Abstract
Quantum dots (QDs) are a novel class of inorganic fluorochromes composed of nanometer-scale crystals made of a semiconductor material. They are resistant to photo-bleaching, have narrow excitation and emission wavelengths that can be controlled by particle size and thus have the potential for multiplexing experiments. Given the remarkable optical properties that quantum dots possess, they have been proposed as an ideal material for use in molecular cytogenetics, specifically the technique of fluorescent in situ hybridisation (FISH). In this review, we provide an account of the current QD-FISH literature, and speculate as to why QDs are not yet optimised for FISH in their current form.
Keywords: quantum dot, nanotechnology, FISH, imaging
Nanotechnology has to date been closely affiliated with engineering since nanomaterials became the major components of computer chips (1). Within the last 10 years or so, however, there has been a growing relationship between nanoscience and fluorescent biological imaging (2). Applications of fluorescent imaging have generated a tremendous drive to develop new probes for tagging molecules, enabling changes in their localisation, concentration and activities to be documented (3). However, traditionally used organic fluorochromes face limitations affecting imaging and multicolour detection.
A novel class of semiconductor nanocrystals, termed quantum dots (QDs) (4,5), are inorganic fluorophores that provide a promising alternative to their organic counterparts. In this review, we will provide a brief account of QD properties and applications, then turn our focus on QDs and their applications for studying chromosomes - principally through the use of the technique ‘FISH’ (fluorescent (or fluorescence) in situ hybridisation). We appraise the current literature and offer possible explanations as to why QDs are not yet optimised for FISH in their current form.
Quantum dots (QDs): core concepts
Synthesis
QDs are composed of a semiconductor core such as cadmium selenide (CdSe), indium phosphate (InP) or lead selenide (PbSe) (6,7). This core is coated with a second semiconductor shell (usually zinc sulphide - ZnS) for the purpose of improving the optical properties of the nanocrystal (7,8). To improve further the utility of QDs, an extra polymer coating is attached that serves as a site for conjugation with biomolecule moieties. This brings the total size of the nanocrystal to 10–20 nm (a few hundred to a few thousand atoms). Fig. 1 shows a diagram of the structural components of a QD conjugate.
fig. 1.
Schematic representation of a QD conjugate.
The core material is chosen with respect to the required emission wavelength range (e.g. CdS for UV-blue, CdSe for the visible spectrum and CdTe for the far red and near infrared - NIR) (9), thus fluorophore colour is size dependent and controlled during synthesis (10). Synthesis occurs by injecting liquid precursors (dimethyl cadmium and selenium powder dissolved in tributylphosphione) in a hot organic solvent (trioctylphosphine oxide - TOPO) at temperatures reaching 300°C (11). Nanocrystals initiate formation immediately and the colourless starting mix becomes coloured. The size of the nanocrystals is adjusted by changing the amount of injected precursors and crystal growth time in the hot TOPO mix (2,12). A variety of core shapes can be synthesised, but they require an extra shell of a high band gap semiconductor material, typically ZnS, to stabilise the core and increase the quantum yield [QY, ratio of the amount of light emitted from a sample to the amount of light absorbed by the sample (13)] up to 80% (10,14). The surface layer of the ZnS shell is, however, hydrophobic and insoluble in aqueous solutions (8).
Optical properties
The most characteristic optical property of the QDs is that their colour is size dependent and thus controlled during synthesis (10). This arises as a result of the quantum confinement phenomenon (15), which refers to the spatial confinement of charge carriers (electrons and holes) within a semiconductor (16).
Because the physical size of the semiconductor nanocrystal is considerably reduced to be much smaller than the natural radius of the electron-hole pair, when a semiconductor is excited to emit light, the energy required to confine this excitation within the nanocrystal is higher, leading to a shift in emission in shorter wavelengths (i.e. towards the blue of emission) (13). To better understand this, an example of two different-sized CdSe QDs of 2.3 and 5.5 nm will be considered (Fig. 2).
fig. 2.
The size-dependent luminescence of quantum dots. Larger QDs have narrow band gaps (red QD, b) comparing to small QDs (blue QD, b). In the example discussed, the 5.5 QD emits orange light (longer wavelength 590 nm), whereas the 2.3 QD emits turquoise light (shorter wavelength 500 nm). Adapted from Jonathan (17).
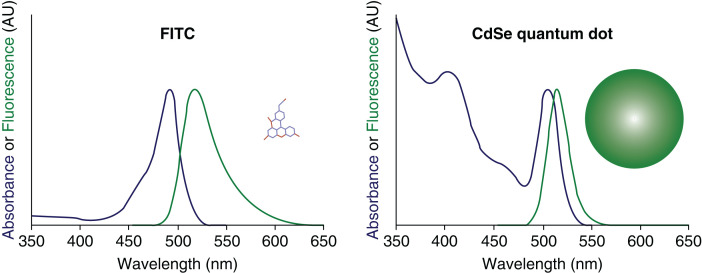
Another unique property of QDs is their broad excitation and narrow symmetric emission spectra. The spectral width of QDs (full width at half maximum is 12 nm) (18) designate that multicolour nanocrystals of different sizes can be excited by a single wavelength (excitation source) that is shorter than their emission wavelength (14,19, 20). This cannot be achieved with classical organic fluorophores because they have narrow excitation and broad emission that often results in spectrum overlap or red tailing (21). Fig. 3 compares excitation and emission spectra between an organic fluorophore and a QD.
fig. 3.
Comparison of absorption and excitation spectra between FITC (Fluorescein isothiocyanate) (blue) and a CdSe QD (green). Adapted from Bailey et al. (22).
QDs are reported to produce two to eleven times brighter fluorescence than organic fluorophores (23) because of the large molar extinction coefficients (10–50 times greater) (24) and, because QDs are inorganic, they are not prone to photo-bleaching (25,26). Moreover, the two-photon action cross-section of QDs (linked to direct measure of brightness) is significantly higher compared with organic fluorophores (approximate value of 45,000 Goeppert-Mayer units, GM) (23). Moreover, QDs have a longer fluorescence lifetime (10–40 ns) (27) than typical organic dyes, which can decay after a few nanoseconds.
The aforementioned optical properties relate mostly to the inorganic nature of QD and provide great potential; however, some photophysical properties can impose limitations on QD use.
Blinking is a phenomenon where the QD alternates between an emitting (on) and non-emitting (off) state (28,29). This behaviour has been interpreted according to an Auger ionisation model (30). Blinking affects single molecule detection applications by saturation of the signal. Hohng and Ha (31) carried out the first demonstration of blinking suppression by passivating the QD surface with thiol groups. Other strategies for blinking suppression are recently reviewed elsewhere (32). Photobrightening, where QD fluorescence intensity increases at the first stage of illumination and then stabilises, can impose limitations on quantitative studies (33). Both these properties are associated with mobile charges on the surface of the QDs (13).
Water solubility
Synthesis of QDs renders hydrophobic nanocrystals as it occurs in non-polar organic solvents (8). However, for QDs to be useful in biological applications, they need to be soluble in aqueous buffers since all experiments involving cells require water-soluble conditions (34,35). This essentially means that the surface of the QD needs to become hydrophilic. Several strategies have been employed to achieve this and most rely on exchanging the hydrophobic surfactant molecules with bifunctional molecules that are hydrophobic towards the ZnS shell of the nanocrystal and hydrophilic on the other end (8,34).
Commonly, thiols (-SH) are used as the hydrophobic anchoring parts to ZnS and carboxyl (-COOH) as the hydrophilic (36,37). The strategy of using mercaptohydrocarbonic acid to solubilise QDs has been applied in DNA immobilisation on the surface of the QD (38), FRET studies (39) and immunolabelling of proteins (40). Alternative approaches include surface silanisation (33,41), coating the QD surface with amphiphilic polymers (42,43), or polysaccharides (44), phospholipid micelles (45), non-charged molecules [i.e. dithiothreitol (36)], dendrons (46), peptides (phytochelatin-related) (47) and oligomeric ligands (oligomeric phosphines - OPs) (48). The effect of surface functionalisation on the optical properties of QDs is difficult to predict. In general, however, QY and decay behaviour respond to this effect whereas shape and spectral position of absorption and emission are hardly affected (49). These strategies allow QDs to be conjugated with a variety of biomolecules, including biotin (41), albumin (50), antibodies (51), avidin (52) and streptavidin (25,53). Covalently linked avidin/streptavidin QDs are very popular amongst companies (e.g. Invitrogen, Evident Technologies); they take advantage of the strong affinity that avidin and streptavidin have for biotin, and the plethora of biotinylated reagents (e.g. antibodies, DNA probes) available (54).
