Abstract
Past genetic studies have indicated that the genes encoding early enzymes of clavulanic acid biosynthesis may be duplicated in Streptomyces clavuligerus. We observed cross-hybridizing bands upon Southern analyses of proclavaminate amidinohydrolase (pah)-defective mutant strains of S. clavuligerus screened with a pah-specific probe. The DNA fragment responsible for this cross hybridization was cloned and sequenced and shown to encode a second copy of the pah gene. The new pah gene (pah1) was 1,056 bp in length, and its sequence was 72% identical to that of the original pah gene (pah2). Disruption mutants with defects in pah1 showed no significant effects on production of clavulanic acid or any of the clavam metabolites with stereochemistries opposite that of clavulanic acid (5S clavams) produced by S. clavuligerus when they were grown on starch asparagine or soy medium. However, double mutants with defects in both pah1 and pah2 were defective in the production of both clavulanic acid and all of the 5S clavam metabolites.
Streptomyces clavuligerus is a filamentous soil bacterium with an unusual facility for the production of β-lactam metabolites. In addition to cephamycin C, this organism produces clavulanic acid and at least four other clavam compounds with stereochemistries opposite that of clavulanic acid (hereafter referred to as 5S clavams) (7). Clavulanic acid is an important industrial product because it irreversibly inactivates a wide range of β-lactamase enzymes. When used in combination with conventional β-lactam antibiotics, it restores their effectiveness against antibiotic-resistant microbes. As a result, the biosynthesis of clavulanic acid and the genes associated with its production have come under intense investigation in recent years.
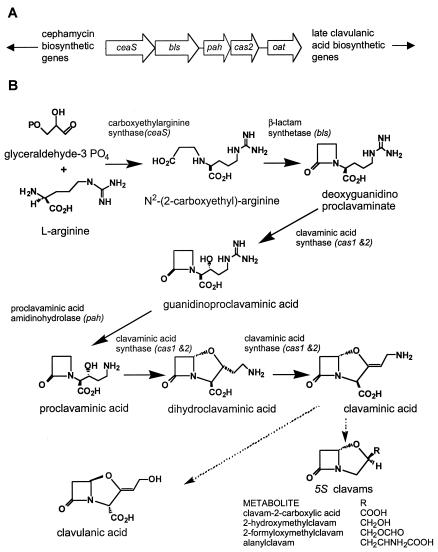
The cluster of genes responsible for clavulanic acid biosynthesis has been found to lie immediately adjacent to the cephamycin C gene cluster on the chromosome of S. clavuligerus, and the two pathways are coregulated by a single pathway-specific transcriptional regulator, CcaR (2, 16). The full details of the clavulanic acid biosynthetic pathway remain to be elucidated, but the early enzymes in the pathway are now known (up to the level of clavaminate), and the genes responsible for their synthesis are grouped at one end of the clavulanic acid gene cluster (Fig. 1A). These include genes encoding carboxyethylarginine synthase (ceaS) (10), β-lactam synthetase (bls) (3, 13), proclavaminate amidinohydrolase (pah) (1, 5, 22), and clavaminate synthase (cas2) (12), as well as a gene encoding ornithine acetyltransferase (oat) (6, 9). The involvement of oat in clavulanic acid synthesis is unclear, but it is grouped with the rest of the early genes because it shows the same pattern of regulation of gene expression (6). The biosynthetic steps catalyzed by these activities are represented in Fig. 1B.
FIG. 1.
Early steps in the biosynthesis of clavulanic acid and the 5S clavams. (A) Diagrammatic representation of the first five genes (encoding enzymes involved in early steps of the pathway) from the clavulanic acid gene cluster; (B) enzyme activities involved in the early steps of clavulanic acid and 5S clavam biosynthesis.
cas, the gene encoding clavaminate synthase, was the first gene of the cluster to be cloned and sequenced (12). Purification of the Cas enzyme as an initial step in the reverse genetic procedure used to clone the gene showed that there were actually two closely related forms of clavaminate synthase in the cells (18). Ultimately, two highly similar genes were identified as encoding the two forms of Cas. The two copies of the cas gene were 85.5% identical at the nucleotide level and encoded proteins showing 88% similarity, including 82% identical residues at the amino acid level. The two genes were named cas1 and cas2, and it has since been shown that cas2 is the gene located within the clavulanic acid cluster (1, 21). In contrast, cas1 resides elsewhere on the chromosome and is apparently unlinked to cas2 (12, 14). Examination of the genetic material flanking cas1 has implicated it in the biosynthesis of the 5S clavam metabolites produced by S. clavuligerus, since a mutation in cvm1, located immediately upstream of cas1, or a deletion which spanned parts of both cvm4 and cvm5, located downstream of cas1, caused a complete loss of production of all of the 5S clavam metabolites with no effect on clavulanic acid production (14).
