Abstract
2-Amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP) is a carcinogenic heterocyclic aromatic amine that is produced in cooked meats. The simultaneous analysis of PhIP and its metabolites in human urine is a challenge, because these biomarkers only occur in urine at parts-per-billion or lower concentrations, and must be selectively purifed from thousands of other urinary constituents. We have developed a facile solid-phase extraction method, employing a mixed-mode reverse phase cation exchange resin, to simultaneously isolate PhIP, its glucuronide conjugates, and the glucuronide conjugates of the genotoxic metabolite 2-hydroxyamino-1-methyl-6-phenylimidazo[4,5-b]pyridine from urine of meat-eaters. PhIP and its metabolites were quantified by liquid chromatography-electrospray ionization tandem mass spectrometry (LC-ESI/MS/MS), using a triple stage quadrupole mass spectrometer in the selected reaction monitoring scan mode (SRM). The lower limit of quantification (LOQ) of PhIP is 5 parts-per-trillion (ppt), and the LOQ values for the glucuronide conjugates are 50 ppt, when 25 μL of urine are employed for assay. The extraction scheme is versatile and has been employed to isolate other ring-hydroxylated and glucuronidated metabolites of PhIP, for characterization by LC-ESI/MS/MS.
INTRODUCTION
Heterocyclic aromatic amines (HAA)1 are a class of carcinogenic compounds that form in cooked meats (1). PhIP is one of the most mass-abundant HAAs formed in red meats and poultry cooked well-done: the concentrations of PhIP can range from several parts-per-billion (ppb) up to 500 ppb (2,3). Putative DNA adducts of PhIP have been detected in human tissues, even though the dietary concentrations of PhIP generally are low (4,5). Thus, PhIP is a health hazard and is reasonably anticipated to be a human carcinogen (6).
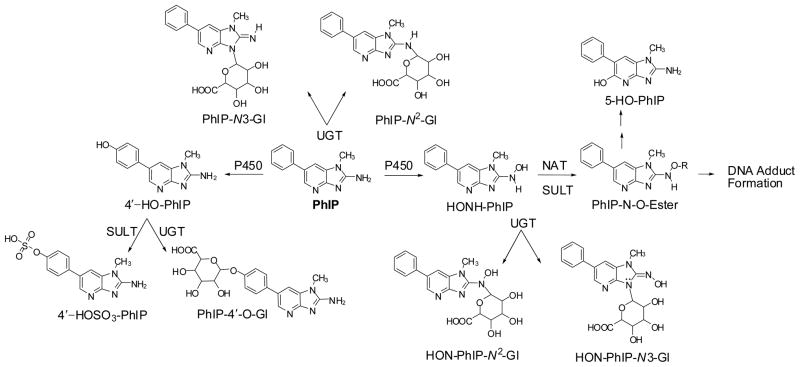
The metabolism of PhIP has been extensively studied in vitro with tissue fractions, purified and recombinant enzymes (7–10), and hepatocytes (11,12), and in vivo in experimental laboratory animals (11–15), and humans (16–22). Cytochrome P450s (P450s 1A1, 1A2 and 1B1) catalyze the oxidation of the exocyclic amine group of PhIP to form the genotoxic metabolite, 2-hydroxyamino-1-methyl-6-phenylimidazo[4,5-b]pyridine (HONH-PhIP) (9,10,23). This metabolite can undergo conjugation by sulfotransferases (SULTs) (24,25) or N-acetyltransferases (NATs) (26), to produce highly reactive esters that bind to DNA (27), or undergo solvolysis to produce 2-amino-1-methyl-6-(5-hydroxy)phenylimidazo[4,5-b]pyridine (5-HO-PhIP) (Figure 1). These same P450s catalyze oxidation at the 4′ position of PhIP to form 2-amino-1-methyl-6-(4′-hydroxy)-phenylimidazo[4,5-b]pyridine (4′-HO-PhIP), a principal detoxication product of PhIP in rodents and non-human primates (11,14). However, human P450s primarily catalyze the formation of HONH-PhIP as the major oxidation product, the formation of 4′-HO-PhIP occurs at considerably lower levels (9,15,23).
Figure 1.
Major pathways of metabolism of PhIP in experimental laboratory animals and humans.
PhIP undergoes extensive metabolism, by uridine diphosphate glucuronosyltransferases (UGTs). Both PhIP and HNOH-PhIP undergo conjugation by UGT1A1 isoforms to produce N2- and N3-glucuronide conjugates (28–30). UGT activity toward PhIP and HONH-PhIP has been detected in human liver and colon microsome samples (28,31–34). The glucuronide conjugates of HONH-PhIP have been viewed as detoxication products (21), although bacterial glucuronidases can hydrolyze HON-PhIP-N3-Gl to liberate HONH-PhIP for futher metabolism and potential DNA adduct formation (35).
Urine is a useful biological matrix for the measurement of PhIP and its metabolites, since large quantities can be obtained noninvasively. Although measurements of PhIP or its metabolites in urine do not shed light on DNA damage, they can reflect the capacity of an individual to bioactivate and detoxicate this procarcinogen (36). PhIP is rapidly absorbed from the gastrointestinal tract and is eliminated in urine as multiple metabolites within 24 hr of consumption of grilled meats (19,37,38). Approximately 70% of the ingested dose of PhIP has been reported to undergo metabolism by P450 1A2 in humans (38), while often < 1% of the ingested dose is eliminated in urine as the unaltered compound.
Several different analytical approaches have been devised to isolate PhIP from human urine: These techniques include solvent extraction (38,39), solid-phase extraction (SPE) (40), molecularly imprinted polymers (22), or immunoaffinity methods (17), followed by quantification by gas chromatography and negative ion chemical ionization mass spectrometry (GC-NICI-MS) (38,39,41), or LC-ESI/MS/MS (22,40), or followed by fluorescence detection (18). [14C]PhIP and several radiolabeled metabolites have been identified in human urine by accelerator mass spectrometry (AMS) (21,42): urinary metabolites have also been detected by LC-ESI/MS/MS (19,20), or indirectly after chemical reduction or acid hydrolysis of HONH-PhIP conjugates, employing either LC-ESI/MS/MS or GC-NICI-MS (43,44).
To understand the interindividual variation in the activities of xenobiotic metabolism enzymes involved in PhIP metabolism, we require a robust, analytical method for simultaneous quantitation of unaltered PhIP and its principal metabolites in urine. To our knowledge, there is no report in the literature of a validated method to conduct measurements of these biomarkers simultaneously, and details about intra-day and inter-day peformance of the method (accuracy and precision) are lacking. Such an analysis is a challenge, because the concentrations of PhIP and its metabolites occur at or below the ppb level in urine. Moreover, the polar and ionic nature of the PhIP metabolites presents difficulties for the selective isolation of them with PhIP, from thousands of other components in the urine matrix (45). In this paper, we describe a facile extraction scheme that employs a mixed-mode reverse phase cation exchange solid phase resin, to isolate PhIP and its glucuronide metabolites from urine of meat-eaters (21,42). Quantification of these biomarkers is achieved by LC-ESI/MS/MS, in the selected reaction monitoring (SRM) scan mode, using the stable isotope dilution method. Moreover, the sensitivity of the triple-stage quadrupole mass spectrometer has allowed us to corroborate the structures of these known PhIP metabolites; it has also enabled us to characterize three other unknown glucuronide conjugates of hydroxylated-PhIP metabolites, in the full product ion scan mode.
EXPERIMENTAL PROCEDURES
Caution: PhIP and several of its derivatives are potential human carcinogens and should be handled with caution in a well-ventilated fume hood with the appropriate protective clothing.
Materials and Methods
PhIP and 1-[2H3C]-PhIP (99% isotopic purity) were purchased from Toronto Research Chemicals (Toronto, ON, Canada). NADPH, NADH, glucose-6-phosphate, uridine-5′-diphosphoglucuronic acid (UDPGA), glucose-6-phosphate dehydrogenase, alamethicin, and NH4OH solution (25%) were from Sigma (St. Louis, MO). All solvents used were high-purity B & J Brand® from Honeywell Burdick and Jackson (Muskegon, MI). ACS reagent grade HCO2H (88%) was purchased from J.T. Baker (Phillipsburg, NJ), Retain CX resins (30 mg) were purchased from ThermoFisher Scientific (Palm Beach, FL) and Baker C18 solid-phase extraction (SPE) resins (500 mg) were purchased through Krackeler Scientific Inc. (Albany, NY). Male SD rat liver microsomes of animals pretreated with polychlorinated biphenyls (PCB, Aroclor-1254) were obtained from Moltox (Boone, NC). Human liver microsomal samples were from Tennessee Donor Services, Nashville, TN, and were kindly provided by Dr. F. P. Guengerich, Vanderbilt University. The P450 1A2 protein expression and metabolic activity were previously characterized (9). All other chemical reagents were ACS grade, and purchased from Sigma Aldrich.
