Abstract
We studied the telithromycin, erythromycin, azithromycin, and clindamycin susceptibilities of serotype III macrolide-resistant group B streptococci, together with genetic mechanisms of resistance and genomic diversity. ermB, ermA, and mefA were found in, respectively, 57, 32, and 9% of isolates. The telithromycin MIC at which 90% of isolates were inhibited was 0.5 μg/ml. Macrolide resistance was associated with dissemination of resistance determinants among isolates of different genetic backgrounds.
Group B Streptococcus (GBS) infection remains a leading cause of neonatal morbidity and mortality, despite major advances in perinatal GBS disease prevention in the 1990s. Serotype III GBS is the most common cause of invasive neonatal infection (9, 18). Intrapartum antibiotic prophylaxis can prevent early-onset GBS infection (26). Penicillin is the drug of choice, but about 10% of pregnant women in the United States are allergic to this agent (24). Erythromycin and clindamycin are recommended as alternatives to penicillin in this setting (25). Widespread implementation of prevention guidelines has increased the use of antimicrobials during labor and has contributed to the emergence of resistant GBS (22). Increasing macrolide resistance among GBS isolates has raised concerns about the use of these antimicrobials in the prophylaxis of early-onset GBS infection. GBS strains expressing the serotype III capsular polysaccharide have been found to have higher rates of erythromycin resistance (15, 21). Telithromycin is a semisynthetic erythromycin A derivative with enhanced activity against macrolide-resistant streptococci (7), but GBS susceptibility to this drug has rarely been studied (3). The aim of this study was to determine the telithromycin susceptibility of macrolide-resistant serotype III GBS clinical strains recently isolated in France and to examine the genetic mechanisms of resistance. We also investigated whether erythromycin resistance among GBS isolates was due to clonal spread of resistant strains.
In 2001 and 2002, 88 unrelated erythromycin-resistant serotype III GBS strains were identified among 430 consecutive isolates obtained from different patients in the Paris (France) area. The isolates were recovered from genital specimens of pregnant women (n = 47), cultures of blood (n = 2) or cerebrospinal fluid (n = 5) from neonates with invasive infections, or gastric fluid or ear specimens of colonized or infected newborns (n = 34). Beta-hemolytic colonies and suspected nonhemolytic colonies were identified as GBS by using a commercial agglutination technique (Murex Diagnostics, Dartford, United Kingdom). Erythromycin-resistant GBS isolates were identified as previously described (14). The MICs of erythromycin, azithromycin, clindamycin, and telithromycin for all isolates were determined by the agar dilution method in Mueller-Hinton medium supplemented with 5% defibrinated sheep blood (10, 23). The plates were incubated overnight at 35°C in air. Pulsed-field gel electrophoresis (PFGE) was performed using the SmaI restriction enzyme as previously described (17). Cluster analysis (unweighted pair group method with arithmetic mean) with whole-band analyzer software (Biogene, Vilber-Lourmat, Marne la Vallée, France) was used to calculate similarity or dissimilarity among GBS isolates. Clonally related PFGE patterns were defined by a similarity coefficient higher than 80% (usually corresponding to a difference of no more than four bands in our study). All erythromycin-resistant isolates were screened for erythromycin resistance genes. The mefA, ermB, and ermA genes were detected by multiplex PCR amplification with previously described primers (4, 15, 27, 28). Streptococcus agalactiae BM 132, S. agalactiae SBI, and Streptococcus pyogenes O2 C1110 were used as positive PCR controls for the ermB, mefA, and ermA genes, respectively (2, 4, 8). Five erythromycin-susceptible GBS isolates were used as negative controls. The positive controls yielded PCR products of the expected sizes (616, 348, and 206 bp for ermB, ermA, and mefA, respectively).