Studies on the genes encoding early enzymes of the clavulanic acid biosynthetic pathway revealed that each of these genes could be disrupted by gene replacement procedures, yet the mutant strains could still synthesize clavulanic acid, albeit at somewhat reduced levels (6, 17). Several of these genes encode enzymes with unusual activities very different from the activities of any enzymes found to be part of primary metabolism, and so it seemed unlikely that the residual clavulanic acid synthesis seen in the disruption mutants could be due to endogenous activity from enzymes with broad specificities. Similarly, since the intermediates of clavulanic acid biosynthesis have unusual structures, these compounds are unlikely to be found to preexist in bacterial growth media. This led to the hypothesis that, as is the case for cas, there might be second copies, or paralogues, for each of the genes encoding the early enzymes of clavulanic acid biosynthesis in S. clavuligerus (6, 17). These paralogues would then enable clavulanic acid production to continue in the face of mutations in seemingly essential structural genes.
Proclavaminate amidinohydrolase (Pah) is the only enzyme from the early part of the pathway, other than Cas, which has been purified from S. clavuligerus. This purified Pah was partially sequenced, and the resulting data were compared with the amino acid sequence deduced from the pah gene sequence (4). Unlike the case for Cas, no evidence suggesting that there are two forms of Pah was found, and only a single copy of the pah gene was identified (1, 5). Subsequent analysis showed that the pah gene is located in the clavulanic acid cluster immediately adjacent to cas2 (1), and no paralogue of pah was found in the corresponding location adjacent to cas1 (14).
In view of these conflicting pieces of evidence regarding the likelihood that a paralogue for pah might exist, a search for a second copy of pah was undertaken. In this study we report on the isolation of a paralogue for the pah gene and examine its involvement in the production of clavulanic acid and the 5S clavams in S. clavuligerus.
MATERIALS AND METHODS
Bacterial strains, plasmids, media, and culture conditions.
S. clavuligerus NRRL 3585 was obtained from the Northern Regional Research Laboratory (Peoria, Ill.). S. clavuligerus was maintained on ISP Medium 4 agar plates (Difco, Detroit, Mich.). Cultures for the isolation of chromosomal DNA were grown on a 2:3 mixture of Trypticase soy broth and yeast extract malt extract (YEME) medium as described by Alexander et al. (2). Cultures for analysis of the production of clavulanic acid and 5S clavam metabolites were grown both on starch asparagine (SA) medium and on soy medium as described previously (15). All liquid cultures were grown at 28°C on a rotary shaker at 250 rpm. Plasmid-containing cultures were supplemented with apramycin (50 μg/ml; Apralan; Provel, Division of Eli Lilly, Indianapolis, Ind.) or thiostrepton (5 μg/ml; Sigma, Oakville, Ontario, Canada), as appropriate.
Manipulation of DNA in Escherichia coli was done by using strain XL-1 Blue (Stratagene, La Jolla, Calif.). E. coli cultures were maintained on Luria-Bertani (LB) agar medium and grown in liquid culture in LB medium at 37°C (19). Plasmid-containing cultures were supplemented with ampicillin (100 μg/ml; Sigma) or apramycin (50 μg/ml), as appropriate.
The plasmids used in this study included pUC120 (20) and pUC120apr (15), a pUC120 derivative carrying an apramycin resistance cassette flanked by NcoI sites. pDA501 is a shuttle vector prepared by fusing Streptomyces plasmid pIJ486 (11) to E. coli plasmid pTZ18R (Stratagene) by means of their EcoRI and BamHI sites (this study). pDA501 is segregationally unstable in S. clavuligerus in the absence of antibiotic selection and so is useful for the introduction of gene disruption constructs. pWE15 is an E. coli cosmid cloning vector (Promega, Madison, Wis.). A library of S. clavuligerus genomic DNA fragments in the cosmid pWE15 was generously provided by W. Jin, Seoul National University.
DNA manipulations.
Standard DNA manipulations, such as E. coli plasmid isolation, restriction endonuclease digestion, generation of blunt-ended fragments, ligation, 32P labeling of DNA probes by nick translation, and E. coli transformation, were carried out as described by Sambrook et al. (19). Isolation of plasmid and genomic DNA from Streptomyces spp. was conducted as described by Kieser et al. (11). Southern analysis of S. clavuligerus DNA fragments for the detection of the pah paralogue was conducted at high stringency, as described by Sambrook et al. (19). The hybridization membranes (Hybond-N; Amersham Pharmacia) were washed twice for 10 min each time in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.1% SDS at room temperature and then once for 15 min in 1.0× SSC-0.1% SDS and twice for 10 min each time in 0.1× SSC-0.1% SDS, all at 65°C.
HPLC analyses of culture filtrates.
Culture filtrates were derivatized with imidazole as described previously before analysis by high-pressure liquid chromatography (HPLC) (14).
Bioassays.
Clavulanic acid was detected in an indirect bioassay for β-lactamase inhibitors by using Klebsiella pneumoniae ATCC 29665 as the indicator organism growing on Nutrient Agar (Difco) containing penicillin G at 6 μg/ml. Cephamycin C was detected in culture filtrates by bioassay with the indicator organism E. coli ESS (8).
Cloning and sequence analysis of the pah1 gene.