Synthesis of HONH-PhIP and 5-HO-PhIP
Briefly, the nitro derivative of PhIP (46) was reduced to HONH-PhIP with hydrazine and Pd/C (46,47), followed by purification of HNOH-PhIP with a Baker C18 SPE resin. The product was eluted with DMSO:C2H5OH (3:1) and stored in liquid nitrogen prior to use. 5-HO-PhIP was prepared by reaction of HONH-PhIP with acetic anhydride in C2H5OH to produce the reactive N-acetoxy-PhIP intermediate, which decomposes to produce 5-HO-PhIP as one of several solvolysis products (48). The product was isolated and characterized by UV, 1H NMR, and MS as previously described (32).
Biosynthesis of 4′HO-PhIP, and PhIP-4′-O-Gl, PhIP-N2-Glu, PhIP-N3-Gl, HON-PhIP-N2-Glu and HON-PhIP-N3-Glu conjugates
The biosyntheses of PhIP-N2-Gl and PhIP-N3-Gl were done with human liver microsomal protein (2 mg/mL) in 100 mM Tris HCl buffer (pH 7.5), containing 10 mM MgC12, PhIP (0.5 mM) and UDPGA (5 mM). The mixture was preincubated with alamethicin (50 μg/mg protein) on ice for 15 min (28). Thereafter, the glucuronidation assay was conducted at 37 °C for 3 h. The reaction was terminated by the addition of 3 vol CH3CN and placed on ice for 30 min, to precipitate proteins. The CH3CN was evaporated under a stream of argon gas. The aqeuous solution was diluted with 10 vol of H2O, and metabolites were partially purified by SPE with a Baker C18 resin (500 mg), and then purified by HPLC (32). The biosynthesis of HON-PhIP-N2-Glu was done as described above except that HONH-PhIP (0.5 mM) was used instead of PhIP. The HON-PhIP-N3-Glu conjugate was prepared in a similar manner, except that microsomes (2 mg protein/mL) of rats pretreated with PCBs were employed. The corresponding isotopically labeled internal standards were prepared in the same manner, employing 1-[2H3C]-PhIP or 1-[2H3C]-HONH-PhIP. The concentrations of the glucuronide metabolites were determined from their UV absorption spectra at their maxima absorbance in CH3OH (HON-PhIP-N2-Glu, 318 nm; HON-PhIP-N3-Glu, 320 nm; PhIP-N2-Gl 305 nm), using the molar extinction coefficient for PhIP (315 nm; ε (M−1cm−1) was determined as 22,220).
For some incubations, NADPH and NADH (1 mM), glucose-6-phopshate (5 mM), and glucose-6-phosphate dehydrogenase (1 unit) were added to the microsomal samples in 100 mM potassium phosphate buffer (pH 7.4) containing 10 mM MgCl2, to catalyze the formation of HONH-PhIP and 4′-HO-PhIP and their glucuronide conjugates in situ. After completion of the reaction, the supernatants were either assayed directly by LC-ESI-MS/MS (vide infra) or purified by solid phase extraction (SPE), followed by HPLC (32). The products were characterized by UV spectroscopy, LC-ESI/MS/MS, and 1H NMR (32).
Human Subjects and Meat Consumption
The analyses of PhIP metabolites were conducted with urine samples from male volunteers who participated in a previous investigation; and full details were reported previously (20,49). In brief, each subject consumed 275 g of cooked minced beef patties that had been fried without added oil or fat for 6 min on each side, using a hot metal griddle at 300 °C, until the meat was well-browned. The average amount of PhIP ingested was estimated to be 4,900 ng (49). A 10-h urine collection was then commenced, and urine samples were stored at −80 °C. A subset of samples were sent blindcoded on dry-ice to the Wadsworth Center for further analyses. This study was approved by the Institutional Review Board at the Wadsworth Center.
Solid-phase Extraction (SPE) of PhIP and its Metabolites from Urine
Urine samples (0.5 mL) were added to chilled CH3OH/Acetone (1:1) (1.5 mL) in Eppendorf tubes. The samples were placed on ice for 15 min, and the precipitated protein and salts were removed by centrifugation at 15,000g for 5 min at 4 °C. The supernatants were transferred into clean Eppendorf tubes, and the organic solvent was removed by vacuum centrifugation at room temperature. The remaining aqueous fraction (~ 0.5 mL) was acidified with HCO2H (10 μL). Then, the urine samples were applied to ThermoFisher HyperSep Retain CX (30 mg resin) cartridges that had been prewashed with CH3OH containing 5% NH4OH (1 mL), followed by 2% HCO2H in H2O (1 mL). The resins were attached to a vacuum manifold, under slight pressure (~5 mm Hg), to achieve a flow rate of the eluent of approximately 1 mL/min. After application of the samples, the cartridges were washed with 2% HCO2H in H2O (1 mL), followed by 2% HCO2H in CH3OH (1 mL), H2O (1 mL), 5% NH4OH (2 × 1 mL), and H2O (1 mL). PhIP and its metabolites were eluted from the resin with CH3OH containing 1% NH4OH (1 mL), and were collected in total recovery cap LC vials (Waters, New Milford, MA). The samples were placed in a ventilated hood for 15 min to allow the NH3 to evaporate. Then, the samples were evaporated to dryness by vacuum centrifugation and resuspended in 1:1 CH3OH:H2O (20 μL).
LC-ESI/MS/MS Analyses
Chromatography was performed with an Agilent 1100 series capillary LC system (Agilent Technologies, Palo Alto, CA) equipped with an Agilent Zorbax-XDB-C18 column (0.3 × 250 mm; 5 μm particle size). Analytes were separated by a gradient. The A solvent contained 0.01% HCO2H and 5% CH3CN in H2O, and the B solvent contained 0.01% HCO2H and 5% H2O in CH3CN. The flow rate was set at 6 μL/min, starting at 100% A and holding for 1 min, followed by a linear gradient to 60% B at 25 min, and then to 100% B at 26 min. The gradient was reversed to the starting conditions over 1 min, and a post-run time of 15 min was required for re-equilibration. The mass-spectral data were acquired on a Finnigan™ Quantum Ultra Triple Stage Quadrupole MS (TSQ/MS) (Thermo Fisher, San Jose, CA), data manipulations were done with Xcalibur version 2.07 software. Analyses were conducted in the positive ionization mode and employed an ADVANCE nanospray source from Michrom Bioresources Inc. (Auburn, CA). The spray voltage was set at 1500 V; the in-source fragmentation was −10 V; and the capillary temperature was 250 °C. There was no sheath or auxiliary gas. The peak and scan widths (in Q1 and Q3) were set at 0.7 Da. The following transitions and collision energies were used for the quantification of PhIP and its metabolites: PhIP and [2H3C]-PhIP: 225.1→ 210.1 and 228.1→ 210.1 @ 33 eV; PhIP-N2-Gl and [2H3C]-PhIP-N2-Gl: 401.1→ 210.1 and 404.1→ 210.1 @ 55 eV; HON-PhIP-N2-Gl, HON-PhIP-N3-Gl, and [2H3C]-HON-PhIP-N2-Gl and [2H3C]-HON-PhIP-N3-Gl: 417.1→ 225.1 and 224.1 and 420.1→ 228.1 and 227.1 @ 32 eV. The dwell time for each transition was 10 ms. Argon was used as the collision gas and was set at 1.5 mTorr. Product ion spectra were acquired on the protonated molecules [M + H]+, scanning from m/z 100 to 500 at a scan speed of 500 amu/s using the same acquisition parameters.
Calibration Curves
Calibration curves were produced in quadruplicate by the addition of a fixed amount of [2H3C]-PhIP (50 pg) and 0, 2.5, 5, 10, 15, or 20 pg of PhIP per 0.5 mL urine from a volunteer who had not consumed cooked meat for at least 24 h. The calibration curves of PhIP metabolites were also constructed in quadruplicate with [2H3C]-PhIP-N2-Gl, [2H3C]-HON-PhIP-N2-Gl, and [2H3C]-HON-PhIP-N3-Gl added at a fixed concentration of 500 pg; the unlabeled analytes were each added at concentrations of 0, 50, 100, 250, 500 or 1000 pg per 0.5 mL urine. The calibration data were fitted to a straight line using the ordinary least-squares method with equal weightings. Each urine sample was subjected to the organic solvent and SPE processing conditions described above. The within- and between-day precisions for PhIP and its metabolites were calculated in quadruplicate as described (50), with urine samples from three different subjects collected over 10 hr, following consumption of cooked meat (20). The measurements were done on four different days over a time period of 2 months.
