Abstract
Purpose
Mapping the genes for age-related macular degeneration (AMD) had not been successful until recent genome-wide association studies revealed Tyr402His in CFH and rs11200638 in HTRA1 as AMD-related genetic variants. This study was conducted to identify other critical factors in HTRA1 that are associated with exudative AMD.
Methods
The promoter, splice regions, and coding exons of HTRA1 were sequenced in 163 patients with exudative AMD and 183 sex- and age-matched control subjects. Also documented were the CFH genotype and smoking status.
Results
Four significant SNPs were found in the promoter and the first exon of HTRA1: rs11200638 (–625G>A), rs2672598 (–487T>C), rs1049331 (102C>T, Ala34Ala), and rs2293870 (108G>T, Gly36Gly) with respective P = 1.7 × 10−14, 3.0 × 10− 10, 3.7 × 10−12, and 3.7 × 10−12. Among them, rs11200638 is the most significant associated SNP with a high odds ratio (OR) of 7.6 (95% CI: 3.94–14.51). One risk haplotype block across the promoter and exon 1, ACCTT, significantly predisposes to AMD (P = 6.68 × 10−14). In both models, significant independent additive effects were identified with smoking and rs800292 (184G>A, Val62Ile) of CFH. Smoking and rs11200638 (HTRA1) combined caused a 15.7-fold increased risk, whereas combined rs800292 and rs11200638 caused a 23.3-fold increased risk. An extremely high population attributable risk (PAR) of 78% was also found.
Conclusions
A high impact of the additive effect of CFH and HTRA1 in the development of exudative AMD was shown. The HTRA1-smoking additive effect found in this study further suggests the importance of this environmental risk factor in AMD.
Age-related macular degeneration (AMD) is a leading cause of severe visual impairment in most developed countries, affecting more than 10 million people worldwide.1,2 Exudative AMD is an advanced form of the disease that poses a serious threat of vision loss. It is characterized by the formation of new blood vessels beneath the retina. Leakage of blood and fluid from the choroidal neovascular membrane causes scarring that leads to permanent damage from the surrounding retinal tissue, destroying central vision. Nearly 90% of severe cases of AMD are exudative.
Population-based association studies have consistently shown that, among the various environmental factors, smoking is associated with a higher risk of advanced AMD.3–9 The population attributable risk (PAR) of AMD in former and current smokers has been reported to be from 20% to 68%.10 Smoking may affect metabolic processes in the retinal pigment epithelium (RPE) by depressing the release of antioxidants and altering the choroidal blood flow. The retina is thus subjected to oxidative stress due to a high consumption of oxygen. The free radicals, the extracellular matrix along Bruch’s membrane, and the increased levels of the angiogenic factors in RPE cells from the insult of oxidative stress then promote neovascularization.11
First-degree relatives of patients with advanced AMD have a higher incidence and earlier age of disease manifestation according to familial aggregation analysis studies.12,13 Twin studies have revealed a higher incidence of of AMD manifestation in monozygotic than in dizygotic twin pairs, which further reinforces the importance of genetic factors in AMD risk.14–16 Genome-wide linkage studies of AMD in large pedigrees, affected sib pairs, and discordant sib pairs have mapped susceptible genetic loci to 1q, 2q, 3p, 6q, 9q, 10q, 16q, 17q, and 22q.17–25 Associations of AMD risk with most loci have been inconsistent in various reports, but the two loci on 1q31 and 10q26 have frequently been implicated in AMD in different studies. A meta-analysis of six independent studies has provided further evidence of linkage on 10q26 and 1q.26 This meta-analysis also showed weak linkage to 2p, 3p, 4q, 12q, and 16q. Yet no AMD-causing gene has been identified, with subsequent traditional positional cloning. After a genome-wide scan, candidate gene screening of extended families and sib pairs was performed on exon 104 of hemicentin-1 and the 11 exons of EFEMP1 residing on 1q31 and 2p16, respectively. No disease-causing allele was identified.21
In 2005, a genome-wide association study revealed the single-nucleotide polymorphism (SNP) Tyr402His in the complement factor H (CFH) gene on 1q32 as an AMD-related variant, but no association was found between CFH Tyr402His and AMD in Chinese and Japanese populations.27–32 We have shown that some sequence variants in CFH, including rs3753394 (P = 0.003), rs800292 (Val62Ile, P = 0.00053), and rs1329428 (P = 0.00092), but not rs1061170 (Tyr402His), have significant associations with exudative AMD in Chinese.32 Recently, we reported the SNP rs11200638, residing in the promoter of HTRA1, to be associated with exudative AMD in a cohort of Chinese patients.33 It was also found to have a PAR of 49.3% of AMD in Caucasians.34 In this study, the whole sequence of HTRA1 was examined to characterize further the critical genetic factors linked to AMD. The effect of smoking was also investigated. Because the CFH SNP rs800292 was found to be significantly associated with exudative AMD in our previous study in a Chinese cohort,32 its interactions with HTRA1 SNPs were examined in this study.
Materials and Methods
Study Subjects
All study subjects underwent a detailed eye examination, including best corrected visual acuity and slit lamp biomicroscopy of the fundus. Stereoscopic color fundus photographs were taken. The standard classification of the International Age-related Maculopathy Epidemiologic Study Group was used.35 Patients with exudative AMD had nondrusenoid retinal pigment epithelium detachment, choroidal neovascularization (CNV), serous or hemorrhagic retinal detachments, subretinal or sub-RPE hemorrhage, or fibrosis. A total of 163 patients with exudative AMD were recruited, including 88 men and 75 women (Table 1). All these patients had been studied for the CFH genotype,32 and 96 were screened in a published association study.33 There were 183 control subjects: 91 men and 92 women. They did not have a family history of AMD, signs of AMD, or any other major eye diseases except mild senile cataracts and low myopia. Fundus examination results were normal, with no drusen, no abnormal retinal pigment epithelium change, and no foveal reflex. The cases and controls were sex matched. In addition, smoking habits were recorded. A smoker was defined as a person who had smoked at least five cigarettes daily for more than 1 year. The study subjects were divided into two groups: those who had never smoked and those who were ex-smokers and current smokers. Data of smoking status was available on 153 cases and 143 controls (Table 1). This study was conducted in accordance to the Declaration of Helsinki, and informed consent was obtained from the participating subjects after explanation of the nature of the study. The study protocol was approved by the Ethics Committee on Human Research of the Chinese University of Hong Kong.
Table 1.