Total genomic DNA from S. clavuligerus was digested to completion with NcoI, and the resulting fragments were separated by electrophoresis on a 0.8% agarose gel. Regions of the gel corresponding to DNA fragments of 4 to 5 kb were excised with a sharp blade, and the DNA was recovered by using a QIAquick gel extraction kit (Qiagen, Mississauga, Ontario, Canada). The purified DNA fragments were inserted into pUC120 digested with NcoI, and the ligation mixture was transformed into E. coli XL-1 Blue. Ampicillin-resistant transformants were screened by colony hybridization with a 0.46-kb SalI fragment from the original pah gene which was 32P labeled by nick translation for use as a probe. Plasmid DNA was isolated from hybridizing transformants and was confirmed to carry a 4.3-kb NcoI fragment. The pah1 gene within the 4.3-kb NcoI fragment was then sequenced by the Molecular Biology Service Unit, University of Alberta, by using the DYEnamic ET Terminator cycle sequencing kit (Amersham Pharmacia, Baie d'Urfe, Quebec, Canada). A 0.46-kb SalI fragment from within the pah1 gene was subcloned into pUC118R and sequenced by the use of universal primers. The remainder of the pah1 gene sequence was obtained by using sequence-specific primers. Sequence information was obtained in full for both strands.
Insertional inactivation of pah1 by gene disruption.
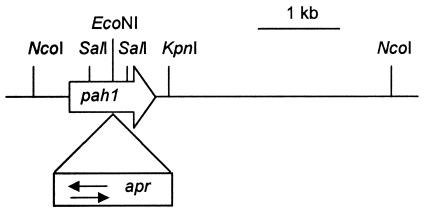
The pah1 gene was initially isolated as a 4.3-kb NcoI fragment of S. clavuligerus genomic DNA cloned into the NcoI site of pUC120. This construct was digested with KpnI (one site within the cloned fragment and one site in the vector) and religated to reduce the size of the insert to 1.65 kb, thereby generating plasmid p4K-1, which still carried the complete pah1 gene. The pah1 gene within p4K-1 was disrupted by digestion at its centrally located EcoNI site and insertion of an apramycin resistance gene cassette from pUC120apr after both fragments had been made blunt by treatment with the Klenow fragment of DNA polymerase I. The KpnI-NcoI insert carrying the disrupted pah1 gene was then inserted into the EcoRI site of pDA501 after blunting of the ends of both the insert and the vector. Constructs with the apr gene in both orientations relative to that of pah1 were obtained.
The disruption constructs were then passaged through Streptomyces lividans and into S. clavuligerus to generate gene replacement mutants, essentially as described by Paradkar and Jensen (15). For each mutant strain a corresponding cured wild-type control was also obtained. The cured control strain originated from the same primary transformant that gave rise to the mutant, but represents the case in which the disruption plasmid was simply lost from the cell rather than recombined with the chromosome.
Production of pah1 and pah2 double mutants.
The method used for the production of mutants with disruptions in pah2 has been described previously (1). Double mutants with disruptions in both pah1 (apr) and pah2 (thio) were produced by using the gene disruption construct originally constructed for the disruption of pah2 (1) and introducing it into one of the newly created pah1 mutants (mutant 5A).
Nucleotide sequence accession number.
The DNA sequence of pah1 has been deposited in GenBank under accession no. AY394923.
RESULTS
Cloning and sequence analysis of pah1.
Previous studies have shown that a mutant strain of S. clavuligerus in which the pah gene was knocked out by a gene replacement procedure was nonetheless still able to produce clavulanic acid (6). One possible interpretation of these results is that a second pah gene may be present and able to compensate for the mutated gene. In the case of cas2, in which a second copy of the gene is known to exist, the cas1 gene is 85.5% identical to cas2, which is sufficient to give extensive cross-hybridization upon Southern analysis unless probes are carefully designed to target regions of dissimilarity between the two genes.
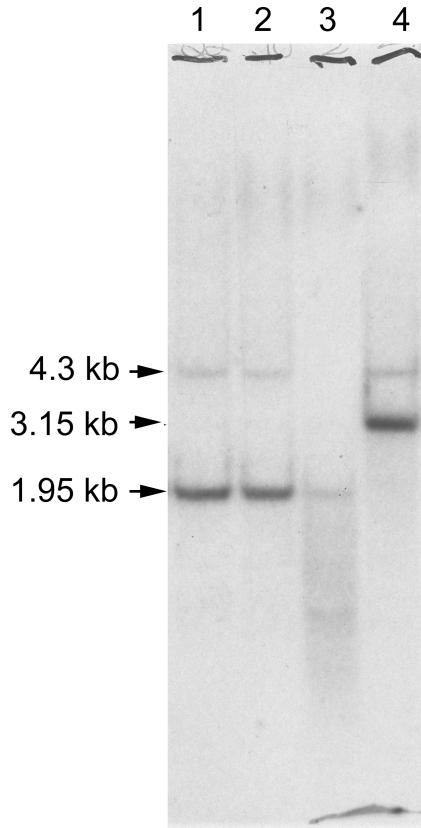
When data from the Southern analyses used in previous studies of pah disruption mutants were reexamined, autoradiograms that had been exposed for longer durations than normal showed evidence of additional weakly hybridizing bands (Fig. 2). In particular, genomic DNA from the wild-type strain of S. clavuligerus (lane 1) digested with NcoI showed a band of about 4.3 kb which hybridized weakly to the pah-specific probe, as well as the 1.95-kb strongly hybridizing band which corresponded to the known pah gene. The pah disruption mutant (lane 4) lacked the 1.95-kb strongly hybridizing band and showed a new 3.15-kb hybridizing band which corresponded to the pah gene disrupted by introduction of a 1.2-kb thiostrepton resistance gene. However, the pah disruption mutant also showed the same 4.3-kb weakly hybridizing band seen in the wild type. Since the weakly hybridizing bands were the same size in both the wild-type and the pah mutant strains, this suggested that they were not just artifacts resulting from incomplete digestion of DNA fragments carrying the known pah gene. On the basis of the assumption that this 4.3-kb NcoI fragment might carry a second copy of the pah gene, a library of NcoI-digested genomic DNA fragments, biased to contain primarily fragments of 4 to 5 kb, was prepared in plasmid vector pUC120 and transformed into E. coli. About 800 transformants were patched onto nylon membranes and then analyzed by colony hybridization with a radiolabeled pah-specific probe. Colonies showing levels of hybridization above the background levels were selected for further analysis.
