Abstract
The interface between biosystems and nanomaterials is emerging for detection of various biomolecules and subtle cellular activities. In particular, the development of cost-effective and sequence-selective DNA detection is urgent for the diagnosis of genetic or pathogenic diseases. Graphene-based nanocarbon materials, such as carbon nanotubes and thin graphene layers, have been employed as biosensors because they are biocompatible, extraordinarily sensitive, and promising for large-area detection. Electrical and label-free detection of DNA can be achieved by monitoring the conductance change of devices fabricated from these carbon materials. Here, the recent advances in this research area are briefly reviewed. The key issues and perspectives of future development are also discussed.
Keywords: carbon nanotubes, graphene, DNA hybridization, biosensors, label-free detection, transistors
The interface between biosystems and nanomaterials is emerging as one of the most diverse and dynamic areas of science and technology. The research of nano/bio interfaces, comprising the dynamic physical or chemical interactions, kinetics, and thermodynamic exchanges between nanomaterial surfaces and the surfaces of biological components such as proteins, membranes, phospholipids, endocytic vesicles, organelles, and DNA is rapidly growing. In particular, the development of sequence-selective DNA sensors for diagnosis of genetic or pathogenic diseases has attracted much attention. Many methods have been adopted to detect the DNA hybridization process including the detection relying on optical (1–10), piezoelectric (11–16), and electrochemical transductions (17–27). However, fluorescent or electrochemical tags are required for these detection methods. Alternative approaches based on the resistance change of semiconductor nanomaterials (e.g. silicon nanowires and carbon nanotubes) have been demonstrated as a potential for label-free electrical detection (28–35). The nanomaterials of graphene (aromatic sp2)-based carbon have been widely used for DNA sensing because they are biocompatible and highly sensitive to environmental perturbations such as electronic doping (36–40) and molecular adsorption (41–46). In this article, we briefly review the recent developments of label-free bioelectronic sensors for detecting DNA hybridization using the field-effect transistors (FETs) based on single-walled carbon nanotubes (SWNTs) and graphene-related materials. The detection mechanisms for various devices are also discussed in detail.
Field-effect transistors (FETs) for signal transduction
Graphene, a one-atom-thick planar sheet of aromatic sp2 carbon crystal, holds great promise for replacing conventional Si semiconductors in applications, including high-speed computer chips and biochemical sensors. It has been recently demonstrated that its intrinsic carrier mobility (∼200,000 cm2/V-s) is higher than other known materials at room temperature (47, 48). The two-dimensional (2-D) graphene is the basic structural element of some carbon allotropes including graphite (3-D), SWNTs (1-D), and fullerenes. Among these allotropes, SWNTs can be easily adopted for device fabrication due to having a high length-to-diameter ratio. The SWNTs can be considered as a cylindrical roll-up of the planar graphene sheet with a sp2 bonding of carbon atoms. These cylindrical carbon molecules have novel properties that make them potentially useful in many applications in nanotechnology, electronics, optics, and other fields of materials science, as well as a potential use in architectural fields. Similar to graphene, they exhibit extraordinary strength, high thermal conductivity, unique electrical properties, and are biocompatible. The diameter size of SWNTs (∼1 nm) is comparable to the size of DNA molecules, which is suitable for revealing the interactions between biomolecules and nanomaterials.
The electronic components, such as resistors and FETs based on 2-D graphene thin layers and 1-D SWNTs, have been successfully demonstrated using nano-/microlithographic fabrication. It is noteworthy that the transport carriers in both 2-D graphene thin layers and 1-D SWNTs flow plentifully on their surfaces as their π electrons are delocalized on the surfaces. Consequently, their conductance is highly sensitive to the environmental perturbations occurring proximate to the carbon surfaces.
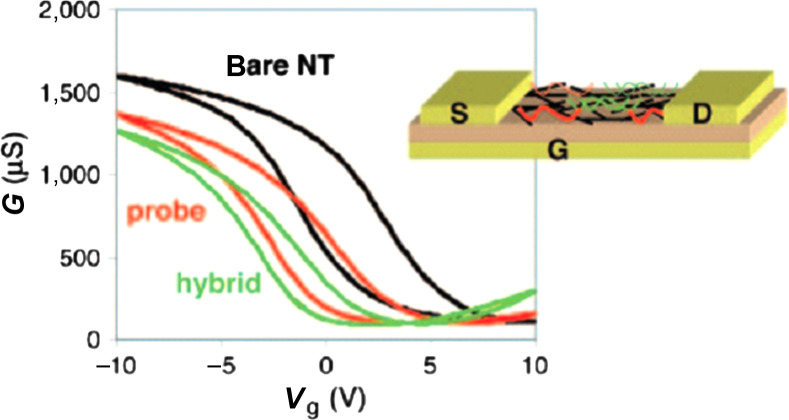
DNA molecules consist of various nucleobases, cytosine, guanine, adenine, and thymine, which can interact with graphene or SWNT surfaces. Theoretical calculations and experiments have suggested that an interaction between nucleobases and graphene (or SWNT) surface can be explained by the van der Waals interaction (π–π interaction) and solvation energy contributed by solvent molecules (49). The interaction between recognition DNA (probe-DNA) and carbon surfaces or the binding of analyte DNA with preconjugated probe-DNA may cause significant changes in the conductivity of devices through different mechanisms such as electrostatic gating (caused by the charges on DNA) (50–53), electronic doping (from DNA to carbon materials) (54), or modification of the junction between electrodes and carbon materials (55, 56). Taking advantage of the sensitive electrical responses from these devices, SWNTs or graphene-related materials configured into FETs have been successfully utilized to electrically differentiate the DNA molecules with single-base specificity. The physical mechanism underlying the sensing varies significantly with the device configuration and operation modes. Fig. 1 shows one of these detection strategies reported by Star et al. (54), and this example illustrates how the label-free electrical detection works for biomolecular detection. The source-drain conductance of the device was recorded before and after incubation with 12-mer oligonucleotide capture probes (5′-CCT AAT AAC AAT-3′), as well as after incubation with the complementary DNA targets. In this experiment, each electrical measurement was performed in dry state, after the device was immersed in desired solutions followed by rinsing and drying. The conductance decreases with the addition of probe- or target-DNA. Typically the percentage decrease in conductance caused by the addition of target-DNA can be correlated to the concentration of the target-DNAs. Also, the addition of mismatched DNA does not result in significant conductance drop. Therefore, the differentiation between the target and mismatched DNAs can be achieved by such a detection strategy.
Fig. 1.
Transfer characteristics, conductance (G) as function of gate voltage (Vg), and schematic drawings of the SWNT FETs used for DNA assays before and after incubation with 12-mer oligonucleotide capture probes (5′-CCT AAT AAC AAT - 3′), as well as after incubation with the complementary DNA targets. (Reproduced with permission from Ref. (54), copyright 2006 National Academy of Sciences of the United States of America.)
