not hydrolyze the peptides cleaved by dd-transpeptidases. In B. subtilis 168, the reverse transcription (RT)-PCR analysis does not detect the presence of mRNA encoding YbxI during the exponential phase of growth, in agreement with the absence of a typical promoter.
MATERIALS AND METHODS
Bacterial strains and plasmids.
B. subtilis 168 was used as source of DNA for PCR amplification of the ybxI gene. E. coli DH5α (Gibco BRL Life Technology) was used for the subcloning experiments, and E. coli BL21(DE3) (Invitrogen) was used as host for YbxI overexpression. A mature YbxI protein expressed in the periplasm of E. coli BL21(DE3) was obtained as follows. PCR was performed using primers 5′TAACCATGGATGCATCATCTGCTCATGAAAAAC3′ (YbxI-up, sense primer) and 5′TAGGATCCTTAGGATGTAATCAGGCC3′ (YbxI-rp, reverse primer) and the purified B. subtilis 168 chromosome as a template. In this way, NcoI and BamHI restriction sites were added at the 5′ and 3′ termini of the ybxI gene. The amplified 754-bp fragment coding for a protein devoid of signal peptide was then cloned in the pGEM-T Easy cloning vector (Promega). Automatic DNA sequencing (ALFexpress) confirmed the correct nucleotide sequence of the PCR product. In the final construct, pCIP 51 (a derivative of pET-26b expression vector [Novagen]), the mature ybxI gene is fused with the pullulanase-encoding signal peptide sequence.
Culture and YbxI production.
E. coli BL21(DE3) was grown in 1 liter of Luria-Bertani medium supplemented with kanamycin (50 μg/ml) at 37°C until the A600 was approximately 0.8. The culture was then induced by addition of IPTG (isopropyl-β-d-thiogalactopyranoside) at a final concentration of 0.5 mM. The culture was centrifuged at 4,000 × g for 20 min, and the harvested cells were washed with 30 mM Tris-HCl buffer (pH 8), centrifuged for 20 min at 4,000 × g, and resuspended in the same buffer containing 27% (wt/vol) sucrose, 10 mM EDTA, and hen egg white lysozyme (0.2 mg/ml). After 30-min of incubation at 4°C, addition of 0.1 M CaCl2, and centrifugation for 20 min at 18,000 × g, the supernatant corresponding to the periplasmic fraction was stored overnight at 4°C.
YbxI purification.
The periplasmic fraction (100 ml) was dialyzed against 50 mM Tris-HCl buffer, pH 8.5, starting buffer, concentrated 10-fold by ultrafiltration on an Amicon membrane (Mr cutoff, 10,000), and deposited onto a Source 15Q column (20 ml) equilibrated with the same buffer. The proteins were eluted with a linear NaCl gradient (0 to 1.0 M) over 100 ml. The enzyme activity was detected by a β-lactamase assay using nitrocefin as the substrate. The active fractions were pooled, dialyzed against 20 mM potassium acetate buffer (pH 5), concentrated to 10 ml, and applied to a Source 15S column (5 ml) equilibrated with 25 ml of the same potassium acetate buffer. The protein was eluted with a linear NaCl gradient (0 to 1.0 M) over 25 ml. The active fractions were pooled and dialyzed against 50 mM phosphate buffer (pH 7). The fractions containing β-lactamase activity were concentrated and then applied to a 100-ml Sephacryl S100 HR column. The active fractions were pooled and concentrated.
SDS-PAGE.
Samples were subjected to 15% polyacrylamide gel electrophoresis (PAGE) (27) in the presence of sodium dodecyl sulfate (SDS) (0.1%, wt/vol). After electrophoresis, the proteins were revealed by Coomassie blue R250 staining.
Penicillin binding assay.
Three micrograms of purified YbxI and 0.5 nmol of fluoresceyl ampicillin (Ampi-flu) (28) were incubated for various times at 37°C in a total volume of 20 μl. The reaction was stopped by addition of 5 μl of SDS-PAGE loading buffer, and the reaction mixture was subjected to SDS-PAGE. As a control protein, 10 μg of the carboxy-terminal domain of the Bacillus licheniformis BlaR penicillin receptor was used. The fluorescent complexes were visualized using a Molecular Imager FX (Bio-Rad).
Measurement of kinetic parameters.
All the spectrophotometric measurements were performed with the help of a UVIKON 860 (KONTRON instrument) or a Beckman DU 500 spectrophotometer interfaced to microcomputers. The time courses of the hydrolysis of the β-lactam antibiotics were recorded at 482 nm for nitrocefin; 260 nm for other cephalosporins, oxacillin, and cloxacillin; 235 nm for other penicillin antibiotics; and 300 nm for imipenem. The kinetic parameters were determined using the Hanes linearization ([S]/v versus [S]) or the integrated form of Michaelis-Menten equation (4). The catalytic activity for all the tested antibiotics was determined with a β-lactam concentration of 100 μM.