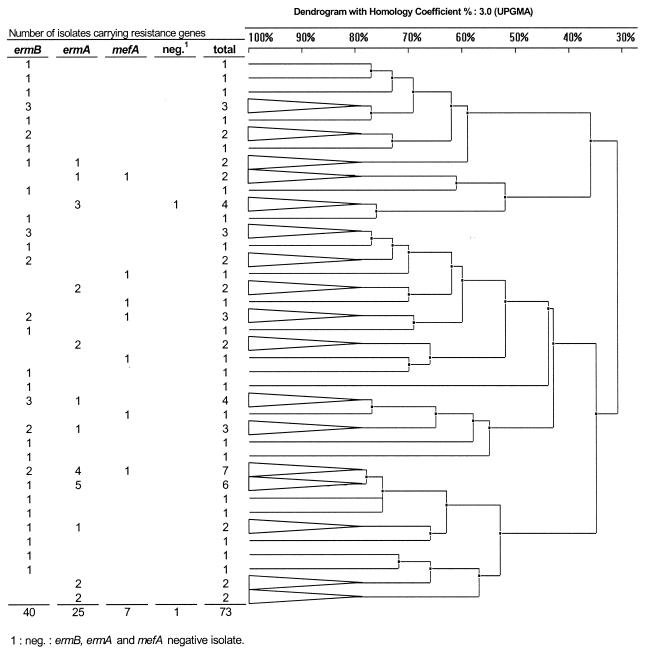
MIC ranges and MICs at which 50% of the isolates were inhibited (MIC50s) and MIC90s are shown in Table 1. The ermB, ermA, and mefA genes were found in, respectively, 57, 32, and 9% of the isolates. Multiplex PCR amplification was unsuccessful with two isolates. For these two isolates, amplification of a housekeeping gene (mreA) (15) was positive, indicating that the failure of our multiplex PCR was not due to a PCR-inhibitory preparation. We did not examine 23S rRNA mutations or ribosomal protein mutations. Table 1 shows MICs according to the erythromycin resistance genotype. Fifteen isolates were repeatedly nontypeable by PFGE because of incomplete total-DNA digestion by SmaI. The remaining 73 isolates displayed extensive genetic diversity. The dendrogram calculated from PFGE patterns identified 39 different clonal lineages (≥80% similarity) (Fig. 1). The 40 ermB isolates gave 37 patterns, the 25 ermA isolates gave 12 patterns, and the 7 mefA isolates gave 7 patterns (Fig. 1). Multiple resistance types were found within some clonal groups (Fig. 1).
TABLE 1.
MICs of macrolides and related agents for 88 erythromycin-resistant GBS isolates according to known mechanisms of resistance
| Group (n) and antimicrobial agent | MICa (μg/ml)
|
||
|---|---|---|---|
| 50% | 90% | Range | |
| All isolates (88)b | |||
| Erythromycin | 4 | >128 | 0.5->128 |
| Azithromycin | 16 | >128 | 1->128 |
| Clindamycin | 64 | >128 | 0.064->128 |
| Telithromycin | 0.125 | 0.5 | 0.032-2 |
| ermB (50) | |||
| Erythromycin | 128 | >128 | 0.5->128 |
| Azithromycin | >128 | >128 | 2->128 |
| Clindamycin | 64 | >128 | 0.25->128 |
| Telithromycin | 0.125 | 1 | 0.064-2 |
| ermA (28) | |||
| Erythromycin | 2 | 8 | 0.5-16 |
| Azithromycin | 8 | 32 | 1-64 |
| Clindamycin | 0.5 | 128 | 0.064->128 |
| Telithromycin | 0.064 | 0.064 | 0.032-0.125 |
| mefA (8) | |||
| Erythromycin | 2 | 4 | 2-4 |
| Azithromycin | 2 | 4 | 2-4 |
| Clindamycin | 0.064 | 0.125 | 0.064-0.125 |
| Telithromycin | 0.25 | 0.25 | 0.125-0.5 |
50 and 90%, MIC50 and MIC90, respectively.