FIG. 2.
Southern analysis of genomic DNA from S. clavuligerus wild-type and pah mutant strains. Genomic DNA was digested with NcoI, separated by electrophoresis on agarose, blotted onto a nylon membrane, and probed with a pah-specific probe. The nylon membrane was exposed to X-ray film for 10 days at −70°C. Lane 1, S. clavuligerus wild type; lanes 2 and 3, thiostrepton-resistant isolates selected as potential pah mutants but shown to carry a wild-type pah gene (not pah mutants); lane 4, thiostrepton-resistant true pah mutant.
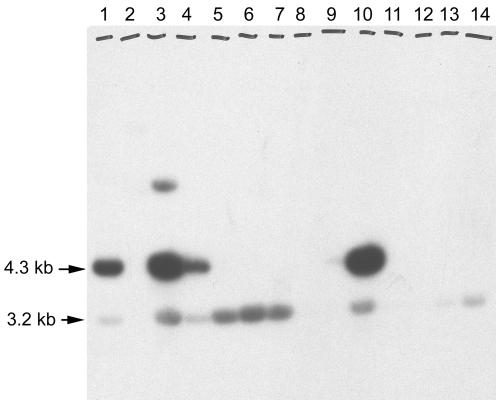
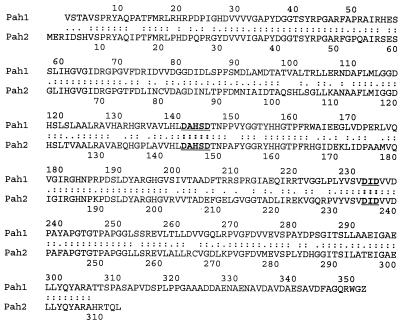
When plasmid DNA was isolated from 13 of these transformants, digested with NcoI, and subjected to Southern analysis, hybridization with the pah-specific probe was associated with the presence of the desired 4.3-kb NcoI insert fragment in four of the plasmids (Fig. 3). The band seen at 3.2 kb represents the pUC120 plasmid and shows some hybridization due to the presence of a small amount of contaminating plasmid in the pah-specific probe. Within the 4.3-kb NcoI fragment, the pah-hybridizing region was further localized to a 0.46-kb SalI fragment. Subcloning and DNA sequence analysis of this 0.46-kb SalI fragment indicated the presence of a gene with significant similarity to pah. Additional DNA sequence analysis with sequence-specific oligonucleotide primers was used to complete the determination of the DNA sequence of this pah paralogue. Since the clavaminate synthase-encoding gene located in the clavulanic acid gene cluster is named cas2, we propose that the original pah gene which lies adjacent to it should be called pah2. The newly isolated pah paralogue would then be named pah1. The newly identified pah1 gene was 1,056 bp in length, whereas the known pah2 gene is 942 bp. Sequence alignment indicated that it showed 72% end-to-end sequence identity with pah2. The corresponding protein predicted by the gene sequence showed 81% similarity, including 71% residues identical to the residues in the Pah2 protein (Fig. 4). The boldface underlined residues in Fig. 4 indicate the locations of two predicted manganese-binding sites that are typically found in proteins belonging to the arginase family, including Pah. The amino acid sequences of the two Pah proteins are identical across both of these sites.
FIG. 3.
Southern analysis of clones hybridizing to a pah-specific probe. S. clavuligerus genomic DNA was digested with NcoI, and fragments of 4 to 5 kb were cloned into pUC120. Transformants were analyzed by colony hybridization with a pah-specific probe, and plasmids from hybridizing colonies were subjected to Southern analysis after NcoI digestion. Lanes 1 and 3 to 14, NcoI-digested plasmids from hybridizing colonies; lane 2, PstI digest of bacteriophage lambda DNA.
FIG. 4.
Similarity between the proteins predicted to be encoded by pah1 and pah2. Identical residues are indicated by colons, and similar residues are indicated by dots. The locations of two motifs associated with binding of manganese in arginase-type enzymes are shown underlined and in boldface.
When the 0.46-kb SalI fragment, which is internal to the pah1 gene, was used as a probe to screen cosmids from a pWE15 library of S. clavuligerus genomic DNA fragments, two recombinant cosmids which hybridized specifically with pah1 were identified. Neither of these recombinant cosmids hybridized to a probe specific for the cas1 gene, nor was there any overlap between the inserts of cas1- and pah1-bearing cosmids (data not shown). Therefore, there is no evidence of a physical linkage between the cas1 gene and the pah1 gene, at least within a range of about 40 kb (the average insert size of cosmids in this library).