Characteristics of the Study Population of HTRA1 Screening
| Exudative AMD | Controls | P | OR (95% CI) | |
|---|---|---|---|---|
| Total | 163 | 183 | ||
| Males, n (%) | 88 (54.0%) | 91 (49.7%) | 0.43 | |
| Females, n (%) | 75 (46.0%) | 92 (50.3%) | ||
| Mean age (±SD) (y) | 75.5 ± 7.5 | 73.3 ± 6.5 | ||
| Age range (y) | 60–94 | 60–99 | 0.84 | |
| Smoker, n (%) | 79 (51.6%) | 54 (37.8%) | 0.017 | 1.76 (1.11–2.80) |
| Nonsmoker, n (%) | 74 (48.4%) | 89 (62.2%) | ||
| Missing smoking status, n | 10 | 40 |
Sequence Analysis
Genomic DNA was extracted (QIAamp DNA Blood Mini kit; Qiagen, Hilden, Germany) from peripheral venous blood, according to the manufacturer’s blood and a body fluid spin protocol. Purified DNA was taken for cycle sequencing (BigDye Terminator Cycle Sequencing Reaction Kit, ver. 3.1; Applied Biosystems, Inc., Foster City, CA), according to the manufacturer’s protocol, on a capillary DNA sequencer (model 3130XL; Applied Biosystems, Inc.). The 5′- and 3′-untranslated regions, coding and noncoding exons, and exon–intron junctions of the HTRA1 gene (NM_002775.3; ENSG00000166033) were sequenced as described.33 All samples were genotyped for CFH-rs800292 (TaqMan assay; Applied Biosystems, Inc.).
Statistical Analysis
All the identified SNPs were assessed for Hardy-Weinberg equilibrium (HWE). Sequence variants were analyzed by χ2 test to eliminate genotyping errors. The less frequently occurring allele in the control group was considered to be the risk allele. The χ2 test or Fisher exact test was used to compare the allele frequencies and genotypes between the cases and controls. The odds ratio (OR) was calculated by comparing those homozygous for the risk allele (RR) with the baseline group (those homozygous for the normal allele, NN) (ORhom) and comparing those heterozygous for the risk allele (RN) with the baseline group (ORhet). The contrasts for dominant (RR and RN versus NN) and recessive (RR versus RN and NN) effects were also evaluated. The Bonferroni method was used for correction of multiple testings (SPSS ver., 11.5; SPSS Inc., Chicago, IL). Common genetic models (recessive, additive, and dominant) were also compared. Pair-wise SNP linkage disequilibrium (LD) coefficients (D′) were calculated by using Haplo-view.36 In addition, subsequent haplotype counts between cases and controls were compared and analyzed accordingly.
Interaction Analysis
The joint contributions of the HTRA1-rs11200638 with smoking and of HTRA1-rs11200638 with CFH-rs800292 to AMD were assessed by logistic regression analysis implemented in the R statistical analysis package (http://www.r-project.org/), modeling the case–control status of the CFH and HTRA1 genotypes. A series of models were fitted to draw inferences about the most likely and most parsimonious model(s). Both additive (coded 0, 1, and 2 for the number of copies of the haplotype) and dominant (coded 1 for possessing exactly one copy of a haplotype and 0 otherwise) effects of each haplotype were considered. Models were compared by using the Akaike information criterion (AIC).37 The best-fit model gives the lowest AIC. Models were considered to be statistically indistinguishable when AIC differed <2 units, and those having fewer parameters were chosen as the best fit and most parsimonious models. The joint ORs of the risk factors were further estimated in this best fit, most parsimonious model. The baseline haplotype is the observed individuals with the lowest-risk genotypes at the two loci studied.
RESULTS
A total of 45 sequence variants were identified (Table 2). Among them, four SNPs—IVS5+76delGTTT, IVS6+111G>A, IVS7+149C>G, and IVS8–36C>T—violated HWE and were excluded from further analysis. For the remaining 41 SNPs, χ2 statistics of both allele and genotype frequencies were assessed for association with AMD. Fifteen variants were found each in one control. Six variants existed each in only one AMD case. These 21 rare variants were excluded in subsequent association analyses. A heterozygous 176G>C (Arg59Pro) was found in one AMD case and one control. Therefore, 18 SNPs remained for further association analysis. Their frequency assessment in a recessive model is shown, with calculated ORs and P, in Table 2 and Figure 1. Among these 18 variants, 7 were significantly associated with exudative AMD (Table 2). A heterozygous 34delCinsTCCT was present in 2 AMD and 16 control subjects. It conferred marginally significant protective effects against AMD (P = 0.001) after Bonferroni correction.
Table 2.
HTRA1 Sequence Variants Identified in Exudative AMD Cases and Controls
| Sequence Variants | SNPs ID | Risk Allele | AMD n = 163 NN/RN/RR |
Control n = 183 NN/RN/RR (MAF) |
P | OR | |
|---|---|---|---|---|---|---|---|
| 1 | –625G>A‡ | rs11200638 | A | 18/51/94 (0.73) | 55/90/38 (0.45) | 1.74 × 10−12‡ | 5.20 (3.24–8.35)‡ |
| 2 | –502C>T | C | 0/10/153 | 0/20/163 | 0.11 | 1.88 (0.85–4.14) | |
| 3 | –497C>T | C | 0/4/159 | 0/8/175 | 0.39 | 1.82 (0.54–6.15) | |
| 4 | –487T>C‡ | rs2672598 | C | 1/24/138 | 18/68/97 (0.28) | 3.03 × 10−10‡ | 4.89 (2.92–8.19)‡ |