To test the possible carboxylation of YbxI Lys-82, a 200-μl portion of 17 μM protein solution was diluted in 2 ml of 6 mM acetic acid to obtain a final pH of 4.5. Buffer exchange was performed by dialysis against degassed 50 mM phosphate buffer, pH 7, under an N2 atmosphere. The kinetic measurements were performed with nitrocefin at 30°C in 50 mM sodium phosphate buffer (pH 7.0) with and without addition of various sodium bicarbonate concentration (0.15 to 50 mM).
The hydrolysis of benzoyl-glycyl-thioglycolate (S2a) was monitored at 250 nm (1).
To probe for a possible dd-peptidase activity, the purified recombinant YbxI was incubated with Ac2-l-Lys-d-Ala-d-Ala (35 mM) in 100 mM Tris-HCl buffer, pH 8. The release of d-Ala was estimated by the d-amino acid oxidase method (11, 20).
The d-amino peptidase activity of YbxI was probed with the help of the chromogenic substrate d-Ala paranitroanilide (39) (10 mM in 20 mM Tris-HCl buffer, pH 8).
To test a possible inactivation or inhibition of YbxI by clavulanic acid, kinetic measurements were performed with 200 μM nitrocefin at 30°C in 50 mM sodium phosphate buffer (pH 7.0), after preincubation of 0.34 μM YbxI with clavulanic acid concentrations ranging from 0 to 1 mM for 10 to 30 min at 37°C. The effect of NaCl was tested with nitrocefin concentrations ranging from 50 to 420 μM at 30°C in 50 mM sodium phosphate buffer (pH 7.0) supplemented with 50 mM NaCl.
Interaction and hydrolysis of peptidoglycan.
An 8-μg sample of purified YbxI protein was incubated with a suspension of Bacillus megaterium peptidoglycan (1.62 mg per ml of 50 mM Tris-HCl buffer, pH 7.5, at 37°C) for different times (30 min or 1, 2, 3, or 20 h). The variation of absorbance was monitored at 450 nm. Hen egg white lysozyme (4 μg in 50 mM Tris-HCl buffer, pH 7.5) was used as a positive control, under the same conditions.
Study of protein expression by RT-PCR.
The isolation of the mRNA produced by B. subtilis 168 cells in the exponential growth phase was carried out by using the SV Total RNA Isolation System kit (Promega). The presence of ybxI mRNA was probed by RT-PCR with the help of the Access RT-PCR System kit (Promega). The PCR fragments, amplified using the YbxI-up and YbxI-rp primers, were subjected to electrophoresis in a 1% agarose gel followed by ethidium bromide staining. The positive control was the B. subtilis yjbJ protein, which is a putative autolysin, using the primers 5′ATAGGATCCTTCGCGTAATAAACTGAAG3′ (BSSLTBH1) and 5′ATACATATGCTGAACAGCGCGAATACAACA3′ (BSSLNDEI1271). The same conditions were used for YbxI.
Sequence comparison.
Comparison of YbxI amino acid sequence (accession number p54427) with the nonredundant EMBL-GenBank-SWISSPROT protein database (http://www.expasy.ch) was performed by using the Blast software. The multiple sequence alignment was made with the help of the Multalin software (5).
RESULTS AND DISCUSSION
Comparison of YbxI with serine PREs.
The polypeptide deduced from the nucleotide sequence of ybxI consists of 268 amino acids, as shown in the sequence alignment presented in Fig. 2. The N-terminal region of the ybxI gene product contains a characteristic prokaryotic signal peptide; the mature protein would begin at Ala-24. According to this feature the ybxI gene product of B. subtilis 168 is expected to be exported outside the cell.
FIG.2.
Sequence alignment of class D β-lactamases and C-terminal domains of penicillin receptors. Alignment of amino acid sequences of the YbxI protein; class D β-lactamases OXA-20 (30), OXA-2 (7), OXA-15 (8), OXA-3 (37), PSE2 (19), OXA-11 (16), OXA-7 (38), OXA-5 (6), LCR1 (6), OXA-1 (32), OXA-18 (34), and OXA-9 (40); and the CTDs of penicillin receptors involved in the induction of β-lactamases (B. licheniformis [B.li] BlaR [26] and S. aureus [S.au] BlaR [35] or methicillin-resistant S. aureus PBP [S. aureus MecR] [18]). The underlined sequence in YbxI corresponds to the signal peptide. In the consensus sequence, residues in uppercase letters are strictly conserved in all aligned sequences. The residues in lowercase letters are those conserved in more than 50% of the sequences. The other consensus symbols are as follows: ! is either I or V; $ is L or M; % is F or Y; and # is N, D, Q, or E. The residues involved in the active-site signatures of serine-PREs are underlined. Dots indicate deletions. Amino acid sequence alignments were performed with the Multalin program of the ExPAsy server (http://us.expasy.org/tools/#align) (5).