Involvement of pah1 in clavulanic acid and clavam metabolite biosynthesis.
Although the pah1 gene showed considerable similarity to pah2, no direct evidence was available to link it to the production of clavulanic acid or the 5S clavams. In order to investigate the role of pah1 in clavulanic acid and 5S clavam metabolite biosynthesis, the gene was disrupted by a gene replacement technique similar to that used to disrupt pah2. The original disruption of pah2 used a thiostrepton resistance gene as a disruption marker, and so an apramycin resistance gene (apr) was used for disruption of pah1 to facilitate the eventual creation of a double mutant. apr was inserted in both orientations into an EcoNI site located centrally within pah1 (Fig. 5). The disrupted forms of pah1 carried on 3.1-kb NcoI-KpnI fragments (originally 1.65 kb before insertion of the 1.45-kb apramycin cassette) were then transferred into E. coli-Streptomyces shuttle plasmid pDA501. The disruption constructs were passaged through S. lividans before transformation into S. clavuligerus. Thiostrepton-resistant, apramycin-resistant transformants were then subjected to two rounds of sporulation in the absence of selection. Thiostrepton-sensitive, apramycin-resistant colonies were subjected to Southern analysis to confirm their status as pah1 mutants.
FIG. 5.
Diagrammatic representation of the 4.3-kb NcoI fragment carrying the pah1 gene. The open arrow indicates the size and direction of the pah1 gene. The open box indicates the size of the apr gene, and the arrows indicate that it was inserted in both directions relative to the orientation of the pah1 gene. Only the restriction sites mentioned in the text are shown.
Southern transfers of NcoI-digested DNA fragments were hybridized with linearized plasmid pUC120apr carrying the apramycin marker cassette. Disruption of pah1 in the mutants was evident because the 4.3-kb NcoI fragment seen in wild-type strains disappeared from the mutants and was replaced by a 5.75-kb hybridizing fragment corresponding to the 4.3-kb NcoI fragment plus the 1.45-kb apramycin resistance cassette (data not shown). No hybridization to the apramycin probe was seen by cured wild-type strains.
Single-flask cultures of eight of the pah1 mutants, including strains carrying the apr gene in both the forward and the reverse orientations relative to the orientation of the pah1 gene, were surveyed for clavulanic acid and 5S clavam metabolite production (Table 1). The level of production volumetrically was compared to that of the cured wild-type controls, and the strains were grown in both soy and SA media. Mutants grown in SA medium showed similar or slightly increased levels of clavulanic acid production compared to the levels of production by wild-type control strains, whereas mutants grown in soy medium showed slightly decreased levels of production relative to the levels of production of the wild-type controls. Changes in the levels of production of the 5S clavam metabolites were highly variable, but no definite pattern related to the mutation was observed. The extent of growth of the mutants (determined by measurement of the DNA content) was similar to that of the wild-type cultures on both media, and so when data were corrected for growth, no change in the production trends was observed (data not shown). Similarly, no differences in whether the disruption cassette was in the forward or the reverse orientation relative to the orientation of the pah1 gene were noted. Cephamycin C production was also unaffected by the mutation in pah1, as judged by a bioassay (data not shown).
TABLE 1.
Production of clavulanic acid and the 5S clavams by pah1 mutants
| Medium and mutant or wild- type straina | Concn (μg/ml)b
|
|||
|---|---|---|---|---|
| Clavulanic acid | Alanyl- clavam | Clavam-2- carboxylic acid | 2-Hydroxy- methylclavam | |
| SA medium | ||||
| M-5A | 13.0 | 0 | 0 | 0 |
| M-5B | 16.0 | 0 | 0 | 0 |
| WT-5 | 5.6 | 0 | 0 | 0 |
| M-6Ac | 10.7 | 0 | 0 | 0 |
| M-6Bc | 13.1 | 0 | 0 | 0 |
| WT-6 | 9.6 | 0 | 0 | 0 |
| M-11A | 18.6 | 0 | 0 | 0 |
| M-11B | 25.4 | 0 | 0 | 0 |
| M-11E | 22.9 | 0 | 0 | 0 |
| M-11F | 19.9 | 0 | 0 | 0 |
| WT-11 | 15.2 | 0 | 0 | 0 |
| Soy medium | ||||
| M-5A | 145 | 42 | 79 | 49 |
| M-5B | 180 | 40 | 60 | 34 |
| WT-5 | 441 | 0.5 | 66 | 121 |
| M-6Ac | 151 | 0.1 | 1 | 0 |
| M-6Bc | 155 | 0.4 | 3 | 0 |
| WT-6 | 132 | 0.9 | 3 | 0 |
| M-11A | 113 | 45 | 114 | 65 |
| M-11B | 161 | 37 | 87 | 44 |
| M-11E | 188 | 19 | 37 | 20 |
| M-11F | 91 | 45 | 121 | 79 |
| WT-11 | 175 | 24 | 75 | 45 |
Mutant strains are designated with the letter M, followed by a number to indicate the primary transformant which gave rise to the mutant, and then the letters A, B, etc., indicate the particular mutant strain. Wild-type strains are plasmid-free (cured) isolates from each primary transformant that gave rise to mutants and are designated WT, followed by a number to indicate the primary transformant. The cultures were grown for 96 h in SA or soy medium before analysis by HPLC.