RESULTS
LC-ESI/MS Characterization of PhIP, HONH-PhIP, 4′-HO-PhIP, 5-HO-PhIP, and their Glucuronide Metabolites Produced by Rat or Human Liver Microsomes
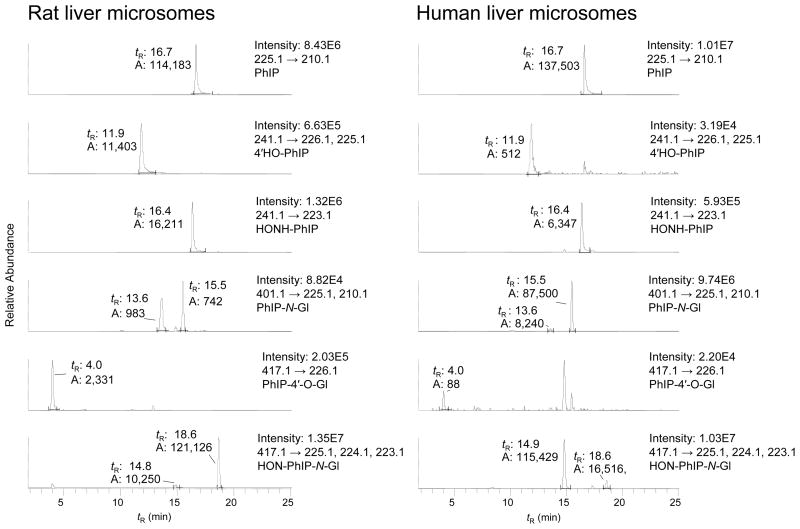
Rat liver (pretreated with PCBs) and human liver microsomal samples containing high levels of PhIP N-oxidation acitivity (rat liver microsomes: 11.3 nmol HONH-PhIP/min/mg protein; human liver microsomes: 2.1 nmol HONH-PhIP/min/mg protein) were used to produce PhIP metabolites (Figure 2). Interspecies differences in regioselectivity of P450-catalyzed oxidation and UGT-catalyzed glucuronidation of PhIP were observed. Both rat and human microsomal preparations produced large quantities of HONH-PhIP. The rat liver microsomes also produced large amounts of 4′-HO-PhIP and its glucuronide conjugate 4′-β-glucosiduronyloxy-2-amino-1-methyl-6-(4′-hydroxy)-phenylimidazo[4,5-b]pyridine (PhIP-4′-O-Gl); however, these PhIP metabolites were produced in low quantities by human liver microsomes. The levels of 5-HO-PhIP and its glucuronide conjugates, if these were formed in either type of microsomal sample, were negligible. The rat liver microsomes catalyzed low levels of glucuronidation of PhIP at both the endocyclic N3 and exocyclic N2 atoms of PhIP. The N3 atom of HONH-PhIP was the predominant position of glucuronidation by rat liver microsomes, whereas the exocyclic N2 atom was the principal site of glucuronidation of PhIP and HONH-PhIP by human liver microsomes. These interspecies differences in regioselectivity of PhIP metabolism have previously been observed for liver microsomes, purified and recombinant P450s, recombinant UGTs, and human and rodent hepatocytes (9,23,28–33).
Figure 2.
LC-ESI/MS/MS chromatograms of PhIP metabolites produced with (A) Rat liver microsomes and (B) Human liver microsomes. Both microsomal samples were fortified with UDPGA and cofactors for P450 catalysis (respective tR: PhIP-N3-Gl 13.6 min; PhIP-N2-Gl 15.5 min; HON-PhIP-N2-Gl 14.9 min; HON-PhIP-N3-Gl 18.6 min). The ion intensity and area counts × 10−3 are reported.
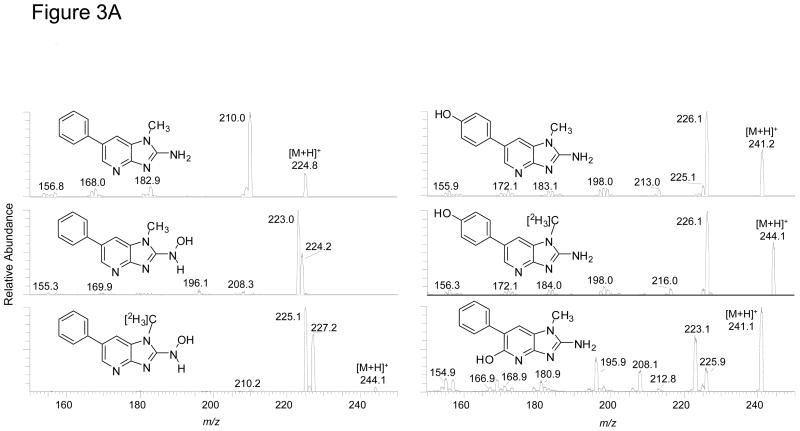
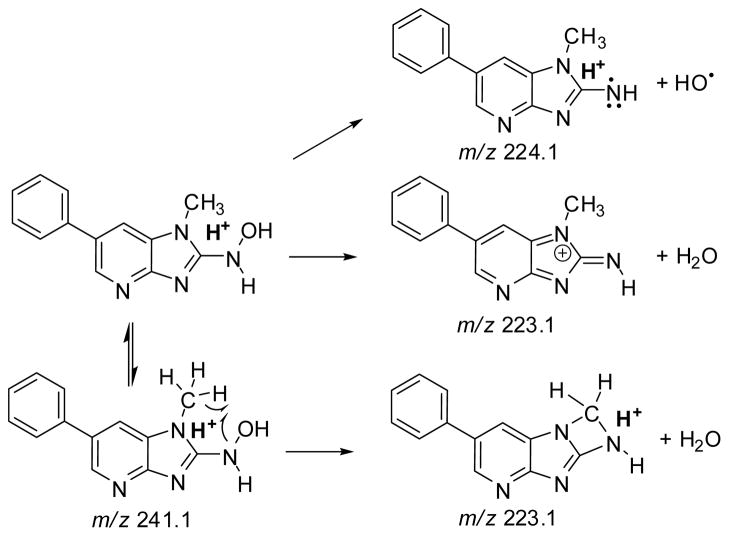
The ESI product ion spectra of the protonated molecules PhIP and its metabolites are shown in Figure 3A, B. The principal fragment ion of PhIP occurs at m/z 210.0 [M + H - 15]+• and is attributed to cleavage of the N1-CH3 bond. The HONH-PhIP ([M + H]+ at m/z 241.1) undergoes fragmentation to form ions at m/z 224.2 [M + H - 17]+• and m/z 223.0 [M + H - 18]+, respectively due to the loss of OH• and of H2O The [M + H - 17]+• is a characteristic fragment ion for N-hydroxy-HAAs, but not for ring-hydroxylated HAAs, under these LC-ESI/MS/MS conditions (32,51). The product ion spectrum of 1-[2H3C]-HONH-PhIP ([M + H]+ at m/z 244.1) displays prominent ions at m/z 227.2 [M + H - 17]+• and 225.1 [M + H - 19]+•, due to losses of OH• and HOD. However, the ion at m/z 226.1 [M + H - 18]+, due to the loss of H2O, is minor. Therefore, the principal mechanism of dehydration of HONH-PhIP occurs by homolytic cleavages at a deuterium atom bonded to the C1 atom and at the HO-N2 bond of 1-[2H3C]-HONH-PhIP, to produce HOD: The loss of 18 (H2O), by solvolysis of HONH-PhIP, is a minor pathway (Scheme 1). The isomeric 4′-HO-PhIP metabolite ([M + H]+ at m/z 241.1) undergoes collision-induced dissociation (CID) to form product ions at m/z 226.1 [M + H]+ and m/z 213.0 [M + H - 28]+, due to losses of CH3• and CO. The isomeric, synthetic 5-HO-PhIP undergoes fragmentation to produce prominent product ions at m/z 225.9, 223.1, 208.1, and 195.9, respectively, attributed to losses of CH3•, H2O, CH3• and H2O, and H2O and HCN.
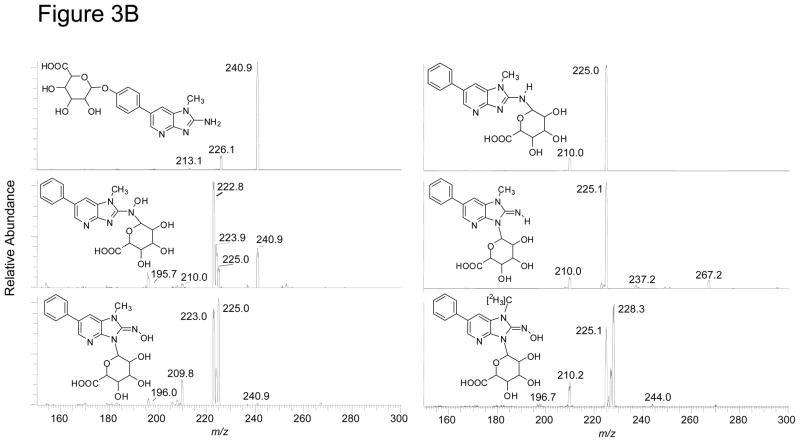
Figure 3.
