Abstract
In eukaryotic cells, the transcription of genes is accurately orchestrated both spatially and temporally by the C-terminal domain of RNA polymerase II (CTD). The CTD provides a dynamic platform to recruit different regulators of the transcription apparatus. Different posttranslational modifications are precisely applied to specific sites of the CTD to coordinate transcription process. Regulators of the RNA polymerase II must identify specific sites in the CTD for cellular survival, metabolism, and development. Even though the CTD is disordered in the eukaryotic RNA polymerase II crystal structures due to its intrinsic flexibility, recent advances in the complex structural analysis of the CTD with its binding partners provide essential clues for understanding how selectivity is achieved for individual site recognition. The recent discoveries of the interactions between the CTD and histone modification enzymes disclose an important role of the CTD in epigenetic control of the eukaryotic gene expression. The intersection of the CTD code with the histone code discloses an intriguing yet complicated network for eukaryotic transcriptional regulation.
Keywords: C-terminal domain, RNA polymerase II, transcription regulation, phosphorylation, CTD code, WW domain, CID domain, SRI domain, FF domain, histone code, epigenetic regulation
Biological systems have long served as a source of inspiration for engineering, resulting in numerous inventions based on biomimetics. During the last few decades, as electronics-based information technology has matured and flourished, our understanding of the biological system has also proceeded to the molecular and informational level. Such understanding has enabled researchers to reprogram cells to undertake unnatural tasks, such as the production of proteins and metabolites for medical and industrial purposes. Recent development of systems and synthetic biology promises the development of cells with even more complex artificial functions that will require the collaboration of a large number of genes. With a similar philosophy, information-based molecular programming has also been established as a new path for nanotechnology. Central to all these new technologies, cellular or acellular, is the encoding and decoding of information at the molecular level, particularly using DNA as the information carrier. Therefore, from both a scientific and engineering point of view, it is important to understand how biological systems read information from DNA.
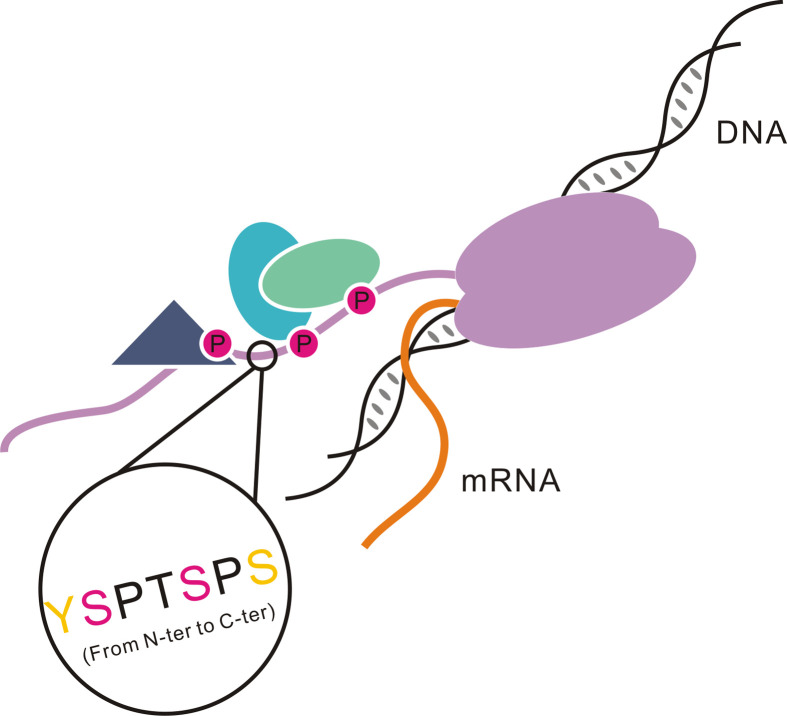
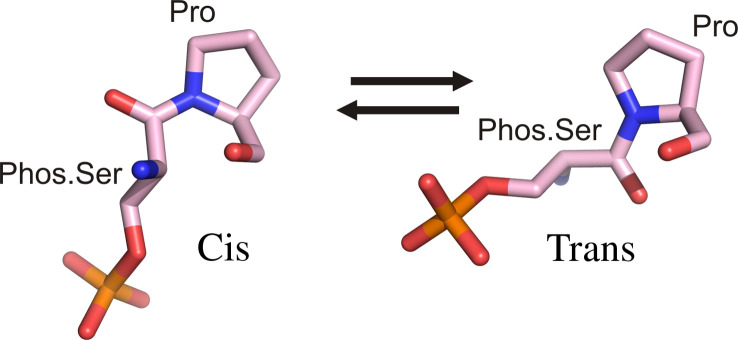
In biology, the major interpretation of this genetic information is through transcription regulation where eukaryotic RNA polymerase II plays the central role of transcribing the genetic information to the expressed protein. A special domain of RNA polymerase II functions as master controller for the transcription process by providing the template to recruit regulatory proteins to nascent mRNA (1). The conformational states of the CTD of RNA polymerase II, termed the CTD code, represent a critical regulatory check point for transcription (2–4). The CTD, found only in eukaryotes, consists of 26–52 tandem heptapeptide repeats generally with the consensus sequence, Tyr1Ser2Pro3Thr4Ser5Pro6Ser7 (1). Alterations of the sequence or the copy number of the heptapeptide may lead to distinguishable phenotypes or cell death (5, 6). The CTD can spatially and temporally recruit different regulatory and processing factors to the transcriptional machinery, reviewed in Corden (1) (Fig. 1) but the domain is disordered in X-ray crystal structures. The CTD phosphorylation is a major mechanism by which cells regulate gene expression, with serines at position 2 and 5 as major phosphorylation sites (7). Recently, Ser7 was also found to be phosphorylated in vivo although its function is still elusive (5). A secondary mechanism for CTD regulation is prolyl-isomerization of the two prolines in the CTD heptapeptide sequence (Fig. 2). By adjusting the cis–trans conformation of a proline adjacent to a phosphorylated serine, interaction of the CTD and binding partners it recruits can be modulated.
Fig 1.
Model of the CTD of RNA polymerase II. The RNA polymerase II is colored with purple. Different shapes bound to the CTD indicate various proteins that are recruited by the CTD. Magenta circles labeled with ‘P’ indicate phosphorylation on the CTD. One repeat in the black circle is zoomed in to show its primary sequence ‘YSPTSPS’. The Ser2 and Ser5 (colored with magenta) are always phosphorylated in each round of transcription, and Tyr1 and Ser7 (colored with yellow) are also detected as phosphorylation sites in vivo.
Fig 2.
Cis and trans conversion of proline in a phos.Ser-Pro motif. One mechanism of CTD phosphorylation is the conversion of proline isomerization states.
Coordinately regulated phosphorylation and dephosphorylation of the CTD plays an essential role not only in the recruitment and assembly of transcription complexes but also in temporal control of transcription and mRNA processing, reviewed in Refs (8, 9). Evidence points to the phosphorylation state of Ser2 and Ser5 as the trigger for transcriptional process modulation. Ser5 phosphorylation is required for assembly of the preinitiation complex (PIC) and facilitates mRNA capping via recruitment of capping enzymes (10, 11). During the transition when the transcription complex moves away from the initiation site, Ser5 gradually becomes dephosphorylated, whereas Ser2 is phosphorylated. Ser2 phosphorylation is the predominant CTD pattern on both elongating and terminating RNA Polymerase II, which ensures efficient 3′-RNA processing by triggering recruitment of 3′-RNA processing machinery (8). At the end of transcription, CTDs are free of phosphate groups; non-phosphorylated CTDs are required for RNA polymerase II to recycle and bind a promoter for the next cycle of transcription (8). Little is known about the timing of Ser7 phosphorylation and how it affects the transcription but it appears to be an essential event specific for snRNA expression (12).
One central question for CTD-directed transcription regulation is how high resolution recognition of different states of Ser2 and Ser5 are identified. Residues flanking Ser2 and Ser5 are highly similar, so it is puzzling how the transcription regulation is managed with such precision in location and timing. The most direct way to visualize and identify molecular elements in binding specificity is X-ray crystallography. A careful examination of the primary sequence of the CTD reveals the possibility that the CTD might have little secondary structure until its association with binding partners. In this review, we will discuss our current understanding of the CTD structure and how the CTD is recognized by its binding partners (Table 1).