| 5 | 34delCinsTCCT§† | 0/2/161 | 0/16/167 | 0.001§† | 7.71 (1.74–34.08)§† | ||
| 6 | 59C>T | C | 3/36/124 | 2/56/125 | 0.11 | 1.47 (0.92–2.37) | |
| 7 | 77G>C | 162/1/0 | 180/0/0 | ||||
| 8 | 102C>T‡ | rs1049331 | T | 18/52/93 | 55/90/38 (0.45) | 3.73 × 10−12‡ | 5.07 (3.16–8.14)‡ |
| 9 | 108G>T‡ | rs2293870 | T | 18/52/93 | 55/90/38 (0.45) | 3.73 × 10−12‡ | 5.07 (3.16–8.14)‡ |
| 10 | 176G>C | 0/1/162 | 0/1/182 | ||||
| 11 | IVS1–176C>G||† | rs12267142 | C | 0/16/147 | 0/25/158 | 0.27||† | 1.45 (0.75–2.83)||† |
| 12 | IVS2+34G>A | 162/1/0 | 183/0/0 | ||||
| 13 | IVS2+81C>T | 163/0/0 | 182/1/0 | ||||
| 14 | ISV2+99T>C | 163/1/0 | 183/0/0 | ||||
| 15 | IVS2+100C>T | C | 0/3/160 | 0/8/175 | 0.22 | 2.44 (0.64–9.35) | |
| 16 | IVS2+172_179del8 | 162/1/0 | 183/0/0 | ||||
| 17 | IVS2+216A>G | 163/0/0 | 182/1/0 | ||||
| 18 | IVS2+317C>T | T | 161/2/0 | 182/1/0 | 0.60 | 2.26 (0.20–25.17) | |
| 19 | 663G>A | 163/0/0 | 182/1/0 | ||||
| 20 | IVS3+93C>T | rs2239586 | C | 15/77/71 | 22/92/69 | 0.27 | 1.27 (0.83–1.96) |
| 21 | IVS3+167G>A | rs2239587 | G | 15/77/71 | 22/92/69 | 0.27 | 1.27 (0.83–1.96) |
| 22 | 834C>T | 163/0/0 | 182/1/0 | ||||
| 23 | IVS4+99C>T | rs2672582 | T | 46/90/27 | 67/86/30 | 0.79 | 1.08 (0.61–1.90) |
| 24 | IVS4–34G>A | 163/0/0 | 182/1/0 | ||||
| 25 | 996A>G | 163/0/0 | 182/1/0 | ||||
| 26 | IVS5+21delG | 163/0/0 | 182/1/0 | ||||
| 27 | IVS5+51G>C | 163/0/0 | 182/1/0 | ||||
| 28 | IVS5+76_79del4* | Del | 104/59/0 | 135/48/0 | |||
| 29 | IVS5+168C>T | 163/0/0 | 182/1/0 | ||||
| 30 | IVS5+169G>A|| | rs2672583 | A | 42/90/31 | 68/87/28 | 0.36|| | 1.30 (0.74–2.28)|| |
| 31 | IVS5–133G>A | 162/1/0 | 183/0/0 | ||||
| 32 | IVS6+90G>T | 163/0/0 | 182/1/0 | ||||
| 33 | IVS6+111G>A* | A | 127/32/4 | 125/57/1 | |||
| 34 | IVS6+115C>G | rs2672585 | G | 45/92/26 | 67/88/28 | 0.87 | 1.05 (0.59–1.88) |
| 35 | IVS7+17C>A | 162/1/0 | 183/0/0 | ||||
| 36 | IVS7+130G>T | 163/0/0 | 182/1/0 | ||||
| 37 | IVS7+149C>G* | G | 126/33/4 | 125/57/1 | |||
| 38 | IVS7–123G>C | 162/1/0 | 183/0/0 | ||||
| 39 | 1221C>T† | rs11538140 | C | 0/3/160 | 0/5/178 | 0.73† | 1.50 (0.35–6.37)† |
| 40 | 1249G>A | 163/0/0 | 182/1/0 | ||||
| 41 | IVS8+14G>A | rs2272599 | G | 43/92/28 | 67/85/31 | 0.95 | 1.02 (0.58–1.78) |
| 42 | IVS8+61A>G | 163/0/0 | 182/1/0 | ||||
| 43 | IVS8–36C>T* | rs2293871 | T | 62/77/24 | 57/102/24 | ||
| 44 | 1487C>T | 163/0/0 | 182/1/0 | ||||
| 45 | 1537C>G | 163/0/0 | 182/1/0 |
Genotypes are ordered NN, NR, RR, where N is the normal allele and R is the risk allele. The risk allele is defined as the least frequent allele in control subjects. ORs and P of 18 SNPs were calculated in the recessive model.
These four SNPs violated HWE and were excluded from further analysis.
These SNPs are calculated in the heterozygous model.
These four SNPs remained significantly associated with AMD after Bonferroni correction (0.05/18/2 = 0.001).
This variant was marginally significant, with two heterozygous carriers in the cases and 16 heterozygous carriers in the controls.
These two SNPs were not significantly associated with AMD after Bonferroni correction.
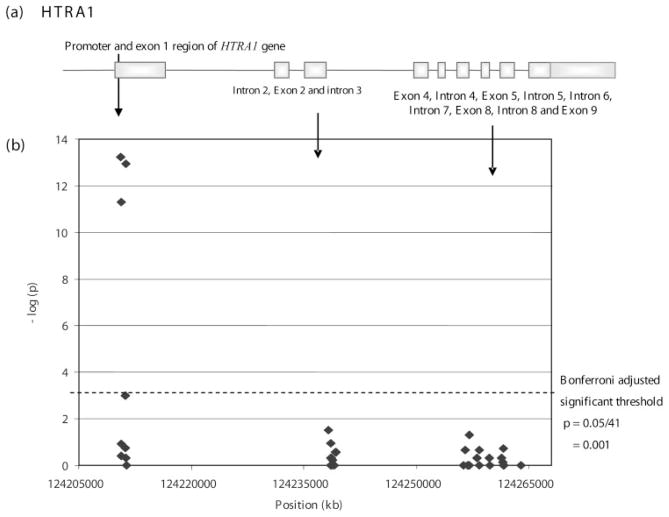
Figure 1.
The association of exudative AMD with 41 variants across a 54-Mb region of chromosome 10 in the HTRA1 gene. The position of each SNP is given on a kilobase scale. (a) The location of exons of HTRA1 that map to the interval. (b) –Log (P) is plotted against the chromosomal location of the 41 variants.
After Bonferroni correction (P = 0.05/41 = 0.001), four variants remained significantly associated with exudative AMD:–625G>A (rs11200638), –487T>C (rs2672598), 102C>T (rs1049331, Ala34Ala), and 108G>T (rs2293870, Gly36Gly) with P = 1.7 × 10−14, 3.0 × 10−10, 3.7 × 10−12, and 3.7 × 10−12 (Table 2), respectively. 102C>T and 108G>T, both in exon 1, are in perfect LD, where D′ = 1 and are synonymous variants with no amino acid change (Ala34Ala and Gly36Gly, respectively). Both dominant and recessive models showed significant association with AMD in all four SNPs (Table 3). In addition, an allele dosage effect appeared to be present. Carriers of two risk alleles were at substantially higher risk (approximately four times higher) for exudative AMD than were carriers of one risk allele. The ORhet versus ORhom for the four SNPs were: 1.73 vs. 7.56, 6.35 vs. 25.61, 1.76 vs. 7.48, and 1.76 vs. 7.48, respectively (Table 3). With the observed increased risk in carriers of the two risk alleles of these SNPs, the PARs were also found to have increased in similar fashion. PAR estimated at –625G>A, 102C>T, and 108G>T increased from 24% to 55% with heterozygous and homozygous genotypes (Table 3). The relative high frequency of the risk (C) genotype at –487T>C (rs2672598) in the study samples (AMD: 84.7%; controls: 53.0%) was observed, suggesting an overestimation of the corresponding attributable risk, which does not provide a good estimate of the risk in the population. The ORs of rs2672598 were also inflated, with ORhet being 6.35 (95% CI: 0.80–50.18) and ORhom being 25.61 (95% CI: 3.36–195.05).
Table 3.