Single-walled carbon nanotubes (SWNTs)
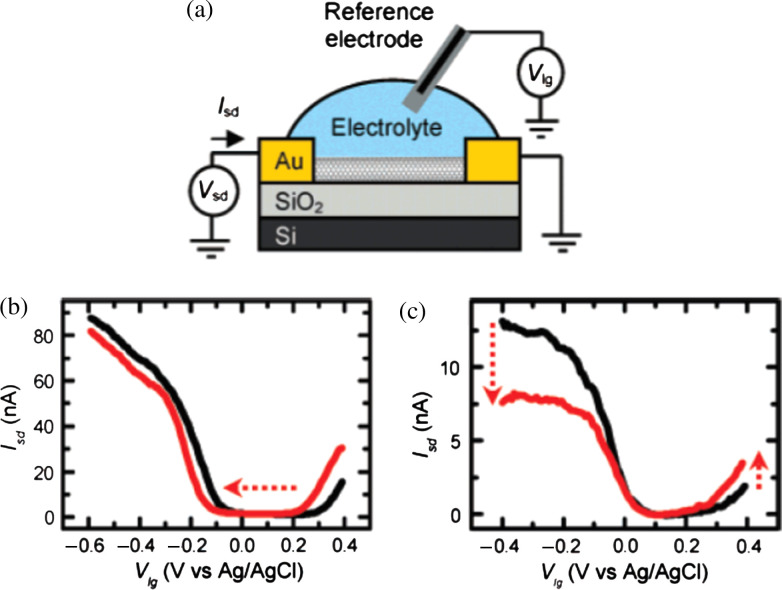
Kong et al. (36) have first successfully demonstrated the electrical detection of gases and chemicals using the FET based on individual SWNTs. Several biosensors based on nanotube FETs have been used for the electronic detection of the following interactions: biotin-streptavidin (57), human immunoglobin (IgG) (58–62), various monoclonal antibodies (63–66), and pig serum albumin (67). The utilization of SWNT networks for DNA detection has been reported by Star et al. (54) and Gui et al. (56). Inspired by these works, Heller et al. (52) have recently discussed the possible sensing mechanisms for the individual SWNT device operated by liquid-gating as schematically illustrated in Fig. 2a, where a source-drain bias potential is applied and the device is gated through a Ag/AgCl reference electrode inserted in the electrolyte. Their study concludes that the electrostatic gating and changes of the Schottky barrier between electrodes and SWNTs are the two competitive detection mechanisms. Fig. 2b and c show two typical changes in liquid gate sweeps of ambipolar devices measured during protein adsorption experiments. Fig. 2b is an example of a strong electrostatic gating (adsorption of 185 nM poly-l-lysine on an ambipolar SWNT device) and Fig. 2c displays the result of a strong Schottky barrier effect in the case of adsorption of 1 µM horse heart cytochrome-c on a short (40 nm) SWNT device. It is noted that these sensors, however, involve a high production cost because nanolithographic facilities are required. Another concern is that these transistors may have significant device-to-device variation due to the difficulty in obtaining desired single-chirality of SWNT species for device fabrication.
Fig. 2.
(a) Measurement setup, where a source-drain bias potential is applied and the device is gated through a Ag/AgCl reference electrode inserted in the electrolyte. (b, c) Changes in liquid gate sweeps of ambipolar devices measured during protein adsorption experiments. (b) Example of strong electrostatic gating (adsorption of 185 nM poly-L-lysine on an ambipolar SWNT device). (c) Example of a strong Schottky barrier effect in the case of adsorption of 1 µM horse heart cytochrome-c on a short (40 nm) SWNT device. (Reproduced with permission from Ref. (52), copyright 2008 American Chemical Society).
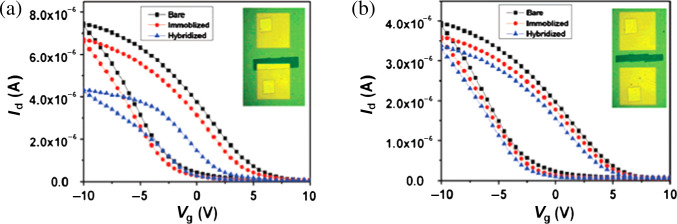
In contrast, SWNT network-based transistors are practically simpler in fabrication and involve lower production cost. Their electrical properties depend on the SWNT percolative path in the conduction channel and thus the device-to-device variations are expected to be small. Label-free electrical detection of DNA hybridization and antibody-antigen binding using FETs fabricated from SWNT networks have been successfully achieved recently (54). We have verified that the sensing of DNA hybridization for the devices with an apparent on/off ratio, the ratio of an on current and off current measured during the gate voltage sweeping, is dominated by the change in electrode-SWNT junctions rather than the conductance change of channel SWNTs (56). Fig. 3 compares the sensing performance for the SWNT FETs devices with (a) one junction exposed, and (b) only channel exposed before immobilization, after immobilization, and upon hybridization with its complementary DNA, where the unexposed area was covered with photoresist. The inset for each graph shows the photoresist pattern as imaged in an optical microscope. The results verify that the SWNT-electrode junction is more sensitive than SWNTs themselves. This can be reasoned that this FET is governed by the electrode-SWNT network junction when the contact resistance is non-trivial, where the contact barrier shall be sensitive to the charges brought in by DNA molecules. Separately, if a metallic tube network is used, the electrode-SWNT contact resistance becomes insignificantly small and therefore the DNA detection will be governed by the electronic doping (from DNA molecules to SWNTs) occurring at the channel area (68).
Fig. 3.
Transfer curves (drain current Id vs gate voltage Vg) for photoresist capped Au-contacted SWNT FETs with (a) one junction exposed, (b) a channel exposed before immobilization, after immobilization, and upon hybridization with its complementary DNA, where the drain voltage was fixed at -0.5V. The inset for each graph shows the photoresist pattern as imaged in an optical microscope. (Reproduced with permission from Ref. (56), copyright 2007 American Chemical Society).