Quantum dot (QD) applications in biology (in-vitro and in-vivo)
The robust optical properties alone of QDs make them powerful substitutes for organic fluorophores for a variety of biological applications. For the purposes of this review, we will refer to some of the in-vitro and in-vivo published applications of QDs. However, in order to provide a broader aspect of their potential applications and limitations, we have summarised them in Table 1.
Table 1.
QD applications and limitations
| QD applications | Target/application | Potential limitations for QDs (all categories) |
|---|---|---|
| In-vitro imaging | Fixed cells, tissues, intracellular organelles | ● Cytotoxicity and how they are metabolised in the body (for use in human medical imaging) |
| In-vivo targeting | Cells, tissues, tumours in animals | ● Size - QDs are bigger from organic fluorophores - imposes limitation on targeting for in-vivo and potentially in situ studies, plus on the success of multicolour experiments |
| Bioanalytical assays | Flow cytometry, microarrays | ● Blinking suppression |
| Other applications (non-life sciences) | LEDs, telecommunications, quantum computers, cryptography, anti-counterfeit technologies | |
| Future applications | Gene/drug delivery, gene expression, biosensors |
The first published study in a biological context was labelling of nanocrystals with F-actin using the biotin-streptavidin bridge (41). Tokumasu and Dvorak (55) used this approach to label human erythrocytes for immunocytochemistry purposes, Wu et al. (25) used QD-streptavidin probes linked with IgG to detect the cancer marker HER2 on the surface of cancer cells, whereas Rosenthal et al. (56) used serotonin-labelled nanocrystals (SNACs) to target the serotonin transporter protein (SERT) in transfected HeLa cells and oocytes in-vitro. The erbB/HER family of transmembrane receptor tyrosine kinases (RTKs) that mediate cellular responses to epidermal growth factor (EGF) were studied using a QD-EGF conjugate that was specific in activating the EGF receptor (57).
Additionally, QDs have found applicability as cellular markers given their inherent ability to be internalised by cells, using either a receptor (18,58), non-specific endocytosis (59) or, for in-vivo injection, under the guidance of peptides (60). A more recent example of a peptide able to carry QDs in living cells is allatostatin, which was conjugated to streptavidin QDs and delivered without aggregation inside 3T3L1 and A431 cells (61). They can be employed for studies of cell-cell interaction by creating unique colour tags for individual cell lines (62), they can be encapsulated in micelles to track embryogenesis in frog or zebra fish embryos (45) for 3D optical sectioning investigations of the vascular endothelium (63), for cell motility assays of actinomyosin function (64) and for phagokinetic tracking of small epithelial cells that cause numerous cancers (65). In all these experiments, labelling of cells with QDs is apparently non-harmful to the cell (59).
The tunable size of QDs has allowed the use of NIR QDs as contrast agents during a surgical procedure to map sentinel lymph nodes (SLN) in pig and mouse (45,66). Using this technique, the surgeon is provided with visual guidance during SLN mapping that minimises incision and dissection inaccuracies, enabling real-time confirmation of complete resection (67). Despite the challenges for QD technology, cancer research has already made extensive use of QD applications for in-vivo tumour cell imaging (68–71), surgical oncology (72) and metastasis detection (73).
Quantum dots (QDs) and their potential for molecular cytogenetics
The term ‘cytogenetics’ refers to the study of chromosomes. For both research and clinical applications, the recognition of specific chromosomal patterns has widespread applications. From the mid-1980s, cytogenetics entered the molecular era through the development of the technique known as FISH (74–76). FISH allowed for direct DNA sequences to be visualised on chromosomes, the principal application being gene mapping, but with many more besides, including chromosome painting, advanced diagnostics and comparative genomics. Most FISH experiments use biotinylated probes and (strept)avidin-fluorochrome conjugates for detection. Moreover, the use of coloured fluorophores allow for the detection of several DNA sequences in the same cell, culminating (with some judicious mixing of colours) with many multicolour applications. FISH techniques have thus continuously been adapted but, as with many fluorescence microscopy applications, face limitations imposed by the use of organic fluorophores. These include the number of available fluorochromes and their broad emission spectra that make multicolour experiments difficult to resolve because of spectrum overlapping and photo-bleaching. Thus, given the aforementioned properties of QDs, they are, potentially, most suitable candidates for the study of chromosomes through adaptations of FISH protocols, particularly as the conjugation of QDs and streptavidin is already widely reported. Indeed, QD-FISH has the potential to revolutionise FISH by overcoming many of the inherent difficulties from the use of organic fluorochromes. It is noteworthy however that a PubMed search using terms such as ‘Quantum Dots FISH’ or ‘Quantum Dots Fluorescent in situ hybridisation’ yields few results, of which only 11 are actually QD-FISH studies. Table 2 lists these studies from February 2004. The purpose of the current paper is to review these studies and provide insight, from our own experience, why they are so few in number, despite the enormous potential of QD-FISH.
Table 2.
The total number of QD-FISH studies to the best of the authors’ knowledge
| Authors | Type of study | Comment | PMID | Published Date |
|---|---|---|---|---|
| Xiao and Barker | Research | First FISH application in human metaphase spreads | 14960711 | February 2004 |
| Xiao and Barker | Review | Review on QD-FISH potential and comments from their previous study | Not indexed for PubMed | December 2004 |
| Chan et al. | Research | First direct labelling of QDs with DNA to detect mRNA targets in mice brain sections | 16224100 | October 2005 |
| Xiao et al. | Correspondence | Importance of pH for QD-FISH | 16179915 | October 2005 |
| Wu et al. | Research | QD-FISH application in E. coli | 16625674 | April 2006 |
| Müller et al. | Research | QD-FISH attempt on plant chromosomes | 16776835 | June 2006 |
| Tholouli et al. | Research | Application of QD-FISH on mRNA targets from clinical biopsies | 16893519 | September 2006 |
| Bentolila and Weiss | Research | Direct labelling and first use of multicolour QD-FISH for mice satellite families | 16679564 | September 2006 |
| Jiang et al. | Research | QD-FISH for the analysis of cancer-related genomic aberrations in basic research and clinical application | 18283800 | December 2007 |
| Knoll | Book chapter | This chapter provided general protocols about slide preparation, probe labelling and a small amount on indirect detection of a chromosome loci using QDs | 17237529 | 2007 |
| Ma et al. | Research | Direct QD-FISH application in maize | 18046569 | December 2007 |
| Choi et al. | Research | QD-DNA probes for direct localization and quantification of gene expression in situ | 19517489 | June 2009 |
| Müller et al. | Research | Concurrent utilisation of QDs and organic fluorochromes for multiplex experiments in 4Pi microscopy | 19556786 | June 2009 |
| Ioannou et al. | Research | An account of QD-FISH experiments (both indirect and direct labelling) with possible reasoning as to why QD-FISH is not fully optimised yet | 19644760 | July 2009 |
A review of the quantum dot-fluorescent in situ hybridisation (QD-FISH) literature
In the initial study, Xiao and Barker (77) made use of biotinylated total genomic DNA as a probe on human metaphase chromosomes. The probe was detected using streptavidin-conjugated QD605 (infrared). Direct comparisons of detection with QDs and organic fluorochromes (Texas Red and Fluorescein) showed that QD-FISH was significantly more photostable and brighter than the more traditional approaches. More specifically, they noticed that after 2 h of continuous illumination there was a moderate loss of the QD signals (30%) compared to the more severe 73% and 89% loss for Texas Red and FITC, respectively. In addition, they made an initial observation regarding the pH and buffer used, as with a more alkaline pH (8.3) for the buffer used to dilute the QD conjugate, there was failure of signal detection in centromeres with QD probes. This did not seem to affect the organic fluorochromes. The importance of pH was further explored in a short correspondence by the authors, where signals from QD-FISH were at an optimum when the buffer pH was between 6 and 7 (78). Furthermore, they applied this technique to detect the clinically important locus of HER2 in low copy human cells and breast cancer cells, demonstrating that QD-FISH has the potential to become a medical diagnostic tool. They underlined the potential of QD probes stating that although expectations were raised, more evaluation of QDs was required entering a clinical setting (79).