Two isolates were negative for ermA, ermB, and mefA genes.
FIG. 1.
Dendrogram constructed from PFGE analysis of 73 typeable erythromycin-resistant serotype III GBS isolates in relation to PCR results for ermB, ermA, and mefA genes. Triangles, collapsed branches gathering isolates with 80% similarity according to the banding patterns. UPGMA, unweighted pair group method with arithmetic mean.
GBS resistance to penicillin or ampicillin has not yet been described (1, 21), while resistance to erythromycin and clindamycin has increased substantially in the last few years (22). The prevalence of GBS resistance ranged from 7 to 25% for erythromycin and from 3 to 15% for clindamycin in reports published between 1998 and 2001 (1, 5, 13, 21). A recent French study showed that 18% of GBS isolates were resistant to erythromycin (15). Macrolide resistance is more frequent among serotype V and serotype III GBS strains than among other serotypes (12, 15, 21). In our institution, the rate of erythromycin resistance among serotype III GBS strains isolated in 2002 was 23%, and a similar level of resistance was found by Lin et al. in six U.S. teaching hospitals (21). This is a matter of concern, as serotype III GBS strains are most frequently associated with neonatal invasive infections (9, 18). Guidelines on intrapartum antimicrobial chemoprophylaxis for penicillin-allergic women were recently updated (25). Vancomycin is recommended for women who are at high risk of β-lactam anaphylaxis and from whom macrolide-resistant GBS is isolated (25). However, vancomycin use has been associated with vancomycin resistance among gram-positive cocci (16).
Here, we determined the telithromycin susceptibility of 88 serotype III macrolide-resistant GBS clinical isolates and the mechanisms of resistance. Telithromycin was active against all the isolates, with MIC50s and MIC90s of 0.125 and 0.5 μg/ml, respectively. Inducible clindamycin or telithromycin resistance was not checked in our study. The telithromycin MIC90s were higher for strains carrying ermB than for strains carrying ermA or mefA. Erythromycin resistance was mainly associated with ermB (57% of erythromycin-resistant isolates), as recently reported by Betriu et al. (3) In contrast, in a Canadian study erythromycin resistance was found to be due mainly to ermA (11). The low prevalence (9%) of the mefA gene among our isolates was comparable to that found in previous studies (3, 11, 20). In contrast to the results of Betriu et al. (3), we never found more than one erythromycin resistance gene in the same isolate. Previous studies have demonstrated genetic heterogeneity among serotype III GBS isolates (6). Likewise, PFGE revealed major genetic diversity among our serotype III GBS isolates. In our study, macrolide resistance among serotype III GBS strains was due to the dissemination of resistance determinants among isolates of identical or different genetic backgrounds, rather than to epidemic spread of a single clone, as described for macrolide-resistant serotype V GBS (12) and group A streptococci (19). Our results suggest that telithromycin is a potential alternative for prophylaxis of perinatal GBS disease when the mother is allergic to penicillin and the local prevalence of macrolide resistance is high.
