Abstract
The action of multidrug efflux pumps in MDR (multidrug resistance) acquisition has been proposed to partially depend on the transport of physiological substrates which may indirectly affect drug partition and transport across cell membranes. In the present study, the PDR18 gene [ORF (open reading frame) YNR070w], encoding a putative PDR (pleiotropic drug resistance) transporter of the ATP-binding cassette superfamily, was found to mediate plasma membrane sterol incorporation in yeast. The physiological role of Pdr18 is demonstrated to affect plasma membrane potential and is proposed to underlie its action as a MDR determinant, conferring resistance to the herbicide 2,4-D (2,4-dichlorophenoxyacetic acid). The action of Pdr18 in yeast tolerance to 2,4-D, which was found to contribute to reduce [14C]2,4-D intracellular accumulation, may be indirect, given the observation that 2,4-D exposure deeply affects the sterol plasma membrane composition, this effect being much stronger in a Δpdr18 background. PDR18 activation under 2,4-D stress is regulated by the transcription factors Nrg1, controlling carbon source availability and the stress response, and, less significantly, Yap1, involved in oxidative stress and MDR, and Pdr3, a key regulator of the yeast PDR network, consistent with a broad role in stress defence. Taken together, the results of the present study suggest that Pdr18 plays a role in plasma membrane sterol incorporation, this physiological trait contributing to an MDR phenotype.
Keywords: ergosterol homoeostasis, herbicide resistance, multidrug resistance, pleiotropic drug resistance, Saccharomyces cerevisiae
Abbreviations: ABC, ATP-binding cassette; CM, cell membrane; 2,4-D, 2,4-dichlorophenoxyacetic acid; 2,4-DCP, 2,4-dichlorophenol; DiOC6(3), 3-3′-dihexyloxacarbocianine iodide; ER, endoplasmic reticulum; MCPA, 2-methyl-4-chlorophenoxyacetic acid; MDR, multidrug resistance; MFS, major facilitator superfamily; ORF, open reading frame; PDR, pleiotropic drug resistance; RT, reverse transcription
INTRODUCTION
MDR (multidrug resistance), the ability to acquire simultaneous resistance to unrelated chemical compounds, is a widespread phenomenon that often results from the activity of multidrug efflux pumps of the ABC (ATP-binding cassette) superfamily and of the MFS (major facilitator superfamily). These transporters are proposed to actively extrude or compartmentalize a wide range of chemically and structurally disparate drugs and other xenobiotics, thus providing protection from these compounds. However, the prevalence and apparent redundancy of so many MDR transporters, which protect the cell against toxic compounds that are not present in its natural environment, has led to speculation concerning a natural physiological function [1–3].
In recent years, a physiological role for many of these transporters in the model eukaryote Saccharomyces cerevisiae has been proposed. Among MFS-MDR transporters, for example, Tpo1-4 and Qdr3 were suggested to mediate the export of polyamines [4,5], whereas Qdr2 was proposed to catalyse K+ influx [6]. These results suggest that the chemoprotection role of these MDR transporters may come, at least in some cases, as a result of their influence on plasma membrane potential and/or ΔpH control, which in turn can alter the partitioning and accumulation of drugs [2]. On the other hand, most of the yeast PDR (pleiotropic drug resistance) transporters of the ABC superfamily, characterized as MDR determinants, have been associated with the control of lipid incorporation into cell membranes [3]. Interestingly, Pdr5 and its homologues from Candida albicans, Cdr1 and Cdr2, were shown to function as phospholipid translocators [7]. Pdr10 and Pdr15 were also implicated in membrane lipid organization [8,9], and Aus1 and Pdr11 were found to mediate non-vesicular movement of plasma membrane sterol to the endoplasmic reticulum in S. cerevisiae, facilitating exogenous sterol uptake [10]. The deletion of yet another S. cerevisiae transporter-encoding gene, PDR16, which is regulated by the PDR network transcription factor Pdr1, also leads to changes in yeast plasma membrane sterol composition [11]. Taken together, these results have been suggested to imply that the MDR phenotype conferred by PDR transporters may occur, to some extent, due to the changes they impose on the composition of cell membranes, which in turn may affect drug partition or transport. This hypothesis is consistent with the fact that ERG2, ERG4 and ERG5 genes, encoding the final enzymes of the ergosterol biosynthetic pathway, were found to be determinants of MDR [12].