Blast results revealed 33.7% amino acid identity with the OXA-2 class D β-lactamase and 33% identities with the C-terminal domains of the BlaR penicillin receptors of Bacillus licheniformis and Staphylococcus aureus (Fig. 2). No significant similarities were found with class A or class C β-lactamases and dd-peptidases.
Heterologous expression of YbxI in E. coli and purification of the recombinant protein.
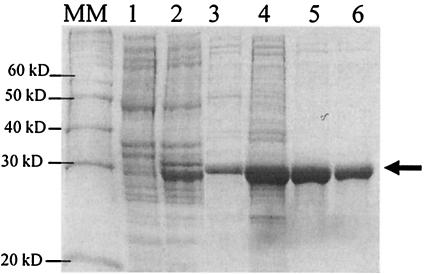
To confirm the biocomputing results that indicated a putative member of the class D β-lactamases, the B. subtilis ybxI gene was expressed in E. coli as described in Materials and Methods. E. coli BL21 cells harboring pCIP 51 were induced with IPTG, and the recombinant mature YbxI protein was produced as a water-soluble periplasmic protein (Fig. 3, lanes 1 to 3), with two additional amino acids, Met-Asp, due to the PelB peptide sequence. The production of the mature YbxI was performed in Luria-Bertani medium (1 liter), and induction was carried out during 4 h by addition of 0.5 mM IPTG when the culture density reached an A600 value of 0.8. After centrifugation and resuspension of the cells in an isotonic buffer, the periplasmic proteins were released by protoplasting the cells with the help of lysozyme. After centrifugation, the supernatant corresponding to the periplasmic extract contained most of the mature YbxI protein. The protein was purified to 95% homogeneity in a three-step procedure (Fig. 3, lanes 4 to 6). At pH 8.5 in 50 mM Tris-HCl buffer, mature YbxI bound to the Source Q ion exchanger and was eluted at 500 mM NaCl in the linear gradient. After dialysis against 20 mM potassium acetate buffer, pH 5, and concentration by ultrafiltration, YbxI was purified by chromatography on a Source S column (elution at 500 mM NaCl) followed by gel filtration on a Sephacryl S100 column. The final yield of YbxI production was 10 mg/liter. Edman degradation of the purified protein yielded the MDASSA sequence. The mass spectrometer analysis showed a main peak (mean ± standard deviation) at 28,420 ± 5 Da (theoretical mass: 28,419.15 Da).
FIG. 3.
SDS-PAGE analysis of crude cellular extracts and at different stages of YbxI purification. Lanes 1 and 2, crude cellular extracts of noninduced and induced (0.5 mM IPTG) E. coli/pCIP 51, respectively; lane 3, periplasmic fraction after cell lysis; lanes 4 to 6, YbxI after Source 15Q, Source 15S, and Sephacryl S100 chromatography steps, respectively; MM, molecular mass markers. The arrow indicates the YbxI position.
Kinetic properties of purified YbxI. (i) β-Lactamase activity.
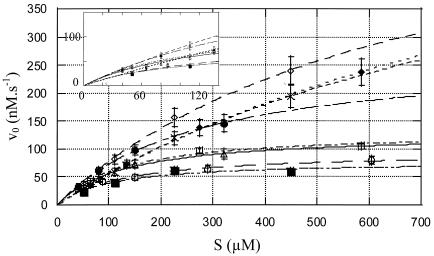
Table 2 summarizes the kinetic parameters for the hydrolysis of nine compounds representing the main families of β-lactam antibiotics. With nitrocefin, initial rates were determined at concentrations ranging from 65 to 600 μM, and the kcat and Km values were obtained with the help of the Hanes linearization method. Similar values were obtained by using complete time courses and the integrated form of the Henri-Michaelis-Menten equation. Incubation of YbxI with increasing concentrations of clavulanic acid did not inactivate the enzyme. With oxacillin, a biphasic reaction was observed, a typical feature of turnover kinetics by a number of class D β-lactamases. Steady-state rates were determined for the second phase at substrate concentrations ranging from 100 to 1,000 μM, and the kcat and Km values were obtained with the help of the Hanes linearization method. The kcat/Km value for cephaloridine was determined on the basis of first-order time courses, with concentrations ranging from 25 to 200 μM. With benzylpenicillin, substrate inhibition was observed. Golemi et al. (13) have shown that the class D OXA-10 β-lactamase activity depends critically on an unusual carboxylation of lysine 70 for both the acylation and deacylation catalytic steps. The carboxylated Lys-70 would act as a general base to activate the serine hydroxyl in the acylation step. The same carboxylated lysine should also be in a suitable position to activate the incoming hydrolytic water for the deacylation event. So the carboxylation of this lysine is necessary for the activity of the OXA-10 class D β-lactamase. To assay the putative activation of the mature YbxI by carboxylation of the active site-lysine residue (Lys-82, corresponding to the class D Lys-70), its activity was assayed without supplemented bicarbonate, under degassed conditions, and in the presence of various bicarbonate concentrations in 50 mM phosphate buffer, pH 7. Initial rates were determined for concentrations of nitrocefin ranging from 40 to 500 μM using 0.07 μM enzyme. As shown in Fig. 4, an activation of the mature YbxI by bicarbonate was recorded.