Chan et al. (80) used direct labelling strategy to target specific mRNAs in mouse brain sections. This study raised the issue of the multiple streptavidin sites on the QD molecule that could interfere with hybridisation efficiency. For this reason, a competitive blocker of streptavidin, biocytin was used, in the presence of which they labelled their oligonucleotide probes. The authors reported that the use of QDs enabled them to observe the details of mRNA expression in the sub-cellular level because of the better image resolution. This study was the first to claim direct labelling of QDs with DNA (specifically oligonucleotides).
Wu et al. (81) were the first to report the successful application of QD-FISH without using the commercial streptavidin-QD conjugates, but by coating naked QDs (synthesised in their laboratory) with mercaptoacetic acid (MAA) to render them water soluble. This was followed by competitive displacing of QD-surface-confined MAA molecules with thiol single-stranded DNA complementary to their plasmid target of interest. By using this technique, they created highly monodisperse QD-DNA probes and because both the single-stranded DNA and the MAA coating were negatively charged, the generated repulsion between those molecules would keep the single-stranded DNA away from the QD surface, facilitating hybridisation in the Escherichia coli bacterium for the first time.
In 2006, Muller et al. (82) made the first attempts on plant chromosomes. An indirect approach to detect non-coding sequences in the plant Allium fistulosum was used, but with limited success. Although different strategies were employed to improve the performance of QDs (slide preparation, pepsin treatment to increase cell permeability), few results were forthcoming with either QD 605 streptavidin conjugate or by a QD 565 anti-Rabbit IgG conjugate. The offered explanation for the intermittent success was the phenomenon of steric hindrance owing to the large size of the nanocrystals (compared to the organic fluorophores).
The wide application of tissue staining by QDs was shown in another study where multiple mRNA targets in formalin-fixed bone marrow biopsies were targeted using QD-streptavidin conjugates, allowing quantitative characterisation of gene expression sites using non-bleaching fluorochromes (83). Testing different molar ratios between QD and oligonucleotide probes, the authors reported the highest signal intensity when a ratio of 1:2 (QD:probe) was used. Furthermore, there was evidence of QD signals still present in the bone marrow tissue even after 18 months of storage. This was not true for the control Cy3-stained tissue.
In September 2006, the first paper describing multicolour FISH using QDs was published by Bentolila and Weiss (84). Using analytical grade QD batches for a variety of QD-streptavidin conjugates, they formed QD-DNA complexes by incubating biotinylated oligonucleotides at various molar ratios at room temperature for 30 min. Complexes were run on an electrophoresis gel and the optimum molar ratio was established. At the same time this assay confirmed binding of the DNA to the nanoparticles because of the motility shift that is caused by the formation of this conjugant. These probes were used to recognise the major (γ) family of mouse satellite DNA. The novel feature in this study was the presentation of a dual colour QD-FISH using QD592 and 655 against centromere-associated sequences (satellites). Reading between the lines of this paper, however, data was presented from only two of the five different QDs that were tried, probably due to technical difficulties or hybridisation failure of the remaining constructs. Nevertheless, this was an important breakthrough for multicolour QD-FISH. Furthermore, QD525 was not used at all in the hybridisation experiments as it showed an irreversible spectral shift. The success of this study in detecting centromeric regions with QDs was in sharp contrast with the study by Xiao and Baker (77), where most of these regions could not be detected. The authors believed that this could be due to the variable steric hindrance effects during the FISH procedure. Another important aspect was the observation of partial loss of QD probes fluorescence over time. However, this was not an irreversible phenomenon as intensity could be fully restored after re-exposure to UV light. The clear message from this study was the great potential of QD-FISH probes to become a sophisticated toolbox that could be applied for high-resolution studies on chromosome binding through the use of spectrally distinguished QDs.
More recently, successful use of QD-FISH was reported by Jiang et al. (85). In this case, selected probes were used in lung cancer specimens to visualise gene amplification, offering another potential diagnostic tool for the study of genomic aberrations in cancer cells. Also in 2007, a methodology book was published entitled ‘Quantum Dots’ Applications in Biology, where Chapter 5 was dedicated to QD-FISH. It provided protocols for the preparation of human metaphase chromosomes, probe labelling by nick translation, standard FISH and indirect detection of a specific region on human chromosome 22 using anti-digoxigenin QD655 (86). Some key points from this chapter to enhance hybridisation efficiency included the importance of cell preparation (good chromosome spreading), formamide quality, temperature, pH and exposure of the probe to the denaturation solution.
In a more specialised investigation, QD-FISH was applied successfully on maize chromosomes (87). In contrast to the Muller et al. (82) study where the conclusion was that QD-streptavidin conjugates could not successfully detect plant chromosomes, successful hybridisation was indeed reported, albeit with QD probes prepared somewhat differently. That is, the nanoparticles were coated with MAA and the oligonucleotide was attached via a metal-thiol bond. The authors tried to address the possible steric hindrance problem by keeping the oligonucleotide probe further away from the QD surface using a homo-polymer of thymidine sequence. By doing this, it was claimed that modification of the hydrodynamic diameter of the bioprobes was small enough to penetrate into maize chromosomes. Moreover, the authors emphasise the improved impact of their own solubilisation strategy on these modified QD probes (MAA-coated) compared to the commercially available polymer-coated QD-streptavidin ones. Mirroring the report by Xiao and Barker (77), this study highlights the importance of pH, ionic strength and formamide to increase the affinity of QD probes to chromosomal targets. Although the report by Ma et al. (87) declared a preference for the MAA coating of QDs compared to the polymer-coated ones, Choi et al. (88) used polymer-coated QDs that maintained high QY and photostability in their FISH experiments. They coupled the DNA oligonucleotides via a 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide hydrochloride (EDC) molecule and were able to visualise gene targets in Drosophila.
The only study that we are aware of to make use of both organic and inorganic fluorochromes in an attempt to increase the number of colours on a single cell was published by Muller et al. (89). One of the objectives of this report was to show the capability of QD probes in 4Pi microscopy, a technique that can push the resolution limits to 100 nm or even less, thereby requiring high photostable fluorophores. Although a combination of QDs and traditional fluorophores could be combined for the visualisation of chromosome painting probes (maximum multiplexing was achieved using three QDs and three traditional fluorochromes), there was some batch variability concerning QD conjugates that manifested as different signal intensity results even in parallel experiments. Thus, the authors argue that further progress is anticipated from the manufacturer's point of view to increase QD robustness and reliability.
Our own experience in quantum dot-fluorescent in situ hybridisation (QD-FISH)
Given the obvious potential of QD-FISH, we have been somewhat puzzled how few studies exist in this area. Around 2006, we began to explore the use of QDs in place of organic fluorochromes, specifically with a view to using QDs in multiplex experiments [i.e. to target multiple regions simultaneously, see Ioannou et al. (90)]. Our own research questions pertain to chromosome copy number and nuclear position of chromosome territories in human sperm (91) and preimplantation embryos (92) and possible links between aberrant nuclear organisation and infertility and/or aneuploidy. In preimplantation embryos specifically, cells are few in number and ethically sensitive; thus as much information as possible should be derived from them. Our other interests relate to genome organisation and evolution in birds (93–96) and fish (97–99), which have large numbers of small chromosomes that are not easily cytologically distinguishable. In all the above, clear bright signals amenable to multiplexing would be of great advantage in advancing our work, particularly if probes could be labelled directly with QDs. Some of our original work was published last year (90) and the following summarises aspects of it.
Our first clear observation was that the emission spectra of the QD samples (from both Invitrogen and Evident Technologies) appeared not to be narrow as the manufacturers claimed them to be. We established this by simply spotting diluted aliquots of the QD-streptavidin conjugates to a slide and observing them under the microscope. Indeed there appeared to be significant emission bleed-through into other filters (Fig. 4).
fig. 4.
QD520 (supplied by Evident) spotted on to a glass slide, excited by a UV filter and then detected with barrier filters at 525, 565, 585 and 605 nm, respectively. Although under the green barrier filter (525 nm) the brightest fluorescence is observed, significant bleed-through is seen on the other filters indicating that the emission spectrum is not as narrow as is usually purported for QDs.
All QDs appeared to show significant bleed-through to other filters but, from visual inspection, QD585 appeared to have the narrowest emission. As a control, the Cy3-streptavidin (organic dye) also showed significant emission bleed-through to other channels, not dissimilar to some of the QDs. We therefore continued experiments mostly using QD585 (7).