In the present study, the functional analysis of PDR18 [ORF (open reading frame) YNR070w], encoding a putative yeast PDR transporter of the plasma membrane, was undertaken. Based on the fact that PDR18 is up-regulated (1.6-fold) in yeast cells challenged by the 2,4-D (herbicide 2,4-dichlorophenoxyacetic acid), as previously revealed by microarray analysis [13], the PDR18 gene was found to confer yeast resistance to the auxin-like herbicides 2,4-D and MCPA (2-methyl-4-chlorophenoxyacetic acid), among other xenobiotic compounds. Our previous studies on the yeast adaptive response mechanisms to the herbicide 2,4-D (reviewed in [14]) indicate that the expression of the drug:H+ antiporter Tpo1, of the ABC transporter Pdr5, and of the Arabidopsis thaliana Tpo1 homologue At5g13750, confers 2,4-D resistance in S. cerevisiae and leads to a lower intracellular accumulation of the herbicide [15,16]. In the present study, the role and regulation of PDR18 expression in yeast resistance to 2,4-D is scrutinized. Given the previous implication of PDR transporters in phospholipid and ergosterol metabolism, the role of Pdr18 in yeast plasma membrane lipid composition was studied. PDR18 expression was found to affect plasma membrane potential and sterol composition, allowing us to propose Pdr18 as mediator of non-vesicular ergosterol transport into the plasma membrane, this physiological role being, at least partially, responsible for the observed MDR phenotype.
EXPERIMENTAL
Strains, plasmids and growth conditions
The parental S. cerevisiae strain BY4741 (MATa, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0) and the derived single deletion mutants Δpdr18, Δnrg1, Δyap1 and Δpdr3 used in the present study were obtained from the EUROSCARF collection. Plasmids pRS416_PDR18 (EUROSCARF), expressing the PDR18 gene from its natural promoter, pRS416_PDR18Δp, and the cloning vector pRS416 (EUROSCARF) were also used. Plasmid pRS416_PDR18Δp was obtained through site-directed mutagenesis of pRS416_PDR18 using the QuikChange® XL site-directed mutagenesis kit (Stratagene), and exhibit a substitution of two nucleotides in the putative Nrg1-binding site in the PDR18 promoter region. The pair of mutagenic oligonucleotides used for this procedure were 5′-GTTTCGTTTCATATGATTGCTTAGGTACCATTC-3′ and its complementary sequence 5′-GAATGGTACCTAAGCAATCATATGAAACGAAAC-3′, in which the replacement nucleotides are underlined.
Yeast cells were cultivated at 30°C with agitation at 250 rev./min in minimal growth medium MM4 [pH 3.5 (adjusted with HCl)], containing 1.7 g/l yeast nitrogen base without amino acids or (NH4)2SO4 (Difco), 20 g/l glucose (Merck), 2.65 g/l (NH4)2SO4 (Merck), 20 mg/l L-histidine (Merck), 20 mg/l L-methionine (Merck), 60 mg/l L-leucine (Sigma) and 20 mg/l L-uracil (Sigma). Solid medium [pH 4.0 (adjusted with HCl)] was prepared by adding 20 g/l agar (Iberagar). Cells harbouring pRS416 or derived plasmids were grown in the same medium conditions and without uracil (MM4-U) to ensure plasmid segregational stability.