TABLE 2.
Kinetic parameters for interaction between YbxI and various β-lactam antibioticsc
| Antibiotic | kcat/Km (M−1 s−1) | kcat (s−1) | Km (μM) | Catalytic activitya [v0/E0 (s−1)] |
|---|---|---|---|---|
| Nitrocefin | 8,054 ± 402 | 1.48 ± 0.07 | 184 ± 9 | 900 × 10−3 ± 180 × 10−3 |
| Penicillin G | 70 × 10−3 ± 1.4 × 10−3b | |||
| Cephalosporin C | 11 ± 2 | 1.63 × 10−3 ± 0.32 × 10−3 | ||
| Cloxacillin | 0.42 × 10−3 ± 0.08 × 10−3 | |||
| Cephaloridin | 240 ± 6 | 5.29 × 10−3 ± 0.9 × 10−3 | ||
| Cefoxitin | 1.34 × 10−3 ± 0.27 × 10−3 | |||
| Oxacillin | 141 ± 28 | 0.09 ± 0.02 | 627 ± 125 | 18 × 10−3 ± 3.6 × 10−3 |
For these experiment the antibiotic and enzyme concentrations were, respectively, 100 and 1.7 μM, except those for nitrocetin and oxacillin, for which the enzyme concentrations were 0.7 and 1.4 μM, respectively. v0, initial rate; E0, total enzyme concentration.
At higher concentrations (200, 300, and 400 μM), inhibition by the substrate was observed (with 400 μM penicillin G the catalytic activity was fivefold lower than that with 100 μM penicillin G).
There was no detectable hydrolysis of 100, 200, or 300 μM imipenem or ampicillin after 5 min at 30°C with 1.7 μM enzyme. Results are presented as means ± standard deviations.
FIG. 4.
Effect of bicarbonate on the rate of nitrocefin hydrolysis. The experiments were carried out in phosphate buffer (pH 7.0) that was degassed (○) or not degassed (□), with addition of 0.150 (▵), 3 (•), 6 (⋄), 25 (×), and 50 (♦) mM bicarbonate and with addition of 50 mM NaCl (▪). Initial rate (v0) values were determined with 0.07 μM YbxI in a total volume of 500 μl. Error bars, standard deviations.
At pH 7.0 and high nitrocefin concentration, the presence of 6 mM HCO3− ions increases the activity versus nitrocefin by about four- to fivefold, and further addition of HCO3− has little or a somewhat-inhibitory influence. By contrast at low substrate concentrations (<Km) the effect of HCO3− is much less significant. This suggests that HCO3− increases both kcat and Km but does not strongly influence kcat/Km. The presence of 50 mM NaCl does not significantly affect the kinetic parameters of nitrocefin hydrolysis (Fig. 4). The presence of 50 mM bicarbonate does not modify the biphasic kinetics observed with oxacillin.
(ii) Trapping of an acyl-enzyme.
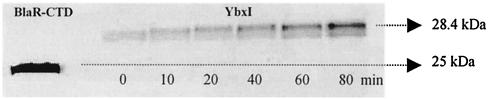
The catalytic pathway of the hydrolysis of a β-lactam antibiotic by a serine β-lactamase involves the formation of an acyl-enzyme between the active serine and the carbonyl of the opened β-lactam ring (Fig. 1). To demonstrate acylation of the mature YbxI by a β-lactam, a time course experiment was carried out by incubating the purified enzyme with a fluorescent derivative of ampicillin (Ampi-flu) (28). In each sample, the reaction was stopped by addition of SDS-PAGE loading buffer, the protein was denatured by heating, and the acyl-enzyme was separated from the excess of antibiotic by electrophoresis. As shown in Fig. 5, a fluorescent band of 28.4 kDa, corresponding to the molecular mass of mature YbxI, was detected, which confirmed the formation of an acyl-enzyme. The B. licheniformis BlaR carboxy-terminal domain (BlaR-CTD) was used as a positive control and molecular mass marker and gave a fluorescent band at 25 kDa.