LC-ESI/MS/MS product ion spectra of PhIP and its metabolites. (A) PhIP, 4′-HO-PhIP and 5-HO-PhIP @ a CID of 32 eV, and HONH-PhIP and 1-[2H3C]-HONH-PhIP @ a CID of 20 eV. (B) Isomeric PhIP-N-Gl and HONH-PhIP-N-Gl conjugates and 1-[2H3C]-HONH-PhIP-N3-Gl @ 32 eV. The ion intensity and area counts × 10−3 are reported.
Scheme 1.
Proposed pathways of fragmentation of HOHN-PhIP [M+H]+ at m/z 241.1, To form the product ions at m/z 223.1 and 224.1.
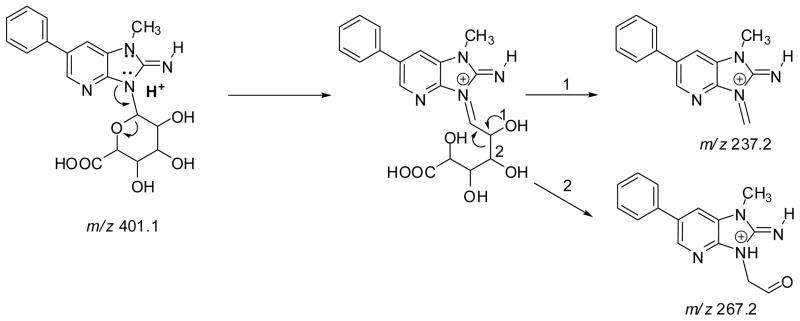
Both PhIP-N2-Gl and PhIP-N3-Gl conjugates ([M + H]+ at m/z 401.1) undergo CID to form ions at m/z 225.1 [M+H–176]+, due to the loss of the glucuronic acid moiety; a secondary fragmentation of the N1-CH3 bond of PhIP occurs to produce the ion at m/z 210.1 [M+H–191]+• Two fragment ions are observed at m/z 237.2 and 267.2 in the product ion spectrum of PhIP-N3-Gl, but are not seen in the spectrum of PhIP-N2-Gl. These ions are proposed to occur via ring opening of the glucuronide, followed by cleavage at the C1–C2 bond (m/z 237.2) or C2–C3 bond (m/z 267.2) of the glucuronide moiety (Scheme 2). The corresponding fragment ions are observed at m/z 240.2 and m/z 270.2, in the product ion spectrum of 1-[2H3C]-PhIP-N3-Gl (data not shown).
Scheme 2.
Proposed pathways of fragmentation of PhIP-N3-Gl To produce the fragment ions at m/z 267.2 and 237.2.
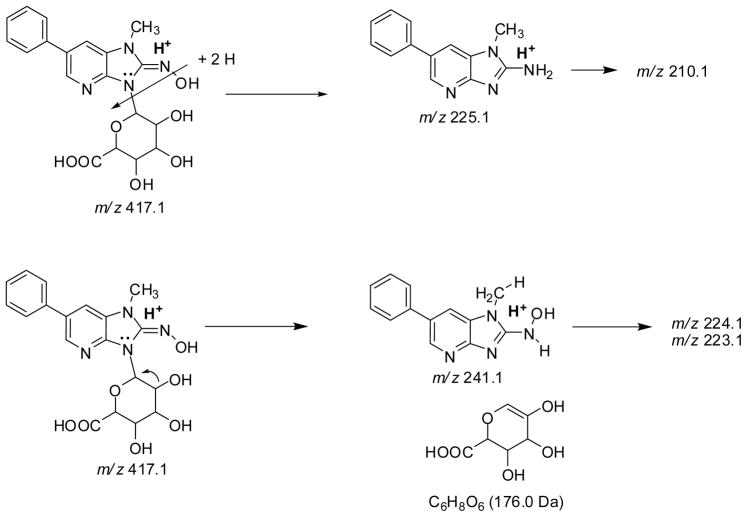
Both HON-PhIP-N-Gl conjugates ([M+H]+ at m/z 417.1) undergo fragmentation of the glycosidic linkage [M+H –176]+, to produce HONH-PhIP at m/z 240.9, as well as other product ions at m/z 225.0, 223.9, and 223.0. The product ion spectra of 1-[2H3C]-HON-PhIP-N-Gl conjugates ([M+H]+ at m/z 420.1) display ions at m/z 244.1 [M+H–176]+ ( the formation of 1-[2H3C]-HONH-PhIP), and ions at m/z 228.1, 227.1, 226.1, and 225.1. The HONH-PhIP, m/z 240.9, (or 1-[2H3C]-HONH-PhIP, m/z 244.1), undergoes fragmentation to lose OH• and H2O (or OH•, H2O, HOD), to form the ions at m/z 224.1 and 223.1 (or m/z 227.1, 226.1, and 225.1). The precursor ion scan mode revealed that the ions at m/z 225.1 and m/z 228.3 in the product ion spectra of the unlabeled and labeled Gl conjugates, respectively, were derived from the parent molecules, indicative or rearrangement of the hydroxyl group of HONH-PhIP to the glucuronide moiety. Subsequent cleavage of the glycosidic linkage produces protonated PhIP (m/z 225.1) (or protonated 1-[2H3C]-PhIP m/z 228.1) (Scheme 3).
Scheme 3.
Proposed pathways of fragmentation of HON-PhIP-N3-PhIP [M+H]+ at m/z 417.1 To produce the product ions at m/z 241.1 and 225.1, and secondary fragmentations to form the product ions at m/z 223.1 and 224.1 (from HONH-PhIP) and m/z 210.1 (from PhIP).
LC-ESI/MS/MS Analysis of PhIP and its Metabolites in Human Urine
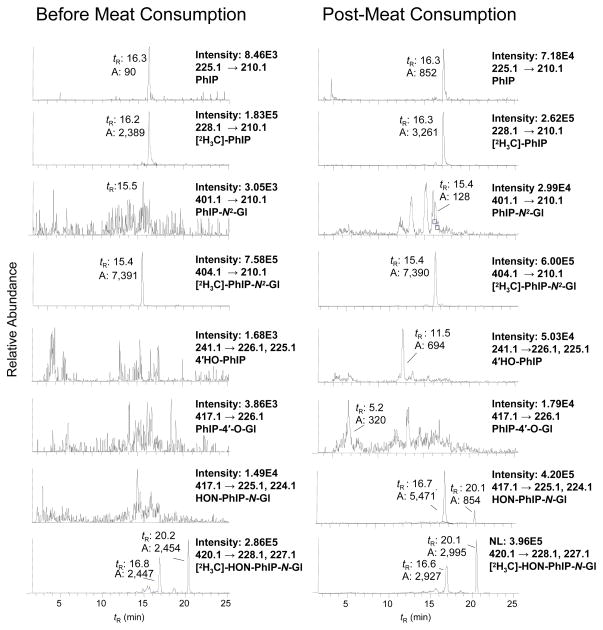
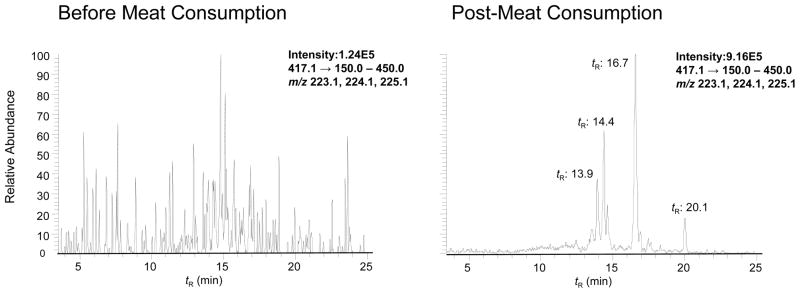
The analysis of urine from a subject before and after consumption of grilled meat is shown in Figure 4. Trace levels of PhIP at (tR 16.3 min, <3 pg/mL) are detected in urine of the subject prior to meat consumption; none of the other metabolites are detected. The LC-ESI/MS/MS chromatogram of metabolites from urine collected over 10 h after meat consumption displays signals for PhIP (tR 16.3 min, 20 pg/mL), HON-PhIP-N2-Gl (tR 16.7 min 1680 pg/mL), and HON-PhIP-N3-Gl (tR 20.1 min 276 pg/mL) (Figure 4). The PhIP-N2-Gl (tR 15.4 min) was difficult to discern when the transition [M+H]+ → [M+H –176]+ (401→225) was employed for monitoring and many isobaric interferences were observed (data not shown). Increasing the CID voltage from 32 to 55 eV resulted in secondary fragmentation of PhIP-N2-Gl [M+H]+→ [M + H–191] +• (401→ 210), attributed to the loss of the glucuronide, followed by loss of the CH3• group of PhIP. This transition greatly improved the signal to noise for monitoring the PhIP-N2-Gl analyte in urine. However, the amount of PhIP-N2-Gl was below 50 pg/mL (the LOQ). There were small quantities of 4′-HO-PhIP (tR 11.5 min). On the basis of ionization efficiency and response similar to those of PhIP, we deduce that 4′-HO-PhIP was present in urine samples at levels of about 16 pg/mL. Trace levels of the PhIP-4′-O-Gl (tR 5.2 min) were also detected.