REFERENCES
- 1.Andrews, J. I., D. J. Diekema, S. K. Hunter, P. R. Rhomberg, M. A. Pfaller, R. N. Jones, and G. V. Doern. 2000. Group B streptococci causing neonatal bloodstream infection: antimicrobial susceptibility and serotyping results from SENTRY centers in the Western Hemisphere. Am. J. Obstet. Gynecol. 183:859-862. [DOI] [PubMed] [Google Scholar]
- 2.Arpin, C., H. Daube, F. Tessier, and C. Quentin. 1999. Presence of mefA and mefE genes in Streptococcus agalactiae. Antimicrob. Agents Chemother. 43:944-946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Betriu, C., E. Culebras, M. Gomez, I. Rodriguez-Avial, B. A. Sanchez, M. C. Agreda, and J. J. Picazo. 2003. Erythromycin and clindamycin resistance and telithromycin susceptibility in Streptococcus agalactiae. Antimicrob. Agents Chemother. 47:1112-1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bingen, E., F. Fitoussi, C. Doit, R. Cohen, A. Tanna, R. George, C. Loukil, N. Brahimi, I. Le Thomas, and D. Deforche. 2000. Resistance to macrolides in Streptococcus pyogenes in France in pediatric patients. Antimicrob. Agents Chemother. 44:1453-1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bland, M. L., S. T. Vermillion, D. E. Soper, and M. Austin. 2001. Antibiotic resistance patterns of group B streptococci in late third-trimester rectovaginal cultures. Am. J. Obstet. Gynecol. 184:1125-1126. [DOI] [PubMed] [Google Scholar]
- 6.Blumberg, H. M., D. S. Stephens, C. Licitra, N. Pigott, R. Facklam, B. Swaminathan, and I. K. Wachsmuth. 1992. Molecular epidemiology of group B streptococcal infections: use of restriction endonuclease analysis of chromosomal DNA and DNA restriction fragment length polymorphisms of ribosomal RNA genes (ribotyping). J. Infect. Dis. 166:574-579. [DOI] [PubMed] [Google Scholar]
- 7.Bryskier, A. 2000. Ketolides-telithromycin, an example of a new class of antibacterial agents. Clin. Microbiol. Infect. 6:661-669. [DOI] [PubMed] [Google Scholar]
- 8.Clancy, J., J. Petitpas, F. Dib-Hajj, W. Yuan, M. Cronan, A. V. Kamath, J. Bergeron, and J. A. Retasema. 1996. Molecular cloning and functional analysis of a novel macrolide-resistance determinant, mefA, from Streptococcus pyogenes. Mol. Microbiol. 22:867-879. [DOI] [PubMed] [Google Scholar]
- 9.Davies, D. H., S. Raj, C. Adair, J. Robinson, and A. M. Geer. 2001. Population-based active surveillance for neonatal group B streptococcal infections in Alberta, Canada: implications for vaccine formulation. Pediatr. Infect. Dis. J. 20:879-884. [DOI] [PubMed] [Google Scholar]
- 10.Davies, T. A., L. M. Ednies, D. M. Hoellmon, G. A. Pankuch, M. R. Jacobs, and P. C. Appelbaum. 2000. Antipneumococcal activity of ABT 773 compared to those of 10 other agents. Antimicrob. Agents Chemother. 44:1894-1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Azavedo, J. C. S., M. McGavin, C. Duncan, D. E. Low, and A. McGeer. 2001. Prevalence and mechanisms of macrolide resistance in invasive and noninvasive group B streptococcus isolates from Ontario, Canada. Antimicrob. Agents Chemother. 45:3504-3508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Diekema, D. J., J. I. Andrews, H. Huynh, P. R. Romberg, S. R. Docktor, J. Beyer, V. D. Shortridge, R. K. Flamm, R. N. Jones, and M. A. Pfaller. 2003. Molecular epidemiology of macrolide resistance in neonatal bloodstream isolates of group B streptococci. J. Clin. Microbiol. 41:2659-2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fernandez, M., M. E. Hickman, and C. J. Baker. 1998. Antimicrobial susceptibilities of group B streptococci isolated between 1992 and 1996 from patients with bacteremia or meningitis. Antimicrob. Agents Chemother. 42:1517-1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fitoussi, F., C. Doit, P. Geslin, N. Brahimi, and E. Bingen. 