Susceptibility assays
The susceptibility of the parental strain BY4741 and Δpdr18 deletion mutant to toxic concentrations of 2,4-D was assessed by comparing their growth curves or growth in spot assays in MM4 medium supplemented or not with inhibitory concentrations of 2,4-D (0.45 mM in liquid medium and 1–2.5 mM in solid medium). Cell suspensions used for the spot assays were prepared as described previously [5]. Besides 2,4-D, other chemical stress inducers (obtained from Sigma) were tested in the specified concentration ranges: the herbicides MCPA (1–1.5 mM) and barban (0.08–0.1 mM); 2,4-DCP (2,4-dichlorophenol; a 2,4-D degradation intermediate) (0.5–1 mM); the fungicides benomyl (17–35 mg/l) and mancozeb (1–1.2 mg/l); and salts of several toxic metal cations, namely, CdSO4 (0.025–0.04 mM), CuSO4 (0.5–1 mM), MnSO4 (5–10 mM), ZnSO4 (5–10 mM), Al2(SO4)3 (0.5–1 mM), TlCl3 (0.1–0.5 mM), CoSO4 (1.5–2 mM) and PbSO4 (1.5–2 mM).
PDR18 gene expression assays
The changes registered in the transcript levels from the PDR18 gene in BY4741 and in derived mutants Δpdr18, Δnrg1, Δyap1 and Δpdr3, upon yeast exposure to 0 mM, 0.3 mM or 0.45 mM 2,4-D, were assessed by real-time RT (reverse transcription)–PCR. RNA extraction from yeast cells was carried out as described previously [15]. The RT–PCR protocol used followed the manufacturer's instructions and has been described previously [5]. Primers for amplification of the PDR18 and ACT1 cDNA were designed using Primer Express Software (Applied Biosystems) and were 3′-TTGGCAAGCCGGATCTGT-5′, 3′-CCACGCGGATTGGGAAT-5′ and 3′-CTCCACCACTGCTGAAAGAGAA-5′, 3′-CCAAGGCGACGTAACATAGTTTT-5′ respectively. The RT–PCR reaction was carried out using a thermal cycler block (7500 Real-Time PCR System; Applied Biosystems). The ACT1 mRNA level was used as the internal control. The relative values obtained for unstressed conditions were set as 1 and the remaining values are relative to that value.
[14C]2,4-D accumulation assays
The intracellular accumulation ratio of 14C-labelled 2,4-D in the parental strain BY4741 and the derived deletion mutant Δpdr18 was assessed as described previously [16].
Plasma membrane potential (ΔΨ) estimation
To estimate the differences in BY4741 and derived mutant Δpdr18 plasma membrane potential, two methods were used: the [14C]methylamine uptake assay [17] and the DiOC6(3) (3-3′-dihexyloxacarbocianine iodide) accumulation assay [18].
The uptake of [14C]methylamine in the parental strain BY4741 and the mutant strain Δpdr18 was monitored as described previously [5,17].
To estimate DiOC6(3) fluorescence, cells were harvested as described above, resuspended in Mes/glucose buffer [10 mM Mes, 0.1 mM MgCl2 and 20 g/l glucose (pH 6)], supplemented with DiOC6(3) (Molecular Probes)] at a final concentration of 0.25 nM and incubated in the dark for 30 min at 30°C with orbital agitation. After centrifugation (8600 g for 5 min at 4°C), cells were immediately observed with a Zeiss Axioplan microscope equipped with adequate epifluorescence filters (BP450-490 and LP520). Fluorescence emission was collected with a CCD (charge-coupled device) camera (Cool SNAPFX, Roper Scientific Photometrics). Bright-field images for determination of ΔΨ were obtained concurrently and recorded at 1 min intervals, each experiment being finished within 15 min. The images were analysed using MetaMorph 3.5. The fluorescence images were background-corrected using dark-current images. The intensity values were calculated for a minimum of 80 cells per experiment. Individual cells were selected using regions of interest obtained from bright-field images recorded before or after the experiment. The value of fluorescence intensity emitted by each cell was obtained pixel-by-pixel in the region of interest. Fluorescence levels given by the software were expressed as a percentage.