Figure 4.
SRM traces of PhIP and its glucuronide metabolites in human urine before and after consumption of cooked beef. Note, 0.01% HCO2H replaced the 0.1% HCO2H that was employed in the analysis of microsomal metabolites presented in Figure 2. The change in acid concentration resulted in shifts of tR values of metabolites: PhIP-N2-Gl 15.2 min; HON-PhIP-N2-Gl 16.7 min; and HON-PhIP-N3-Gl 20.1 min). The ion intensity and area counts × 10−3 are reported.
The quantities of HON-PhIP-N2-Gl and HON-PhIP-N3-Gl were sufficient that product ion spectra could be acquired for corroboration of their identities. The product ion spectra are in excellent agreement with the reference standards (Supporting Information, Figure S-1, S-2). There was evidence for three other glucuronide conjugates of hydroxylated PhIP metabolites [M+H]+ at m/z 417.1, (tR 13.3–14.5 min), when the transition [M+H]+→ [M+H –194]+ (417.1→ 223.1), due to loss of glucuronide and H2O. These analytes were not seen in urine samples of subjects prior to consumption of cooked meat (Figure 5). The product ion spectra of these metabolites differ from those spectra of the glucuronide conjugates of HONH-PhIP and 4′-HO-PhIP. Prominent fragment ions are observed at m/z 241.1, 223.1, and 167.1 (Supporting Information, Figure S-3).
Figure 5.
SRM traces of novel glucuronides of hydroxylated PhIP metabolites in human urine before and after consumption of cooked beef. The novel glucuronide metabolites elute at tR 13.9 – 14.6 min, the tR of HON-PhIP-N2-Gl is 16.7 min; and tR of HON-PhIP-N3-Gl is 20.1 min.
Performance of the Analytical Method
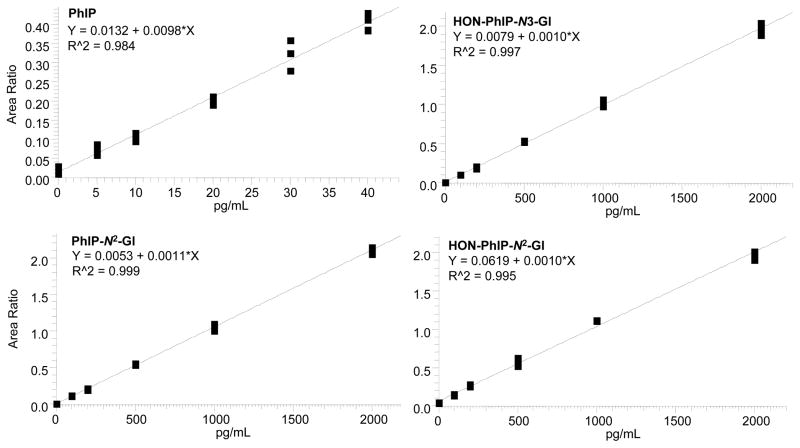
The recoveries of each internal standard [2H3C]-PhIP (100 pg/mL), or [2H3C]-PhIP-N2-Gl, [2H3C]-HON-PhIP-N2-Gl and [2H3C]-HON-PhIP-N3-Gl (each at 1000 pg/mL), added to urine prior to sample processing were consistently between 50 and 80%, based on the response of the signals to those of pure standards measured by LC-ESI/MS/MS. The response of the signals of the processed internal standards is a function of the recoveries of the compounds and the potential ion suppression effects of the urine matrix (40). The calibration curves for PhIP, PhIP-N2-Gl, HON-PhIP-N2-Gl and HON-PhIP-N3-Gl (4 independent replicates per calibrant level), constructed in urine from a subject who refrained from eating cooked meat for 48 h, are depicted in Figure 6. The LOQ for PhIP was estimated at approximately 5 pg/ml, and the LOQs for the glucuronide conjugates of PhIP and HONH-PhIP were estimated at ~50 pg/mL.
Figure 6.
Calibration curves for PhIP, PhIP-N2-Gl, HONH-PhIP-N2-Gl, and HONH-PhIP-N3-Gl.
The isomeric glucuronide conjugates of HONH-PhIP and their isotopically labelled internal standards were monitored by use of the transitions [M+H]+→ [M+H–192]+ and [M+H–193]+, which are attributed to the loss of the glucuronide moiety and an oxygen atom or loss of the glucuronide moiety and the hydroxyl group. These ions were chosen because isobaric interferences were observed in the most abundant transition for the internal standard ([M+H]+→ [M+H–194]+ (loss of glucuronide and H2O), resulting in less reliable estimates of the urinary analytes (data not shown). The precision (CV(%)) of the estimates in the calibration curve at the lowest calibrant levels of PhIP (5 pg/mL) was ≤20%, and the precisions (CV(%)) for PhIP-N2-Gl, HONH-PhIP-N2-Gl, and HONH-PhIP-N3-Gl metabolites at the lowest calibrant levels (100 pg/mL) were ≤11%; these precisions improved at the higher calibrant levels.
The peformance of the method was assessed by the within-day and between-day estimates and precision of measurements of PhIP and the glucuronide conjugates of NHOH-PhIP, in urine of three volunteers determined over 4 days (n = 4 independent measurements per day), within a time period of two months. The results are summarized in Table 1. The within-day and between-day precision values (CV(%)) in estimates of PhIP were <8.8% and 14.0%, respectively. The within-day and between-day precision values (CV(%)) in estimates of the N3- and N2-glucuronide metabolites of HONH-PhIP were <10.2% and 10.8%, respectively. Thus, the analytical method is precise, and inter-day estimates for the quantification of PhIP and its metabolites are highly reproducible.
Table 1.
Performance of the Analytical Method to Measure PhIP and Its Metabolites in Urine
| Amount (pg/mL) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Subject | Metabolite | Day 1 | Day 2 | Day 3 | Day 4 | Overall | CV(%) within-day | CV(%) between-day | |
| 1 | HON-PhIP- N 2-Gl | Mean | 970 | 1040 | 1080 | 1110 | 1050 | ||
| SD | 63 | 177 | 24.5 | 105 | 112 | ||||
| RSD(%) | 6.5 | 17.1 | 2.3 | 9.4 | 10.6 | 10.2 | 10.6 | ||
| HON-PhIP- N3 -Gl | Mean | 145 | 135 | 149 | 136 | 142 | |||
| SD | 15.8 | 12.4 | 16.2 | 10.2 | 13.9 | ||||
| RSD(%) | 10.9 | 9.2 | 10.8 | 7.5 | 9.9 | 10.2 | 10.2 | ||
| PhIP | Mean | 27 | 25 | 28 | 24 | 26 | |||
| SD | 3.6 | 1.3 | 0.6 | 0.8 | 2.3 | ||||
| RSD(%) | 13.2 | 5.0 | 2.1 | 3.4 | 8.7 | 8.8 | 9.3 | ||
| 2 | HON-PhIP- N 2-Gl | Mean | 990 | 950 | 1150 | 1120 | 1054 | ||
| SD | 27 | 24 | 96 | 96 | 105 | ||||
| RSD(%) | 2.7 | 2.5 | 8.4 | 8.6 | 9.9 | 6.7 | 10.8 | ||
| HON-PhIP- N3 -Gl | Mean | 168 | 154 | 163 | 159 | 161 | |||
| SD | 6.5 | 18 | 16 | 5.0 | 12.8 | ||||
| RSD(%) | 3.9 | 11.9 | 10.1 | 3.1 | 7.9 | 8.0 | 7.8 | ||
| PhIP | Mean | 14 | 18 | 17 | 15 | 16 | |||
| SD | 1.0 | 2.3 | 0.5 | 0.4 | 2.0 | ||||
| RSD(%) | 6.7 | 12.7 | 2.9 | 2.7 | 12.6 | 8.2 | 14.0 | ||
| 3 | HON-PhIP- N 2-Gl | Mean | 1610 | 1450 | 1450 | 1490 | 1490 | ||
| SD | 87 | 105 | 62 | 89 | 103 | ||||
| RSD(%) | 5.4 | 7.2 | 4.3 | 6.0 | 6.9 | 5.5 | 7.0 | ||
| HON-PhIP- N3 -Gl | Mean | 258 | 227 | 231 | 222 | 235 | |||
| SD | 22 | 15 | 17 | 22 | 21 | ||||
| RSD(%) | 8.5 | 6.6 | 7.4 | 6.3 | 8.9 | 6.5 | 8.8 | ||
| PhIP | Mean | 18 | 19 | 23 | 19 | 20 | |||
| SD | 1.0 | 0.4 | 1.5 | 0.4 | 1.9 | ||||
| RSD(%) | 5.5 | 1.8 | 6.5 | 2.1 | 9.7 | 2.8 | 10.3 | ||
n = 4 replicates per day
DISCUSSION
A robust analytical method has been established to simultaneously quantitate PhIP, PhIP-N2-Gl, and the isomeric N2 and N3-glucuronide conjugates of HONH-PhIP. The extraction method is facile: It entails treatment of the urine with an organic solvent, to promote precipitation of proteins and salts, followed by SPE of the supernatant with a mixed-mode reverse phase cation exchange resin. The LC-ESI/MS/MS analyses of glucuronide conjugates of HONH-PhIP in human urine have been previously reported, in two different studies. The first study employed a labor-intensive extraction scheme prior to LC-ESI/MS/MS: the analysis was done with a Finnigan™ LCQ (Thermo Electron), a 3-dimensional ion trap mass spectrometer (19). A second study measured these urinary metabolites by triple stage quadrupole MS, employing a Quattro LC (Micromass Ltd.). In the latter study, urine was assayed without any prior purification of the metabolites (20). In our current study, we detected numerous isobaric interferences in urine samples without prior purification of PhIP and its metabolites, when monitored with the Finnigan™ Quantum TSQ/MS. Apparently, the different designs of the ion sources of MS instruments can influence the degree of analyte purification that is required prior to MS analysis. Moreover, we observed that the performance of the analytical LC column deteriorated rapidly, when unprocesssed urine was assayed. Our estimates of PhIP and its N2- and N3-glucuronide metabolites of HONH-PhIP are in excellent agreement with the values previously determined on the same urine samples that were purified and measured by different techniques (20,49): The inter-laboratory estimates of PhIP and these metabolites are within 25%.