2001. Mechanisms of macrolide resistance in clinical pneumococcal isolates in France. Antimicrob. Agents Chemother. 45:636-638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fitoussi, F., C. Loukil, I. Gros, O. Clermont, P. Mariani-Kurkdjian, S. Bonacorsi, I. Le Thomas, D. Deforche, and E. Bingen. 2001. Mechanisms of macrolide resistance in clinical group B streptococci isolated in France. Antimicrob. Agents Chemother. 45:1889-1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fridkin, S. K., J. R. Edwards, J. M. Courval, H. Hill, F. C. Tenover, and R. Lawton. 2001. The effect of vancomycin and third-generation cephalosporins on prevalence of vancomycin-resistant enterococci in 126 U.S. adult intensive care units. Ann. Intern. Med. 135:175-183. [DOI] [PubMed] [Google Scholar]
- 17.Gordillo, M. E., K. V. Singh, C. J. Baker, and B. E. Murray. 1993. Typing of group B streptococci: comparison of pulsed-field gel electrophoresis and conventional electrophoresis. J. Clin. Microbiol. 31:1430-1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harrison, L. H., J. A. Elliott, D. M. Dwyer, J. P. Libonati, P. Ferrieri, L. Billmann, et al. 1998. Serotype distribution of invasive group B streptococcal isolates in Maryland: implications for vaccine formulation. J. Infect. Dis. 177:998-1002. [DOI] [PubMed] [Google Scholar]
- 19.Kataja, J., P. Huovinen, A. Muotiala, J. Vuopio-Varkila, A. Efstratiou, G. Hallas, and H. Seppala. 1998. Clonal spread of group A streptococcus with the new type of erythromycin resistance. J. Infect. Dis. 177:786-789. [DOI] [PubMed] [Google Scholar]
- 20.Leclercq, R. 2002. Mechanisms of resistance to macrolides and lincosamides: nature of the resistance elements and their clinical application. Clin. Infect. Dis. 34:482-492. [DOI] [PubMed] [Google Scholar]
- 21.Lin, F. Y., P. H. Azami, L. E. Weisman, J. B. Philips, J. Regan, P. Clark, G. G. Rhoads, J. Clemens, J. Troendle, E. Pratt, R. A. Brenner, and V. Gill. 2000. Antibiotic susceptibility profiles for group B streptococci isolated from neonates, 1995-1998. Clin. Infect. Dis. 31:76-79. [DOI] [PubMed] [Google Scholar]
- 22.Moore, M. R., S. J. Schrag, and A. Schuchat. 2003. Effects of intrapartum antimicrobial prophylaxis for prevention of group-B-streptococcal disease on the incidence and ecology of early-onset neonatal sepsis. Lancet Infect. Dis. 3:201-213. [DOI] [PubMed] [Google Scholar]
- 23.National Committee for Clinical Laboratory Standards. 2000. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard M7-A5, 5th ed. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 24.Pearlman, M. D., C. L. Pierson, and R. G. Faix. 1998. Frequent resistance of clinical group B streptococcus isolates to clindamycin and erythromycin. Obstet. Gynecol. 92:258-261. [DOI] [PubMed] [Google Scholar]
- 25.Schrag, S. J., R. Gorwitz, K. Fultz-Butts, and A. Schuchat. 2002. Prevention of perinatal group B streptococcal disease. Revised guidelines from CDC. Morb. Mortal. Wkly. Rep. Recomm. Rep. 51:1-22. [PubMed] [Google Scholar]
- 26.Schrag, S. J., S. Zywicki, M. M. Farley, A. L. Reingold, L. H. Harrisson, and L. B. Lefkowitz. 2000. Group B streptococcal disease in the era of intrapartum antibiotic prophylaxis. N. Engl. J. Med. 342:15-20. [DOI] [PubMed]
- 27.Seppäla, H., M. Skurnik, H. Soini, M. C. Roberts, and P. Huovinen. 1998. A novel erythromycin resistance methylase gene (ermTR) in Streptococcus pyogenes. Antimicrob. Agents Chemother. 42:257-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sutcliffe, J., T. Grebe, A. Tait-Kamradt, and L. Wondrack. 1996. Detection of erythromycin-resistant determinants by PCR. Antimicrob. Agents Chemother. 40:2562-2566. [DOI] [PMC free article] [PubMed] [Google Scholar]