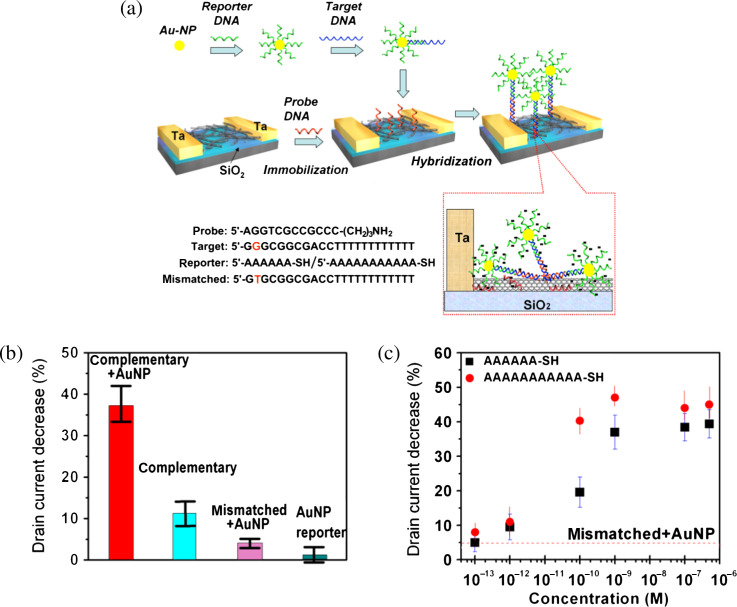
The reported detection limit of distinguishing complementary and one-base mismatched DNA for the FETs based on networks is on the order of ca. 1 nM of DNA. It has been demonstrated that the sensitivity can be improved by using a threading intercalacter attached with a redox-active functional group, where the intercalacter selectively inserts into hybridized DNA strands and the redox functional groups enhance the changes in device conductance (69). Another more efficient method is to bring more electrostatic charges to the proximity of electrode-SWNT contacts through introducing reporter-DNA-Au nanoparticle conjugates (70) as schematically illustrated in Fig. 4a. Each target DNA can bind to a Au-nanoparticle that has been previously attached with many reporter DNA molecules and consequently the change of conductance upon each hybridization event can be greatly magnified. Fig. 4b clearly demonstrates that the sensitivity of reporter DNA-Au nanoparticle assisted detection is significantly higher than those without the cooperation of reporter DNA. Fig. 4c shows that the sensitivity of this approach allows us to differentiate the complementary and one-base mismatched DNA with the sensitivity as good as ∼100 fM of DNA.
Fig. 4.
(a) Schematic illustration of DNA detection enhancement by reporter DNA-Au nanoparticle conjugates. The bottom right panel illustrates the possible molecular binding on SWNTs. (b) Statistics showing the percentage decrease of Id in SWNT FETs for various sensing experiments. (c) Percentage Id decrease versus DNA concentration in the sensing of complementary DNA, enhanced by 6A and 11A reporter DNA-AuNP conjugates. The dashed line shows the limit of selective detection, which is based on the Id response to the mismatched DNA (1 nM) + 11A reporter DNA-AuNP conjugates. (Reproduced with permission from Ref. 70, copyright 2008 Wiley-VCH).
An alternative method, by monitoring the shift of threshold voltage during the gate voltage sweep, reflecting on the increased density of charges trapped around SWNTs upon DNA addition or hybridization (71–74), has also been adopted to perform label-free detection of DNA hybridization (75). Due to that, the change in threshold voltage involves the trapping of moisture and strongly depends on the substrate surface qualities, the detection reliability, and sensitivity for this approach still require more investigations.
Graphene and related materials
Graphene and graphene derivatives, such as graphene oxide (GO), reduced graphene oxide (rGO) and n-doped graphene, are also potentially useful for the biosensing application. The GO is an oxidized form of graphene produced in solution and has negative charges when dispersed in water (76). It is strongly hydrophilic and has a brown/dark-brown color in aqueous solution. The GO can be reduced by chemical methods (77–81), thermal methods (82, 83), and ultraviolet-assisted methods (84) to form rGO. The conductivity can be increased by up to 4 orders of magnitude by reduction of GO to rGO (85–87). However, the conductivity of rGO is still less than that of the pristine graphene by a factor of 10–100 (88, 89). The rGO has been found to contain a considerable amount of topological defects (90) and this is probably one of the main reasons for the limited conductivity. Graphene could also be n-doped by introducing n-dopants (40, 43). Wang's group (91) has also recently reported that graphene nanoribbons could be covalently functionalized by nitrogen species on ribbon edges through high-power electrical joule heating in ammonia gas. All these approaches open up the routes for tailoring electrical properties of graphene sheets for various applications.
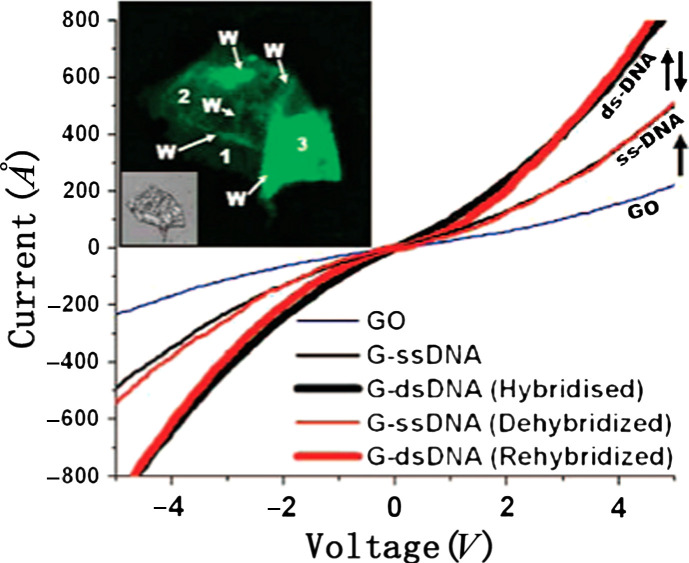
For the DNA hybridization study, GO has been used as a template to host a fluorescence quencher for the detection of DNA hybridization based on the fluorescence from the fluorophore-conjugated probe DNAs (92, 93). Electrical detection of DNA hybridization by monitoring the conductance change of GO sheets has also been reported (94) (shown in Fig. 5), where the authors observe that the single-stranded (ss)-DNA tethering on the GO device (termed as G-DNA) increases its conductivity. Successive hybridization and dehybridization of DNA on the G-DNA device result in a completely reversible increase and restoration of conductivity. The conductance increase (restoration) was explained by the increased (decreased) electrostatic gating induced by the negative charges from DNA molecules. The electrical properties of GO films largely vary with the fabrication process of GO (oxidation processes of graphite); hence, the sensitivity of the devices produced from GO sheets shall strongly depend on the size and the shape of GO, the presence of wrinkles on a GO surface, the degree of oxidation in GO, and the defect density of the GO sheet. Therefore, to use GO as the sensing component, more efforts are needed to control the quality of GO sheets.
Fig. 5.
Single-stranded (ss)-DNA tethering on GO (G-DNA) increases the conductivity of the device. Successive hybridization and dehybridization of DNA on the G-DNA device results in completely reversible increase and restoration of conductivity. Inset shows a fluorescent image for the GO device hybridized G-DNA (ds) sheet with wrinkles and folds clearly visible. (Reproduced with permission from Ref. 78, copyright 2008 American Chemical Society).