Our initial results were very encouraging when biotinylated probes were detected using the QD585-strepavidin conjugate (7). Fig. 5 demonstrates this in chromosome painting experiment compared to a Cy3 control.
fig. 5.
Successful FISH experiments on human chromosome 1 using biotinylated chromosome 1 paint with Cy3-streptavidin conjugate control (upper) and QD585-streptavidin conjugate (lower). QD585 signals were brighter, though more ‘patchy’ and with a greater amount of background. Adapted from Ioannou et al. (90).
When results were successful, the reported properties of QDs were plain to see. In particular, preparations were noticeably brighter than Cy3 preparations and did not fade upon inspection. That is, when Cy3-labelled preparations were exposed continually to the fluorescent lamp, photo-bleaching occurred after about 5 min. By contrast, when QD preparations were exposed to UV light, no noticeable loss of signal was seen, even following 1 h of exposure. We also noticed that, in several chromosome painting experiments, the QD signal was brighter around the periphery of the chromosome - a sort of fluorescent ‘sheath’ (Fig. 6). Moreover, in two or three cases, a bright signal was observed in the less condensed interphase nuclei of the cell, but not in the highly coiled metaphase chromosomes.
fig. 6.
Successful chromosome painting experiment (chromosome 2, tetraploid cell) in chicken, but with signals predominantly around the periphery of the chromosome, giving an impression of a fluorescent ‘sheath’. Adapted from Ioannou et al. (90).
On the negative side, in general terms, QD preparations in these experiments had more non-specific background than were observed for Cy3 preparations and we can confirm a similar observation by Muller et al. (89) on identical experiments giving different levels of hybridisation efficiency. Even more confusingly, our experiment would regularly work on one slide but not the other identically processed in parallel. In general terms, indirect QD experiments were successful approximately 25–35% of the time, compared to Cy3 controls that worked reliably and consistently.
In attempts to improve the efficacy and reliability of our experiments, various FISH conditions were systematically altered. These included removal of a ‘blocking’ step prior to the addition of the conjugate and changing the temperature, pH and time of the post-hybridisation washes. These did not usually improve QD experiments and the same applied when controlled experiments were performed in the presence or absence of dextran sulphate (a component of hybridisation buffer used to chelate the hybridised probe and make the signal stronger). In an attempt to minimise steric hindrance, a longer carbon chain (biotin-21-dUTP) was used instead of 16-dUTP, and different ratios of biotin labelled and unlabelled probes were assessed. No noticeable difference was observed between the two biotins and there was no indication of more efficient hybridisation in any of the different ratios tested.
Several more alternative strategies were attempted with no increased efficacy of QD-FISH; these included trying numerous batches of chromosome preparations, labelling probes with digoxigenin (and attempting detection with anti-digoxigenin) and methods to increase cell permeability (fixation, pepsin). The only intervention that we did observe that had a degree of success was the use of silicon-coated plastic tubes and sonication of the conjugate prior to use. In both conditions, we observed an (albeit temporary) improvement in the reliability of the results. Notwithstanding the repeated efforts to increase the robustness of our approach, on the whole, outcomes were temperamental or unsuccessful. Fig. 7 shows some of our inglorious attempts.
fig. 7.
(A) Chromosome painting attempt in human lymphocytes using QD520. No specific signal was seen and the area surrounding the chromosomes had a very high background (left), moreover the background signal bled through into the red channel (right). (B) Attempts to visualise the centromeres of human chromosome 12. There is some evidence of hybridisation and detection but the preparation has a very high background. (C) A bright red signal is seen on every part of the slide apart from the chromosomes! This was another attempt at human chromosome painting for chromosomes 1 and 2.
This limited degree of success was, however, relatively encouraging compared to our attempts to conjugate QDs directly to FISH probes. Our direct conjugation strategy of DNA to QDs was based on recently published material (84) and, with the direct help of the authors, we were confident that we had made successful conjugates (established by mobility shifts on agarose gels). Such conjugates were generated for chromosome paints and oligonucleotide probes recognising the centromeres of chromosomes, however repeated attempts at subsequent FISH experiments (employing a range of different conditions of stringency, hybridisation buffer, QD:DNA concentration ratios and incubation times) without exception ended in failure (despite considerable success with Cy3 conjugate controls).
Quantum dot-fluorescent in situ hybridisation (QD-FISH): where does this leave us?
The message through our comprehensive appraisal of the utility of QDs for FISH has been that, in their current form, QDs are not suitable materials for FISH applications. If further evidence were needed, it can be found in the fact that traditional fluorochromes have not, for any application, been replaced by QDs, despite their great potential. There are few peer-reviewed studies pertaining to QD-FISH and we are unaware of any company marketing QD-labelled FISH.
In our experience (and following discussions with colleagues from other groups), lack of reproducibility appears to be a distinguishing feature of QD-FISH in contrast to the more robust applications with organic fluorophore-streptavidin conjugates. That is, while we would not claim that we have explored every possible avenue with respect to QD-FISH, we have nonetheless extensive experience in FISH over many years and have been (for the last three to four years) running parallel QD-based experiments (mostly in avian and human cells). Our collective experience paints a general picture of a non-reproducible approach when QDs are used in place of organic fluorochromes.
The unreliable nature of QDs (at least for FISH) is perhaps not totally unexpected as other colleagues have had similar experiences to our own (89,100). There is clearly a challenging set of conditions pertaining to intracellular delivery of QDs and, since there are no reliable FISH protocols for this, individual adaptations need empirical establishment (49). If this was achieved then the reliability may well improve and the benefits of QDs observed in this and other studies (e.g. increased brightness, resistance to photo-bleaching) may be properly realised. With all this in mind, we can speculate about reasons for the lack of reproducibility of QD-FISH results. Clues about QD size and chemistry during synthesis may be a starting point.
QDs vary in size (this is the basis of the fluorescent colour that they emit) from 2 to 10 nm. A Cy3 molecule on the other hand is <2 nm in size (22). This may explain in part why our successful FISH experiments gave the impression of larger fluorescent particles and why there was a greater degree of background for most experiments. It might also explain an observed fluorescent ‘sheath’ effect seen on some metaphases (90) and why certain preparations gave bright signals in decondensed interphase nuclei, but not highly coiled metaphase chromosomes. That is, steric hindrance may have led to signals being brighter in areas where the chromatin is less compact (e.g. at the edge of the chromosomes and/or in the interphase nucleus), indeed steric hindrance has been an issue reported in many studies (82,84, 87,88). If this were true, we might have expected to see an improvement when we reduced the ratio of labelled to unlabelled dUTPs and/or when we made use of a ‘longer-arm’ biotin dUTP. This was not the case. Again, however, a general background of intermittent success may have masked any appreciable difference seen in any given experiment. Furthermore, as QD-streptavidin conjugates were used throughout these experiments, it is worth pointing out that it is not entirely clear how streptavidin binds on the actual polymer site of the QD. For this reason, the number of free streptavidin sites varies per individual QD (10–15). Incidentally, these sites can break off from the nanoparticle (for no reported reason) rendering the probe unstable or even detached, with immediate effect on the hybridisation signal (Bentolila, L personal communication). We are also informed that QD streptavidin conjugates can easily degrade (a batch-specific attribute) and this can be due to barely discernable temperature changes during storage. Additionally, we are given to understand that QDs are prone to adhere to tubes sides and tips (Chan, P personal communication). Our attempts to reduce this problem using siliconised tubes and regular sonication met with a degree of success; however it did not eliminate our technical issues completely.
Another confounding issue was that the emission spectra of the QDs did not appear to be as narrow as the manufacturers claimed, in that we observed ‘bleed-through’ between channels, despite making use of narrow band-pass filters. Apparently, this phenomenon is not as uncommon as the literature might suggest (Bentolila, L personal communication) and could vary from batch to batch. Controlling the size of the core during synthesis (that will tune the colour that the QD will emit) requires high technical skills and sometimes nanoparticles are larger than expected. Addressing the size control is critical in particular for multicolour detection or imaging and could hold the key to the success of multicolour experiments in QD-FISH. Also, abnormalities in their shape could result in the same effect (Bentolila, L personal communication). An additional possible explanation for this emission bleed-through to other channels was that QDs were not monodisperse. Simple spotting experiments confirmed this statement. Fig. 8 shows a QD605-conjugate dissolved in hybridisation mix where different QD populations could be observed under the different band-pass filters.
fig. 8.