Estimated ORs, Ps, PARs, and Corresponding 95% CI of HTRA1 Gene Variants
| Gene Variants | Dominant (RN+RR vs. NN)
|
Recessive (RR vs. RN+NN)
|
Heterozygote (RN vs. NN)
|
Homozygote (RR vs. NN)
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ORdom (95% CI) | P | PAR% | ORree (95% CI) | P | PAR% | ORbet (95% CI) | P | PAR% | ORhom (95% CI) | P | PAR% | |
| –625G>A | ||||||||||||
| rs11200638 | 3.46 (1.93–6.20) | 1.51 × 10−5 | 47.7 | 5.20 (3.24–8.35) | 1.74 × 10−12 | 31.6 | 1.73 (0.92–3.26) | 0.088 | 23.5 | 7.56 (3.94–14.51) | 1.44 × 10−10 | 54.9 |
| –487T>C | ||||||||||||
| rs2672598 | 17.67 (2.33–133.93) | 7.75 × 10−5 | 88.8 | 4.89 (2.92–8.20) | 3.03 × 10−10 | 52.2 | 6.35 (0.80–50.18) | 0.068 | 76.6 | 25.61 (3.36–195.05) | 3.38 × 10−6 | 90.4 |
| 102C>T | ||||||||||||
| rs1049331 | 3.46 (1.93–6.20) | 1.52 × 10−5 | 47.7 | 5.07 (3.16–8.14) | 3.73 × 10−12 | 30.9 | 1.76 (0.94–3.32) | 0.076 | 24.3 | 7.48 (3.89–14.36) | 1.89 × 10−10 | 54.7 |
| 108G>T | ||||||||||||
| rs2293870 | 3.46 (1.93–6.20) | 1.52 × 10−5 | 47.7 | 5.07 (3.16–8.14) | 3.73 × 10−12 | 30.9 | 1.76 (0.94–3.32) | 0.076 | 24.3 | 7.48 (3.89–14.36) | 1.89 × 10−10 | 54.7 |
Genotypes are ordered NN, NR, RR, where N is the normal allele, and R is the risk allele. The risk allele is defined as the least frequent allele in control subjects. NN, homozygous normal alleles; RR, homozygous risk alleles; RN, heterozygous genotype.
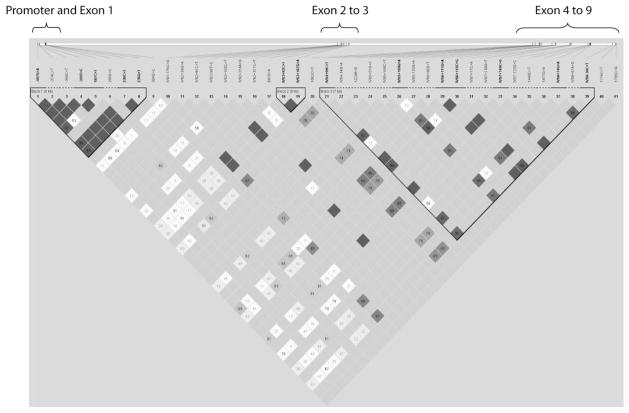
LD analysis revealed three regions of LD in HTRA1 (Fig. 2). Only the first haplotype block that lies in the promoter and exon 1 significantly predisposed to AMD. There are five SNPs within this block: –625G>A (rs11200638), –487T>C (rs2672598), 59C>T (Ala20Val), 102C>T (rs1049331; Ala34Ala), and 108G>T (rs2293870; Gly36Gly) (Table 4). The haplotypes ACCTT and GTCCG significantly predisposed to AMD, with P = 6.68 × 10−14 and 4.97 × 10−12, respectively. After permutation, these two haplotypes remained exceptionally significant, further supporting that this region is associated with exudative AMD as in single SNP analysis. Haplotype block 2 contains only two SNPs in intron 3. Haplotype block 3 spans introns 4 to 8. Both haplotype blocks 2 and 3 were not associated with AMD risk.
Figure 2.
Haploview plot depicting the haplotype block structure of the exudative AMD-associated region. Top diagram: the relative chromosomal position of each SNP.
Table 4.
Haplotype Analysis for the Association of HTRA1 with Risk of Exudative AMD
| Haplotype Block | Haplotype Frequency | AMD:Control Ratios | χ2 | P | Pcorr | OR |
|---|---|---|---|---|---|---|
| Block 1: –625G>A, –487T>C, 59C>T, 102C>T, 108G>T | ||||||
| ACCTT* | 0.584 | 240:88, 165:201 | 56.16 | 6.68 × 10−14* | <0.000001* | 3.32 (2.41–4.57) |
| GTCCG* | 0.187 | 16:302, 104:262 | 47.699 | 4.97 × 10−12* | <0.000001* | 0.13 (0.077–0.23) |
| GCTCG | 0.146 | 41:287, 60:306 | 2.109 | 0.15 | 0.94 | |
| GCCCG | 0.079 | 20:308, 35:331 | 2.847 | 0.09 | 0.74 | |
| Block 2: IVS3+93C>T, IVS3+167G>A | ||||||
| CG | 0.647 | 219:109, 230:136 | 1.168 | 0.28 | 0.99 | |
| TA | 0.353 | 109:219, 136:230 | 1.168 | 0.28 | 0.99 | |
| Block 3: IVS4+99C>T, IVS5+169G>A, IVS6+111G>A, IVS6+115C>G, IVS7 +149C>G, IVS8+14G>A, IVS8–36C>T | ||||||
| TAGGCGC | 0.409 | 142.9:185.1, 140.9:225.1 | 1.839 | 0.1751 | 0.9598 | |
| CGGCCAT | 0.240 | 76.9:251.1, 89.9:276.1 | 0.119 | 0.7296 | 1.0000 | |
| CGGCCAC | 0.184 | 56.4:271.6, 71.1:294.9 | 0.582 | 0.4457 | 1.0000 | |
| CGACGAT | 0.132 | 36.8:291.2, 55:311 | 2.198 | 0.1382 | 0.9297 | |
These two genotypes of haplotype block 1 were significantly associated with risk of AMD. Pcorr is the corrected P after permutation.
Table 5 summarizes the best-fit and most parsimonious model analyzed by logistic regression, which was used to explore the joint contributions of HTRA1 and cigarette smoking and of HTRA1 and CFH. The results suggest that the joint effects of smoking and rs11200638 of HTRA1 were best described as independent multiplicative effects, without significant dominance or interacting effects. An independent multiplicative effect is the additive effect on the log scale. This model yielded the lowest AIC (367.5).
Table 5.
Results of Fitting Two-Factor Models by Logistic Regression
| Two-Factor Model | AIC | AIC Difference |
|---|---|---|
| HTRA1:rs11200638 (factor 1) and smoking (ever vs. never) | ||
| Single | ||
| ADD1† | 369.9 | 2.4‡ |
| SMOKE | 408.2 | 40.7 |
| DOM1 | 405.9 | 38.4 |
| Joint | ||
| ADD1-SMOKE† | 367.5 | 0† |
| DOM1-SMOKE | 402.7 | 35.2 |
| Interaction | ||
| ADD* SMOKE-INT | 400.1 | 32.6 |
| DOM* SMOKE-INT | 413.8 | 46.3 |
| ADD-DOM-SMOKE-INT | 402.1 | 34.6 |
| HTRA1:rs11200638 (factor 1) and CFH:rs800292 (factor 2) | ||
| Single | ||
| ADD1 | 432.2 | 13.2 |
| ADD2 | 466.7 | 47.7 |
| DOM1 | 471.0 | 52.0 |
| DOM2 | 476.1 | 57.1 |
| Joint | ||
| ADD1* ADD2† | 419.0 | 0† |
| DOM1* DOM2 | 465.5 | 46.5 |
| Interaction | ||
| ADD1* ADD2-INT | 461.5 | 42.5 |
| ADD* DOM-INT | 448.9 | 29.9 |
| ALL-INT | 448.7 | 29.7 |
ADD, additive model; DOM, dominance model; INT, interaction model; AIC: Akaike information criterion (the AIC difference is the difference from the AIC of the best fitting model).