Plasma membrane sterol composition assessment
Total CMs (cell membranes) were extracted and prepared from yeast cells grown in MM4 medium (pH 3.5) and harvested in the exponential growth phase. For studying the effect of 2,4-D, exponential cells grown in MM4 medium were transferred to fresh medium supplemented with 0.45 mM 2,4-D, and grown for 1 h at 30°C. Cells were harvested by centrifugation, resuspended in homogenization buffer [50 mM Tris/HCl (pH 7.5), 2.5 mM EDTA and a protease inhibitor cocktail (1 mM PMSF and 1 μg/ml each of leupeptin, pepstatin A and aprotinin)] and broken by vortex mixing with glass beads (Glaperlon 0.40–0.60 mm). The CMs were recovered by centrifugation at 1000 g to remove unbroken cells and finally the CMs were pelleted by ultracentifugation at 25000 rev./min (rotor type SW41Ti, Beckman Coulter) for 1 h. The CMs were resuspended in a buffer containing 20 mM Tris/HCl (pH 7.5), 150 mM NaCl, 20% glycerol and protease inhibitors at the concentrations mentioned above. Plasma membrane fractions were obtained from CM fractions by sucrose gradient centrifugation as described by Monk et al. [19]. The CM and plasma membrane protein concentrations obtained, measured using the BCA (bicinchoninic acid) test, ranged from 8 to 19 μg/μl and 3 to 9 μg/μl respectively. Equal amounts of plasma membranes were used for lipid extraction from each of the yeast strains using the method described by Bligh and Dyer [20] with slight modifications. Sterols were extracted and analysed using a method described previously with slight modifications [21]. The saponified lipids were re-extracted using 5 ml of hexane and vortex mixed several times. Water (1 ml) was added to separate the organic phase, which was then dried in nitrogen at 60°C. The extracted sterols were derivatized using 100 μl of BSTFA/TMCS [N,O-Bis(trimethylsilyl) trifluoroacetamide with trimethyl-chlorosilane; Sigma] at 80°C for 1 h in nitrogen. The derivatized sterols were then analysed using GC-MS (Schimadzu QP2010 Plus, Japan) and a DB5-MS column 60 m×0.2 mm with a film thickness of 0.20 μm. The carrier gas was helium with a flow rate of 1 ml/min and a pressure of 80.8 kPa. The initial column temperature of 120°C was held for 1 min and then programmed at 120–250°C at 5°C/min where it was held for 30 min. A 1 μl injection was made using a split ratio of 10. The injection temperature was 300°C. The total ion mass spectra were recorded in the mass range m/z 40–650 at a scan rate of 1 s/scan. The interface and detector temperature was 300°C. Peak identification was based on relative retention time and total ion mass spectral comparison with an external standard. The sterol standards were obtained from Sigma–Aldrich.
RESULTS
The ABC transporter Pdr18, encoded by ORF YNR070w, confers yeast resistance to 2,4-D and other chemical stresses
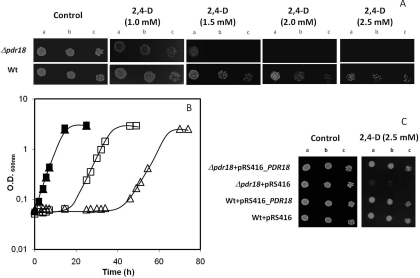
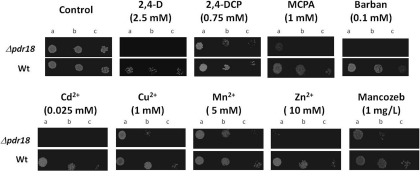
The susceptibility towards 2,4-D-imposed stress of the single deletion mutant Δpdr18 was found to be higher compared with the parental strain, based on spot assays and liquid growth (Figure 1). In the absence of Pdr18, yeast cells become susceptible to 2,4-D, even when supplemented at concentrations that hardly affected wild-type viability (Figure 1A). For higher concentrations of the herbicide, the expression of this transporter become essential for survival under stress (Figure 1A). PDR18 deletion was also found to lead to a longer 2,4-D-induced lag-phase, and also a reduced value of final biomass concentration (Figure 1B). The expression of PDR18 from a centromeric plasmid was found to rescue the 2,4-D-susceptibility phenotype of Δpdr18 cells, to levels comparable with the parental strain, whereas no effect was detected in the control cells harbouring the corresponding cloning vector (Figure 1C). The ability of this gene expression to confer resistance to other pesticides and chemical compounds of agroeconomic importance was further analysed, and PDR18 was also found to be a determinant of yeast resistance to the herbicides MCPA and barban, to the 2,4-D degradation intermediate 2,4-DCP, to the agricultural fungicide mancozeb, and to the metal cations Zn2+, Mn2+, Cu2+ and Cd2+ (Figure 2). No protection was conferred by PDR18 expression towards benomyl, Co2+, Pb2+, Al3+ or Tl3+ (results not shown).