QD605 dissolved in hybridisation mix and viewed directly under the microscope using four barrier filters: 525 nm (blue), 565 nm, 585 nm (red) and 605 nm (far red but pseudo-coloured purple for the purposes of this figure). The image represents a merge of all four filters. The QDs are predominantly purple (as would be expected), but a smaller number of green, blue and red QDs are seen. The discrete appearance of QDs of one or other of the colours indicates there is a mixed population of QDs. Adapted from Ioannou et al. (90).
The different colours seen in Fig. 8 represent different-sized QDs that emit at longer (towards the red - large QDs) or shorter (towards the blue - small QDs) wavelengths. These findings are consistent with those of Murray and colleagues, who have tried to address the monodispersity of QD preparations (101). All these technical features that were attributed to the chemical synthesis of the QDs may require more experimental attention in order to improve QD synthesis. Of course, we cannot rule out the possibility that bleed-through and monodispersity are batch-specific problems; after all, we did not test more than three or four batches for each QD. However, we saw no evidence of batch-specific variance.
A further QD feature that we observed was ‘blinking’ - a phenomenon unknown in conventional FISH where the QD alternates between an emitting (on) and non-emitting (off) state (28,29). Blinking has been explained according to an Auger ionisation model (30) and affects single molecule detection applications by saturation of the signal. It may, however, be suppressed by using thiol groups to passivate the QD surface (31,84). A second phenomenon, photo-brightening, where the fluorescence intensity increases rapidly at the first stage of illumination and then stabilises, can limit on quantitative studies (33). Both these properties are associated with mobile charges on the surface of the QDs (13).
A likely reason to explain the positive results arising from groups that have published in this area (79,84, 87) is that their laboratories were equipped with the ability to synthesise and batch-test their own conjugates (a luxury not afforded to most groups). Ma et al. (87) suggested that the QDs that they used were significantly smaller than those available commercially and may thus have reduced steric hindrance and increased hybridisation ability. Several laboratories (79,84, 87), however, have generated QD-oligonucleotide conjugates and report that, during the time of annealing, steric hindrance has little effect but it may limit the QDs access to the target at the time of detection (84,87). This may provide a possible explanation for our lack of success in generating usable conjugates. Furthermore, negative hybridisation was potentially caused by unbound QD left over after the incubation between QD and DNA (to generate a conjugant) that prevented the complex entering cells and hybridising (acted as a competitor). Excess cytoplasm around the chromosomes cannot solely be blamed as pepsin treatments were introduced to reduce it.
Taking all this into consideration, further research is essential. Advances in nanomaterials synthesis (regarding uniformity and size control) and solubility will assist conjugation to biomolecules. Moreover, a new generation of nanocrystals (FloDots, C-dots) has already been mentioned in the literature (102,103). There may well be a future for a marriage between nanotechnology and molecular cytogenetics. Like all good marriages, however, a little patience may be required.
Acknowledgements
The authors would like to note the efforts of Dr Helen Tempest (formerly of our own group and now of the Florida International University) for her attempts on QD-FISH. We would also like to thank Mr. Michael Ellis (Digital Scientific UK) for his initial enthusiasm and funding for the project, and for assembling the image analysis hardware.
Biographies
 Dimitris Ioannou is a final year PhD student in the laboratory of Professor Griffin. He is a graduate of the University of Wales (BSc) and Nottingham (MPhil), and has performed original research work on applications of FISH including QD-FISH.
Dimitris Ioannou is a final year PhD student in the laboratory of Professor Griffin. He is a graduate of the University of Wales (BSc) and Nottingham (MPhil), and has performed original research work on applications of FISH including QD-FISH.
 Prof. Darren Griffin holds the chair in genetics at the University of Kent, Canterbury, UK. He is a graduate of the University of Manchester (BSc and DSc) and University College London (PhD). He is a Fellow of the Royal College of Pathology and of the Society of Biology. He has published over 100 papers on aspects related to chromosome research and runs a busy research laboratory.
Prof. Darren Griffin holds the chair in genetics at the University of Kent, Canterbury, UK. He is a graduate of the University of Manchester (BSc and DSc) and University College London (PhD). He is a Fellow of the Royal College of Pathology and of the Society of Biology. He has published over 100 papers on aspects related to chromosome research and runs a busy research laboratory.
Conflict of interest and funding
There is no conflict of interest in the present study for any of the authors.
References
- 1.Chan WC. Bionanotechnology progress and advances. Biol Blood Marrow Transplant. 2006;12:87–91. doi: 10.1016/j.bbmt.2005.10.004. [DOI] [PubMed] [Google Scholar]
- 2.Parak WJ, Gerion D, Pellegrino T, Zanchet D, Micheel C, Williams SC, et al. Biological applications of colloidal nanocrystals. Nanotechnology. 2003;14:R15–R27. [Google Scholar]
- 3.Jaiswal JK, Simon SM. Potentials and pitfalls of fluorescent quantum dots for biological imaging. Trends Cell Biol. 2004;14:497–504. doi: 10.1016/j.tcb.2004.07.012. [DOI] [PubMed] [Google Scholar]
- 4.Reed MA, Bate RT, Bradshaw WM, Duncan WR, Frensley JWL, Shih HD. Spatial quantization in GaAs-AlGaAs multiple quantum dots. J Vac Sci Technol B. 1986;4:358–60. [Google Scholar]
- 5.Miller DAB, Chemla DS, Schmittrink S. Absorption saturation of semiconductor quantum dots. J Opt Soc Am B. 1986;3:42. [Google Scholar]
- 6.Lipovskii A, Kolobkova E, Petrikov V, Kang I, Olkhovets A, Krauss T, et al. Synthesis and characterization of PbSe quantum dots in phosphate glass. Appl Phys Lett. 1997;71:3406–8. [Google Scholar]
- 7.Invitrogen. vol. 2006. Carlsbad, CA: Invitrogen Corporation; 2006. Qdot nanocrystal technology. [Google Scholar]
- 8.Michalet X, Pinaud FF, Bentolila LA, Tsay JM, Doose S, Li JJ, et al. Quantum dots for live cells, in vivo imaging, and diagnostics. Science. 2005;307:538–44. doi: 10.1126/science.1104274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Quantum Dot Corporation. Qdot nanocrystals. Anatomy. vol. 2006. Hayward, CA: Quantum Dot Corporation (QDC); 2006. [Google Scholar]
- 10.Chan WC, Maxwell DJ, Gao X, Bailey RE, Han M, Nie S. Luminescent quantum dots for multiplexed biological detection and imaging. Curr Opin Biotechnol. 2002;13:40–6. doi: 10.1016/s0958-1669(02)00282-3. [DOI] [PubMed] [Google Scholar]