The levels of production of the 5S clavam metabolites are expressed in clavulanic acid equivalents. Peak areas from HPLC analyses were converted to concentrations by using a clavulanic acid standard with a known concentration.
Mutants M-6A and M-6B both carried the apramycin cassette in the opposite orientation relative to that in the pah1 gene. All other mutants carried the disruption cassette in the same orientation as that in the pah1 gene.
Creation of a pah1 and pah2 double mutant.
Mutation of the pah1 gene had little effect on the level of production of clavulanic acid or any of the 5S clavams. This suggested that the pah2 gene is able to fulfill the requirement for Pah enzyme activity in a pah1 mutant. To test this theory, we created a pah1 and pah2 double mutant. Initial attempts to create this double mutant used the pah1 disruption construct as described above, but the construct was transformed into protoplasts of a pah2 mutant rather than into the wild type. For reasons which are not clear, we were unable to obtain double mutants by this approach, even though apramycin-resistant primary transformants were obtained. When these transformants were subjected to sporulation under nonselective conditions, only apramycin-sensitive cured isolates were obtained, even though many thousands of colonies were screened. In contrast, when the pah2 disruption construct used to create the original pah2 mutant (1) was introduced into one of the newly created pah1 mutants (mutant 5A), primary transformants were obtained, giving rise to numerous double mutants upon sporulation in the absence of selection.
Confirmation by Southern analysis.
Five double mutants were examined by Southern analysis to confirm the identities of the mutations. Genomic DNA preparations from each of the mutants and from the corresponding pah1 parental strain were digested with ApaI and analyzed by Southern hybridization with a radiolabeled 0.8-kb KpnI fragment comprising part of pah2 and part of cas2 as a probe. The pah1 parental strain gave a single strongly hybridizing band of 1.3 kb that corresponded to the wild-type pah2 gene. In contrast, each of the presumed mutant strains gave a strongly hybridizing band of 2.5 kb, and the 1.3-kb band was absent, confirming that the pah2 disruption had been introduced into the strains which already carried the pah1 disruption. Hybridization with a thiostrepton resistance gene probe also confirmed the identities of the mutants.
Production of β-lactam metabolites by the pah1 and pah2 double mutants.
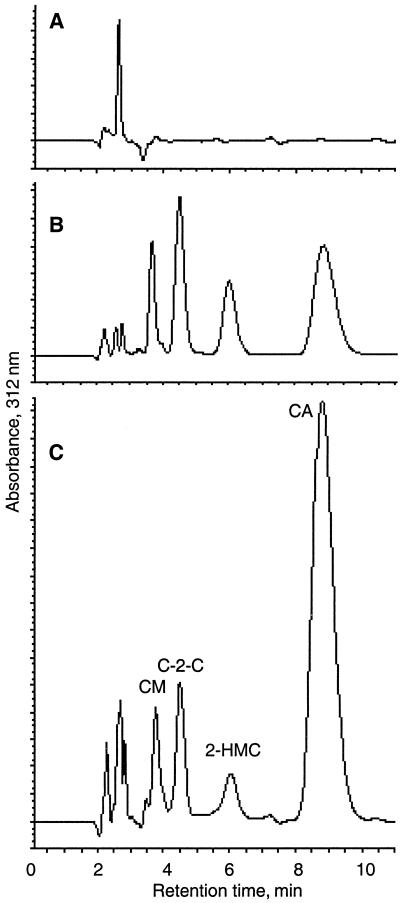
When mutants defective for both copies of the pah genes were grown in soy medium and SA medium and their levels of production of β-lactam metabolites were compared to that for wild-type S. clavuligerus, the production of clavulanic acid (retention time, 9.0 min) and the 5S clavam metabolites was found to be abolished, as determined by HPLC analyses (Fig. 6) and bioassays for clavulanic acid.
FIG. 6.
Analysis of culture filtrates from the wild type, the pah1 mutant, and the pah1 and pah2 double mutants of S. clavuligerus by HPLC. Culture filtrates from 96-h cultures grown on soy medium were analyzed by HPLC after derivatization with imidazole. The elution profiles for the pah1 and pah2 double mutant culture (A), the pah1 mutant culture (mutant 5A) (B), and the wild type (C), with detection at 311 nm, are shown. CM, clavaminic acid; C-2-C, clavam-2-carboxylic acid; 2-HMC, 2-hydroxymethylclavam; CA, clavulanic acid.
DISCUSSION
Past genetic studies with disruption mutants with defects in the early genes encoding clavulanic acid biosynthetic enzymes suggested the possibility that second functionally equivalent copies may exist for each of these genes. The existence of such a second copy was already known for cas, and the Cas enzyme was shown to be a mixture of two closely related isozymes. In contrast, Pah, another of the early enzymes, had been purified to homogeneity with no indication that it was a mixture of two isozymes. However, we had observed in past studies that digests of genomic DNA from S. clavuligerus showed evidence of a weakly hybridizing band when they were probed with a pah-specific probe, in addition to the expected strongly hybridizing band due to the known pah gene. When the DNA fragment representing this weakly hybridizing band was cloned and sequenced, the genetic material responsible for this hybridization was shown to be a second copy of the pah gene. This second copy of the pah gene, now called pah1, showed a high degree of similarity to the original pah gene, now called pah2, at both the nucleotide (72% identical) and amino acid (71% identical) sequence levels. Although this level of similarity is lower than that seen between the two forms of the cas gene, it is nonetheless high enough to suggest that the two Pah enzymes catalyze the same reaction.