Figure 1. Comparison of the susceptibility to 2,4-D of S. cerevisiae BY4741 and the derived deletion mutant Δpdr18, through cultivation in liquid medium (B) or spot assays (A and C).
(A) Spot assays were carried out as described in the Experimental section. The cell suspensions used to prepare the spots in lanes b and c were 1:5 and 1:25 serial dilutions respectively of the suspensions with a D600=0.05±0.005 spotted in lane a. Pictures were taken after 2 days of incubation. (B) Growth curves of wild-type (wt) (■,□), and Δpdr18 (▲,Δ) cells in MM4 liquid medium, pH 3.5 (■,▲) or in this medium supplemented with 0.45 mM 2,4-D (□,Δ). Cells of the inocula were grown in the absence of the herbicide and the growth curves are representative of at least three independent experiments. (C) Spot assays were carried as described in the Experimental section. The cell suspensions used to prepare the spots in lanes b and c were 1:5 and 1:25 serial dilutions respectively of the suspensions with an D600=0.05±0.005 spotted in lane a. Pictures were taken after 4 days of incubation.
Figure 2. Susceptibility to the herbicides 2,4-D, MCPA and barban, to 2,4-DCP, to the agricultural fungicide mancozeb, and to the metal ions Zn2+, Mn2+, Cu2+ and Cd2+ induced stress of the deletion mutant Δpdr18 compared with the wild-type, assessed by spot assays.
Cells used for the spot assays were prepared as described in the Experimental section. The cell suspensions used to prepare the spots in lanes b and c were 1:5 and 1:25 serial dilutions respectively, of the suspensions with an D600=0.05±0.005 spotted in lane a. Pictures were taken after 2 or 3 days of incubation. wt, wild-type.
PDR18 transcription is activated in response to 2,4-D-imposed stress in an Nrg1-, Yap1- and Pdr3-dependent manner
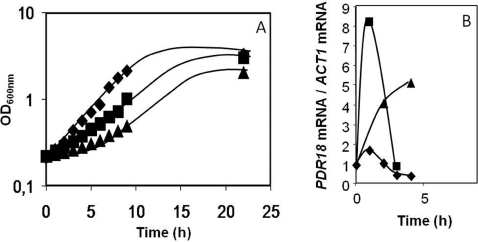
A strong increase in the transcript levels of PDR18 (~8-fold) was registered after 1 h of exposure of an unadapted S. cerevisiae cell population to 0.3 mM 2,4-D. This strong, but transient, increase was followed by a rapid decrease of transcript levels to basal values as cells adapted to growth in the presence of the herbicide. When exposed to a higher concentration of 2,4-D (0.45 mM), PDR18 transcriptional activation reached a maximum of up to 5-fold after 4 h of stress exposure, correlating with the longer duration of the lag-phase imposed by this higher 2,4-D concentration (Figure 3). The fact that a higher herbicide concentration apparently leads to a lower activation of PDR18 may be due to the fact that 0.45 mM 2,4-D induces viability loss, thus reducing the number of cells in the population with the ability to generate a stress response [22].
Figure 3. PDR18 transcript levels in yeast cells exposed to 2,4-D-imposed stress.
(A) Comparison of the susceptibility to 2,4-D-induced stress of S. cerevisiae parental strain BY4741 exposed to 0 (◆), 0.3 (■) or 0.45 (▲) mM 2,4-D through cultivation in MM4 liquid medium (pH 3.5). Cells used to prepare the inocula were previously grown in the absence of 2,4-D until mid-exponential phase. Growth curves are representative of at least three independent growth experiments. (B) Comparison of the relative transcript values of PDR18 mRNA/ACT1 mRNA, in cells of parental strain BY4741 during the period of adaptation to 2,4-D by RT–PCR. The PDR18 mRNA value for the control conditions (0 h, unsupplemented medium) was set as 1 and the remaining values were relative values. Values are the means±S.D. for at least three independent experiments.