Table 1.
Summary of CTD interacting proteins/domains
| Species | Protein | Method | Preference | PDB code | References |
|---|---|---|---|---|---|
| Homo sapiens | Pin1 | X-ray | Phos.Ser5 CTD | 1f8a | (22) |
| Saccharomyces cerevisiae | Pcf11 CID | X-ray | Unphosphorylated CTD; phos.Ser2 CTD | 1sza | (39) |
| Saccharomyces cerevisiae | Nrd CID | X-ray | Phos.Ser5 CTD | 3clj | (43) |
| Homo sapiens | SCAF8 CID | X-ray | Phos.Ser2 CTD | 3d9i | (46) |
| Homo sapiens | Scp1 | X-ray | Phos.Ser5 CTD | 2ght | (65) |
| Schizosaccharomyces pombe | Fcp1 | X-ray | Phos.Ser2 CTD | 3ef0 | (59) |
| Candida albicans | Cgt1 | X-ray | Phos.Ser5 CTD | 1p16 | (67) |
| Homo sapiens | Set2 SRI | NMR | Phos.Ser2 CTD | 2a7o | (77) |
| Saccharomyces cerevisiae | Set2 SRI | NMR | Phos.Ser2 CTD | 2c5z | (78) |
| Homo sapiens | CA150 FF | NMR | ?a | 2kis | (82) |
| Saccharomyces cerevisiae | Prp40 FF | NMR | ? | 2b7e | (81) |
Not determined.
Pin1 and the C-terminal domain (CTD)
The first glimpse of the CTD structure was through its interaction with human Pin1, a unique prolyl isomerase that catalyzes cis/trans isomerization of specific phos.Ser/Thr-Pro motifs in signaling proteins (13–16). Identification and characterization of this novel peptidyl-prolyl cis/trans isomerase (PPIase), Pin1, led to discovery of a novel postphosphorylation regulatory mechanism, in which regulation is achieved by conformational changes of a phosphorylated Ser/Thr-Pro peptide bond upon proline isomerization (Fig. 2). This change in the configuration of the polypeptide has a profound effect on Pin1 targets and therefore modulates various signaling pathways at both transcriptional and posttranslational levels. Specifically, activity of prolyl-isomerization of Pin1 can interconvert the cis/trans conformation of the phos.Ser/Thr-Pro motif of target proteins and make them better or worse substrates for conformation-specific signaling kinases (such as cyclin-dependent protein kinase, glycogen synthetase kinase 3 β, and mitogen-activated protein kinase) and phosphatases (such as PP2A and Cdc25). Recent studies also provide substantial evidence implicating Pin1 in progression of malignant tumor cells (17) and development of Alzheimer's disease (18). High affinity inhibitory unnatural peptides have been developed as a good template for chemical compounds targeting Pin1 for antineoplastic effects (19).
Compelling data have implicated human Pin1 as a key modulator in the transcription mechanism. The yeast homologue of Pin1, Ess1, interacts physiologically and genetically with the CTD (20). Furthermore, hyperphosphorylated RNA polymerase II appears to be the dominant binding target in yeast extracts (21). Considering the high local concentration of the phos.Ser/Thr-Pro motif in the hyperphosphorylated CTD, it is plausible that the CTD is the major substrate of Pin1 in vivo. The binding of a single CTD repeat that is phosphorylated at Ser5 was reported to be 30 µM (22). Presumably, the CTD tail containing 26–52 such repeats localizes a substantial amount of Pin1.
Pin1 is a 163 amino acid polypeptide that can be divided into two domains based on topology and function, a C-terminal PPIase domain and an N-terminal WW domain. Structure of the WW domain reveals three antiparallel β-strands forming a shallow interface with the PPIase domain for substrate peptide binding (23, 24). It has long been realized that WW domains recognize proline-containing sequences but it was not clear until recently that WW domains join a group of modules that bind to protein ligands in a phosphorylation-dependent manner (25, 26); these domains include SH2, PTB, 14-3-3, WD40, FHA, and FF domains. The modular nature of WW domain interactions leads to a classification into four distinct groups based on binding specificity (27). Group I WW domains, such as dystrophin and the Yes-associated protein YAP65, recognize ‘PpxY’ motifs (28). Group II, such as FE65 and forming binding proteins (FBPs), bind the ‘PPLP’ motif (29). A subset of FBPs interacts with ‘PGM’ motifs (30). Group III WW domains select polyproline motifs flanked by arginine or lysine (31, 32). Group IV WW domain, including human Pin1 and Nedd4, specifically recognize a phos.Ser/Thr-Pro motif (22, 26, 33).
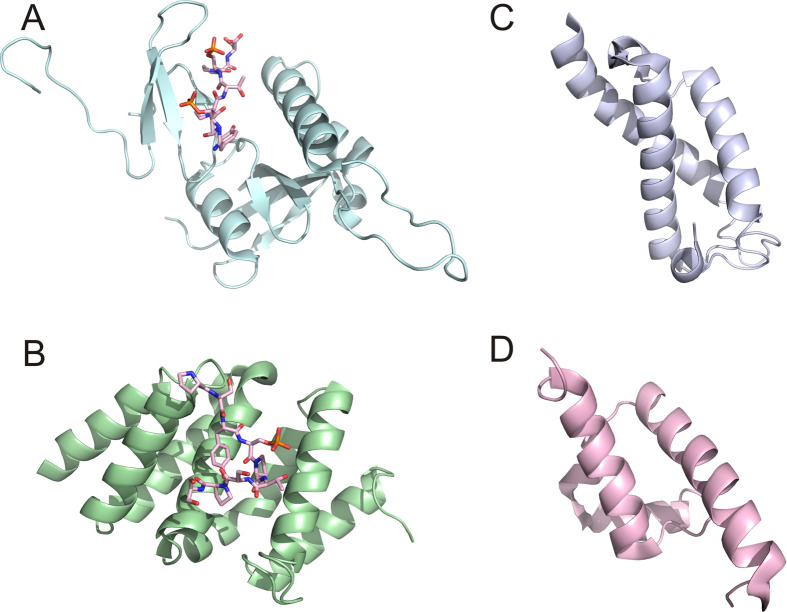
The molecular detail of recognition of the CTD by Pin1 was elucidated by a structure of human Pin1 in complex with one doubly phosphorylated CTD repeat (22) (Fig. 3A). So far, this is the only structure available for a full-length Pin1 binding to its target at the recognition module WW domain. The structure is consistent with thermodynamic data, which the phosphorylated Ser5 in the CTD repeat is the major binding element in recognition. Loop 1 of the WW domain has been shown to be highly flexible in apo Pin1 (34) but this loop is essential for specificity recognition (35). In the complex structure, this loop warps toward the substrate peptide to ensure binding and results in an exaggerated twist in the triple-stranded β-sheet (22) (Fig. 3A). This twist is coupled to a contraction of the WW domain ligand binding surface formed between the WW and PPIase domains. The two essential elements for peptide recognition include binding of phosphate by loop 1 residues and hydrophobic stacking of proline by Tyr23 and Trp34 (22). The binding of Pin1 to the CTD can modulate the regulatory effect of other CTD-binding proteins. In vitro experiments showed that Pin1 can influence the phosphorylation status of the CTD by inhibiting the transcription factor IIF-interacting CTD phosphatase 1 (Fcp1) and stimulating CTD phosphorylation by cdc2/cyclinB (36, 37). The Rsp5, a ubiquitin ligase that binds to the CTD, also functions to oppose Pin1 effects on RNA polymerase II (38). The biological and structural results of Pin1 effects on RNA polymerase II function support a model that Pin1 works in a processive manner on the CTD with the WW domain acting as a binding element restricting movement to an efficient one-dimensional walk and with the PPIase acting much like a reading head to processively isomerize the peptide bonds. The binding of Pin1 prolongs the phosphorylated state of the CTD by suppressing dephosphorylation, thereby enhancing the regulatory effect of CTD binding proteins.