FIG. 5.
Time course analysis of YbxI acylation by fluorescent ampicillin (Ampi-flu). SDS-PAGE and fluorography of YbxI (85 pmol) after incubation (0, 10, 20, 40, 60, and 80 min) with Ampi-flu (500 pmol). BlaR-CTD (25 kDa) (400 pmol) was used as a positive control and was incubated with Ampi-flu for 15 min.
When YbxI was incubated with Ampi-flu for 90 min, added with the TEM1 β-lactamase, and further incubated, no degradation of the fluorescent complex was observed after 24 h (data not shown).
dd-Peptidase and d-aminopeptidase activities.
The dd-carboxypeptidase activity was assayed by incubating mature YbxI (0.2 μM) with Ac2-Lys-d-Ala-d-Ala (18) and S2a thioester (1) substrates. No hydrolysis of these compounds was observed. Similarly, no peptidoglycan hydrolyzing activity was found. The same negative result was obtained with d-Ala-paranitroanilide (39), a substrate used for the kinetic study of the Ochrobacterium anthropi d-aminopeptidase (2), another member of the serine PRE family.
RT-PCR analysis.
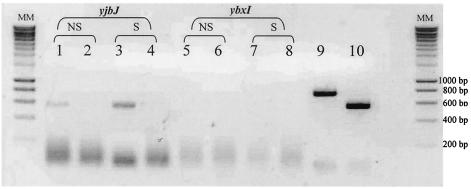
The expression of the ybxI gene was analyzed by RT-PCR. Total mRNAs were isolated from B. subtilis cells in exponential growth, and attempts to amplify the mRNA were made with the help of YbxI-up and YbxI-rp primers. After amplification and electrophoresis on agarose gel, no YbxI transcript could be detected in the absence or presence of β-lactam stress (cephalosporin C, 2.5 μg/ml) (Fig. 6). As a positive control the yjbJ gene was used, which codes for a putative autolysin constitutively expressed by the strain.
FIG. 6.
RT-PCR analysis of ybxI transcripts in B. subtilis. RT-PCR analysis by agarose gel electrophoresis of ybxI transcripts in B. subtilis in the presence or in absence of a β-lactam stress compound. Abbreviations: NS, nonstressed cells; S, cells stressed by the presence of 2.5 μg of cephalosporin C; MM,: molecular mass marker. Lanes 1 to 4, yjbJ control; lane 1, RT-PCR NS; lane 2, PCR NS; lane 3, RT-PCR S; lane 4, PCR S; Lanes 5 to 8, ybxI; lane 5, RT-PCR NS; lane 6, PCR NS; lane 7, RT PCR S; lane 8, PCR S; Lanes 9 and 10 correspond to the PCR products of ybxI and yjbJ, respectively, obtained with the genomic DNA of B. subtilis 168.
Sequence alignments.
Figure 2 shows the comparison of sequences of YbxI, the BlaR-CTD domain of B. licheniformis (26), the BlaR and MecR equivalent domains of Staphylococcus aureus (18, 35), and several class D β-lactamases: OXA-2 (7) and PSE-2 (OXA-10) (19) of Salmonella enterica serovar Typhimurium; OXA-11 (16), OXA-5 (6), OXA-3 (37), OXA-15 (8), OXA-18 (34), OXA-20 (30), and LCR-1 (6) of Pseudomonas aeruginosa; OXA-7 (38) and OXA-1 (32) of E. coli; and OXA-9 (40) of Klebsiella pneumoniae. The major differences between these proteins result from either deletions or insertions. The consensus shows several conserved residues, including the amino acids forming the active-site elements.
Conclusion.