The prominent role of UGTs in the metabolism of HONH-PhIP by humans is well-known (20,21,32,37,44). Together, the amount of the HON-PhIP-N2-Gl and HON-PhIP-N3-Gl conjugates have been reported to account for 50 to 70%, and the levels of PhIP-N2-Gl ranged between 4 and 11% of the ingested dose that were excreted in urine within 24 h, when assayed by AMS (21). The other major urinary metabolite detected by AMS was the 4′-sulfate conjugate of 4′-HO-PhIP, which accounted for as much as 10% of the ingested dose that was excreted in urine (21). We did not detect either the N2 or the N3-Gl conjugate of PhIP (<50 pg/mL), by LC-ESI/MS/MS, in urine of subjects from our study. Glucuronidation has been reported to be less important to the metabolism of PhIP than is the glucuronidation of HONH-PhIP in vivo (21), as well as in human hepatocytes (32). Possibly, the amounts of the PhIP-N-Gl isomers present in these urine specimens were just below the LOQ in our analysis. We did detect trace levels of 4′-HO-PhIP; however, the 4′-sulfate conjugate of 4′HO-PhIP was not found (data not shown). This sulfate metabolite exists as a zwitterion under our LC solvent conditions, and may have escaped detection. Increased coverage of this PhIP metabolite can be achieved by pre-treatment of urine with arylsulfatase, to cleave this sulfate conjugate to form 4′-HO-PhIP (22).
Three other minor glucuronide conjugates of hydroxylated PhIP metabolites were detected; their structures are unknown. Frandsen (22) tentatively identified three glucuronide conjugates of 5-HO-PhIP, a solvolysis product of the reactive N-acetoxy-PhIP intermediate. We have seen that a prominent fragment ion occurs at m/z 223.1 in the product ion spectra of all three of these glucuronide metabolites (Supporting Information, Figure S-3), and is also prominent in the product ion spectrum of 5-HO-PhIP (Figure 3A). Further studies are required to determine whether these three novel glucuronide metabolites are derived from 5-HO-PhIP or other uncharacterized hydroxylated PhIP metabolites.
In summary, our new method for the measurement of PhIP and its urinary metabolites is an effective, non-invasive approach, by which to determine exposure to this carcinogen during the previous 24 h (21). Although the measurements of PhIP urinary metabolites do not shed light on DNA damage, which occurs at a distant target tissues, such as the colon (21), urinary metabolite analyses can aid in identifying genetic polymorphisms in xenobiotic metabolism enzymes that influence the genotoxic potency of this procarcinogen, and they can be used to assess chemoprotective agents that modulate the metabolism and the potential deleterious effects of PhIP (20).
Supplementary Material
SYNOPSIS.
Biomonitoring of PhIP and metabolites in urine.
Acknowledgments
The project was supported by grant R01CA-122320 from the National Cancer Institute (JMF, APT, and RJT).
Footnotes
Abbreviations: PhIP, 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine; 5-HO-PhIP, 2-amino-1-methyl-6-(5-hydroxy)phenylimidazo[4,5-b]pyridine; 4′-HO-PhIP, 2-amino-1-methyl-6-(4′-hydroxy)-phenylimidazo[4,5-b]pyridine; HONH-PhIP, 2-hydroxyamino-1-methyl-6-phenylimidazo[4,5-b]pyridine; HON-PhIP-N2-G1, N2-β-D-glucosidurony1-2-(hydroxyamino)-1-methy1-6-phenylimidazo[4,5-b]pyridine; PhIP-N2-G1, N2-β-D-glucosidurony1-2-amino-1-methy1-6-phenylimidazo[4,5-b]pyridine; HON-PhIP-N3-G1, N3-β-D-glucosidurony1-2-(hydroxyamino)-1-methy1-6-phenylimidazo[4,5-b]pyridine; PhIP-N3-G1, N3-β-D-glucosidurony1-2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine; PhIP-4′-O-Gl; 4′-β-D-glucosiduronyloxy-2-amino-4′-hydroxy1-1-methy1-6-phenylimidazo[4,5-b]pyridine; CID, collision induced dissociation; HAA, heterocyclic aromatic amine; LC-ESI/MS/MS, liquid chromatography-electrospray ionization/ tandem mass spectrometry; LOQ, limit of quantification; MS, mass spectrometry; GC-NICIMS, negative ion chemical ionization mass spectrometry; ppb, parts-per-billion; ppt, parts-per-trillion; SPE, solid-phase extraction; SRM, selected reaction monitoring; TSQ, Finnigan TSQ Quantum Ultra triple stage quadrupole mass spectrometer; UDPGA; uridine-5′-diphosphoglucuronic acid; UGTs, diphosphate glucuronosyltransferases.
Supporting Information Available: Product ion spectrum of urinary HON-PhIP-N3-Gl and synthetic standard (Figure S-1), product ion spectrum of urinary HON-PhIP-N2-Glu and synthetic standard (Figure S-2), and product ion spectrum of novel glucuronide conjugates of hydroxylated PhIP metabolite (Figure S-3). This material is available free of charge via the Internet at http://pubs.acs.org.