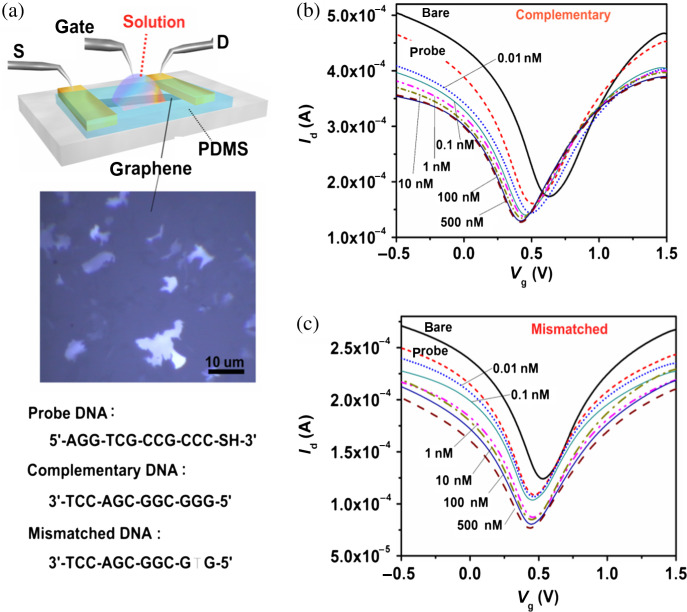
In contrast to GO, large-size graphene films are considered more reliable and achievable from the device fabrication viewpoint. Developing graphene-based biosensors becomes practical with the recent advances of chemical vapor deposition (CVD) of large-sized graphene film (up to wafer size) (95–97). We have fabricated large-sized graphene transistors by transferring the as-grown CVD graphene films from Ni (or Cu) to glass substrates. These films can be configured as liquid-gated FETs (Fig. 6a) and they are able to detect the DNA hybridization with a detection sensitivity of 0.01 nM and the capability to distinguish single-base mismatch (98). As shown in Fig. 6b and c, the conductance of graphene devices exhibited amipolar behaviors subjecting to the gate voltage applied to the bath solution. The Vg,min that gives the minimum graphene conductance can be identified from the transfer curve and be used to monitor the Fermi energy state of the graphene film. Fig. 6b shows that the Vg,min is sensitive to the immobilization of probe DNAs and hybridization of the complementary target DNAs. Specifically, Vg,min is significantly left-shifted with the addition of DNA molecules, suggesting that the electrostatic gating from the DNA is not dominating and that DNA molecules n-dopes the graphene film. The interaction between DNA molecules and graphitic structure has been studied theoretically and experimentally (92, 93, 99–101). It has been demonstrated that the binding between graphene and nucleotides is dominated by the non-electrostatic interaction (99, 100, 102–104).
Fig. 6.
(a) Schematic illustration of the graphene device operated by liquid gating. The middle is an optical microscopy image of the graphene films. The bottom shows the DNA sequences used in the experiments. (b, c) Transfer characteristics for the graphene transistors before adding DNA, after immobilization with probe DNA, and after reaction with (b) complementary or (c) one-base mismatched DNA molecules with the concentration ranging from 0.01 to 500 nM. (Reproduced with permission from Ref 82, copyright 2010 Wiley-VCH).
Meanwhile, the left-shift of Vg,min after DNA hybridization suggests that the complementary DNAs can also effectively interact with graphene and impose the n-doping effect based on the graphene-nucleotide interaction. The shift in Vg,min increased with the increasing concentration of the complementary DNA, specifically, 0.01 nM and 10 nM DNA solutions caused the >10 meV and >55 meV shift, respectively. The Vg,min did not shift further at the higher complementary DNA concentrations (>10 nM) likely due to saturation in hybridization with the limited number of probe DNAs. Furthermore, Fig. 6 demonstrates that the complementary DNA and the one-base mismatched DNA can be easily differentiated because Vg,min is much less sensitive to the mismatched DNA, which only caused a ∼20 meV shift at a high concentration (500 nM). The electrostatic gating effect by adsorbed charge species has been adopted to explain the shift of the Id-Vg curve of carbon nanotube transistors (52, 53). If the same mechanism applies to our graphene devices, a positive shift of Vg,min would be expected because the highly negative charges of the adsorbed DNAs can only be balanced by positive gate voltage. Thus, electrostatic gating is not dominating the electrical characteristics in our testing conditions. It is noted that the electrostatic gating by electrolytes may sometimes become a competing mechanism when the electrolyte concentration is too high or when the gate voltage scanning range is too large.
Future of carbon-based electrical detection of DNA hybridization
Label-free electrical detection of DNA hybridization using carbon-based materials has been carried out for years. High sensitivity and high specificity have been achieved. However, there are still numerous challenges for detecting low concentration of DNA analytes (< femto mole). Along with the decrease of DNA concentration, improvement of signal-to-noise ratio of devices shall become an important issue. More efforts are required to optimize operation conditions of carbon-based devices. It is also attractive to seek the device that allows the detection of a single hybridization event. In practical cases, detection of a longer DNA chain will also be necessary. The electrical detection of DNA hybridization largely depends on the device structure and the forms of these carbon materials. For example, an increase of semi-to-metallic ratio in the SWNT ensemble, controlling chirality of carbon nanotubes, and proper modification of graphene or graphene nanoribbon could potentially enhance the DNA detection sensitivity. The rapid, specific, and low-cost electrical detection of DNA hybridization could speed up the realization of the next generation smart homecare sensor system.
Conclusions
In this review, we have discussed the recent advances and key issues for the development of label-free detection of DNA hybridization using carbon nanotubes and graphene. Sensing mechanisms are also discussed for each type of device. The detection of a single-base polymorphorism or mutation is thought to be the key for diagnosis of genetic diseases and realization of personalized medicine. Carbon nanotube and graphene-based devices have shown great potential for the future application of DNA biosensors in terms of the high sensitivity and selectivity. From the fundamental viewpoint, more research efforts are needed to understand the interface (contact) effects, including nanotube-nanotube and graphene-electrode contacts, on sensing behaviors. In addition, intensive studies to bridge the knowledge to clinical detection of biomarkers are emergent.
Acknowledgements
We acknowledge the financial support from the Taiwan National Science Council (99-2112-M-001-021-MY3 and 99-2738-M-001-001) and from the Academia Sinica Taiwan.
Biographies
 Dongliang Fu received a BSc from Zhejiang University, China in 2006. He is currently a final year PhD student of Nanyang Technological University. He also joined the University of Illinois at Urbana-Champaign and Academia Sinica Taiwan for the overseas attachments during his PhD study. His research interests include carbon nanotube and graphene-based sensors.
Dongliang Fu received a BSc from Zhejiang University, China in 2006. He is currently a final year PhD student of Nanyang Technological University. He also joined the University of Illinois at Urbana-Champaign and Academia Sinica Taiwan for the overseas attachments during his PhD study. His research interests include carbon nanotube and graphene-based sensors.
 Lain-Jong Li received a BSc and an MSc in chemistry at the National Taiwan University. After 5 years of research and development experience at the Taiwan Semiconductor Manufacturing Company, he obtained his PhD in condensed matter physics from Oxford University, United Kingdom in 2006. He is an associate research fellow at Academia Sinica Taiwan. His research interests include carbon nanotubes, boron nitride, graphene, and graphene-derivatives.
Lain-Jong Li received a BSc and an MSc in chemistry at the National Taiwan University. After 5 years of research and development experience at the Taiwan Semiconductor Manufacturing Company, he obtained his PhD in condensed matter physics from Oxford University, United Kingdom in 2006. He is an associate research fellow at Academia Sinica Taiwan. His research interests include carbon nanotubes, boron nitride, graphene, and graphene-derivatives.
Conflict of interest and funding
There is no conflict of interest in the present study for any of the authors.