This study describes the production in E. coli, the purification, and the kinetic study of the putative β-lactamase encoded by the B. subtilis 168 ybxI gene. The predicted sequence of the encoded protein contains an N-terminal signal peptide, and the mature protein shows a high degree of identity to class D β-lactamases and the C-terminal domain of penicillin receptors. In addition, multiple alignments of these sequences confirmed that mature YbxI possesses all the conserved sequence motifs of the serine PRE family. This feature was well highlighted by the ability of YbxI to form an acyl-enzyme with Ampi-flu. The kinetic survey shows that the ybxI gene, previously thought to encode a class D β-lactamase, does not encode a real β-lactamase and is devoid of dd-peptidase and d-aminopeptidase activities, at least with the tested substrates assayed. Indeed, from the kinetic parameters presented in Table 2, YbxI seems to be a PBP with low β-lactamase activity on a few compounds. The rate of hydrolysis of either cephalosporin C, cloxacillin or cefoxitin is very low. The highest values were obtained with nitrocefin, for which the kcat/Km value was ∼8,054 M−1 s−1. If we compare these values with those of the class D β-lactamase OXA10 (PSE2) from S. enterica serovar Typhimurium and PBP5 of Enterococcus hirae ATCC 9790, which exhibit kcat/Km values of ∼6.6 × 106 and ∼20 M−1 s−1, respectively, YbxI has an activity intermediate between those of a PBP and a β-lactamase.
The BlaR and MecR C-terminal domains are penicillin receptors whose sequences are related to those of class D enzymes, but they form stable adducts with β-lactams and do not hydrolyze depsipeptides (9). The activity of several class D enzymes has been shown to significantly increase upon carboxylation of the active-site Lys-70 side chain. For the BlaR receptors, contradictory results have been obtained (14, 23). In the presence of HCO3− ions, the rate of acylation of S. aureus BlaR protein by β-lactams seems to increase at pH 7.0, but under the same conditions that of B. licheniformis BlaR does not appear sensitive to the presence of HCO3− ions and the X-ray structure clearly highlights a normal Lys residue. The results obtained here with YbxI remain ambiguous. The influence of HCO3− on the activity of YbxI versus nitrocefin might be due to carboxylation of Lys-82, but this remains to be established by determining the 3D structure of the protein in the absence and presence of HCO3−. One major difference between the class D β-lactamase, YbxI, and BlaR-CTD is the nature of the third residue of the second element, Ile for YbxI, Val for class D enzymes, and a polar residue (Thr or Asn) for the BlaR/MecR group of penicillin sensor proteins. It has been suggested that the class D Val residue might contribute to the substrate specificity of the class D enzymes by providing interactions with the isoxazoyl moiety of oxacillins. However, the efficient hydrolysis of penicillins catalyzed by the class D β-lactamases would rest on the carboxylation of the Lys residue of the active-site serine element. One can tentatively explain the poor activity of YbxI by the fact that in the second element the bulkier Ile residue would interfere with an efficient binding of the antibiotic side chain. However, despite these considerations it is still difficult at this stage to determine with certainty why YbxI and BlaR-CTD are not β-lactamases. The determination of their 3D structures acylated by, or complexed with, a β-lactam antibiotic may answer this question. The physiological role of YbxI in B. subtilis 168 remains unknown. The RT-PCR experiments showed that the mRNA corresponding to a ybxI transcript is not detectable in the bacterial cell in the exponential growth phase, simultaneously confirming the absence of a promoter upstream of ybxI and that the product of this gene is not essential for cell growth and sporulation (25). The members of the serine PRE superfamily show a great diversity in their catalytic activities. However, they share a common feature: the chemical bond hydrolyzed is always linked to a chiral D center. For this reason, it can be hypothesized that YbxI might catalyze a reaction involving a D center.
Acknowledgments
This work was supported by the Belgian Program of Interuniversity Poles of Attraction initiated by the Federal office for Scientific, Technical and Cultural Affaires (PAI no. P5/33) and the Fond National de la Recherche Scientifique (FNRS), crédit aux chercheurs no. 1.5201.02 and FRFC no. 2.4508.01.
B.J. is an FNRS associate researcher. M.-L.C., S.H., S.L.B., and C.B. are fellows of the Fonds pour la Formation à la Recherche dans l'Industrie et l'Agriculture (FRIA, Brussels, Belgium).