Mutants with disruptions in pah2 showed some modest reduction in the levels of production of clavulanic acid, and the disruptions had no effect on the levels of production of the 5S clavam metabolites when the mutants were grown on a soy medium but resulted in the complete loss of metabolite production on SA medium (6). When mutants with defects in the pah1 gene were created and analyzed for their levels of clavulanic acid and 5S clavam metabolite production, they showed an even less clear-cut phenotype than the pah2 mutants. Neither clavulanic acid nor 5S clavam metabolite production was greatly affected by the pah1 mutation, regardless of whether the cultures were grown on SA medium or soy medium. From this result we concluded that the pah2 gene alone is able to produce sufficient Pah enzyme under the growth conditions used to support the full level of clavulanic acid and 5S clavam biosynthesis seen in the wild-type strain. These results are different from those seen when disruption mutants with defects in cas1 were prepared. In that case the levels of clavulanic acid production were little affected during growth on SA medium but were substantially decreased during growth on soy medium (14). For Pah, it appears that there is sufficient biosynthetic capability in the enzyme output from the pah2 gene alone to provide the intermediates needed for metabolite biosynthesis under the growth conditions used. However, since the Cas enzyme catalyzes three separate reactions in clavulanic acid biosynthesis, a single copy of the gene may be insufficient to meet all of the metabolic needs for cultures growing on soy medium.
Since disruption of pah1 had so little effect on the level of production of clavulanic acid or the other 5S clavam metabolites, it called into question the assumption that pah1 encoded an enzyme functionally equivalent to pah2. Creation of a pah1 and pah2 double mutant was undertaken to establish that pah1 was dispensable only under circumstances in which there was an intact copy of pah2. When these double mutants were prepared and analyzed, they were found to be defective in clavulanic acid and 5S clavam metabolite production on both SA and soy media, thereby establishing the functional equivalence of Pah1 and Pah2.
The discovery of a paralogue for pah, taken together with the known existence of the pair of cas genes, lends strong support to the theory that all of the genes encoding early enzymes for clavulanic acid biosynthesis are duplicated in S. clavuligerus. The reason for this duplication is far less evident. A sizeable number of Streptomyces species are known to produce various mixtures of 5S clavam metabolites, and a smaller number of species are known to produce clavulanic acid only (7). However, S. clavuligerus is the only species in this group of clavam producers that makes both clavulanic acid and 5S clavam metabolites. We originally postulated that S. clavuligerus may accomplish the production of both of these types of clavam metabolites by having evolved or otherwise acquired two complete clusters of biosynthetic genes, one for clavulanic acid production and one for the production of the 5S clavam metabolites. Since the early steps in the biosynthesis of both clavulanic acid and the 5S clavams up to the level of clavaminic acid are common, the organism would therefore carry duplicate genes for these early steps in the pathway. Other species would have one cluster or the other but not both.
However, when the newly discovered pah1 gene was used as a probe to screen a library of cosmids carrying fragments of chromosomal DNA from S. clavuligerus, two cosmids carrying the pah1 gene were identified. Neither of these cosmids hybridized to a cas1-specific probe, and no evidence was obtained to indicate that these pah1-bearing cosmids could cross-react with cosmids carrying cas1. Therefore, although two gene clusters, one for clavulanic acid production and one for production of the 5S clavam metabolites, were predicted, at present there is no evidence that pah1 and cas1 form a cluster. Studies are in progress to establish the organizational relationship between the clavulanic acid gene cluster, the cas1 gene cluster associated with production of the 5S clavam metabolites, and the newly described pah1 gene.
Acknowledgments
This study was supported by GlaxoSmithKline, the Natural Sciences and Engineering Research Council of Canada, and the Canadian Institutes of Health Research.