References
- 1.Piunno P, Krull U, Hudson R, Damha M, Cohen H. Fiber optic biosensor for fluorimetric detection of DNA hybridization. Anal Chim Acta. 1994;288:205–14. [Google Scholar]
- 2.Watts H, Yeung D, Parkes H. Real-time detection and quantification of DNA hybridization by an optical biosensor. Anal Chem. 1995;67:4283–9. doi: 10.1021/ac00119a013. [DOI] [PubMed] [Google Scholar]
- 3.Ferguson J, Boles T, Adams C, Walt D. A fiber-optic DNA biosensor microarray for the analysis of gene expression. Nat Biotechnol. 1996;14:1681–4. doi: 10.1038/nbt1296-1681. [DOI] [PubMed] [Google Scholar]
- 4.Liu X, Tan W. A fiber-optic evanescent wave DNA biosensor based on novel molecular beacons. Anal Chem. 1999;71:5054–9. doi: 10.1021/ac990561c. [DOI] [PubMed] [Google Scholar]
- 5.Reichert J, Csaki A, Kohler J, Fritzsche W. Chip-based optical detection of DNA hybridization by means of nanobead labeling. Anal Chem. 2000;72:6025–9. doi: 10.1021/ac000567y. [DOI] [PubMed] [Google Scholar]
- 6.Hansen K, Ji H, Wu G, Datar R, Cote R, Majumdar A, et al. Cantilever-based optical deflection assay for discrimination of DNA single-nucleotide mismatches. Anal Chem. 2001;73:1567–71. doi: 10.1021/ac0012748. [DOI] [PubMed] [Google Scholar]
- 7.Dubertret B, Calame M, Libchaber A. Single-mismatch detection using gold-quenched fluorescent oligonucleotides. Nat Biotechnol. 2001;19:365–70. doi: 10.1038/86762. [DOI] [PubMed] [Google Scholar]
- 8.Nelson B, Grimsrud T, Liles M, Goodman R, Corn R. Surface plasmon resonance imaging measurements of DNA and RNA hybridization adsorption onto DNA microarrays. Anal Chem. 2001;73:1–7. doi: 10.1021/ac0010431. [DOI] [PubMed] [Google Scholar]
- 9.Peterson A, Heaton R, Georgiadis R. The effect of surface probe density on DNA hybridization. Nucleic Acids Res. 2001;29:5163–8. doi: 10.1093/nar/29.24.5163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ho H, Dore K, Boissinot M, Bergeron M, Tanguay R, Boudreau D, et al. Direct molecular detection of nucleic acids by fluorescence signal amplification. J Am Chem Soc. 2005;127:12673–676. doi: 10.1021/ja053417j. [DOI] [PubMed] [Google Scholar]
- 11.Okahata Y, Kawase M, Niikura K, Ohtake F, Furusawa H, Ebara Y. Kinetic measurements of DNA hybridization on an oligonucleotide-immobilized 27-MHz quartz crystal microbalance. Anal Chem. 1998;70:1288–96. doi: 10.1021/ac970584w. [DOI] [PubMed] [Google Scholar]
- 12.Tombelli S, Mascini M, Sacco C, Turner A. A DNA piezoelectric biosensor assay coupled with a polymerase chain reaction for bacterial toxicity determination in environmental samples. Anal Chim Acta. 2000;418:1–9. [Google Scholar]
- 13.Tombelli S, Mascini M, Turner A. Improved procedures for immobilisation of oligonucleotides on gold-coated piezoelectric quartz crystals. Biosens Bioelectron. 2002;17:929–36. doi: 10.1016/s0956-5663(02)00084-2. [DOI] [PubMed] [Google Scholar]
- 14.Duman M, Saber R, Piskin E. A new approach for immobilization of oligonucleotides onto piezoelectric quartz crystal for preparation of a nucleic acid sensor for following hybridization. Biosen Bioelectron. 2003;18:1355–63. doi: 10.1016/s0956-5663(03)00087-3. [DOI] [PubMed] [Google Scholar]
- 15.Hur Y, Han J, Seon J, Pak Y, Roh Y. Development of an SH-SAW sensor for the detection of DNA hybridization. Sen Actuators A. 2005;120:462–7. [Google Scholar]
- 16.Rijal K, Mutharasan R. PEMC-based method of measuring DNA hybridization at femtomolar concentration directly in human serum and in the presence of copious noncomplementary strands. Anal Chem. 2007;79:7392–400. doi: 10.1021/ac0712042. [DOI] [PubMed] [Google Scholar]
- 17.Wang J, Cai X, Rivas G, Shiraishi H, Farias P, Dontha N. DNA electrochemical biosensor for the detection of short DNA sequences related to the human immunodeficiency virus. Anal Chem. 1996;68:2629–34. doi: 10.1021/ac9602433. [DOI] [PubMed] [Google Scholar]
- 18.Steel A, Herne T, Tarlov M. Electrochemical quantitation of DNA immobilized on gold. Anal Chem. 1998;70:4670–77. doi: 10.1021/ac980037q. [DOI] [PubMed] [Google Scholar]
- 19.Wang J. Towards genoelectronics: electrochemical biosensing of DNA hybridization. Chem Eur J. 1999;5:1681–5. [Google Scholar]
- 20.Takenaka S, Yamashita K, Takagi M, Uto Y, Kondo H. DNA sensing on a DNA probe-modified electrode using ferrocenylnaphthalene diimide as the electrochemically active ligand. Anal Chem. 2000;72:1334–41. doi: 10.1021/ac991031j. [DOI] [PubMed] [Google Scholar]
- 21.Wang J, Xu D, Kawde A, Polsky R. Metal nanoparticle-based electrochemical stripping potentiometric detection of DNA hybridization. Anal Chem. 2001;73:5576–81. doi: 10.1021/ac0107148. [DOI] [PubMed] [Google Scholar]
- 22.Gooding J. Electrochemical DNA hybridization biosensors. Electroanalysis. 2002;14:1149–56. [Google Scholar]
- 23.Wang J. Electrochemical nucleic acid biosensors. Anal Chim Acta. 2002;469:63–71. [Google Scholar]
- 24.Fan C, Plaxco K, Heeger A. Electrochemical interrogation of conformational changes as a reagentless method for the sequence-specific detection of DNA. Proc Natl Acad Sci USA. 2003;100:9134–7. doi: 10.1073/pnas.1633515100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li J, Ng H, Cassell A, Fan W, Chen H, Ye Q, et al. Carbon nanotube nanoelectrode array for ultrasensitive DNA detection. Nano Lett. 2003;3:597–602. [Google Scholar]
- 26.He P, Xu Y, Fang Y. A review: electrochemical DNA biosensors for sequence recognition. Anal Lett. 2005;38:2597–623. [Google Scholar]