- 11.Murray CB, Norris DJ, Bawendi MG. Synthesis and characterization of nearly monodisperse CdE (E=S, Se, Te) semiconductor nanocrystallites. J Am Chem Soc. 1993;115:8706–15. [Google Scholar]
- 12.Michalet X, Pinaud F, Thilo DL, Dahan M, Bruchez MP, Alivisatos AP, et al. Properties of fluorescent semiconductor nanocrystals and their application to biological labeling. Single Mol. 2001;2:261–76. [Google Scholar]
- 13.Fu A, Alivisatos AP, Gu W, Larabell C. Semiconductor nanocrystals for biological imaging. Curr Opin Neurobiol. 2005;15:568–75. doi: 10.1016/j.conb.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 14.Alivisatos AP, Gu W, Larabell C. Quantum dots as cellular probes. Annu Rev Biomed Eng. 2005;7:55–76. doi: 10.1146/annurev.bioeng.7.060804.100432. [DOI] [PubMed] [Google Scholar]
- 15.Alivisatos AP. Perspectives on the physical chemistry of semiconductor nanocrystals. J Phys Chem B. 1996;100:13226–39. [Google Scholar]
- 16.Alivisatos P. The use of nanocrystals in biological detection. Nat Biotechnol. 2004;22:47–52. doi: 10.1038/nbt927. [DOI] [PubMed] [Google Scholar]
- 17.Jonathan C. A quantum paintbox. Chemistry World. 2003. pp. 1–8. Available from: http://www.rsc.org/chemistryworld/Issues/2003/September/paintbox.asp [cited 4 March 2010]
- 18.Chan WC, Nie S. Quantum dot bioconjugates for ultrasensitive nonisotopic detection. Science. 1998;281:2016–8. doi: 10.1126/science.281.5385.2016. [DOI] [PubMed] [Google Scholar]
- 19.Arya H, Kaul Z, Wadhwa R, Taira K, Hirano T, Kaul SC. Quantum dots in bio-imaging: revolution by the small. Biochem Biophys Res Commun. 2005;329:1173–7. doi: 10.1016/j.bbrc.2005.02.043. [DOI] [PubMed] [Google Scholar]
- 20.Green M. Semiconductor quantum dots as biological imaging agents. Angew Chem Int Ed Engl. 2004;43:4129–31. doi: 10.1002/anie.200301758. [DOI] [PubMed] [Google Scholar]
- 21.Dabbousi BO, Rodriguez-Viejo J, Mikulec FV, Heine JR, Mattoussi H, Ober R, et al. (CdSe)ZnS core-shell quantum dots: synthesis and characterization of a size series of highly luminescent nanocrystallites. J Phys Chem B. 1997;101:9463–75. [Google Scholar]
- 22.Bailey RE, Smith AM, Nie S. Quantum dots in biology and medicine. Physica E. 2004;25:1–12. [Google Scholar]
- 23.Larson DR, Zipfel WR, Williams RM, Clark SW, Bruchez MP, Wise FW, et al. Water-soluble quantum dots for multiphoton fluorescence imaging in vivo. Science. 2003;300:1434–6. doi: 10.1126/science.1083780. [DOI] [PubMed] [Google Scholar]
- 24.Gao X, Yang L, Petros JA, Marshall FF, Simons JW, Nie S. In vivo molecular and cellular imaging with quantum dots. Curr Opin Biotechnol. 2005;16:63–72. doi: 10.1016/j.copbio.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 25.Wu X, Liu H, Liu J, Haley KN, Treadway JA, Larson JP, et al. Immunofluorescent labeling of cancer marker HER2 and other cellular targets with semiconductor quantum dots. Nat Biotechnol. 2003;21:41–6. doi: 10.1038/nbt764. [DOI] [PubMed] [Google Scholar]
- 26.Jaiswal JK, Mattoussi H, Mauro JM, Simon SM. Long-term multiple color imaging of live cells using quantum dot bioconjugates. Nat Biotechnol. 2003;21:47–51. doi: 10.1038/nbt767. [DOI] [PubMed] [Google Scholar]
- 27.Lounis B, Bechtel HA, Gerion D, Alivisatos PA, Moerner WE. Photon antibunching in single CdSe/ZnS quantum dot fluorescence. Chem Phys Lett. 2000;329:399–404. [Google Scholar]
- 28.Michler P, Imamoglu A, Mason MD, Carson PJ, Strouse GF, Buratto SK. Quantum correlation among photons from a single quantum dot at room temperature. Nature. 2000;406:968–70. doi: 10.1038/35023100. [DOI] [PubMed] [Google Scholar]
- 29.Pinaud F, Michalet X, Bentolila LA, Tsay JM, Doose S, Li JJ, et al. Advances in fluorescence imaging with quantum dot bio-probes. Biomaterials. 2006;27:1679–87. doi: 10.1016/j.biomaterials.2005.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Efros AL, Rosen M. Random telegraph signal in the photoluminescence intensity of a single quantum dot. Phys Rev Lett. 1997;78:1110–3. [Google Scholar]
- 31.Hohng S, Ha T. Near-complete suppression of quantum dot blinking in ambient conditions. J Am Chem Soc. 2004;126:1324–5. doi: 10.1021/ja039686w. [DOI] [PubMed] [Google Scholar]
- 32.Lee SF, Osborne MA. Brightening, blinking, bluing and bleaching in the life of a quantum dot: friend or foe? Chemphyschem. 2009;10:2174–91. doi: 10.1002/cphc.200900200. [DOI] [PubMed] [Google Scholar]
- 33.Gerion D, Pinaud F, Williams SC, Parak WJ, Zanchet D, Weiss S, et al. Synthesis and properties of biocompatible water-soluble silica-coated CdSe/ZnS semiconductor quantum dots. J Phys Chem B. 2001;105:8861–71. [Google Scholar]
- 34.Parak WJ, Pellegrino T, Plank C. Labelling of cells with quantum dots. Nanotechnology. 2005;16:R9–R25. doi: 10.1088/0957-4484/16/2/R01. [DOI] [PubMed] [Google Scholar]
- 35.Yu WW, Chang E, Drezek R, Colvin VL. Water-soluble quantum dots for biomedical applications. Biochem Biophys Res Commun. 2006;348:781–6. doi: 10.1016/j.bbrc.2006.07.160. [DOI] [PubMed] [Google Scholar]
- 36.Pathak S, Choi SK, Arnheim N, Thompson ME. Hydroxylated quantum dots as luminescent probes for in situ hybridization. J Am Chem Soc. 2001;123:4103–4. doi: 10.1021/ja0058334. [DOI] [PubMed] [Google Scholar]
- 37.Gerion D, Parak WJ, Williams SC, Zanchet D, Micheel CM, Alivisatos AP. Sorting fluorescent nanocrystals with DNA. J Am Chem Soc. 2002;124:7070–4. doi: 10.1021/ja017822w. [DOI] [PubMed] [Google Scholar]
- 38.Mitchell GP, Mirkin CA, Letsinger RL. Programmed assembly of DNA functionalized quantum dots. J Am Chem Soc. 1999;121:8122–3. [Google Scholar]
- 39.Willard DM, Carillo LL, Jung J, van Orden A. CdSe-ZnS quantum dots as resonance energy transfer donors in a model protein-protein binding assay. Nano Lett. 2001;1:469–74. [Google Scholar]
- 40.Sukhanova A, Devy J, Venteo L, Kaplan H, Artemyev M, Oleinikov V, et al. Biocompatible fluorescent nanocrystals for immunolabeling of membrane proteins and cells. Anal Biochem. 2004;324:60–7. doi: 10.1016/j.ab.2003.09.031. [DOI] [PubMed] [Google Scholar]
- 41.Bruchez M, Jr, Moronne M, Gin P, Weiss S, Alivisatos AP. Semiconductor nanocrystals as fluorescent biological labels. Science. 1998;281:2013–6. doi: 10.1126/science.281.5385.2013. [DOI] [PubMed] [Google Scholar]
- 42.Pellegrino T, Manna L, Kudera S, Liedl T, Koktysh D, Rogach AL, et al. Hydrophobic nanocrystals coated with an amphiphilic polymer shell: a general route to water soluble nanocrystals. Nano Lett. 2004;4:703–7. [Google Scholar]
- 43.Gao X, Cui Y, Levenson RM, Chung LW, Nie S. In vivo cancer targeting and imaging with semiconductor quantum dots. Nat Biotechnol. 2004;22:969–76. doi: 10.1038/nbt994. [DOI] [PubMed] [Google Scholar]
- 44.Osaki F, Kanamori T, Sando S, Sera T, Aoyama Y. A quantum dot conjugated sugar ball and its cellular uptake. On the size effects of endocytosis in the subviral region. J Am Chem Soc. 2004;126:6520–1. doi: 10.1021/ja048792a. [DOI] [PubMed] [Google Scholar]