Fig 3.
Ribbon representation of four CTD recognition modules. (A) WW domain of Pin1 in complex with a short CTD peptide (1f8a); (B) CID domain of Pcf11 in complex with a short CTD peptide (1sza); (C) SRI domain of Set2 (2a7o); and (D) FF domain of CA150 (2kis).
CTD-interacting domain (CID) domain and the C-terminal domain (CTD)
Recognition of the phosphorylated CTD by Pin1 is mediated by its WW domain, a modular domain of around 40 residues that is essential for recognition of proline-rich motifs by the PPIase domain. The Pin1 WW domain also recognizes other substrates in addition to the CTD. A more specific recognition domain for the CTD is the CTD-interacting domain (CID) identified in multiple RNA processing and termination factors in eukaryotes (39, 40) (Fig. 4).
Fig 4.
Superimposition of CID domains from Pcf11 (light blue, 1sza), Nrd1 (light pink, 3clj), and SCAF8 (white, 3d9i).
In yeast, the effective termination of transcription relies on the recruitment of cleavage factors by Pcf11. The Pcf11 is a yeast protein of 70 kD with a CID domain directly targeted to the CTD of RNA polymerase II, which gives us the first glance of how a CID recognizes heptad repeats of the CTD. Interestingly, the CID domain of Pcf11 can bind to both unphosphorylated or phos.Ser2 CTD in biophysical binding assays (39). Consistent with the biochemical data for such preferences, the co-crystal structure of the Pcf11 CID and a CTD peptide shows no direct interaction between the phosphate group of phos.Ser2 and Pcf11, indicating no prerequirement of phosphorylation of the CTD for protein binding (39) (Fig. 3B). The CID domain, as an eight-helical bundle, recognizes a span of two heptad repeats in the CTD with a conserved groove, whereas the phosphate group of phos.Ser2 actually forms an intramolecular hydrogen bond with Thr4 of the CTD and stabilizes the sharp β-turn formed by CTD, presenting the side chain to the binding groove (Fig. 3B). Hydrogen bondings between the CID domain and the CTD peptide are distributed between the CID side chain and the main chain amide and carbonyl group of the CTD peptide. Importantly, hydrophobic interaction by Tyr1 of the CTD to CID might contribute greatly for the specificity for phos.Ser2 over phos.Ser5. Meinhart and Cramer (39) conclude that Ser2 phosphorylation stabilizes the CTD β-spiral that is incorporated into the transcription complex. Ser5 phosphorylation would unwind the spiral resulting in an extended region that binds the capping enzyme. Overall, the binding of Pcf11 is not particularly strong but is consistent with its dynamic role involved in dismantling the elongation complex (41, 42).
Another CID containing protein Nrd1 is an essential player in the termination pathway for RNA polymerase II-mediated transcription for snoRNA, snRNA as well as cryptic unstable transcripts (CUT). The complex of Nrd1-Nab3-Sen1 is recruited to the transcription machinery through the CTD by the CID domain of Nrd1 of the complex. The CID domain derived from Nrd1 (Fig. 4) has a very similar overall fold and a strong conservation of residues involved in CTD peptide binding with Pcf11, but Nrd1 shows a different specificity profile in which a much stronger preference for phos.Ser5 over phos.Ser2 in the CTD sequence is detected for the CID from Nrd1 by in vitro binding assays (43). Using yeast-two hybrid and fluorescence anisotropy, Vasiljeva showed a tenfold improvement in binding affinity for singly phosphorylated CTD double repeats at Ser5 over Ser2 (Kd=40 µM for phos.Ser5 vs. 390 µM for phos.Ser2) and a slight improvement when the peptide is doubly phosphorylated (Kd=16 µM upon double phosphorylation for both Ser2 and Ser5 sites). Specificity for phos.Ser5 on the protein is essential to understand how different termination pathways are selected. A potential different phosphate binding site was proposed but more insightful information about specificity will only be available with a structure of the Nrd1–CID complexed with the CTD peptide. Since the Nrd1-dependent termination pathway is usually associated with much shorter transcripts (a few hundred base-pairs), upon which point dephosphorylation of Ser5 is not complete, a logical hypothesis is that phosphorylation at Ser5 favors the selection of Nrd1 complex as a termination pathway in yeast. Indeed, Gudipati et al. (44) showed that reducing phosphorylation of Ser5 using a mutant of kin28, a CTD Ser5 kinase, will hamper Nrd-dependent termination. The selectivity for phos.Ser5 over phos.Ser2 might be the determining factor for the selection of termination pathways. A functional model for how the phosphorylation pattern of the CTD determines the transcriptional pathways suggests that termination by the Nrd1-Nab3-Sen1 complex is enforced when the CTD is still highly phosphorylated at Ser5 (45). The structure of a complex of the CID of Nrd1 with the CTD domain will help provide a molecular explanation of transcription termination.
Another CID containing protein human SCAF8 was crystallized with different phosphorylated forms of CTD peptides (Fig. 4), providing more clues about how registration of a phosphate group on the CTD is encoded in target recognition for CID domains (46). The SCAF8 is implicated in splicing with a preference of phos.Ser2, similar to mRNA 3′-processing factor Pcf11. Indeed, a similar conformation of beta-turn adapted by the CTD is observed in SCAF8 upon peptide binding but with one major distinction: phos.Ser2 is directly recognized by a basic residue, Arg112, through salt bridge interaction (46). At a similar position in Pcf11, a methionine residue was placed with no direct interaction with the CTD peptide. The replacement of methionine by arginine in SCAF8 distinguishes the phos.Ser2 CTD from the unphosphorylated form and might explain a tighter interaction for SCAF8 toward phosphoryl-peptide (68 µM). In the study of Becker et al. (46), an issue was raised whether phosphorylated Ser7 contributes to recognition by the CID domain of SCAF8. Binding affinity measured by fluorescence anisotropy showed no advantage with additional Ser7 phosphorylation (68 µM for phos.Ser2 and 90 µM for doubly phosphorylated Ser2 and Ser7). Consistent with the binding interaction measurement, the structure of the complex with the SCAF8 CID presents no interaction between the phosphate group of Ser7 with the protein.
Small C-terminal domain phosphatases (Scp)/Fcp phosphatases and the C-terminal domain (CTD)
A wide range of enzymes participate in dynamic modifications of the CTD, including kinases and phosphatases responsible for addition and removal of phosphates. The CTD is principally phosphorylated by cyclin-dependent kinases (CDKs) with their associated cyclins. Specifically, Ser5 phosphorylation is mainly catalyzed by Cdk7/cyclin H subunits of TFIIH (47–49); Ser2 phosphorylation is mainly catalyzed by P-TEFb, which contains Cdk9/cyclin T subunits (50, 51). Intriguingly, it is suggested that Cdk9 also makes a contribution to Ser5 phosphorylation and the relative contribution of TFIIH-associated Cdk7 varies between different genes based on experimental observations (52). Moreover, the CTD can also be phosphorylated at both Ser2 and Ser5 by Cdk8 as part of the mediator complex, and preferentially phosphorylated at Ser5 by MAPK2/ERK2 (53). Recently, TFIIH-associated Cdk7 kinase has also been shown to phosphorylate Ser7 in vivo (52, 54). Even though structural information has been obtained for Cdk7 (55) and Cdk9/cycln T (56), how they recognize the CTD peptide and label the phosphorylation mark on CTD is still elusive. It is assumed that specificity is achieved by other associated proteins in the multiprotein complexes they are involved (Cdk7 in TFIIH, Cdk8 in mediator, and Cdk9 in P-TEFb).