- 27.Kelley S, Boon E, Barton J, Jackson N, Hill M. Single-base mismatch detection based on charge transduction through DNA. 1999;27:4830–7. doi: 10.1093/nar/27.24.4830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hahm J, Lieber C. Direct ultrasensitive electrical detection of DNA and DNA sequence variations using nanowire nanosensors. Nano Lett. 2004;4:51–4. [Google Scholar]
- 29.Gao Z, Agarwal A, Trigg A, Singh N, Fang C, Tung C, et al. Silicon nanowire arrays for label-free detection of DNA. Anal Chem. 2007;79:3291–7. doi: 10.1021/ac061808q. [DOI] [PubMed] [Google Scholar]
- 30.Zhang G, Zhang G, Chua J, Chee R, Wong E, Agarwal A, et al. DNA sensing by silicon nanowire: charge layer distance dependence. Nano Lett. 2008;8:1066–70. doi: 10.1021/nl072991l. [DOI] [PubMed] [Google Scholar]
- 31.Zhang G, Chua J, Chee R, Agarwal A, Wong S, Buddharaju K, et al. Highly sensitive measurements of PNA-DNA hybridization using oxide-etched silicon nanowire biosensors. Biosen Bioelectron. 2008;23:1701–7. doi: 10.1016/j.bios.2008.02.006. [DOI] [PubMed] [Google Scholar]
- 32.Cattani-Scholz A, Pedone D, Dubey M, Neppl S, Nickel B, Feulner P, et al. Organophosphonate-based PNA-functionalization of silicon nanowires for label-free DNA detection. ACS Nano. 2008;2:1653–60. doi: 10.1021/nn800136e. [DOI] [PubMed] [Google Scholar]
- 33.Curreli M, Zhang R, Ishikawa F, Chang H, Cote R, Zhou C, et al. Real-time label-free detection of biological entities using nanowire-based FETs. IEEE Trans Nanotechnol. 2008;7:651–67. [Google Scholar]
- 34.Ishikawa F, Stauffer B, Caron D, Zhou C. Rapid and label-free cell detection by metal-cluster-decorated carbon nanotube biosensors. Biosen Bioelectron. 2009;24:2967–72. doi: 10.1016/j.bios.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 35.Basuray S, Senapati S, Aijian A, Mahon A, Chang H. Shear and AC field enhanced carbon nanotube impedance assay for rapid sensitive, and mismatch-discrimination DNA hybridization. ACS Nano. 2009;3:1823–30. doi: 10.1021/nn9004632. [DOI] [PubMed] [Google Scholar]
- 36.Kong J, Franklin N, Zhou C, Chapline M, Peng S, Cho K, et al. Nanotube molecular wires as chemical sensors. Science. 2000;287:622–5. doi: 10.1126/science.287.5453.622. [DOI] [PubMed] [Google Scholar]
- 37.Collins P, Bradley K, Ishigami M, Zettl A. Extreme oxygen sensitivity of electronic properties of carbon nanotubes. Science. 2000;287:1801–4. doi: 10.1126/science.287.5459.1801. [DOI] [PubMed] [Google Scholar]
- 38.Snow E, Perkins F, Houser E, Badescu S, Reinecke T. Chemical detection with a single-walled carbon nanotube capacitor. Science. 2005;307:1942–5. doi: 10.1126/science.1109128. [DOI] [PubMed] [Google Scholar]
- 39.Li L, Nicholas R. Bandgap-selective chemical doping of semiconducting single-walled carbon nanotubes. Nanotechnology. 2004;15:1844–7. [Google Scholar]
- 40.Shi Y, Dong X, Chen P, Wang J, Li L. Effective doping of single-layer graphene from underlying SiO2 substrates. Phys Rev B. 2009;79:115402. [Google Scholar]
- 41.Schedin F, Geim A, Morozov S, Hill E, Blake P, Katsnelson M, et al. Detection of individual gas molecules adsorbed on graphene. Nat Mater. 2007;6:652–5. doi: 10.1038/nmat1967. [DOI] [PubMed] [Google Scholar]
- 42.Wehling T, Novoselov K, Morozov S, Vdovin E, Katsnelson M, Geim A, et al. Molecular doping of graphene. Nano Lett. 2008;8:173–7. doi: 10.1021/nl072364w. [DOI] [PubMed] [Google Scholar]
- 43.Dong X, Fu D, Fang W, Shi Y, Chen P, Li L. Doping single-layer graphene with aromatic molecules. Small. 2009;5:1422–6. doi: 10.1002/smll.200801711. [DOI] [PubMed] [Google Scholar]
- 44.Dan Y, Lu Y, Kybert N, Luo Z, Johnson A. Intrinsic response of graphene vapor sensors. Nano Lett. 2009;9:1472–5. doi: 10.1021/nl8033637. [DOI] [PubMed] [Google Scholar]
- 45.Bradley K, Briman M, Star A, Gruner G. Charge transfer from adsorbed proteins. Nano Lett. 2004;4:253–6. [Google Scholar]
- 46.Li L, Khlobystov A, Wiltshire J, Briggs G, Nicholas R. Diameter-selective encapsulation of metallocenes in single-walled carbon nanotubes. Nat Mater. 2005;4:481–5. doi: 10.1038/nmat1396. [DOI] [PubMed] [Google Scholar]
- 47.Morozov S, Novoselov K, Katsnelson M, Schedin F, Elias D, Jaszczak J, et al. Giant intrinsic carrier mobilities in graphene and its bilayer. Phys Rev Lett. 2008;100:016602. doi: 10.1103/PhysRevLett.100.016602. [DOI] [PubMed] [Google Scholar]
- 48.Chen J, Jang C, Xiao S, Ishigami M, Fuhrer M. Intrinsic and extrinsic performance limits of graphene devices on SiO2. Nat Nanotechnol. 2008;3:206–9. doi: 10.1038/nnano.2008.58. [DOI] [PubMed] [Google Scholar]
- 49.Varghese N, Mogera U, Govindaraj A, Das A, Maiti P, Sood A, et al. Binding of DNA nucleobases and nucleosides with graphene. ChemPhysChem. 2009;10:206–10. doi: 10.1002/cphc.200800459. [DOI] [PubMed] [Google Scholar]
- 50.Pandana H, Aschenbach K, Lenski D, Fuhrer M, Khan J, Gomez R. A versatile biomolecular charge-based sensor using oxide-gated carbon nanotube transistor arrays. IEEE Sensors J. 2008;8:655–60. [Google Scholar]
- 51.Maehashi K, Matsumoto K, Kerman K, Takamura Y, Tamiya E. Ultrasensitive detection of DNA hybridization using carbon nanotube field-effect transistors. Jpn J Appl Phys. 2004;43:1558–60. [Google Scholar]
- 52.Heller I, Janssens A, Mnnik J, Minot E, Lemay S, Dekker C. Identifying the mechanism of biosensing with carbon nanotube transistors. Nano Lett. 2008;8:591–5. doi: 10.1021/nl072996i. [DOI] [PubMed] [Google Scholar]
- 53.Artyukhin A, Stadermann M, Friddle R, Stroeve P, Bakajin O, Noy A. Controlled electrostatic gating of carbon nanotube FET devices. Nano Lett. 2006;6:2080–5. doi: 10.1021/nl061343j. [DOI] [PubMed] [Google Scholar]