Reference List
- 1.Sugimura T, Wakabayashi K, Nakagama H, Nagao M. Heterocyclic amines: Mutagens/carcinogens produced during cooking of meat and fish. Cancer Sci. 2004;95:290–299. doi: 10.1111/j.1349-7006.2004.tb03205.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Felton JS, Knize MG, Bennett LM, Malfatti MA, Colvin ME, Kulp KS. Impact of environmental exposures on the mutagenicity/carcinogenicity of heterocyclic amines. Toxicology. 2004;198:135–145. doi: 10.1016/j.tox.2004.01.024. [DOI] [PubMed] [Google Scholar]
- 3.Ni W, McNaughton L, LeMaster DM, Sinha R, Turesky RJ. Quantitation of 13 heterocyclic aromatic amines in cooked beef, pork, and chicken by liquid chromatography-electrospray iIonization/tandem mass spectrometry. J Agric Food Chem. 2008;56:68–78. doi: 10.1021/jf072461a. [DOI] [PubMed] [Google Scholar]
- 4.Friesen MD, Kaderlik K, Lin D, Garren L, Bartsch H, Lang NP, Kadlubar FF. Analysis of DNA adducts of 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine in rat and human tissues by alkaline hydrolysis and gas chromatography/electron capture mass spectrometry: validation by comparison with 32P-postlabeling. Chem Res Toxicol. 1994;7:733–739. doi: 10.1021/tx00042a004. [DOI] [PubMed] [Google Scholar]
- 5.Magagnotti C, Pastorelli R, Pozzi S, Andreoni B, Fanelli R, Airoldi L. Genetic polymorphisms and modulation of 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP)-DNA adducts in human lymphocytes. Int J Cancer. 2003;107:878–884. doi: 10.1002/ijc.11492. [DOI] [PubMed] [Google Scholar]
- 6.National Toxicology Program Report on Carcinogenesis. 11. 2005 U.S. Department of Health and Human Services, Public Health Service; Research Triangle Park, N.C: 2005. [Google Scholar]
- 7.Zhao K, Murray S, Davies DS, Boobis AR, Gooderham NJ. Metabolism of the food derived mutagen and carcinogen 2-amino-1-methyl-6-phenylimidazo(4,5-b)pyridine (PhIP) by human liver microsomes. Carcinogenesis. 1994;15:1285–1288. doi: 10.1093/carcin/15.6.1285. [DOI] [PubMed] [Google Scholar]
- 8.Hammons GJ, Milton D, Stepps K, Guengerich FP, Kadlubar FF. Metabolism of carcinogenic heterocyclic and aromatic amines by recombinant human cytochrome P450 enzymes. Carcinogenesis. 1997;18:851–854. doi: 10.1093/carcin/18.4.851. [DOI] [PubMed] [Google Scholar]
- 9.Turesky RJ, Constable A, Richoz J, Varga N, Markovic J, Martin MV, Guengerich FP. Activation of heterocyclic aromatic amines by rat and human liver microsomes and by purified rat and human cytochrome P450 1A2. Chem Res Toxicol. 1998;11:925–936. doi: 10.1021/tx980022n. [DOI] [PubMed] [Google Scholar]
- 10.Crofts FG, Sutter TR, Strickland PT. Metabolism of 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine by human cytochrome P4501A1, P4501A2 and P4501B1. Carcinogenesis. 1998;19:1969–1973. doi: 10.1093/carcin/19.11.1969. [DOI] [PubMed] [Google Scholar]
- 11.Alexander J, Heidenreich B, Reistad R, Holme JA. Metabolism of the food carcinogen 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP) in the rat and other rodents. In: Adamson RH, Gustafsson J-A, Ito N, Nagao M, Sugimura T, Wakabayashi K, Yamazoe Y, editors. Heterocyclic amines in cooked foods: Possible human carcinogens. 23rd Proceedings of the Princess Takamatusu Cancer Society. Princeton Scientific Publishing Co., Inc; New Jersey: 1995. pp. 59–68. [Google Scholar]
- 12.Turesky RJ, Guengerich FP, Guillouzo A, Langouet S. Metabolism of heterocyclic aromatic amines by human hepatocytes and cytochrome P4501A2. Mutat Res. 2002;506–507:187–195. doi: 10.1016/s0027-5107(02)00165-3. [DOI] [PubMed] [Google Scholar]
- 13.Kaderlik KR, Minchin RF, Mulder GJ, Ilett KF, Daugaard-Jenson M, Teitel CH, Kadlubar FF. Metabolic activation pathway for the formation of DNA adducts of the carcinogen 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP) in rat extrahepatic tissues. Carcinogenesis. 1994;15:1703–1709. doi: 10.1093/carcin/15.8.1703. [DOI] [PubMed] [Google Scholar]
- 14.Snyderwine EG, Turesky RJ, Turteltaub KW, Davis CD, Sadrieh N, Schut HA, Nagao M, Sugimura T, Thorgeirsson UP, Adamson RH, Thorgeirsson SS. Metabolism of food-derived heterocyclic amines in nonhuman primates. Mutat Res. 1997;376:203–210. doi: 10.1016/s0027-5107(97)00044-4. [DOI] [PubMed] [Google Scholar]
- 15.Chen C, Ma X, Malfatti MA, Krausz KW, Kimura S, Felton JS, Idle JR, Gonzalez FJ. A comprehensive investigation of 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP) metabolism in the mouse using a multivariate data analysis approach. Chem Res Toxicol. 2007;20:531–542. doi: 10.1021/tx600320w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.King RS, Kadlubar FF, Turesky RJ. In vivo Metabolism of Heterocyclic Amines. In: Nagao M, Sugimura T, editors. Food Borne Carcinogens: Heterocyclic Amines. John Wiley & Sons, Ltd; Chichester, England: 2000. pp. 90–111. [Google Scholar]
- 17.Stillwell WG, Kidd L-CKS-B, Wishnok JW, Tannenbaum SR, Sinha R. Urinary excretion of unmetabolized and phase II conjugates of 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine and 2-amino-3,8-dimethylimidazo[4,5-f]quinoxaline in humans: Relationship to cytochrome P450 1A2 and N-acetyltransferase actvity. Cancer Res. 1997;57:3457–3464. [PubMed] [Google Scholar]
- 18.Strickland PT, Qian Z, Friesen MD, Rothman N, Sinha R. Metabolites of 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP) in human urine after consumption of charbroiled or fried beef. Mutat Res. 2002;506–507:163–173. doi: 10.1016/s0027-5107(02)00163-x. [DOI] [PubMed] [Google Scholar]
- 19.Kulp KS, Knize MG, Fowler ND, Salmon CP, Felton JS. PhIP metabolites in human urine after consumption of well-cooked chicken. J Chromatogr B Analyt Technol Biomed Life Sci. 2004;802:143–153. doi: 10.1016/j.jchromb.2003.09.032. [DOI] [PubMed] [Google Scholar]
- 20.Walters DG, Young PJ, Agus C, Knize MG, Boobis AR, Gooderham NJ, Lake BG. Cruciferous vegetable consumption alters the metabolism of the dietary carcinogen 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP) in humans. Carcinogenesis. 2004;25:1659–1669. doi: 10.1093/carcin/bgh164. [DOI] [PubMed] [Google Scholar]
- 21.Malfatti MA, Dingley KH, Nowell-Kadlubar S, Ubick EA, Mulakken N, Nelson D, Lang NP, Felton JS, Turteltaub KW. The urinary metabolite profile of the dietary carcinogen 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine is predictive of colon DNA adducts after a low-dose exposure in humans. Cancer Res. 2006;66:10541–10547. doi: 10.1158/0008-5472.CAN-06-1573. [DOI] [PubMed] [Google Scholar]
- 22.Frandsen H. Biomonitoring of urinary metabolites of 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP) following human consumption of cooked chicken. Food Chem Toxicol. 2008;46:3200–3205. doi: 10.1016/j.fct.2008.07.008. [DOI] [PubMed] [Google Scholar]
- 23.Wallin H, Mikalsen A, Guengerich FP, Ingelman-Sundberg I, Solberg KE, Rossland OJ, Alexander J. Differential rates of metabolic activation and detoxification of the food mutagen 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine by different cytochrome P450 enzymes. Carcinogenesis. 1990;11:489–492. doi: 10.1093/carcin/11.3.489. [DOI] [PubMed] [Google Scholar]
- 24.Nowell S, Ambrosone CB, Ozawa S, MacLeod SL, Mrackova G, Williams S, Plaxco J, Kadlubar FF, Lang NP. Relationship of phenol sulfotransferase activity (SULT1A1) genotype to sulfotransferase phenotype in platelet cytosol. Pharmacogenetics. 2000;10:789–797. doi: 10.1097/00008571-200012000-00004. [DOI] [PubMed] [Google Scholar]
- 25.Glatt H. Metabolic factors affecting the mutagenicity of heteroyclic amines. In: Skog K, Alexander J, editors. Acrylamide and Other Hazardous Compounds in Heat-Treated fFoods. Woodhead Publishing Ltd; Cambridge, England: 2006. pp. 358–404. [Google Scholar]