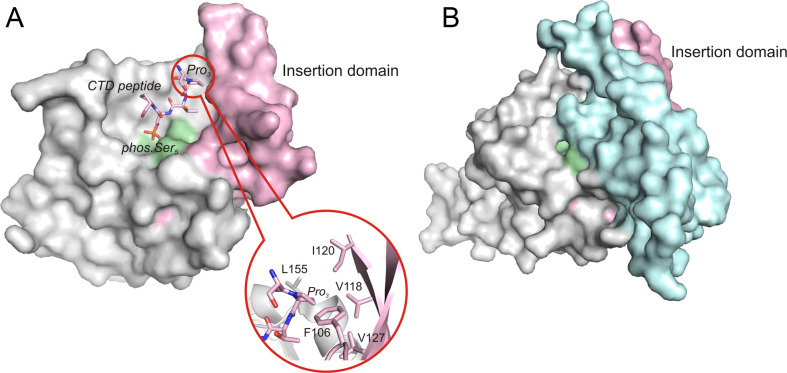
Dephosphorylation is essential for recycling RNA polymerase II, because after each round of transcription, the CTD has to be dephosphorylated in order to actively restart a new round of transcription. In humans, Fcp1, which is required for general transcription and cell viability, was the first discovered CTD-specific phosphatase with a catalytic preference for phos.Ser2. The Fcp1 is conserved among eukaryotes and was shown to be essential for cell survival in budding and fission yeast (57, 58). The conserved region of Fcp1 is composed of two domains: an N-terminal FCP homology (FCPH) domain with phosphatase activity and a C-terminal breast cancer protein related C-terminal (BRCT) domain (59) (Fig. 5).
Fig 5.
Surface representation of Scp1 and Fcp1 FCPH domains. (A) Scp1 in complex with a short CTD peptide (2ght). The zoom-in picture shows the Pro3 binds to the hydrophobic pocket with the aromatic residues shown in stick. (B) Fcp1 FCPH domain (3ef0). In both structures, the active site signature motif is colored with pale green, and the insertion domain is colored with light pink. Notably, the additional helical domain (light cyan) covers the ‘insertion domain’ in Fcp1 and makes it much less accessible for substrates.
Recently, a family of small CTD phosphatases (Scps) with activities preferential for phos.Ser5 was identified (60, 61). This family includes three highly similar proteins designated Scp1, Scp2, and Scp3. The Scps also contain the FCPH catalytic domain that includes the DXDX(T/V) motif, the signature of a superfamily of phosphotransferases and phosphohydrolases called the haloacid dehalogenase (HAD) superfamily (62). Therefore, Fcp/Scp family members are classified as HAD superfamily enzymes. Interestingly, outside the signature motif, Scps share very little sequence similarity with the other enzymes in the HAD superfamily (63). In humans, Scp1 has more than 20% sequence identity to Fcp1 in the FCPH domain but lacks the C-terminal BRCT domain that exists in Fcp1. Moreover, Scp2 and Scp3, which also lack the BRCT domain, share more than 90% similarity with Scp1 in the FCPH domain (60) (Fig. 5).
The apo structure of Scp1 solved by Kamenski et al. (64) showed a central parallel β sheet flanked by two α helices, a two-stranded β sheet, and a short 310 helix. The conserved DXDX(T/V) signature motif lines part of a central crevice, which forms the active site and coordinates the Mg2+ ion that is essential to Scp1 phosphatase activity. The first aspartate in the signature motif is involved in Mg2+-assisted phosphoryl transfer and acts as the phosphoryl acceptor. Mutation of this residue (Asp96 in Scp1) to alanine or asparagine abolished the activity of Scp1. The second aspartate (Asp98 in Scp1) also contributes to metal ion-binding and could possibly function as a general acid/base (64). The proposed phosphoryl transfer mechanism for the Scp/Fcp family involves a phosphoryl-aspartate intermediate. Existence of this phosphoryl-enzyme intermediate was confirmed in a recent structural and functional study when we successfully trapped the phosphoryl-aspartate intermediate in the crystal structure of an Scp1D206A mutant soaked with para-nitrophenyl phosphate (pNPP) (63). The steady-state kinetic analysis of a variety of Scp1 mutants revealed the importance of Asp206 in Mg2+ coordination mediated by a water molecule. Moreover, snapshots of the phosphoryl transfer reaction at each stage of Scp1-mediated catalysis were also captured in this study.
In order to understand the discrimination of phos.Ser5 over phos.Ser2 as a substrate by Scp1, the complex structure of a dominant negative form of human Scp1 (Scp1D96N) bound with Ser2/Ser5-phosphorylated CTD peptide was obtained by crystal soaking (65). The defined complex structure revealed a unique binding mode of the peptide in which Ser2Pro3Thr4(phos.Ser5) forms a β-turn. The phos.Ser5 binds to the active site groove through Mg2+ coordination. The Pro3 is recognized by an aromatic-rich hydrophobic pocket near the active site that further confers substrate specificity (65). Notably, Scp1 shows remarkable specificity toward the trans peptide-bond configuration of the two prolines in the CTD repeat, which can adopt both cis and trans configurations. Such configuration switching is known to modulate the structure of the CTD and its accessibility to kinases or phosphatases (21, 22). An insertion domain formed by a three-stranded β sheet directly follows the signature motif (Fig. 5). This insertion domain is unique to Fcp1/Scp1 family phosphatases and may assist in substrate recognition. Consistent with the known specificity of Scp1 toward phos.Ser5 instead of phos.Ser2, in the complex structure the phos.Ser2 flips out of the active site, making no direct interaction with the protein.
Even though Scp and Fcp share similar phosphatase active sites, their strategy for substrate recognition might be different, as suggested by the recent crystal structure of apo Schizosaccharomyces pombe Fcp1 (SpFcp1) (59) (Fig. 5). The minimal effective CTD substrate for SpFcp1 is a single heptad CTD peptide: Ser5Pro6Ser7Tyr1(phos.Ser2)Pro3Thr4, among which the Tyr1 and Pro3 flanking the phos.Ser2 are critical determinants of Fcp1 activity (66). The SpFcp1 structure revealed that it is a Y-shaped protein composed of three structural domains (59). The stem of the Y is the FCPH domain that contains a globular catalytic phosphatase core similar to that of Scp1. One major difference is that the three-stranded β sheet in Scp1 (insertion domain) is accessible for substrate recognition, whereas in Fcp1 it is buried by a helical insertion domain, suggesting a different binding interface between Fcp and the CTD with phos.Ser2 (Fig. 5).
mRNA capping enzyme Cgt1 and the C-terminal domain (CTD)
Even though 26–52 heptad repeats exist in the CTD primary sequence, recognition of the CTD by proteins in all the complex structures discussed above only show a spanning of one or two repeats. This is understandable since a balance between a favorable interaction of the CTD with its binding partners versus the entropy cost for binding a disordered peptide needs to be achieved. This leads to the proposed mechanism that a double repeat of the CTD sequence is the functional unit for transcription. To explore if such rule is consistent with the mRNA capping enzyme Cgt1, four heptad repeats each with their Ser5 phosphorylated were used in a cocrystallization experiment (67). Interestingly, a long span of CTD peptide consisting of 17 amino acids was modeled in the density of one of the two monomers with an extensive buried surface of 1,600 Å2 (67). Three of the four phosphorylated Ser5 residues were visible in this structure with both the first and third phosphate group recognized by positively charged patches on the Cgt1 surface. Consistent with a previous study of Cgt1, the Tyr1 position in the CTD sequence is essential for recognition by the protein (68). On the other hand, a single mutation of the Cgt protein for the recognition interface did not show obvious deleterious effects during yeast mutation screening (67), possibly due to the extended binding surface without one interaction dominating binding. Since the other monomer in the asymmetric unit shows a much smaller interface, it suggests that multiple phosphorylation sites are not a prerequisite for ligand binding. Further affinity measurements with a different length and registration of the phos.Ser5 would elucidate whether interaction with multiple repeats of the CTD is essential for the effective binding by Cgt1.