Interaction.
The most parsimonious and best-fit model in both interaction analyses. Model with best fit (lowest AIC) has an AIC difference of 0.
The second best-fit model, giving an AIC difference of 2.4. This model is based on the multiplicative effect of HTRA1 only.
Given the evidence of independent multiplicative joint effects of smoking and HTRA1, the combined effect of smoking and HTRA1-rs11200638 was further illustrated by the joint ORs shown in Table 6a, b. The risk of AMD due to the genotype was increased with smoking. Estimates from this model demonstrated a 15.71-fold increased risk of exudative AMD in homozygous carriers of the HTRA1 risk allele who had ever smoked compared with no risk alleles at HTRA1-rs11200638 in those who had never smoked. The heterozygous ever-smoker who carried the HTRA1 risk allele was 4.80 times more likely to have exudative AMD. The adjusted OR of a smoker with the nonrisk allele was calculated to be 3.67. This indicated approximately a fivefold increased risk of AMD in smokers with the homozygous risk allele than in smokers with no risk allele (Table 6b). Corresponding ORs for smoker and nonsmoker homozygous risk allele carriers of HTRA1-rs11200638 were similar (15.71 and 14.33, respectively). This indicated that the risk allele HTRA1-rs11200638 conferred a stronger risk of AMD than did smoking.
Table 6.
Interaction Analysis between HTRA1-rs11200638 and Smoking in Exudative AMD
| a. Genotype Distribution | ||||
|---|---|---|---|---|
| HTRA1-rs11200638 | Smoking Status
|
|||
| Controls (n = 143)
|
Exudative AMD (n = 153)
|
|||
| Never | Ever | Never | Ever | |
| GG | 30 (2.1%) | 15 (10.5%) | 6 (3.9%) | 11 (7.2%) |
| GA | 44 (30.8%) | 25 (17.5%) | 25 (16.3%) | 24 (15.7%) |
| AA | 15 (10.5%) | 14 (9.8%) | 43 (28.1%) | 44 (28.8%) |
| b. Joint Odds Ratios and 95% CI | |||
|---|---|---|---|
| HTRA1-rs11200638 | Smoking Status
|
||
| Never | Ever | ||
| Main effects | |||
| ORsmoke | 1.00 (Ref) | 1.76 (1.11–2.80) | |
| PARsmoke | 12.2 | ||
| PARHTRA1 | 53.1 | ||
| PARjoint | 68.7 | ||
|
| |||
|
ORHTRA1
|
Joint Effects
|
||
| GG | 1.00 (Ref) | 1.00 (Ref) | 3.67 (1.14–11.84) |
| GA | 1.88 (0.96–3.66) | 2.84 (1.04–7.76) | 4.80 (1.70–13.58) |
| AA | 7.94 (3.95–15.97) | 14.33 (4.99–41.18) | 15.71 (5.43–45.49) |
Data in a are number (%). Data in b are the OR (95% CI). ncontrols = 143; nAMD = 153. GG, homozygous GG alleles; GA, heterozygous GA alleles; AA, homozygous AA alleles.
The PAR was calculated to be 12.2% for smoking, 53.1% for HTRA1, and 68.7% for joint smoking and HTRA1 effect (Table 6b). There was an observable increase in PAR (~5.5-fold increase) of the joint effect of smoking and the HTRA1 risk allele, indicating that smokers with the homozygous risk allele had a substantially higher likelihood of the development of exudative AMD. Gene–gene interaction analysis on different models of the HTRA1 and CFH variants showed no evidence of interaction that enhanced the risk of exudative AMD. These variants in the two genes contributed independently to disease risk. The best fit and most parsimonious model for the joint effect was best described by an independent multiplicative model (independent log-additive effect) with no dominance or interactive effects, yielding the lowest AIC of 419.0 (Table 5).
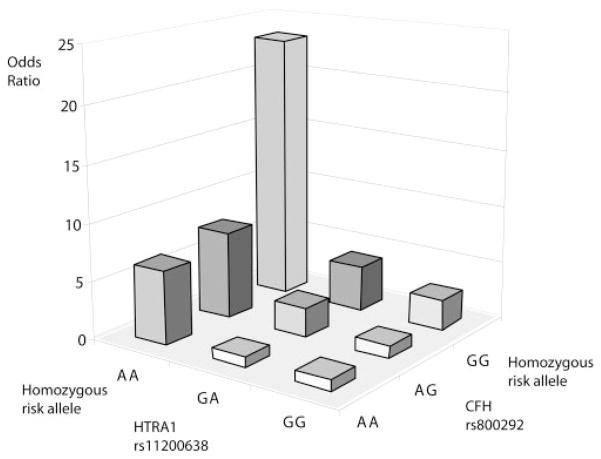
Based on the independent multiplicative effects of the HTRA1 and CFH variants as observed in the model comparison, corresponding ORs of AMD for each possible combination of the genotypes from the two variants were estimated (Table 7a, b). The risk allele of HTRA1-rs11200638 was the A allele and of CFH-rs800292 was the G allele. The ORs were compared with the baseline genotype of the two genes that showed the lowest frequency of the disease risk alleles (homozygous G carrier of HTRA1 with homozygous A carrier of CFH). The frequency of the homozygous risk genotype at both loci was five times lower in the controls (6.56%) than in the cases (34.36%; Table 7a). A joint disease OR of 23.3 in individuals with homozygous risk alleles at both loci was observed when compared with the baseline wild-type (nonrisk) genotype (Table 7), with a wide range of calculated 95% CI from 2.5 to 218.2. Individuals homozygous for the HTRA1 risk (A) allele and any genotype for the CFH risk allele (for the AA-GA joint genotype, OR 7.63; AA-AA joint genotype, OR 6.43) were at higher risk of AMD than those who were homozygous for the CFH risk allele and any genotype for the HTRA1 risk allele (for the GA-GG joint genotype, OR = 3.95; and GG-GG joint genotype, OR = 2.63; Table 7b). The homozygous risk genotype of HTRA1 exerted a twofold higher risk than CFH in the joint multiplicative model of risk for exudative AMD. The HTRA1 risk allele revealed a stronger impact on the development of exudative AMD than did the CFH risk allele. A plot of the two-locus genotype-specific AMD risks further illustrates the stronger impact of HTRA1 on AMD (Fig. 3). PAR was estimated to be 41.2% for CFH-rs800292, 54.7% for HTRA1-rs11200638, and 78.4% for the joint effect (Table 7b).