- 26.Minchin RF, Reeves PT, Teitel CH, McManus ME, Mojarrabi B, Ilett KF, Kadlubar FF. N- and O-acetylation of aromatic and hetercyclic amine carcinogens by human monomorphic and polymorphic acetyltransferases expressed in COS-1 cells. Biochem and Biophys Res Comm. 1992;185:839–844. doi: 10.1016/0006-291x(92)91703-s. [DOI] [PubMed] [Google Scholar]
- 27.Schut HA, Snyderwine EG. DNA adducts of heterocyclic amine food mutagens: implications for mutagenesis and carcinogenesis. Carcinogenesis. 1999;20:353–368. doi: 10.1093/carcin/20.3.353. [DOI] [PubMed] [Google Scholar]
- 28.Nowell SA, Massengill JS, Williams S, Radominska-Pandya A, Tephly TR, Cheng Z, Strassburg CP, Tukey RH, MacLeod SL, Lang NP, Kadlubar FF. Glucuronidation of 2-hydroxyamino-1-methyl-6-phenylimidazo[4,5-b]pyridine by human microsomal UDP-glucuronosyltransferases: identification of specific UGT1A family isoforms involved. Carcinogenesis. 1999;20:1107–1114. doi: 10.1093/carcin/20.6.1107. [DOI] [PubMed] [Google Scholar]
- 29.Malfatti MA, Felton JS. Human UDP-glucuronosyltransferase 1A1 is the primary enzyme responsible for the N-glucuronidation of N-hydroxy-PhIP in vitro. Chem Res Toxicol. 2004;17:1137–1144. doi: 10.1021/tx049898m. [DOI] [PubMed] [Google Scholar]
- 30.Dellinger RW, Chen G, Blevins-Primeau AS, Krzeminski J, Amin S, Lazarus P. Glucuronidation of PhIP and N-OH-PhIP by UDP-glucuronosyltransferase 1A10. Carcinogenesis. 2007;28:2412–2418. doi: 10.1093/carcin/bgm164. [DOI] [PubMed] [Google Scholar]
- 31.Styczynski PB, Blackmon RC, Groopman JD, Kensler TW. The direct glucuronidation of 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP) by human and rabbit liver microsomes. Chem Res Toxicol. 1993;6:846–851. doi: 10.1021/tx00036a014. [DOI] [PubMed] [Google Scholar]
- 32.Langouët S, Paehler A, Welti DH, Kerriguy N, Guillouzo A, Turesky RJ. Differential metabolism of 2-amino-1-methyl-6-phenylimidazo[4,5- b]pyridine in rat and human hepatocytes. Carcinogenesis. 2002;23:115–122. doi: 10.1093/carcin/23.1.115. [DOI] [PubMed] [Google Scholar]
- 33.Girard H, Thibaudeau J, Court MH, Fortier LC, Villeneuve L, Caron P, Hao Q, von Moltke LL, Greenblatt DJ, Guillemette C. UGT1A1 polymorphisms are important determinants of dietary carcinogen detoxification in the liver. Hepatology. 2005;42:448–457. doi: 10.1002/hep.20770. [DOI] [PubMed] [Google Scholar]
- 34.Strassburg CP, Vogel A, Kneip S, Tukey RH, Manns MP. Polymorphisms of the human UDP-glucuronosyltransferase (UGT) 1A7 gene in colorectal cancer. Gut. 2002;50:851–856. doi: 10.1136/gut.50.6.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alexander J, Wallin H, Rossland OJ, Solberg KE, Holme JA, Becher G, Andersson R, Grivas S. Formation of a glutathione conjugate and a semistable transportable glucuronide conjugate of N2-oxidized species of 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP) in rat liver. Carcinogenesis. 1991;12:2239–2245. doi: 10.1093/carcin/12.12.2239. [DOI] [PubMed] [Google Scholar]
- 36.Hecht SS. Human urinary carcinogen metabolites: biomarkers for investigating tobacco and cancer. Carcinogenesis. 2002;23:907–922. doi: 10.1093/carcin/23.6.907. [DOI] [PubMed] [Google Scholar]
- 37.Lynch AM, Knize MG, Boobis AR, Gooderham N, Davies DS, Murray S. Intra- and interindividual variability in systemic exposure in humans to 2-amino-3,8-dimethylimidazo[4,5-f]quinoxaline and 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine, carcinogens present in food. Cancer Res. 1992;52:6216–6223. [PubMed] [Google Scholar]
- 38.Boobis AR, Lynch AM, Murray S, de la Torre R, Solans A, Farré M, Segura J, Gooderham NJ, Davies DS. CYP1A2-catalyzed conversion of dietary heterocyclic amines to their proximate carcinogens is their major route of metabolism in humans. Cancer Res. 1994;54:89–94. [PubMed] [Google Scholar]
- 39.Alexander J, Reistad R, Hegstad S, Frandsen H, Ingebrigtsen K, Paulsen JE, Becher G. Biomarkers of exposure to heterocyclic amines: approaches to improve the exposure assessment. Food Chem Toxicol. 2002;40:1131–1137. doi: 10.1016/s0278-6915(02)00053-4. [DOI] [PubMed] [Google Scholar]
- 40.Holland RD, Taylor J, Schoenbachler L, Jones RC, Freeman JP, Miller DW, Lake BG, Gooderham NJ, Turesky RJ. Rapid biomonitoring of heterocyclic aromatic amines in human urine by tandem solvent solid phase extraction liquid chromatography electrospray ionization mass spectrometry. Chem Res Toxicol. 2004;17:1121–1136. doi: 10.1021/tx049910a. [DOI] [PubMed] [Google Scholar]
- 41.Stillwell WG, Turesky RJ, Sinha R, Skipper PL, Tannenbaum SR. Biomonitoring of heterocyclic aromatic amine metabolites in human urine. Cancer Lett. 1999;143:145–148. doi: 10.1016/s0304-3835(99)00144-5. [DOI] [PubMed] [Google Scholar]
- 42.Malfatti MA, Kulp KS, Knize MG, Davis C, Massengill JP, Williams S, Nowell S, MacLeod S, Dingley KH, Turteltaub KW, Lang NP, Felton JS. The identification of [2-14C]2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine metabolites in humans. Carcinogenesis. 1999;20:705–713. doi: 10.1093/carcin/20.4.705. [DOI] [PubMed] [Google Scholar]
- 43.Frandsen H. Deconjugation of N-glucuronide conjugated metabolites with hydrazine hydrate--biomarkers for exposure to the food-borne carcinogen 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP) Food Chem Toxicol. 2007;45:863–870. doi: 10.1016/j.fct.2006.11.006. [DOI] [PubMed] [Google Scholar]
- 44.Stillwell WG, Sinha R, Tannenbaum SR. Excretion of the N(2)-glucuronide conjugate of 2-hydroxyamino-1-methyl-6-phenylimidazo[4,5-b]pyridine in urine and its relationship to CYP1A2 and NAT2 activity levels in humans. Carcinogenesis. 2002;23:831–838. doi: 10.1093/carcin/23.5.831. [DOI] [PubMed] [Google Scholar]
- 45.Wishart DS, Tzur D, Knox C, Eisner R, Guo AC, Young N, Cheng D, Jewell K, Arndt D, Sawhney S, Fung C, Nikolai L, Lewis M, Coutouly MA, Forsythe I, Tang P, Shrivastava S, Jeroncic K, Stothard P, Amegbey G, Block D, Hau DD, Wagner J, Miniaci J, Clements M, Gebremedhin M, Guo N, Zhang Y, Duggan GE, Macinnis GD, Weljie AM, Dowlatabadi R, Bamforth F, Clive D, Greiner R, Li L, Marrie T, Sykes BD, Vogel HJ, Querengesser L. HMDB: the Human Metabolome Database. Nucleic Acids Res. 2007;35:D521–D526. doi: 10.1093/nar/gkl923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Turesky RJ, Lang NP, Butler MA, Teitel CH, Kadlubar FF. Metabolic activation of carcinogenic heterocyclic aromatic amines by human liver and colon. Carcinogenesis. 1991;12:1839–1845. doi: 10.1093/carcin/12.10.1839. [DOI] [PubMed] [Google Scholar]
- 47.Westra JG. A rapid and simple synthesis of reactive metabolites of carcinogenic aromatic amines in high yield. Carcinogenesis. 1981;2:355–357. doi: 10.1093/carcin/2.4.355. [DOI] [PubMed] [Google Scholar]
- 48.Frandsen H, Frederiksen H, Alexander J. 2-Amino-1-methyl-6-(5-hydroxy-)phenylimidazo[4,5-b]pyridine (5-OH-PhIP), a biomarker for the genotoxic dose of the heterocyclic amine, 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP) Food Chem Toxicol. 2002;40:1125–1130. doi: 10.1016/s0278-6915(02)00033-9. [DOI] [PubMed] [Google Scholar]
- 49.Murray S, Lake BG, Gray S, Edwards AJ, Springall C, Bowey EA, Williamson G, Boobis AR, Gooderham NJ. Effect of cruciferous vegetable consumption on heterocyclic aromatic amine metabolism in man. Carcinogenesis. 2001;22:1413–1420. doi: 10.1093/carcin/22.9.1413. [DOI] [PubMed] [Google Scholar]
- 50.Mottier P, Hammel YA, Gremaud E, Guy PA. Quantitative high-throughput analysis of 16 (fluoro)quinolones in honey using automated extraction by turbulent flow chromatography coupled to liquid chromatography-tandem mass spectrometry. J Agric Food Chem. 2008;56:35–43. doi: 10.1021/jf072934d. [DOI] [PubMed] [Google Scholar]
- 51.Turesky RJ, Parisod V, Huynh-Ba T, Langouet S, Guengerich FP. Regioselective differences in C(8)- and N-oxidation of 2-amino-3,8- dimethylimidazo[4,5-f]quinoxaline by human and rat liver microsomes and cytochromes P450 1A2. Chem Res Toxicol. 2001;14:901–911. doi: 10.1021/tx010035s. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.