- 45.Dubertret B, Skourides P, Norris DJ, Noireaux V, Brivanlou AH, Libchaber A. In vivo imaging of quantum dots encapsulated in phospholipid micelles. Science. 2002;298:1759–62. doi: 10.1126/science.1077194. [DOI] [PubMed] [Google Scholar]
- 46.Wang YA, Li JJ, Chen H, Peng X. Stabilization of inorganic nanocrystals by organic dendrons. J Am Chem Soc. 2002;124:2293–8. doi: 10.1021/ja016711u. [DOI] [PubMed] [Google Scholar]
- 47.Pinaud F, King D, Moore HP, Weiss S. Bioactivation and cell targeting of semiconductor CdSe/ZnS nanocrystals with phytochelatin-related peptides. J Am Chem Soc. 2004;126:6115–23. doi: 10.1021/ja031691c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim S, Bawendi MG. Oligomeric ligands for luminescent and stable nanocrystal quantum dots. J Am Chem Soc. 2003;125:14652–3. doi: 10.1021/ja0368094. [DOI] [PubMed] [Google Scholar]
- 49.Resch-Genger U, Grabolle M, Cavaliere-Jaricot S, Nitschke R, Nann T. Quantum dots versus organic dyes as fluorescent labels. Nat Methods. 2008;5:763–75. doi: 10.1038/nmeth.1248. [DOI] [PubMed] [Google Scholar]
- 50.Gao X, Chan WC, Nie S. Quantum-dot nanocrystals for ultrasensitive biological labeling and multicolor optical encoding. J Biomed Opt. 2002;7:532–7. doi: 10.1117/1.1506706. [DOI] [PubMed] [Google Scholar]
- 51.Goldman ER, Balighian ED, Kuno MK, Labrenz S, Tran PT, Anderson GP, et al. Luminescent quantum dot-adaptor protein-antibody conjugates for use in fluoroimmunoassays. Phys Stat Sol B. 2002;229:407–14. [Google Scholar]
- 52.Goldman ER, Balighian ED, Mattoussi H, Kuno MK, Mauro JM, Tran PT, et al. Avidin: a natural bridge for quantum dot-antibody conjugates. J Am Chem Soc. 2002;124:6378–82. doi: 10.1021/ja0125570. [DOI] [PubMed] [Google Scholar]
- 53.Mason JN, Tomlinson ID, Rosenthal SJ, Blakely RD. Labeling cell-surface proteins via antibody quantum dot streptavidin conjugates. Methods Mol Biol. 2005;303:35–50. doi: 10.1385/1-59259-901-X:035. [DOI] [PubMed] [Google Scholar]
- 54.Dahan M, Levi S, Luccardini C, Rostaing P, Riveau B, Triller A. Diffusion dynamics of glycine receptors revealed by single-quantum dot tracking. Science. 2003;302:442–5. doi: 10.1126/science.1088525. [DOI] [PubMed] [Google Scholar]
- 55.Tokumasu F, Dvorak J. Development and application of quantum dots for immunocytochemistry of human erythrocytes. J Microsc. 2003;211:256–61. doi: 10.1046/j.1365-2818.2003.01219.x. [DOI] [PubMed] [Google Scholar]
- 56.Rosenthal SJ, Tomlinson A, Adkins EM, Schroeter S, Adams S, Swafford L, et al. Targeting cell surface receptors with ligand-conjugated nanocrystals. J Am Chem Soc. 2002;124:4586–94. doi: 10.1021/ja003486s. [DOI] [PubMed] [Google Scholar]
- 57.Lidke DS, Nagy P, Heintzmann R, Arndt-Jovin DJ, Post JN, Grecco HE, et al. Quantum dot ligands provide new insights into erbB/HER receptor-mediated signal transduction. Nat Biotechnol. 2004;22:198–203. doi: 10.1038/nbt929. [DOI] [PubMed] [Google Scholar]
- 58.Zheng J, Ghazani AA, Song Q, Mardyani S, Chan WC, Wang C. Cellular imaging and surface marker labeling of hematopoietic cells using quantum dot bioconjugates. Lab Hematol. 2006;12:94–8. [PubMed] [Google Scholar]
- 59.Parak WJ, Boudreau R, Le Gros MA, Gerion D, Zanchet D, Micheel CM, et al. Cell motility and metastatic potential studies based on quantum dot imaging of phagokinetic tracks. Adv Mater. 2002;14:882–5. [Google Scholar]
- 60.Akerman ME, Chan WC, Laakkonen P, Bhatia SN, Ruoslahti E. Nanocrystal targeting in vivo. Proc Natl Acad Sci USA. 2002;99:12617–21. doi: 10.1073/pnas.152463399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Biju V, Itoh T, Anas A, Sujith A, Ishikawa M. Semiconductor quantum dots and metal nanoparticles: syntheses, optical properties, and biological applications. Anal Bioanal Chem. 2008;391:2469–95. doi: 10.1007/s00216-008-2185-7. [DOI] [PubMed] [Google Scholar]
- 62.Mattheakis LC, Dias JM, Choi YJ, Gong J, Bruchez MP, Liu J, et al. Optical coding of mammalian cells using semiconductor quantum dots. Anal Biochem. 2004;327:200–8. doi: 10.1016/j.ab.2004.01.031. [DOI] [PubMed] [Google Scholar]
- 63.Rieger S, Kulkarni RP, Darcy D, Fraser SE, Koster RW. Quantum dots are powerful multipurpose vital labeling agents in zebrafish embryos. Dev Dyn. 2005;234:670–81. doi: 10.1002/dvdy.20524. [DOI] [PubMed] [Google Scholar]
- 64.Ferrara DE, Weiss D, Carnell PH, Vito RP, Vega D, Gao X, et al. Quantitative 3D fluorescence technique for the analysis of en face preparations of arterial walls using quantum dot nanocrystals and two-photon excitation laser scanning microscopy. Am J Physiol Regul Integr Comp Physiol. 2006;290:R114–23. doi: 10.1152/ajpregu.00449.2005. [DOI] [PubMed] [Google Scholar]
- 65.Mansson A, Sundberg M, Balaz M, Bunk R, Nicholls IA, Omling P, et al. In vitro sliding of actin filaments labelled with single quantum dots. Biochem Biophys Res Commun. 2004;314:529–34. doi: 10.1016/j.bbrc.2003.12.133. [DOI] [PubMed] [Google Scholar]
- 66.Bruchez MP. Turning all the lights on: quantum dots in cellular assays. Curr Opin Chem Biol. 2005;9:533–7. doi: 10.1016/j.cbpa.2005.08.019. [DOI] [PubMed] [Google Scholar]
- 67.Kim S, Lim YT, Soltesz EG, De Grand AM, Lee J, Nakayama A, et al. Near-infrared fluorescent type II quantum dots for sentinel lymph node mapping. Nat Biotechnol. 2004;22:93–7. doi: 10.1038/nbt920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Takeda M, Tada H, Higuchi H, Kobayashi Y, Kobayashi M, Sakurai Y, et al. In vivo single molecular imaging and sentinel node navigation by nanotechnology for molecular targeting drug-delivery systems and tailor-made medicine. Breast Cancer. 2008;15:145–52. doi: 10.1007/s12282-008-0037-0. [DOI] [PubMed] [Google Scholar]
- 69.Ciarlo M, Russo P, Cesario A, Ramella S, Baio G, Neumaier CE, et al. Use of the semiconductor nanotechnologies ‘quantum dots’ for in vivo cancer imaging. Recent Pat Anticancer Drug Discov. 2009;4:207–15. doi: 10.2174/157489209789206841. [DOI] [PubMed] [Google Scholar]
- 70.Ballou B, Ernst LA, Andreko S, Fitzpatrick JA, Lagerholm BC, Waggoner AS, et al. Imaging vasculature and lymphatic flow in mice using quantum dots. Methods Mol Biol. 2009;574:63–74. doi: 10.1007/978-1-60327-321-3_6. [DOI] [PubMed] [Google Scholar]
- 71.Kang WJ, Chae JR, Cho YL, Lee JD, Kim S. Multiplex imaging of single tumor cells using quantum-dot-conjugated aptamers. Small. 2009;5:2519–22. doi: 10.1002/smll.200900848. [DOI] [PubMed] [Google Scholar]
- 72.Singhal S, Nie S, Wang MD. Nanotechnology applications in surgical oncology. Annu Rev Med. 2010;61:359–73. doi: 10.1146/annurev.med.60.052907.094936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mahmoud W, Sukhanova A, Oleinikov V, Rakovich YP, Donegan JF, Pluot M, et al. Emerging applications of fluorescent nanocrystals quantum dots for micrometastases detection. Proteomics. 2009;10:700–16. doi: 10.1002/pmic.200900540. [DOI] [PubMed] [Google Scholar]
- 74.Pinkel D, Straume T, Gray JW. Cytogenetic analysis using quantitative, high-sensitivity, fluorescence hybridization. Proc Natl Acad Sci USA. 1986;83:2934–8. doi: 10.1073/pnas.83.9.2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ekong R, Wolfe J. Advances in fluorescence in situ hybridization. Curr Opin Biotechnol. 1998;9:19–24. doi: 10.1016/s0958-1669(98)80079-7. [DOI] [PubMed] [Google Scholar]