Nuclear magnetic resonance (NMR) structure of Set2
The identification of the histone methyltransferase Set2 as a novel CTD binding partner bridges the CTD code to the histone code by implicating the regulatory effect of the CTD on histone modification and epigenetic control (69, 70) (Fig. 3C). In eukaryotes, large genomes are efficiently organized into nucleosomes that are fundamental repeat units of chromatin. The nucleosome is composed of a histone octamer consisting of two copies of each of the core histones H2A, H2B, H3, and H4, around which 147 bp of DNA is wrapped. Such a structure is not only a strategy to compress large genomic DNA, but also provides a potential solution for tight regulation of DNA replication, repair, and transcription. During transcription, for example, the binding sites for a variety of transcription factors can be occluded by histones, leading to transcriptional silencing (71). Access to specific loci on the nucleosomal DNA is dynamically regulated by many factors including chromatin modifiers and chromatin remodelers. Histones can be covalently modified by chromatin modifiers at particular loci, most of which are concentrated in the relatively unstructured N-terminal tails of histones. These modifications include acetylation, methylation, phosphorylation, ubiquitination, and sumoylation (72). The histone code, which is defined by covalent modifications, represents a fundamental regulatory mechanism of gene expression and repression (73). Furthermore, the histone code can be interpreted by different modules in a modification-dependent manner to decide whether a gene is to be transcribed.
A novel Rpb1-binding domain of Set2, called Set2–Rpb1 interacting (SRI) domain, mediates the recognition and interaction with the phosphoryl CTD (Fig. 3C). Lys4 of histone 3 is methylated by the proteins of the Set1 family, while Lys36 is methylated by proteins of the Set2 family (69). Both Set1 and Set2 associate with the CTD but at different stages of transcription: Set1 binds to phosphorylated CTD enriched with Ser5 at the promoter region through the mediation of the Paf complex (74, 75); whereas the recognition of the CTD by Set2 relies on the phosphorylation of Ser2 in heptad repeats (76). The SRI domains from both human (77) and yeast (78) Set2 were determined by NMR and the binding interface mapped by phosphoryl-peptide titration. The study of affinity with different length and phosphorylation registration showed that single repeats are not sufficient for SRI recognition and a strong preference for doubly phosphorylated peptide was observed. Five residues were identified as essential for the effective association between Set2 SRI and the CTD using NMR and mutagenesis (74). One can assume the positively charged residues, Lys54, Arg58, and His62 of the Set2 SRI, are involved in phosphate binding whereas Val31 and Phe53 are important for hydrophobic interaction with the side chain residues of the CTD, possibly tyrosine or proline.
In addition, the FF domain is a protein-protein interaction module existing in several CTD interaction proteins such as human transcription factor CA150 (79) and splicing factor Prp40 (80). Like the SRI domain, the FF domain exhibits a structure of three-helical bundle (Fig. 3D). The FF domains usually are arranged as tandem arrays in proteins and such architecture might account for interaction with the CTD with each module contributing very weakly to binding. Identification of the specificity of phosphorylation states of the CTD recognized by FF domains has been challenging due to the weak binding. In vitro binding assays and NMR titration experiments did not detect interaction with a CTD peptide with the FF domain arrays derived from Prp40 (81) or CA150 (82). A binding constant of 50 µM was reported for the mammalian homologue of Prp40, FBP11, using isothermal titration calorimetry (83) but detailed information about the interaction between molecules is yet to be elucidated.
Concluding remarks
Recognition of the phosphorylation states of the CTD is essential for mediation of the expression of genetic information. Proteins that are highly selective toward specific phosphorylated forms of the CTD decipher the ‘CTD code’ and synchronize the transcriptional events accordingly. Communication of the CTD and histone codes provides an exquisite regulatory network for the precise control of transcription at multiple levels. Some CTD interacting proteins function as global transcriptional regulators non-discriminatingly. The inactivation of such molecules will lead to the shutdown of transcription machinery and eventually, cell death. For example, the deletion of the CTD Ser2 phosphatase Fcp1 is lethal for yeast due to the abolishment of RNA polymerase II recycling (58, 84). Similar effects on yeast are observed when the CTD Ser5 phosphatase Ssu72 is eliminated (85). On the other hand, evidence has indicated that CTD-interacting proteins can control gene expression at an epigenetic level and regulate expression of specific genes based on timing and the developmental needs. Human Scps are only found in higher eukaryotes and their expression is limited to neuronal stem cells or non-neural tissues. Consistent with their unique expression profile, Scps are identified as a component of the neuronal chromatin remodeling complex, REST (61). The inactivation of Scp activity leads to the inappropriate differentiation of neuronal stem cells (61). Another example of epigenetic regulation of a CTD-interacting protein is the Cdk8/cyclin C pair, which has been linked to transcriptional repression (48). Histone methyltransferases Set1 (75) and Set2 (74) can identify the different phosphorylation stages of CTD. The phosphorylation of Ser7 of CTD itself also occurs in a gene-specific fashion (12). The apparent simple primary sequence of CTD integrates genetic and epigenetic information and plays a pivotal role in transcriptional activation or repression. Understanding how such coding is deciphered at the molecular level is essential to the interpretation of the central role of RNA polymerase II and its regulatory domains. The application of CTD code provides a unique opportunity to engineer the transcriptional process in an epigenetic level in tissue-specificity manner.
Biographies
 Mengmeng Zhang is a second year PhD student in the laboratory of Professor Zhang. She is a graduate from Shanghai Jiao Tong University, China (BE) and Clemson University (MS). She has performed several research works on the Scp1 phosphatase.
Mengmeng Zhang is a second year PhD student in the laboratory of Professor Zhang. She is a graduate from Shanghai Jiao Tong University, China (BE) and Clemson University (MS). She has performed several research works on the Scp1 phosphatase.
 Dr. Gordon Gill, is Professor Emeritus in the Departments of Medicine and Cellular and Molecular Medicine at the University of California, San Diego and actively serves as Dean for Scientific Affairs for the School of Medicine. Dr. Gill received his BA and MD from Vanderbilt University. After serving a residency and a National Institutes of Health postdoctoral fellowship at the Yale-New Haven Hospital, he continued his NIH-sponsored postdoctoral research in metabolism and endocrinology at UCSD. During his tenure there, he served as chief of the Division of Endocrinlogy and Metabolism in the Department of Medicine (1971–1995) and was associate chair for scientific affairs in the Department of Medicine (1992–1995).
Dr. Gordon Gill, is Professor Emeritus in the Departments of Medicine and Cellular and Molecular Medicine at the University of California, San Diego and actively serves as Dean for Scientific Affairs for the School of Medicine. Dr. Gill received his BA and MD from Vanderbilt University. After serving a residency and a National Institutes of Health postdoctoral fellowship at the Yale-New Haven Hospital, he continued his NIH-sponsored postdoctoral research in metabolism and endocrinology at UCSD. During his tenure there, he served as chief of the Division of Endocrinlogy and Metabolism in the Department of Medicine (1971–1995) and was associate chair for scientific affairs in the Department of Medicine (1992–1995).
 Dr. Yan Jessie Zhang is currently a tenure-track assistant professor in the Department of Chemistry and Biochemistry at the University of Texas, Austin. She obtained her BS at the Tsinghua University in China in the area of medicinal chemistry and later came to the United States for crystallographic training from Dr. Brian W. Matthews of the University of Oregon. Dr. Zhang received her PhD from The Scripps Research Institute mentored by Dr. Ian A. Wilson and completed her postdoctoral training with Dr. Joseph P. Noel at The Salk Institute for Biological Studies. Her independent laboratory is now focusing on the elucidation of the fundamental principles of transcriptional regulation and their impact on tumor and stem cell differentiation using X-ray crystallography and other biophysical methods.
Dr. Yan Jessie Zhang is currently a tenure-track assistant professor in the Department of Chemistry and Biochemistry at the University of Texas, Austin. She obtained her BS at the Tsinghua University in China in the area of medicinal chemistry and later came to the United States for crystallographic training from Dr. Brian W. Matthews of the University of Oregon. Dr. Zhang received her PhD from The Scripps Research Institute mentored by Dr. Ian A. Wilson and completed her postdoctoral training with Dr. Joseph P. Noel at The Salk Institute for Biological Studies. Her independent laboratory is now focusing on the elucidation of the fundamental principles of transcriptional regulation and their impact on tumor and stem cell differentiation using X-ray crystallography and other biophysical methods.
Conflict of interest and funding
There is no conflict of interest in the present study for any of the authors.