Table 7.
Interaction Analysis between HTRA1-rs11200638 and CFH-rs800292 in Exudative AMD
| a. Genotype Distribution | ||||||
|---|---|---|---|---|---|---|
| Genotype at HTRA1-rs11200638 | Genotype at CFH-rs800292
|
|||||
| Controls (n = 183)
|
AMD (n = 163)
|
|||||
| AA | AG | GG | AA | AG | GG | |
| GG | 5 (2.73%) | 31 (16.9%) | 19 (10.38%) | 1 (0.61%) | 7 (4.29%) | 10 (6.13%) |
| GA | 17 (9.29%) | 35 (19.13%) | 38 (20.77%) | 3 (1.84%) | 18 (11.04%) | 30 (18.40%) |
| AA | 7 (3.83%) | 19 (10.38%) | 12 (6.56%) | 9 (5.52%) | 29 (17.79%) | 56 (34.36%) |
| b. Joint Odds Ratios and 95% Confidence | ||||
|---|---|---|---|---|
| HTRA1-rs11200638 |
CFH-rs800292
|
|||
| AA | AG | GG | ||
| Main effects | ||||
| ORCFH | 1.0 (Ref) | 1.09 (0.60–1.97) | 2.14 (1.18–3.86) | |
| PARCFH | 41.2 | |||
| PARHTRA1 | 54.7 | |||
| PARJoint | 78.4 | |||
|
| ||||
| ORHTRA1 |
Joint Effects
|
|||
| AA | AG | GG | ||
|
| ||||
| GG | 1.00 (Ref) | 1.00 (Ref) | 1.13 (0.11–11.24) | 2.63 (0.27–25.71) |
| GA | 1.88 (0.96–3.66) | 0.88 (0.074–10.46) | 2.57 (0.28–23.70) | 3.95 (0.44–35.62) |
| AA | 7.94 (3.95–15.97) | 6.43 (0.61–68.31) | 7.63 (0.83–70.52) | 23.3 (2.5–218.2) |
Data in a are number (%). Data in b are the OR (95% CI). ncontrols = 183; nAMD = 163. GG, homozygous GG alleles; GA, heterozygous GA alleles; AA, homozygous AA alleles.
Figure 3.
Two-locus (HTRA1 and CFH) genotype-specific AMD risk.
Discussion
In this study of the HTRA1 sequence, two variants in the promoter and two variants in exon 1 were identified to be associated with exudative AMD. Carriers of the risk genotype (AA) of the most significant SNP, rs11200638 at the HTRA1 promoter increased the risk of exudative AMD with an OR of 7.6. A similar association has recently been reported in the Japanese population (OR = 10.1).38 A consistently significant association of rs11200638 with the risk of the two advanced forms of AMD, geographic atrophy (GA) and exudative, was also identified in Caucasians.39 The HTRA1-rs11200638 risk allele is common in the Chinese population, with 21% of control subjects having the homozygous risk allele (Table 2). According to the high PAR, individuals carrying two copies of the HTRA1 risk allele are at high risk of the development of exudative AMD at a later age, even though they may be asymptomatic at the time of genetic testing. Hence, they require close monitoring of any signs of AMD for early detection and prompt treatment, which may help to delay presymptomatic progression of the disease. rs11200638 is located 625 bp upstream of the HTRA1 transcription start site and resides within the putative binding sites for the transcription factors adaptor-related protein complex 2α (AP2α) and serum response factor (SRF).33 The A allele is expected to disturb the expression of HTRA1 by altering the binding affinity of AP2α and SRF. This genotype-driven expression pattern was demonstrated by an increased trend of luciferase expression in HTRA1 promoter–AA constructs in transfected RPE and HeLa S3 cells.33 Another immunohistochemical study of the drusen and along Bruch’s membrane in patients with AMD showed an elevated expression of HTRA1 mRNA and protein associated with AMD patients carrying the AA risk genotype.34,39 These data support our finding that the AA carrier of rs11200638 in the HTRA1 promoter had a risk of exudative AMD caused by upregulation of the expression of HTRA1. However, the role of genotype-driven, increased expression of HTRA1 leading to the development of exudative AMD remains unclear. A recent paper reported the mitochondrial protein LOC387715/ARMS2, rather than HTRA1, to be strongly associated with AMD.40 Their data gave evidence that SNP rs10490924 alone accounts for the AMD susceptibility associated with the AMD locus at chromosome 10q26. Since the phenotype of their patients with AMD was not specified, whether rs10490924 also has prominent association with specific subtypes of AMD warrants further investigation. The LOC387715/ARMS2 genotypes of their patients were unknown, but it would be interesting to examine the genotypes’ interactions with HTRA1 variants. In a previous study on a Chinese cohort with exudative AMD, rs10490924 was slightly more associated with AMD than HTRA1-rs11200638, and they were in the same LD block.33 Although the biological role of the LOC387715/ARMS2 gene in humans is unclear, all these studies further show the complexity of the pathogenesis of AMD. It is possible that rs10490924 and rs11200638 contribute to AMD, or different subtypes of AMD, through diverse mechanisms, the former due to disruption of mitochondrial oxidation reduction40 and the latter to proteolytic effects.34
The frequencies of risk allele A of HTRA1-rs11200638 associated with AMD in smokers (36.6%) and nonsmokers (36.2%) were very similar, whereas that in control smokers (18.5%) and nonsmokers (25.9%) was two times lower (Table 6a). Smoking appeared to be independent of the rs11200638 variant in HTRA1 with no significant interaction. Numerous studies had reported smoking to be associated with the risk of advanced AMD independently, with no interaction with the other susceptibility genes studied: LOC387715, APOE, and CFH.41–44 Only one recent study reported smoking as a modifier of LOC387715.45
In this study, the large increase in PAR estimates in smoking (12%) and HTRA1 (53%) to the joint effect of 68.7% (Table 6b) indicates that a smoker carrying the risk allele is at high risk of AMD. A high prevalence of the risk allele A at HTRA1-rs11200638 was found in patients with exudative AMD (MAF [minor allele frequency] = 73%) and control subjects (MAF = 45%). This reinforces and motivates the necessity of modifying our lifestyle by quitting smoking and having regular eye examinations, especially for those high-risk control subjects who are homozygous carriers of the variant and are at higher risk of the later development of AMD.