- 76.Levsky JM, Singer RH. Fluorescence in situ hybridization: past, present and future. J Cell Sci. 2003;116:2833–8. doi: 10.1242/jcs.00633. [DOI] [PubMed] [Google Scholar]
- 77.Xiao Y, Barker PE. Semiconductor nanocrystal probes for human metaphase chromosomes. Nucleic Acids Res. 2004;32:e28. doi: 10.1093/nar/gnh024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Xiao Y, Telford WG, Ball JC, Locascio LE, Barker PE. Semiconductor nanocrystal conjugates, FISH and pH. Nat Methods. 2005;2:723. doi: 10.1038/nmeth1005-723. [DOI] [PubMed] [Google Scholar]
- 79.Xiao Y, Barker PE. Semiconductor nanocrystal probes for human chromosomes and DNA. Minerva Biotec. 2004;16:281–8. doi: 10.1093/nar/gnh024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chan P, Yuen T, Ruf F, Gonzalez-Maeso J, Sealfon SC. Method for multiplex cellular detection of mRNAs using quantum dot fluorescent in situ hybridization. Nucleic Acids Res. 2005;33:1–8. doi: 10.1093/nar/gni162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wu SM, Zhao X, Zhang ZL, Xie HY, Tian ZQ, Peng J, et al. Quantum-dot-labeled DNA probes for fluorescence in situ hybridization (FISH) in the microorganism Escherichia coli . Chem Phys Chem. 2006;7:1062–7. doi: 10.1002/cphc.200500608. [DOI] [PubMed] [Google Scholar]
- 82.Muller F, Houben A, Barker PE, Xiao Y, Kas JA, Melzer M. Quantum dots - a versatile tool in plant science? J Nanobiotechnol. 2006;4:5. doi: 10.1186/1477-3155-4-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tholouli E, Hoyland JA, Di Vizio D, O'Connell F, Macdermott SA, Twomey D, et al. Imaging of multiple mRNA targets using quantum dot based in situ hybridization and spectral deconvolution in clinical biopsies. Biochem Biophys Res Commun. 2006;348:628–36. doi: 10.1016/j.bbrc.2006.07.122. [DOI] [PubMed] [Google Scholar]
- 84.Bentolila LA, Weiss S. Single-step multicolor fluorescence in situ hybridization using semiconductor quantum dot-DNA conjugates. Cell Biochem Biophys. 2006;45:59–70. doi: 10.1385/CBB:45:1:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Jiang Z, Li R, Todd NW, Stass SA, Jiang F. Detecting genomic aberrations by fluorescence in situ hybridization with quantum dots-labeled probes. J Nanosci Nanotechnol. 2007;7:4254–9. doi: 10.1166/jnn.2007.920. [DOI] [PubMed] [Google Scholar]
- 86.Knoll JH. Human metaphase chromosome FISH using quantum dot conjugates. In: Bruchez MP, Hotz CZ, editors. Quantum dots applications in biology. vol. 374. Totowa, NJ: Humana Press; 2007. pp. 55–66. [DOI] [PubMed] [Google Scholar]
- 87.Ma L, Wu SM, Huang J, Ding Y, Pang DW, Li L. Fluorescence in situ hybridization (FISH) on maize metaphase chromosomes with quantum dot-labeled DNA conjugates. Chromosoma. 2008;117:181–17. doi: 10.1007/s00412-007-0136-2. [DOI] [PubMed] [Google Scholar]
- 88.Choi Y, Kim HP, Hong SM, Ryu JY, Han SJ, Song R. In situ visualization of gene expression using polymer-coated quantum-dot-DNA conjugates. Small. 2009;5:2085–91. doi: 10.1002/smll.200900116. [DOI] [PubMed] [Google Scholar]
- 89.Muller S, Cremer M, Neusser M, Grasser F, Cremer T. A technical note on quantum dots for multi-color fluorescence in situ hybridization. Cytogenet Genome Res. 2009;124:351–9. doi: 10.1159/000218138. [DOI] [PubMed] [Google Scholar]
- 90.Ioannou D, Tempest HG, Skinner BM, Thornhill AR, Ellis M, Griffin DK. Quantum dots as new-generation fluorochromes for FISH: an appraisal. Chromosome Res. 2009;17:519–30. doi: 10.1007/s10577-009-9051-0. [DOI] [PubMed] [Google Scholar]
- 91.Finch KA, Fonseka KG, Abogrein A, Ioannou D, Handyside AH, Thornhill AR, et al. Nuclear organization in human sperm: preliminary evidence for altered sex chromosome centromere position in infertile males. Hum Reprod. 2008;23:1263–70. doi: 10.1093/humrep/den112. [DOI] [PubMed] [Google Scholar]
- 92.Finch KA, Fonseka G, Ioannou D, Hickson N, Barclay Z, Chatzimeletiou K, et al. Nuclear organisation in totipotent human nuclei and its relationship to chromosomal abnormality. J Cell Sci. 2008;121:655–63. doi: 10.1242/jcs.025205. [DOI] [PubMed] [Google Scholar]
- 93.Masabanda JS, Burt DW, O'Brien PC, Vignal A, Fillon V, Walsh PS, et al. Molecular cytogenetic definition of the chicken genome: the first complete avian karyotype. Genetics. 2004;166:1367–73. doi: 10.1534/genetics.166.3.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Robertson LB, Griffin DK, Tempest HG, Skinner BM. The evolution of the avian genome as revealed by comparative molecular cytogenetics. Cytogenet Genome Res. 2007;117:64–77. doi: 10.1159/000103166. [DOI] [PubMed] [Google Scholar]
- 95.Skinner BM, Volker M, Ellis M, Griffin DK. An appraisal of nuclear organisation in interphase embryonic fibroblasts of chicken, turkey and duck. Cytogenet Genome Res. 2009;126:156–64. doi: 10.1159/000245915. [DOI] [PubMed] [Google Scholar]
- 96.Griffin DK, Haberman F, Masabanda J, O'Brien PCM, Bagga M, Smith J, et al. Micro-and macro chromosome painting probes generated by flow cytometry and chromosome microdissection: tools for mapping the chicken genome. Cytogenet Cell Genet. 1999;87:278–81. doi: 10.1159/000015449. [DOI] [PubMed] [Google Scholar]
- 97.Campos-Ramos R, Harvey SC, Masabanda JS, Carrasco LA, Griffin DK, McAndrew BJ, et al. Identification of putative sex chromosomes in the blue tilapia, Oreochromis aureus, through synaptonemal complex and FISH analysis. Genetica. 2001;111:143–53. doi: 10.1023/a:1013707818534. [DOI] [PubMed] [Google Scholar]
- 98.Harvey SC, Masabanda J, Carrasco LA, Bromage NR, Penman DJ, Griffin DK. Molecular-cytogenetic analysis reveals sequence differences between the sex chromosomes of Oreochromis niloticus: evidence for an early stage of sex-chromosome differentiation. Cytogenet Genome Res. 2002;97:76–80. doi: 10.1159/000064036. [DOI] [PubMed] [Google Scholar]
- 99.Griffin DK, Harvey SC, Campos-Ramos R, Ayling LJ, Bromage NR, Masabanda JS, et al. Early origins of the X and Y chromosomes: lessons from tilapia. Cytogenet Genome Res. 2002;99:157–63. doi: 10.1159/000071588. [DOI] [PubMed] [Google Scholar]
- 100.Bruchez M. Quantum dots for ultra-sensitive multicolor detection of proteins and genes. In: Jong H, Tanke H, Fransz P, editors. 16th International Chromosome Conference (16th ICC) Amsterdam, the Netherlands: Springer; 2007. pp. 1–108. [Google Scholar]
- 101.Murray CB, Kagan CR, Bawendi M. Synthesis and characterization of monodisperse nanocrystals and close-packed nanocrystal assemblies. Annu Rev Mater Sci. 2000;30:545–610. [Google Scholar]
- 102.Yao G, Wang L, Wu Y, Smith J, Xu J, Zhao W, et al. FloDots: luminescent nanoparticles. Anal Bioanal Chem. 2006;385:518–24. doi: 10.1007/s00216-006-0452-z. [DOI] [PubMed] [Google Scholar]
- 103.Choi J, Burns AA, Williams RM, Zhou Z, Flesken-Nikitin A, Zipfel WR, et al. Core-shell silica nanoparticles as fluorescent labels for nanomedicine. J Biomed Opt. 2007;12:064007. doi: 10.1117/1.2823149. [DOI] [PubMed] [Google Scholar]