References
- 1.Corden JL. Tails of RNA polymerase II. Trends Biochem Sci. 1990;15:383–7. doi: 10.1016/0968-0004(90)90236-5. [DOI] [PubMed] [Google Scholar]
- 2.Dahmus ME. Reversible phosphorylation of the C-terminal domain of RNA polymerase II. J Biol Chem. 1996;271:19009–12. doi: 10.1074/jbc.271.32.19009. [DOI] [PubMed] [Google Scholar]
- 3.Meinhart A, Kamenski T, Hoeppner S, Baumli S, Cramer P. A structural perspective of CTD function. Genes Dev. 2005;19:1401–15. doi: 10.1101/gad.1318105. [DOI] [PubMed] [Google Scholar]
- 4.Palancade B, Bensaude O. Investigating RNA polymerase II carboxyl-terminal domain (CTD) phosphorylation. Eur J Biochem. 2003;270:3859–70. doi: 10.1046/j.1432-1033.2003.03794.x. [DOI] [PubMed] [Google Scholar]
- 5.Chapman RD, Heidemann M, Albert TK, Mailhammer R, Flatley A, Meisterernst M, et al. Transcribing RNA polymerase II is phosphorylated at CTD residue serine-7. Science. 2007;318:1780–2. doi: 10.1126/science.1145977. [DOI] [PubMed] [Google Scholar]
- 6.Rogers C, Guo Z, Stiller JW. Connecting mutations of the RNA polymerase II C-terminal domain to complex phenotypic changes using combined gene expression and network analyses. PLoS One. 2010;5:e11386. doi: 10.1371/journal.pone.0011386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Phatnani HP, Greenleaf AL. Phosphorylation and functions of the RNA polymerase II CTD. Genes Dev. 2006;20:2922–36. doi: 10.1101/gad.1477006. [DOI] [PubMed] [Google Scholar]
- 8.Fuda NJ, Ardehali MB, Lis JT. Defining mechanisms that regulate RNA polymerase II transcription in vivo. Nature. 2009;461:186–92. doi: 10.1038/nature08449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buratowski S. Progression through the RNA polymerase II CTD cycle. Mol Cell. 2009;36:541–46. doi: 10.1016/j.molcel.2009.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cho EJ, Takagi T, Moore CR, Buratowski S. mRNA capping enzyme is recruited to the transcription complex by phosphorylation of the RNA polymerase II carboxy-terminal domain. Genes Dev. 1997;11:3319–26. doi: 10.1101/gad.11.24.3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Komarnitsky P, Cho EJ, Buratowski S. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev. 2000;14:2452–60. doi: 10.1101/gad.824700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Egloff S, O'Reilly D, Chapman RD, Taylor A, Tanzhaus K, Pitts L, et al. Serine-7 of the RNA polymerase II CTD is specifically required for snRNA gene expression. Science. 2007;318:1777–9. doi: 10.1126/science.1145989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lu KP, Hanes SD, Hunter T. A human peptidyl-prolyl isomerase essential for regulation of mitosis. Nature. 1996;380:544–7. doi: 10.1038/380544a0. [DOI] [PubMed] [Google Scholar]
- 14.Ranganathan R, Lu KP, Hunter T, Noel JP. Structural and functional analysis of the mitotic rotamase Pin1 suggests substrate recognition is phosphorylation dependent. Cell. 1997;89:875–86. doi: 10.1016/s0092-8674(00)80273-1. [DOI] [PubMed] [Google Scholar]
- 15.Yaffe MB, Schutkowski M, Shen M, Zhou XZ, Stukenberg PT, Rahfeld JU, et al. Sequence-specific and phosphorylation-dependent proline isomerization: a potential mitotic regulatory mechanism. Science. 1997;278:1957–60. doi: 10.1126/science.278.5345.1957. [DOI] [PubMed] [Google Scholar]
- 16.Lu PJ, Wulf G, Zhou XZ, Davies P, Lu KP. The prolyl isomerase Pin1 restores the function of Alzheimer-associated phosphorylated tau protein. Nature. 1999;399:784–8. doi: 10.1038/21650. [DOI] [PubMed] [Google Scholar]
- 17.Lu KP. Pinning down cell signaling, cancer and Alzheimer's disease. Trends Biochem Sci. 2004;29:200–209. doi: 10.1016/j.tibs.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 18.Etzkorn FA. Pin1 flips Alzheimer's switch. ACS Chem Biol. 2006;1:214–6. doi: 10.1021/cb600171g. [DOI] [PubMed] [Google Scholar]
- 19.Zhang Y, Daum S, Wildemann D, Zhou XZ, Verdecia MA, Bowman ME, et al. Structural basis for high-affinity peptide inhibition of human Pin1. ACS Chem Biol. 2007;2:320–8. doi: 10.1021/cb7000044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu X, Wilcox CB, Devasahayam G, Hackett RL, Arevalo-Rodriguez M, Cardenas ME, et al. The Ess1 prolyl isomerase is linked to chromatin remodeling complexes and the general transcription machinery. Embo J. 2000;19:3727–38. doi: 10.1093/emboj/19.14.3727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morris DP, Phatnani HP, Greenleaf AL. Phospho-carboxyl-terminal domain binding and the role of a prolyl isomerase in pre-mRNA 3'-end formation. J Biol Chem. 1999;274:31583–7. doi: 10.1074/jbc.274.44.31583. [DOI] [PubMed] [Google Scholar]
- 22.Verdecia MA, Bowman ME, Lu KP, Hunter T, Noel JP. Structural basis for phosphoserine-proline recognition by group IV WW domains. Nat Struct Biol. 2000;7:639–43. doi: 10.1038/77929. [DOI] [PubMed] [Google Scholar]
- 23.Macias MJ, Hyvonen M, Baraldi E, Schultz J, Sudol M, Saraste M, et al. Structure of the WW domain of a kinase-associated protein complexed with a proline-rich peptide. Nature. 1996;382:646–9. doi: 10.1038/382646a0. [DOI] [PubMed] [Google Scholar]
- 24.Huang K, Johnson KD, Petcherski AG, Vandergon T, Mosser EA, Copeland NG, et al. A HECT domain ubiquitin ligase closely related to the mammalian protein WWP1 is essential for Caenorhabditis elegans embryogenesis. Gene. 2000;252:137–45. doi: 10.1016/s0378-1119(00)00216-x. [DOI] [PubMed] [Google Scholar]
- 25.Lu PJ, Zhou XZ, Liou YC, Noel JP, Lu KP. Critical role of WW domain phosphorylation in regulating phosphoserine binding activity and Pin1 function. J Biol Chem. 2002;277:2381–4. doi: 10.1074/jbc.C100228200. [DOI] [PubMed] [Google Scholar]
- 26.Lu PJ, Zhou XZ, Shen M, Lu KP. Function of WW domains as phosphoserine- or phosphothreonine-binding modules. Science. 1999;283:1325–8. doi: 10.1126/science.283.5406.1325. [DOI] [PubMed] [Google Scholar]
- 27.Sudol M, Hunter T. NeW wrinkles for an old domain. Cell. 2000;103:1001–4. doi: 10.1016/s0092-8674(00)00203-8. [DOI] [PubMed] [Google Scholar]
- 28.Chen HI, Einbond A, Kwak SJ, Linn H, Koepf E, Peterson S, et al. Characterization of the WW domain of human yes-associated protein and its polyproline-containing ligands. J Biol Chem. 1997;272:17070–7. doi: 10.1074/jbc.272.27.17070. [DOI] [PubMed] [Google Scholar]
- 29.Ermekova KS, Zambrano N, Linn H, Minopoli G, Gertler F, Russo T, et al. The WW domain of neural protein FE65 interacts with proline-rich motifs in Mena, the mammalian homolog of Drosophila enabled. J Biol Chem. 1997;272:32869–77. doi: 10.1074/jbc.272.52.32869. [DOI] [PubMed] [Google Scholar]
- 30.Bedford MT, Reed R, Leder P. WW domain-mediated interactions reveal a spliceosome-associated protein that binds a third class of proline-rich motif: the proline glycine and methionine-rich motif. Proc Natl Acad Sci USA. 1998;95:10602–7. doi: 10.1073/pnas.95.18.10602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bedford MT, Sarbassova D, Xu J, Leder P, Yaffe MB. A novel pro–Arg motif recognized by WW domains. J Biol Chem. 2000;275:10359–69. doi: 10.1074/jbc.275.14.10359. [DOI] [PubMed] [Google Scholar]