The risk of exudative AMD conferred by CFH-rs800292 (OR = 2.14; 95% CI 1.18–3.86) was similar to the risk of smoking (OR = 1.76; 95% CI 1.11–2.80), whereas the risk of exudative AMD associated with HTRA1-rs11200638 was substantially higher (OR = 7.94; 95% CI 3.95–15.97; as shown in Table 7b). These data show a stronger influence of HTRA1 on the development of exudative AMD than that of smoking and CFH. The joint risk for exudative AMD of CFH and HTRA1 was 23.33 (95% CI 2.49–218.24 (Table 7). CFH and HTRA1 appear to be involved in different pathways in the development of exudative AMD. A recent study reported immunohistochemical detection of HtrA1 in drusen of patients with GA.39 Evidence has shown that Y402H on CFH is associated with the risk of both GA and exudative AMD. These genotype data, together with the observed expression of HTRA1 in drusen of patients with GA, suggest that CFH and HTRA1 contribute to the development and progression of advanced AMD.
The high joint prevalence of the CFH-rs800292 and HTRA1-rs11200638 variants (prevalence of carriers homozygous for either or both variants = 78.4%; Table 7b) and their strong association with exudative AMD (joint OR = 23.3) raises the possibility of using a population genetic test for the joint genotype to predict risk for exudative AMD. The potential benefits of identifying high-risk presymptomatic subjects would allow insight into the earliest manifestations of the disease and allow trials of preventive treatment in high-risk groups. This insight would help in early detection of manifestations of the disease and immediate treatment to delay disease progression.
Acknowledgments
The authors thank all the participants in the study and Chan Kwok Ping for help in the management of the subjects’ samples.
Supported by The Chinese University of Hong Kong and the Lim Por Yen Eye Foundation Endowment Fund.
Footnotes
Disclosure: P.O.S. Tam, None; T.K. Ng, None; D.T.L. Liu, None; W.M. Chan, None; S.W.Y. Chiang, None; L.J. Chen, None; A. DeWan, None; J. Hoh, None; D.S.C. Lam, None; C.P. Pang, None
References
- 1.Friedman DS, O’Colmain BJ, Muñoz B, et al. Prevalence of age-related macular degeneration in the United States. Arch Ophthalmol. 2004;122:564–572. doi: 10.1001/archopht.122.4.564. [DOI] [PubMed] [Google Scholar]
- 2.la Cour M, Kiilgaard JF, Nissen MH. Age-related macular degeneration: epidemiology and optimal treatment. Drugs Aging. 2002;19:101–133. doi: 10.2165/00002512-200219020-00003. [DOI] [PubMed] [Google Scholar]
- 3.Seddon JM, Willett WC, Speizer FE, Hankinson SE. A prospective study of cigarette smoking and age-related macular degeneration in women. JAMA. 1996;276:1141–1146. [PubMed] [Google Scholar]
- 4.Vingerling JR, Hofman A, Grobbee DE, de Jong PT. Age-related macular degeneration and smoking. The Rotterdam Study. Arch Ophthalmol. 1996;114:1193–1196. doi: 10.1001/archopht.1996.01100140393005. [DOI] [PubMed] [Google Scholar]
- 5.DeAngelis MM, Ji F, Kim IK, et al. Cigarette smoking, CFH, APOE, ELOVL4, and risk of neovascular age-related macular degeneration. Arch Ophthalmol. 2007;125:49–54. doi: 10.1001/archopht.125.1.49. [DOI] [PubMed] [Google Scholar]
- 6.Oneill C, Jamison J, McCulloch D, Smith D. Age-related macular degeneration: cost-of-illness issues. Drugs Aging. 2001;18:233–241. doi: 10.2165/00002512-200118040-00001. [DOI] [PubMed] [Google Scholar]
- 7.Fraser-Bell S, Wu J, Klein R, Azen SP, Varma R. Smoking, alcohol intake, estrogen use, and age-related macular degeneration in Latinos: the Los Angeles Latino Eye Study. Am J Ophthalmol. 2006;141:79–87. doi: 10.1016/j.ajo.2005.08.024. [DOI] [PubMed] [Google Scholar]
- 8.Conley YP, Jakobsdottir J, Mah T, et al. CFH, ELOVL4, PLEKHA1 and LOC387715 genes and susceptibility to age-related maculopathy: AREDS and CHS cohorts and meta-analyses. Hum Mol Genet. 2006;15:3206–3218. doi: 10.1093/hmg/ddl396. [DOI] [PubMed] [Google Scholar]
- 9.Smith W, Assink J, Klein R, et al. Risk factors for age-related macular degeneration: pooled findings from three continents. Ophthalmology. 2001;108:697–704. doi: 10.1016/s0161-6420(00)00580-7. [DOI] [PubMed] [Google Scholar]
- 10.Mitchell P, Chapman S, Smith W. Smoking is a major cause of blindness. Med J Aust. 1999;16:173–174. doi: 10.5694/j.1326-5377.1999.tb123591.x. [DOI] [PubMed] [Google Scholar]
- 11.Mousa SA, Lorelli W, Campochiaro PA. Role of hypoxia and extra-cellular matrix-integrin binding in the modulation of angiogenic growth factors secretion by retinal pigmented epithelial cells. J Cell Biochem. 1999;74:135–143. [PubMed] [Google Scholar]
- 12.Seddon JM, Ajani UA, Mitchell BD. Familial aggregation of age-related maculopathy. Am J Ophthalmol. 1997;123:199–206. doi: 10.1016/s0002-9394(14)71036-0. [DOI] [PubMed] [Google Scholar]
- 13.Klaver CC, Wolfs RC, Assink JJ, van Duijn CM, Hofman A, de Jong PT. Genetic risk of age-related maculopathy: population-based familial aggregation study. Arch Ophthalmol. 1998;116:1646–1651. doi: 10.1001/archopht.116.12.1646. [DOI] [PubMed] [Google Scholar]
- 14.Hammond CJ, Webster AR, Snieder H, Bird AC, Gilbert CE, Spector TD. Genetic influence on early age-related maculopathy: a twin study. Ophthalmology. 2002;109:730–736. doi: 10.1016/s0161-6420(01)01049-1. [DOI] [PubMed] [Google Scholar]
- 15.Meyers SM, Greene T, Gutman FA. A twin study of age-related macular degeneration. Am J Ophthalmol. 1995;120:757–766. doi: 10.1016/s0002-9394(14)72729-1. [DOI] [PubMed] [Google Scholar]
- 16.Seddon JM, Cote J, Page WF, Aggen SH, Neale MC. The US twin study of age-related macular degeneration: relative roles of genetic and environmental influences. Arch Ophthalmol. 2005;123:321–327. doi: 10.1001/archopht.123.3.321. [DOI] [PubMed] [Google Scholar]
- 17.Weeks DE, Conley YP, Tsai HJ, et al. Age-related maculopathy: an expanded genome-wide scan with evidence of susceptibility loci within the 1q31 and 17q25 regions. Am J Ophthalmol. 2001;132:682–692. doi: 10.1016/s0002-9394(01)01214-4. [DOI] [PubMed] [Google Scholar]