REFERENCES
- 1.Adam, M., C. Damblon, B. Plaitin, L. Christiaens, and J. M. Frère. 1990. Chromogenic depsipeptide substrates for β-lactamases and penicillin-sensitive DD-peptidases. Biochem. J. 270:525-529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Asano, Y., A. Nakasawa, Y. Kato, and K. Kondo. 1989. Properties of a novel D-stereospecific aminopeptidase from Ochrobactrum anthropi. J. Biol. Chem. 264:14233-14239. [PubMed] [Google Scholar]
- 3.Barlow, M., and B. G. Hall. 2002. Phylogenetic analysis shows that the OXA β-lactamase genes have been on plasmids for millions of years. J. Mol. Evol. 55:314-321. [DOI] [PubMed] [Google Scholar]
- 4.Bush, K., and R. B. Sykes. 1986. Methodology for the study of β-lactamases. Antimicrob. Agents Chemother. 30:6-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Corpet, F. 1988. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 16:10881-10890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Couture, F., J. Lachapelle, and R. C. Levesque. 1992. Phylogeny of LCR-1 and OXA-5 with class A and class D β-lactamases. Mol. Microbiol. 6:1693-1705. [DOI] [PubMed] [Google Scholar]
- 7.Dale, J. W., D. Godwin, D. Mossakowska, P. Stephenson, and S. Wall. 1985. Sequence of the OXA-2 β-lactamase: comparison with other penicillin-reactive enzyme. FEBS Lett. 191:39-44. [DOI] [PubMed] [Google Scholar]
- 8.Danel, F., L. M., Hall, D. Gur, and D. M. Livermore. 1997. OXA-15, an extended-spectrum variant of OXA-2 β-lactamase, isolated from a Pseudomonas aeruginosa strain. Antimicrob. Agents Chemother. 41:785-790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duval, V., M. Swinnen, S. Lepage, A. Brans, B. Garnier, C. Franssen, J. M. Frère, and B. Joris. 2003. The kinetic properties of the carboxy terminal domain of the Bacillus licheniformis 749/I BlaR penicillin-receptor shed a new light on the derepression of beta-lactamase synthesis. Mol. Microbiol. 48:1553-1564. [DOI] [PubMed] [Google Scholar]
- 10.Frère, J. M., and B. Joris. 1985. Penicillin-sensitive enzymes in peptidoglycan biosynthesis. Crit. Rev. Microbiol. 11:299-396. [DOI] [PubMed] [Google Scholar]
- 11.Frère, J. M., M. Leyh-Bouille, J. M. Ghuysen, M. Nieto, and H. R. Perkins. 1976. Exocellular DD-carboxypeptidases-transpeptidases from Streptomyces. Methods Enzymol. 45:610-636. [DOI] [PubMed] [Google Scholar]
- 12.Frère, J. M. 1989. Quantitative relationship between sensitivity to β-lactam antibiotics and β-lactamase production in gram-negative bacteria-I. Steady-state treatment. Biochem. Pharmacol. 38:1415-1426. [DOI] [PubMed] [Google Scholar]
- 13.Golemi, D., L. Maveyraud, S. Vakulenko, J. P. Samama, and S. Mobashery. 2001. Critical involvement of a carbamylated lysine in catalytic function of class D beta-lactamases. Proc. Natl. Acad. Sci. USA 98:14280-14285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Golemi-Kotra, D., J. Y. Cha, S. O. Meroueh, S. B. Vakulenko, and S. Mobashery. 2003. Resistance to beta-lactam antibiotics and its mediation by the sensor domain of the transmembrane BlaR signaling pathway in Staphylococcus aureus. J. Biol. Chem. 278:18419-18425. [DOI] [PubMed] [Google Scholar]
- 15.Gordon, E., N. Mouz, E. Duée, and O. Dideberg. 2000. The crystal structure of the penicillin-binding protein 2x from Streptococcus pneumoniae and its acyl-enzyme form: implication in drug resistance. J. Mol. Biol. 299:477-485. [DOI] [PubMed] [Google Scholar]
- 16.Hall, L. M., D. M. Livermore, D. Gur, M. Akova, and H. E. Akalin. 1993. OXA-11, an extended-spectrum variant of OXA-10 (PSE-2) β-lactamase from Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 37:1637-1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Herzberg, D., and J. Moult. 1987. Bacterial resistance to β-lactam antibiotics crystal structure of the β-lactamase from Staphylococcus aureus PC1 at 2.5 A resolution. Science 236:694-701. [DOI] [PubMed] [Google Scholar]
- 18.Hiramatsu, K., K. Asada, E. Suzuki, K. Okonogi, and T. Yokota. 1992. Molecular cloning and nucleotide sequence determination of the regulator region of mecA gene in methicillin-resistant Staphylococcus aureus (MRSA). FEBS Lett. 298:133-136. [DOI] [PubMed] [Google Scholar]
- 19.Huovinen, P., S. Huovinen, and G. A. Jacoby. 1988. Sequence of the PSE-2 β-lactamase. Antimicrob. Agents Chemother. 32:134-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnson, K., C. Duez, J. M. Frère, and J. M. Ghuysen. 1975. Beta-lactamases (Actinomycetes species). Methods Enzymol. 43:687-698. [DOI] [PubMed] [Google Scholar]