- 32.Komuro A, Saeki M, Kato S. Npw38, a novel nuclear protein possessing a WW domain capable of activating basal transcription. Nucleic Acids Res. 1999;27:1957–65. doi: 10.1093/nar/27.9.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Myers JK, Morris DP, Greenleaf AL, Oas TG. Phosphorylation of RNA polymerase II CTD fragments results in tight binding to the WW domain from the yeast prolyl isomerase Ess1. Biochemistry. 2001;40:8479–86. doi: 10.1021/bi0027884. [DOI] [PubMed] [Google Scholar]
- 34.Kowalski JA, Liu K, Kelly JW. NMR solution structure of the isolated Apo Pin1 WW domain: comparison to the X-ray crystal structures of Pin1. Biopolymers. 2002;63:111–21. doi: 10.1002/bip.10020. [DOI] [PubMed] [Google Scholar]
- 35.Jager M, Zhang Y, Bieschke J, Nguyen H, Dendle M, Bowman ME, et al. Structure–function-folding relationship in a WW domain. Proc Natl Acad Sci USA. 2006;103:10648–53. doi: 10.1073/pnas.0600511103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu YX, Hirose Y, Zhou XZ, Lu KP, Manley JL. Pin1 modulates the structure and function of human RNA polymerase II. Genes Dev. 2003;17:2765–76. doi: 10.1101/gad.1135503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Palancade B, Marshall NF, Tremeau-Bravard A, Bensaude O, Dahmus ME, Dubois MF. Dephosphorylation of RNA polymerase II by CTD-phosphatase FCP1 is inhibited by phospho-CTD associating proteins. J Mol Biol. 2004;335:415–24. doi: 10.1016/j.jmb.2003.10.036. [DOI] [PubMed] [Google Scholar]
- 38.Wu X, Chang A, Sudol M, Hanes SD. Genetic interactions between the ESS1 prolyl-isomerase and the RSP5 ubiquitin ligase reveal opposing effects on RNA polymerase II function. Curr Genet. 2001;40:234–42. doi: 10.1007/s00294-001-0257-8. [DOI] [PubMed] [Google Scholar]
- 39.Meinhart A, Cramer P. Recognition of RNA polymerase II carboxy-terminal domain by 3'-RNA-processing factors. Nature. 2004;430:223–6. doi: 10.1038/nature02679. [DOI] [PubMed] [Google Scholar]
- 40.Noble CG, Hollingworth D, Martin SR, Ennis-Adeniran V, Smerdon SJ, Kelly G, et al. Key features of the interaction between Pcf11 CID and RNA polymerase II CTD. Nat Struct Mol Biol. 2005;12:144–51. doi: 10.1038/nsmb887. [DOI] [PubMed] [Google Scholar]
- 41.Hollingworth D, Noble CG, Taylor IA, Ramos A. RNA polymerase II CTD phosphopeptides compete with RNA for the interaction with Pcf11. RNA. 2006;12:555–60. doi: 10.1261/rna.2304506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang Z, Fu J, Gilmour DS. CTD-dependent dismantling of the RNA polymerase II elongation complex by the pre-mRNA 3'-end processing factor, Pcf11. Genes Dev. 2005;19:1572–80. doi: 10.1101/gad.1296305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vasiljeva L, Kim M, Mutschler H, Buratowski S, Meinhart A. The Nrd1-Nab3-Sen1 termination complex interacts with the Ser5-phosphorylated RNA polymerase II C-terminal domain. Nat Struct Mol Biol. 2008;15:795–804. doi: 10.1038/nsmb.1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gudipati RK, Villa T, Boulay J, Libri D. Phosphorylation of the RNA polymerase II C-terminal domain dictates transcription termination choice. Nat Struct Mol Biol. 2008;15:786–94. doi: 10.1038/nsmb.1460. [DOI] [PubMed] [Google Scholar]
- 45.Rondon AG, Mischo HE, Proudfoot NJ. Terminating transcription in yeast: whether to be a ‘nerd’ or a ‘rat’. Nat Struct Mol Biol. 2008;15:775–6. doi: 10.1038/nsmb0808-775. [DOI] [PubMed] [Google Scholar]
- 46.Becker R, Loll B, Meinhart A. Snapshots of the RNA processing factor SCAF8 bound to different phosphorylated forms of the carboxyl-terminal domain of RNA polymerase II. J Biol Chem. 2008;283:22659–69. doi: 10.1074/jbc.M803540200. [DOI] [PubMed] [Google Scholar]
- 47.Lu H, Zawel L, Fisher L, Egly JM, Reinberg D. Human general transcription factor IIH phosphorylates the C-terminal domain of RNA polymerase II. Nature. 1992;358:641–5. doi: 10.1038/358641a0. [DOI] [PubMed] [Google Scholar]
- 48.Hengartner CJ, Myer VE, Liao SM, Wilson CJ, Koh SS, Young RA. Temporal regulation of RNA polymerase II by Srb10 and Kin28 cyclin-dependent kinases. Mol Cell. 1998;2:43–53. doi: 10.1016/s1097-2765(00)80112-4. [DOI] [PubMed] [Google Scholar]
- 49.Fouillen L, Abdulrahman W, Moras D, Van Dorsselaer A, Poterszman A, Sanglier-Cianferani S. Analysis of recombinant phosphoprotein complexes with complementary mass spectrometry approaches. Anal Biochem. doi: 10.1016/j.ab.2010.07.006. (in press) [DOI] [PubMed] [Google Scholar]
- 50.Zhou M, Halanski MA, Radonovich MF, Kashanchi F, Peng J, Price DH, et al. Tat modifies the activity of CDK9 to phosphorylate serine 5 of the RNA polymerase II carboxyl-terminal domain during human immunodeficiency virus type 1 transcription. Mol Cell Biol. 2000;20:5077–86. doi: 10.1128/mcb.20.14.5077-5086.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shim EY, Walker AK, Shi Y, Blackwell TK. CDK-9/cyclin T (P-TEFb) is required in two postinitiation pathways for transcription in the C. elegans embryo. Genes Dev. 2002;16:2135–46. doi: 10.1101/gad.999002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Glover-Cutter K, Larochelle S, Erickson B, Zhang C, Shokat K, Fisher RP, et al. TFIIH-associated Cdk7 kinase functions in phosphorylation of C-terminal domain Ser7 residues, promoter-proximal pausing, and termination by RNA polymerase II. Mol Cell Biol. 2009;29:5455–64. doi: 10.1128/MCB.00637-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Trigon S, Serizawa H, Conaway JW, Conaway RC, Jackson SP, Morange M. Characterization of the residues phosphorylated in vitro by different C-terminal domain kinases. J Biol Chem. 1998;273:6769–75. doi: 10.1074/jbc.273.12.6769. [DOI] [PubMed] [Google Scholar]
- 54.Akhtar MS, Heidemann M, Tietjen JR, Zhang DW, Chapman RD, Eick D, et al. TFIIH kinase places bivalent marks on the carboxy-terminal domain of RNA polymerase II. Mol Cell. 2009;34:387–93. doi: 10.1016/j.molcel.2009.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lolli G, Lowe ED, Brown NR, Johnson LN. The crystal structure of human CDK7 and its protein recognition properties. Structure. 2004;12:2067–79. doi: 10.1016/j.str.2004.08.013. [DOI] [PubMed] [Google Scholar]