- 54.Star A, Tu E, Niemann J, Gabriel J, Joiner C, Valcke C. Label-free detection of DNA hybridization using carbon nanotube network field-effect transistors. Proc Natl Acad Sci USA. 2006;103:921–6. doi: 10.1073/pnas.0504146103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tang X, Bansaruntip S, Nakayama N, Yenilmez E, Chang Y, Wang Q. Carbon nanotube DNA sensor and sensing mechanism. Nano Lett. 2006;6:1632–6. doi: 10.1021/nl060613v. [DOI] [PubMed] [Google Scholar]
- 56.Gui E, Li L, Zhang K, Xu Y, Dong X, Ho X, et al. DNA sensing by field-effect transistors based on networks of carbon nanotubes. J Am Chem Soc. 2007;129:14427–32. doi: 10.1021/ja075176g. [DOI] [PubMed] [Google Scholar]
- 57.Star A, Gabriel J, Bradley K, Gruner G. Electronic detection of specific protein binding using nanotube FET devices. Nano Lett. 2003;3:459–63. [Google Scholar]
- 58.Maehashi K, Katsura T, Kerman K, Takamura Y, Matsumoto K, Tamiya E. Label-free protein biosensor based on aptamer-modified carbon nanotube field-effect transistors. Anal Chem. 2007;79:782–7. doi: 10.1021/ac060830g. [DOI] [PubMed] [Google Scholar]
- 59.Maehashi K, Matsumoto K, Takamura Y, Tamiya E. Aptamer-based label-free immunosensors using carbon nanotube field-effect transistors. Electroanalysis. 2009;21:1285–90. [Google Scholar]
- 60.Chen R, Choi H, Bangsaruntip S, Yenilmez E, Tang X, Wang Q, et al. An investigation of the mechanisms of electronic sensing of protein adsorption on carbon nanotube devices. J Am Chem Soc. 2004;126:1563–8. doi: 10.1021/ja038702m. [DOI] [PubMed] [Google Scholar]
- 61.Cid C, Riu J, Maroto A, Rius F. Detection of human immunoglobulin G at physiological conditions with chemically functionalized carbon nanotube field effect transistors. Curr Nanosci. 2008;4:314–7. [Google Scholar]
- 62.Kim J, Lee B, Hong S, Sim S. Ultrasensitive carbon nanotube-based biosensors using antibody-binding fragments. Anal Biochem. 2008;381:193–8. doi: 10.1016/j.ab.2008.06.040. [DOI] [PubMed] [Google Scholar]
- 63.Chen R, Bangsaruntip S, Drouvalakis K, Wong N, Shim M, Li Y, et al. Noncovalent functionalization of carbon nanotubes for highly specific electronic biosensors. Proc Natl Acad Sci USA. 2003;100:4984–9. doi: 10.1073/pnas.0837064100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Byon H, Choi H. Network single-walled carbon nanotube-field effect transistors with increased schottky contact area for highly sensitive biosensor applications. J A Chem Soc. 2006;128:2188–9. doi: 10.1021/ja056897n. [DOI] [PubMed] [Google Scholar]
- 65.Takeda S, Sbagyo A, Sakoda Y, Ishii A, Sawamura M, Sueoka K, et al. Application of carbon nanotubes for detecting anti-hemagglutinins based on antigen-antibody interaction. Biosen Bioelectron. 2005;21:201–5. doi: 10.1016/j.bios.2004.08.030. [DOI] [PubMed] [Google Scholar]
- 66.Li C, Curreli M, Lin H, Lei B, Ishikawa F, Datar R, et al. Complementary detection of prostate-specific antigen using In2O3 nanowires and carbon nanotubes. J Am Chem Soc. 2005;127:12484–5. doi: 10.1021/ja053761g. [DOI] [PubMed] [Google Scholar]
- 67.Kojima A, Hyon C, Kamimura T, Maeda M, Matsumoto K. Protein sensor using carbon nanotube field effect transistor. Jpn J Appl Phys. 2005;44:1596–8. [Google Scholar]
- 68.Dong X, Fu D, Xu Y, Wei J, Shi Y, Chen P, et al. Label-free electronic detection of DNA using simple double-walled carbon nanotube resistors. J Phys Chem C. 2008;112:9891–5. [Google Scholar]
- 69.Gui E, Li L, Lee P, Lohani A, Mhaisalkar S, Cao Q, et al. Electrical detection of DNA hybridization and threading intercalation using carbon nanotubes network field-effect transistors. Appl Phys Lett. 2006;89:232104. [Google Scholar]
- 70.Dong X, Lau C, Lohani A, Mhaisalkar S, Kasim J, Shen Z, et al. Electrical detection of femtomolar DNA via gold-nanoparticle enhancement in carbon-nanotube-network field-effect transistors. Adv Mater. 2008;20:2389–93. [Google Scholar]
- 71.Kim W, Javey A, Vermesh O, Wang Q, Li Y, Dai H. Hysteresis caused by water molecules in carbon nanotube field-effect transistors. Nano Lett. 2003;3:193–8. [Google Scholar]
- 72.Sung D, Hong S, Kim Y, Park N, Kim S, Maeng S, et al. Ab initio study of the effect of water adsorption on the carbon nanotube field-effect transistor. Appl Phys Lett. 2006;89:243110. [Google Scholar]
- 73.Lee J, Ryu S, Yoo K, Choi I, Yun W, Kim J. Origin of gate hysteresis in carbon nanotube field-effect transistors. J Phys Chem C. 2007;111:12504–7. [Google Scholar]
- 74.Lee C, Dong X, Goh S, Wang J, Wei J, Li L. Illumination-enhanced hysteresis of transistors based on carbon nanotube networks. J. Phys. Chem. C. 2009;113:4745–7. [Google Scholar]
- 75.Martinez M, Tseng Y, Ormategui N, Loinaz I, Eritja R, Bokor J. Label-free DNA biosensors based on functionalized carbon nanotube field effect transistors. Nano Lett. 2009;9:530–6. doi: 10.1021/nl8025604. [DOI] [PubMed] [Google Scholar]
- 76.Li D, Muller M, Gilje S, Kaner R, Wallace G. Processable aqueous dispersions of graphene nanosheets. Nat Nanotech. 2008;3:101–5. doi: 10.1038/nnano.2007.451. [DOI] [PubMed] [Google Scholar]
- 77.Stankovich S, Dikin D, Dommett G, Kohlhaas K, Zimney E, Stach E, et al. Graphene-based composite materials. Nature. 2006;442:282–6. doi: 10.1038/nature04969. [DOI] [PubMed] [Google Scholar]
- 78.Stankovich S, Dikin D, Piner R, Kohlhaas K, Kleinhammes A, Jia Y, et al. Synthesis of graphene-based nanosheets via chemical reduction of exfoliated graphite oxide. Carbon. 2007;45:1558–65. [Google Scholar]