- 21.Joris, B., J. M. Ghuysen, G. Dive, A. Renard, O. Dideberg, P. Charlier, J. M. Frère, J. A. Kelly, J. C. Boyington, P. C. Moews, et al. 1988. The active-site-serine penicillin-recognizing enzymes as members of the Streptomyces R61 DD-peptidase family. Biochem. J. 250:313-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kelly, J. A., O. Dideberg, P. Charlier, J. P. Wery, M. Libert, P. C. Moews, J. R. Knox, C. Duez, C. Fraipont, B. Joris, et al. 1986. On the origin of bacterial resistance to penicillin: comparison of a beta-lactamase and a penicillin target. Science 231:1429-1431. [DOI] [PubMed] [Google Scholar]
- 23.Kerff, F., P. Charlier, M. L. Colombo, E. Sauvage, A. Brans, J. M. Frère, B. Joris, and E. Fonzé. 2003. Crystal structure of the sensor domain of BlaR penicillin-receptor from Bacillus licheniformis. Biochemistry 42:12835-12843. [DOI] [PubMed] [Google Scholar]
- 24.Knox, J. R., P. C. Moews, and J. M. Frère. 1996. Molecular evolution of bacterial β-lactam resistance. Chem. Biol. 3:937-947. [DOI] [PubMed] [Google Scholar]
- 25.Kobayashi, K., S. D. Ehrlich, and. N. Ogasawara. 2003. Essential Bacillus subtilis genes. Proc. Natl. Acad. Sci. USA 100:4678-4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kobayashi, T., Y. F. Zhu, N. J. Nicholls, and J. O. Lampen. 1987. A second regulatory gene, blaR1, encoding a potential penicillin-binding protein required for induction of β-lactamase in Bacillus licheniformis. J. Bacteriol. 169:3873-3878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 28.Lakaye, B., C. Damblon, M. Jamin, M. Galleni, S. Lepage, B. Joris, J. Marchand-Brynaert, C. Frydrych, and J. M. Frère. 1994. Synthesis, purification and kinetic properties of fluorescein-labelled penicillins. Biochem. J. 300:141-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Naas, T., and P. Nordmann. 1999. OXA-type β-lactamases. Curr. Pharm. Design 5:865-879. [PubMed] [Google Scholar]
- 30.Naas, T., W. Sougakoff, A. Casetta, and P. Nordmann. 1998. Molecular characterization of OXA-20, a novel class D β-lactamase, and its integron from Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 42:2074-2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nikaido, H., and S. Normark. 1987. Sensitivity of Escherichia coli to various β-lactams is determined by the interplay of outer membrane permeability and degradation by periplasmic β-lactamases: a quantitative predictive treatment. Mol. Microbiol. 1:29-36. [DOI] [PubMed] [Google Scholar]
- 32.Ouellette, M., L. Bissonnette, and P. H. Roy. 1987. Precise insertion of antibiotic resistance determinants into Tn21-like transposons: nucleotide sequence of the OXA-1 β-lactamase gene. Proc. Natl. Acad. Sci. USA 84:7378-7382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Paetzel, M., F. Danel, L. de Castro, S. C. Misimann, M. G. P. Page, and N. C. J. Strynadka. 2000. Crystal structure of the class D β-lactamase OXA-10. Nat. Struct. Biol. 7:918-925. [DOI] [PubMed] [Google Scholar]
- 34.Philippon, L. N., T. Naas, A. T. Bouthors, V. Barakett, and P. Nordmann. 1997. OXA-18, a class D clavulanic acid-inhibited extended-spectrum β-lactamase from Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 41:2188-2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rowland, S. J., and K. G. Dyke. 1990. Tn552, a novel transposable element from Staphylococcus aureus. Mol. Microbiol. 4:961-975. [DOI] [PubMed] [Google Scholar]
- 36.Samraoui, B., B. J. Sutton, R. J. Todd, P. J. Artymiuk, S. G. Waley, and D. C. Phillips. 1986. Tertiary structural similarity between a class A beta-lactamase and a penicillin-sensitive D-alanyl carboxypeptidase-transpeptidase. Nature 320:378-380. [DOI] [PubMed] [Google Scholar]
- 37.Sanschagrin, F., F. Couture, and R. C. Levesque. 1995. Primary structure of OXA-3 and phylogeny of oxacillin-hydrolyzing class D β-lactamases. Antimicrob. Agents Chemother. 39:887-893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Scoulica, E., A. Aransay, and Y. Tselentis. 1995. Molecular characterization of the OXA-7 β-lactamase gene. Antimicrob. Agents Chemother. 39:1379-1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shibata, K., and T. Watanabe. 1987. Purification and characterization of an aminopeptidase from Mycoplasma salivarium. J. Bacteriol. 169:3409-3413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tolmasky, M. E. 1990. Sequencing and expression of aadA, bla, and tnpR from the multiresistance transposon Tn1331. Plasmid 24:218-226. [DOI] [PubMed] [Google Scholar]