- 56.Baumli S, Lolli G, Lowe ED, Troiani S, Rusconi L, Bullock AN, et al. The structure of P-TEFb (CDK9/cyclin T1), its complex with flavopiridol and regulation by phosphorylation. Embo J. 2008;27:1907–18. doi: 10.1038/emboj.2008.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Archambault J, Chambers RS, Kobor MS, Ho Y, Cartier M, Bolotin D, et al. An essential component of a C-terminal domain phosphatase that interacts with transcription factor IIF in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1997;94:14300–5. doi: 10.1073/pnas.94.26.14300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kimura M, Suzuki H, Ishihama A. Formation of a carboxy-terminal domain phosphatase (Fcp1)/TFIIF/RNA polymerase II (pol II) complex in Schizosaccharomyces pombe involves direct interaction between Fcp1 and the Rpb4 subunit of pol II. Mol Cell Biol. 2002;22:1577–88. doi: 10.1128/mcb.22.5.1577-1588.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ghosh A, Shuman S, Lima CD. The structure of Fcp1, an essential RNA polymerase II CTD phosphatase. Mol Cell. 2008;32:478–90. doi: 10.1016/j.molcel.2008.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yeo M, Lin PS, Dahmus ME, Gill GN. A novel RNA polymerase II C-terminal domain phosphatase that preferentially dephosphorylates serine 5. J Biol Chem. 2003;278:26078–85. doi: 10.1074/jbc.M301791200. [DOI] [PubMed] [Google Scholar]
- 61.Yeo M, Lee SK, Lee B, Ruiz EC, Pfaff SL, Gill GN. Small CTD phosphatases function in silencing neuronal gene expression. Science. 2005;307:596–600. doi: 10.1126/science.1100801. [DOI] [PubMed] [Google Scholar]
- 62.Collet JF, Stroobant V, Pirard M, Delpierre G, Van Schaftingen E. A new class of phosphotransferases phosphorylated on an aspartate residue in an amino-terminal DXDX(T/V) motif. J Biol Chem. 1998;273:14107–12. doi: 10.1074/jbc.273.23.14107. [DOI] [PubMed] [Google Scholar]
- 63.Zhang M, Liu J, Kim Y, Dixon JE, Pfaff SL, Gill GN, et al. Structural and functional analysis of the phosphoryl transfer reaction mediated by the human small C-terminal domain phosphatase, Scp1. Protein Sci. 2010;19:974–86. doi: 10.1002/pro.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kamenski T, Heilmeier S, Meinhart A, Cramer P. Structure and mechanism of RNA polymerase II CTD phosphatases. Mol Cell. 2004;15:399–407. doi: 10.1016/j.molcel.2004.06.035. [DOI] [PubMed] [Google Scholar]
- 65.Zhang Y, Kim Y, Genoud N, Gao J, Kelly JW, Pfaff SL, et al. Determinants for dephosphorylation of the RNA polymerase II C-terminal domain by Scp1. Mol Cell. 2006;24:759–70. doi: 10.1016/j.molcel.2006.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hausmann S, Erdjument-Bromage H, Shuman S. Schizosaccharomyces pombe carboxyl-terminal domain (CTD) phosphatase Fcp1: distributive mechanism, minimal CTD substrate, and active site mapping. J Biol Chem. 2004;279:10892–900. doi: 10.1074/jbc.M312513200. [DOI] [PubMed] [Google Scholar]
- 67.Fabrega C, Shen V, Shuman S, Lima CD. Structure of an mRNA capping enzyme bound to the phosphorylated carboxy-terminal domain of RNA polymerase II. Mol Cell. 2003;11:1549–61. doi: 10.1016/s1097-2765(03)00187-4. [DOI] [PubMed] [Google Scholar]
- 68.West ML, Corden JL. Construction and analysis of yeast RNA polymerase II CTD deletion and substitution mutations. Genetics. 1995;140:1223–33. doi: 10.1093/genetics/140.4.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gerber M, Shilatifard A. Transcriptional elongation by RNA polymerase II and histone methylation. J Biol Chem. 2003;278:26303–6. doi: 10.1074/jbc.R300014200. [DOI] [PubMed] [Google Scholar]
- 70.Hampsey M, Reinberg D. Tails of intrigue: phosphorylation of RNA polymerase II mediates histone methylation. Cell. 2003;113:429–32. doi: 10.1016/s0092-8674(03)00360-x. [DOI] [PubMed] [Google Scholar]
- 71.Kornberg RD, Lorch Y. Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell. 1999;98:285–94. doi: 10.1016/s0092-8674(00)81958-3. [DOI] [PubMed] [Google Scholar]
- 72.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 73.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–5. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 74.Krogan NJ, Kim M, Tong A, Golshani A, Cagney G, Canadien V, et al. Methylation of histone H3 by Set2 in Saccharomyces cerevisiae is linked to transcriptional elongation by RNA polymerase II. Mol Cell Biol. 2003;23:4207–18. doi: 10.1128/MCB.23.12.4207-4218.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ng HH, Robert F, Young RA, Struhl K. Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol Cell. 2003;11:709–19. doi: 10.1016/s1097-2765(03)00092-3. [DOI] [PubMed] [Google Scholar]
- 76.Xiao T, Hall H, Kizer KO, Shibata Y, Hall MC, Borchers CH, et al. Phosphorylation of RNA polymerase II CTD regulates H3 methylation in yeast. Genes Dev. 2003;17:654–63. doi: 10.1101/gad.1055503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Li M, Phatnani HP, Guan Z, Sage H, Greenleaf AL, Zhou P. Solution structure of the Set2–Rpb1 interacting domain of human Set2 and its interaction with the hyperphosphorylated C-terminal domain of Rpb1. Proc Natl Acad Sci USA. 2005;102:17636–41. doi: 10.1073/pnas.0506350102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Vojnic E, Simon B, Strahl BD, Sattler M, Cramer P. Structure and carboxyl-terminal domain (CTD) binding of the Set2 SRI domain that couples histone H3 Lys36 methylation to transcription. J Biol Chem. 2006;281:13–5. doi: 10.1074/jbc.C500423200. [DOI] [PubMed] [Google Scholar]
- 79.Carty SM, Goldstrohm AC, Sune C, Garcia-Blanco MA, Greenleaf AL. Protein-interaction modules that organize nuclear function: FF domains of CA150 bind the phosphoCTD of RNA polymerase II. Proc Natl Acad Sci USA. 2000;97:9015–20. doi: 10.1073/pnas.160266597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Morris DP, Greenleaf AL. The splicing factor, Prp40, binds the phosphorylated carboxyl-terminal domain of RNA polymerase II. J Biol Chem. 2000;275:39935–43. doi: 10.1074/jbc.M004118200. [DOI] [PubMed] [Google Scholar]
- 81.Gasch A, Wiesner S, Martin-Malpartida P, Ramirez-Espain X, Ruiz L, Macias MJ. The structure of Prp40 FF1 domain and its interaction with the crn-TPR1 motif of Clf1 gives a new insight into the binding mode of FF domains. J Biol Chem. 2006;281:356–64. doi: 10.1074/jbc.M508047200. [DOI] [PubMed] [Google Scholar]
- 82.Murphy JM, Hansen DF, Wiesner S, Muhandiram DR, Borg M, Smith MJ, et al. Structural studies of FF domains of the transcription factor CA150 provide insights into the organization of FF domain tandem arrays. J Mol Biol. 2009;393:409–24. doi: 10.1016/j.jmb.2009.08.049. [DOI] [PubMed] [Google Scholar]
- 83.Allen M, Friedler A, Schon O, Bycroft M. The structure of an FF domain from human HYPA/FBP11. J Mol Biol. 2002;323:411–6. doi: 10.1016/s0022-2836(02)00968-3. [DOI] [PubMed] [Google Scholar]
- 84.Kobor MS, Archambault J, Lester W, Holstege FC, Gileadi O, Jansma DB, et al. An unusual eukaryotic protein phosphatase required for transcription by RNA polymerase II and CTD dephosphorylation in S. cerevisiae . Mol Cell. 1999;4:55–62. doi: 10.1016/s1097-2765(00)80187-2. [DOI] [PubMed] [Google Scholar]
- 85.Sun ZW, Hampsey M. Synthetic enhancement of a TFIIB defect by a mutation in SSU72, an essential yeast gene encoding a novel protein that affects transcription start site selection in vivo. Mol Cell Biol. 1996;16:1557–66. doi: 10.1128/mcb.16.4.1557. [DOI] [PMC free article] [PubMed] [Google Scholar]