REFERENCES
- 1.Aidoo, K. A., A. Wong, D. C. Alexander, R. A. Rittammer, and S. E. Jensen. 1994. Cloning, sequencing and disruption of a gene from Streptomyces clavuligerus involved in clavulanic acid biosynthesis. Gene 147:41-46. [DOI] [PubMed] [Google Scholar]
- 2.Alexander, D. C., and S. E. Jensen. 1998. Investigation of the Streptomyces clavuligerus cephamycin C gene cluster and its regulation by the CcaR protein. J. Bacteriol. 180:4068-4079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bachmann, B. O., R. Li, and C. A. Townsend. 1998. β-Lactam synthetase: a new biosynthetic enzyme. Proc. Natl. Acad. Sci. USA 95:9082-9086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Elson, S. W., K. H. Baggaley, M. Davison, M. Fulston, N. H. Nicholson, G. D. Risbridger, and J. W. Tyler. 1993. The identification of three new biosynthetic intermediates and one further biosynthetic enzyme in the clavulanic acid pathway. J. Chem. Soc. Chem. Commun. 1993:1212-1214. [Google Scholar]
- 5.Hodgson, J. E., A. P. Fosberry, N. S. Rawlinson, H. N. M. Ross, R. J. Neal, J. C. Arnell, A. J. Earl, and E. J. Lawlor. 1995. Clavulanic acid biosynthesis in Streptomyces clavuligerus: gene cloning and characterization. Gene 166:49-55. [DOI] [PubMed] [Google Scholar]
- 6.Jensen, S. E., K. J. Elder, K. A. Aidoo, and A. S. Paradkar. 2000. Enzymes catalyzing the early steps of clavulanic acid biosynthesis are encoded by two sets of paralogous genes in Streptomyces clavuligerus. Antimicrob. Agents Chemother. 44:720-726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jensen, S. E., and A. S. Paradkar. 1999. Biosynthesis and molecular genetics of clavulanic acid. Antonie Leeuwenhoek 75:125-133. [DOI] [PubMed] [Google Scholar]
- 8.Jensen, S. E., D. W. Westlake, and S. Wolfe. 1982. Cyclization of δ-(l-α-aminoadipyl)-l-cysteinyl-d-valine to penicillins by cell-free extracts of Streptomyces clavuligerus. J. Antibiot. (Tokyo) 35:483-490. [DOI] [PubMed] [Google Scholar]
- 9.Kershaw, N. J., H. J. McNaughton, K. S. Hewitson, H. Hernandez, J. Griffen, C. Hughes, P. Greaves, B. Barton, C. V. Robinson, and C. J. Schofield. 2002. ORF6 from the clavulanic acid gene cluster of Streptomyces clavuligerus has ornithine acetyltransferase activity. Eur. J. Biochem. 269:2052-2059. [DOI] [PubMed] [Google Scholar]
- 10.Khaleeli, N., R. F. Li, and C. A. Townsend. 1999. Origin of the β-lactam carbons in clavulanic acid from an unusual thiamine pyrophosphate-mediated reaction. J. Am. Chem. Soc. 121:9223-9224. [Google Scholar]
- 11.Kieser, T., M. J. Bibb, M. J. Buttner, K. F. Chater, and D. A. Hopwood. 2000. Practical Streptomyces genetics. John Innes Foundation, Norwich, England.
- 12.Marsh, E. N., M. D. Chang, and C. A. Townsend. 1992. Two isozymes of clavaminate synthase central to clavulanic acid formation: cloning and sequencing of both genes from Streptomyces clavuligerus. Biochemistry 31:12648-12657. [DOI] [PubMed] [Google Scholar]
- 13.McNaughton, H. J., J. E. Thirkettle, Z. H. Zhang, C. J. Schofield, S. E. Jensen, B. Barton, and P. Greaves. 1998. β-Lactam synthetase: implications for β-lactamase evolution. Chem. Commun. 1998:2325-2326. [Google Scholar]
- 14.Mosher, R. H., A. S. Paradkar, C. Anders, B. Barton, and S. E. Jensen. 1999. Genes specific for the biosynthesis of clavam metabolites antipodal to clavulanic acid are clustered with the gene for clavaminate synthase 1 in Streptomyces clavuligerus. Antimicrob. Agents Chemother. 43:1215-1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Paradkar, A. S., and S. E. Jensen. 1995. Functional analysis of the gene encoding the clavaminate synthase 2 isoenzyme involved in clavulanic acid biosynthesis in Streptomyces clavuligerus. J. Bacteriol. 177:1307-1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Perez-Llarena, F. J., P. Liras, A. Rodriguez-Garcia, and J. F. Martin. 1997. A regulatory gene (ccaR) required for cephamycin and clavulanic acid production in Streptomyces clavuligerus: amplification results in overproduction of both β-lactam compounds. J. Bacteriol. 179:2053-2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Perez-Redondo, R., A. Rodriguez-Garcia, J. F. Martin, and P. Liras. 1999. Deletion of the pyc gene blocks clavulanic acid biosynthesis except in glycerol-containing medium: evidence for two different genes in formation of the C3 unit. J. Bacteriol. 181:6922-6928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Salowe, S. P., E. N. Marsh, and C. A. Townsend. 1990. Purification and characterization of clavaminate synthase from Streptomyces clavuligerus: an unusual oxidative enzyme in natural product biosynthesis. Biochemistry 29:6499-6508. [DOI] [PubMed] [Google Scholar]
- 19.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 20.Vieira, J., and J. Messing. 1987. Production of single-stranded plasmid DNA. Methods Enzymol. 153:3-11. [DOI] [PubMed] [Google Scholar]
- 21.Ward, J. M., and J. E. Hodgson. 1993. The biosynthetic genes for clavulanic acid and cephamycin production occur as a ′super-cluster' in three Streptomyces. FEMS Microbiol. Lett. 110:239-242. [DOI] [PubMed] [Google Scholar]
- 22.Wu, T. K., R. W. Busby, T. A. Houston, D. B. McIlwaine, L. A. Egan, and C. A. Townsend. 1995. Identification, cloning, sequencing, and overexpression of the gene encoding proclavaminate amidino hydrolase and characterization of protein function in clavulanic acid biosynthesis. J. Bacteriol. 177:3714-3720. [DOI] [PMC free article] [PubMed] [Google Scholar]