- 79.Lomeda J, Doyle C, Kosynkin D, Hwang W, Tour J. Diazonium functionalization of surfactant-wrapped chemically converted graphene sheets. J Am Chem Soc. 2008;130:16201–6. doi: 10.1021/ja806499w. [DOI] [PubMed] [Google Scholar]
- 80.Tung V, Allen M, Yang Y, Kaner R. High-throughput solution processing of large-scale graphene. Nat Nanotech. 2008;4:25–9. doi: 10.1038/nnano.2008.329. [DOI] [PubMed] [Google Scholar]
- 81.Wang G, Yang J, Park J, Gou X, Wang B, Liu H, et al. Facile synthesis and characterization of graphene nanosheets. J Phys Chem C. 2008;112:8192–5. [Google Scholar]
- 82.Schniepp H, Li J, McAllister M, Sai H, Herrera-Alonso M, Adamson D, et al. Functionalized single graphene sheets derived from splitting graphite oxide. J Phys Chem B. 2006;110:8535–9. doi: 10.1021/jp060936f. [DOI] [PubMed] [Google Scholar]
- 83.McAllister M, Li J, Adamson D, Schniepp H, Abdala A, Liu J, et al. Single sheet functionalized graphene by oxidation and thermal expansion of graphite. Chem Mater. 2007;19:4396–404. [Google Scholar]
- 84.Williams G, Serger B, Kamat P. TiO2-graphene nanocomposites. UV-assisted photocatalytic reduction of graphene oxide. ACS Nano. 2008;2:1487–91. doi: 10.1021/nn800251f. [DOI] [PubMed] [Google Scholar]
- 85.Jung I, Dikin D, Piner R, Ruoff R. Tunable electrical conductivity of individual graphene oxide sheets reduced at “low” temperatures. Nano Lett. 2008;8:4283–7. doi: 10.1021/nl8019938. [DOI] [PubMed] [Google Scholar]
- 86.Gilje S, Han S, Minsheng W, Kang L, Kaner R. A chemical route to graphene for device applications. Nano Lett. 2007;7:3394–8. doi: 10.1021/nl0717715. [DOI] [PubMed] [Google Scholar]
- 87.Gomez-Navarro C, Weitz R, Bittner A, Scolari M, Mews A, Burghard M, et al. Electronic transport properties of individual chemically reduced graphene oxide sheets. Nano Lett. 2007;7:3499–503. doi: 10.1021/nl072090c. [DOI] [PubMed] [Google Scholar]
- 88.Lopez V, Sundaram R, Gomez-Navarro C, Olea D, Burghard M, Gomez-Herrero J, et al. Chemical vapor deposition repair of graphene oxide: a route to highly conductive graphene monolayers. Adv Mater. 2009;21:4683–6. [Google Scholar]
- 89.Tung V, Allen M, Yang Y, Kaner R. High-throughput solution processing of large-scale graphene. Nat Nanotechnol. 2009;4:25–9. doi: 10.1038/nnano.2008.329. [DOI] [PubMed] [Google Scholar]
- 90.Gomez-Navarro C, Meyer J, Sundaram R, Chuvilin A, Kurasch S, Burghard M, et al. Atomic Structure of reduced graphene oxide. Nano Lett. 2010;10:1144–8. doi: 10.1021/nl9031617. [DOI] [PubMed] [Google Scholar]
- 91.Wang X, Li X, Zhang L, Yoon Y, Weber P, Wang H, et al. N-doping of graphene through electrothermal reactions with ammonia. Science. 2009;324:768–71. doi: 10.1126/science.1170335. [DOI] [PubMed] [Google Scholar]
- 92.He S, Song B, Li D, Zhu C, Qi W, Wen Y, et al. A graphene nanoprobe for rapid, sensitive, and multicolor fluorescent DNA analysis. Adv Funct Mater. 2010;20:453–9. [Google Scholar]
- 93.Lu C, Yang H, Zhu C, Chen X, Chen G. A graphene platform for sensing biomolecules. Angew Chem Int Ed. 2009;48:4785–7. doi: 10.1002/anie.200901479. [DOI] [PubMed] [Google Scholar]
- 94.Mohanty N, Berry V. Graphene-based single-bacterium resolution biodevice and DNA transistor: interfacing graphene derivatives with nanoscale and microscale biocomponents. Nano Lett. 2008;8:4469–76. doi: 10.1021/nl802412n. [DOI] [PubMed] [Google Scholar]
- 95.Reina A, Jia X, Ho J, Nezich D, Son H, Bulovic V, et al. Large area, few-layer graphene films on arbitrary substrates by chemical vapor deposition. Nano Lett. 2009;9:30–5. doi: 10.1021/nl801827v. [DOI] [PubMed] [Google Scholar]
- 96.Li X, Cai W, An J, Kim S, Nah J, Yang D, et al. Large-area synthesis of high-quality and uniform graphene films on copper foils. Science. 2009;324:1312–4. doi: 10.1126/science.1171245. [DOI] [PubMed] [Google Scholar]
- 97.Kim K, Zhao Y, Jang H, Lee S, Kim J, Kim K, et al. Large-scale pattern growth of graphene films for stretchable transparent electrodes. Nature. 2009;457:706–10. doi: 10.1038/nature07719. [DOI] [PubMed] [Google Scholar]
- 98.Dong X, Shi Y, Huang W, Chen P, Li L. Electrical detection of DNA hybridization with single-base specificity using transistors based on CVD-grown graphene sheets. Adv Mater. 2010;22:1649–53. doi: 10.1002/adma.200903645. [DOI] [PubMed] [Google Scholar]
- 99.Manohar S, Mantz A, Bancroft K, Hui C, Jagota A, Vezenov D. Peeling single-stranded DNA from graphite surface to determine oligonucleotide binding energy by force spectroscopy. Nano Lett. 2008;8:4365–72. doi: 10.1021/nl8022143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Varghese N, Mogera U, Govindaraj A, Das A, Maiti P, Sood A, et al. Binding of DNA nucleobases and nucleosides with graphene. ChemPhysChem. 2009;10:206–10. doi: 10.1002/cphc.200800459. [DOI] [PubMed] [Google Scholar]
- 101.Liu Z, Robinson J, Sun X, Dai H. PEGylated nanographene oxide for delivery of water-insoluble cancer drugs. J Am Chem Soc. 2008;130:10876–7. doi: 10.1021/ja803688x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Antony J, Grimme S. Structures and interaction energies of stacked graphene-nucleobase complexes. Phys Chem Chem Phys. 2008;10:2722–9. doi: 10.1039/b718788b. [DOI] [PubMed] [Google Scholar]
- 103.Gowtham S, Scheicher R, Ahuja R, Pandey R, Karna S. Physisorption of nucleobases on graphene: density-functional calculations. Phys Rev B. 2007;76:033401–4. [Google Scholar]
- 104.Ortmann F, Schmidt W, Bechstedt F. Attracted by long-range electron correlation: adenine on graphite. Phys Rev Lett. 2005;95:186101–4. doi: 10.1103/PhysRevLett.95.186101. [DOI] [PubMed] [Google Scholar]