- 18.Weeks DE, Conley YP, Tsai HJ, et al. Age-related maculopathy: a genomewide scan with continued evidence of susceptibility loci within the 1q31, 10q26, and 17q25 regions. Am J Hum Genet. 2004;75:174–189. doi: 10.1086/422476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Abecasis GR, Yashar BM, Zhao Y, et al. Age-related macular degeneration: a high-resolution genome scan for susceptibility loci in a population enriched for late-stage disease. Am J Hum Genet. 2004;74:482–494. doi: 10.1086/382786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schmidt S, Scott WK, Postel EA, et al. Ordered subset linkage analysis supports a susceptibility locus for age-related macular degeneration on chromosome 16p12. BMC Genet. 2004;5:18. doi: 10.1186/1471-2156-5-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Iyengar SK, Song D, Klein BE, et al. Dissection of genomewide-scan data in extended families reveals a major locus and oligogenic susceptibility for age-related macular degeneration. Am J Hum Genet. 2004;74:20–39. doi: 10.1086/380912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kenealy SJ, Schmidt S, Agarwal A, et al. Linkage analysis for age-related macular degeneration supports a gene on chromosome 10q26. Mol Vis. 2004;10:57–61. [PubMed] [Google Scholar]
- 23.Majewski J, Schultz DW, Weleber RG, et al. Age-related macular degeneration: a genome scan in extended families. Am J Hum Genet. 2003;73:540–550. doi: 10.1086/377701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schick JH, Iyengar SK, Klein BE, et al. A whole-genome screen of a quantitative trait of age-related maculopathy in sibships from the Beaver Dam Eye Study. Am J Hum Genet. 2003;72:1412–1424. doi: 10.1086/375500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Seddon JM, Santangelo SL, Book K, Chong S, Cote J. A genome-wide scan for age-related macular degeneration provides evidence for linkage to several chromosomal regions. Am J Hum Genet. 2003;73:780–790. doi: 10.1086/378505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fisher SA, Abecasis GR, Yashar BM, et al. Meta-analysis of genome scans of age-related macular degeneration. Hum Mol Genet. 2005;14:2257–2264. doi: 10.1093/hmg/ddi230. [DOI] [PubMed] [Google Scholar]
- 27.Fuse N, Miyazawa A, Mengkegale M, et al. Polymorphisms in Complement Factor H and hemicentin-1 genes in a Japanese population with dry-type age-related macular degeneration. Am J Ophthalmol. 2006;142:1074–1076. doi: 10.1016/j.ajo.2006.07.030. [DOI] [PubMed] [Google Scholar]
- 28.Gotoh N, Yamada R, Hiratani H, et al. No association between complement factor H gene polymorphism and exudative age-related macular degeneration in Japanese. Hum Genet. 2006;120:139–143. doi: 10.1007/s00439-006-0187-0. [DOI] [PubMed] [Google Scholar]
- 29.Grassi MA, Fingert JH, Scheetz TE, et al. Ethnic variation in AMD-associated complement factor H polymorphism p.Tyr402His. Hum Mutat. 2006;27:921–925. doi: 10.1002/humu.20359. [DOI] [PubMed] [Google Scholar]
- 30.Uka J, Tamura H, Kobayashi T, et al. No association of complement factor H gene polymorphism and age-related macular degeneration in the Japanese population. Retina. 2006;26:985–987. doi: 10.1097/01.iae.0000244068.18520.3e. [DOI] [PubMed] [Google Scholar]
- 31.Okamoto H, Umeda S, Obazawa M, et al. Complement factor H polymorphisms in Japanese population with age-related macular degeneration. Mol Vis. 2006;12:156–158. [PubMed] [Google Scholar]
- 32.Chen LJ, Liu DT, Tam PO, et al. Association of complement factor H polymorphisms with exudative age-related macular degeneration. Mol Vis. 2006;12:1536–1542. [PubMed] [Google Scholar]
- 33.Dewan A, Liu M, Hartman S, et al. HTRA1 promoter polymorphism in wet age-related macular degeneration. Science. 2006;314:989–992. doi: 10.1126/science.1133807. [DOI] [PubMed] [Google Scholar]
- 34.Yang Z, Camp NJ, Sun H, et al. A variant of the HTRA1 gene increases susceptibility to age-related macular degeneration. Science. 2006;314:992–993. doi: 10.1126/science.1133811. [DOI] [PubMed] [Google Scholar]
- 35.Bird AC, Bressler NM, Bressler SB, et al. An international classification and grading system for age-related maculopathy and age-related macular degeneration: The International ARM Epidemiological Study Group. Surv Ophthalmol. 1995;39:367–374. doi: 10.1016/s0039-6257(05)80092-x. [DOI] [PubMed] [Google Scholar]
- 36.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 37.North BV, Curtis D, Sham PC. Application of logistic regression to case-control association studies involving two causative loci. Hum Hered. 2005;59:79–87. doi: 10.1159/000085222. [DOI] [PubMed] [Google Scholar]
- 38.Yoshida T, DeWan A, Zhang H, et al. HTRA1 promoter polymorphism predisposes Japanese to age-related macular degeneration. Mol Vis. 2007;13:545–548. [PMC free article] [PubMed] [Google Scholar]
- 39.Cameron DJ, Yang Z, Gibbs D, et al. HTRA1 variant confers similar risks to geographic atrophy and neovascular age-related macular degeneration. Cell Cycle. 2007;6:1122–1125. doi: 10.4161/cc.6.9.4157. [DOI] [PubMed] [Google Scholar]
- 40.Kanda A, Chen W, Othman M, et al. A variant of mitochondrial protein LOC387715/ARMS2, not HTRA1, is strongly associated with age-related macular degeneration. Proc Natl Acad Sci USA. 2007;104:16227–16232. doi: 10.1073/pnas.0703933104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Francis PJ, George S, Schultz DW, et al. The LOC387715 gene, smoking, body mass index, environmental associations with advanced age-related macular degeneration. Hum Hered. 2007;63:212–218. doi: 10.1159/000100046. [DOI] [PubMed] [Google Scholar]
- 42.Scott WK, Schmidt S, Hauser MA, et al. Independent effects of complement factor H Y402H polymorphism and cigarette smoking on risk of age-related macular degeneration. Ophthalmology. 2007;114:1151–1156. doi: 10.1016/j.ophtha.2006.08.054. [DOI] [PubMed] [Google Scholar]
- 43.Seddon JM, George S, Rosner B, Klein ML. CFH gene variant, Y402H, and smoking, body mass index, environmental associations with advanced age-related macular degeneration. Hum Hered. 2006;61:157–165. doi: 10.1159/000094141. [DOI] [PubMed] [Google Scholar]
- 44.Schmidt S, Haines JL, Postel EA, et al. Joint effects of smoking history and APOE genotypes in age-related macular degeneration. Mol Vis. 2005;11:941–949. [PubMed] [Google Scholar]
- 45.Schmidt S, Hauser MA, Scott WK, et al. Cigarette smoking strongly modifies the association of LOC387715 and age-related macular degeneration. Am J Hum Genet. 2006;78:852–864. doi: 10.1086/503822. [DOI] [PMC free article] [PubMed] [Google Scholar]