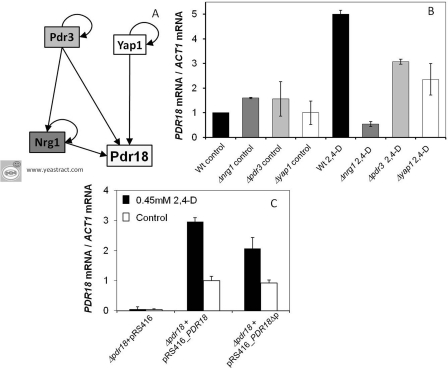
The YEASTRACT database (http://www.yeastract.com) [23,24], was used to guide the analysis of the transcriptional control underlying PDR18 activation. Five transcription factors were identified as documented PDR18 regulators previously proven to bind to its promoter region, and, thus, were selected as candidates for, directly or indirectly, controlling 2,4-D-induced PDR18 up-regulation (Figure 4A). Of these, only Nrg1, Pdr3 and Yap1 were examined in the present study because mutants deleted for the other two genes were either unviable (Δrap1) or exhibited marked growth defects (Δswi4) [SGD (Saccharomyces Genome Database); http://www.yeastgenome.com]. The three mutant strains tested, devoid of PDR3, YAP1 or NRG1, exhibit nearly identical PDR18 mRNA basal levels, as registered in unstressed parental strain cells. However, the PDR18 up-regulation detected in the wild-type cells after 4 h of incubation with 2,4-D was abrogated in the Δnrg1 mutant or reduced in Δyap1 and Δpdr3 mutants (Figure 4B), suggesting that both Pdr3 and Yap1 transcription factors are necessary to ensure full activation of PDR18.
Figure 4. PDR18 expression regulation under 2,4-D-imposed stress.
(A) Representation of the putative regulatory network controlling PDR18 transcription, according to the information in the YEASTRACT database (http://www.yeastract.com). (B) Relative values of PDR18 mRNA in wild-type strain (wt) and Δnrg1, Δpdr3 and Δyap1 mutant cells before and 4 h following a yeast cell population exposure to 0.45 mM 2,4-D. The relative value of mRNA for the wild-type strain immediately before exposure to the herbicide (control) was set as 1. Values are means±S.D. for at least three independent experiments. (C) Relative values of PDR18 mRNA in Δpdr18 cells transformed with pRS416_PDR18, pRS416_PDR18Δp or the corresponding empty vector before and 4 h following a yeast cell population exposure to 0.45 mM 2,4-D. The relative value of mRNA for the Δpdr18 strain, harbouring the pRS416_PDR18 plasmid, immediately before exposure to the herbicide was set as 1. Values are means±S.D. for at least three independent experiments.
Since Nrg1 has been described as a transcriptional repressor, its action as an activator of PDR18 was hypothesized to be indirect. Nonetheless, this transcription factor was previously demonstrated, through genome-wide screenings [25,26], to bind to the PDR18 promoter region. Furthermore, according to the YEASTRACT database, a potential Nrg1-binding site can be found in the PDR18 promoter at position −567. To evaluate whether or not the action of Nrg1 on PDR18 transcriptional up-regulation might be direct, site-directed mutagenesis was used to abrogate the putative Nrg1-binding site found in the pRS416_PDR18 expression plasmid. Both pRS416_PDR18 and pRS416_PDR18Δp plasmids were transformed into Δpdr18 cells, so that the genomic expression of PDR18 from its natural promoter would not be accounted for. RT–PCR was used to measure the PDR18 transcript levels in these cells upon exposure to 0.45 mM 2,4-D. The abrogation of the Nrg1-binding site was seen to have only a moderate effect on herbicide-induced PDR18 up-regulation (Figure 4C) when compared with the full effect observed upon Nrg1 deletion. Taken together, these results suggest that the action of Nrg1 on PDR18 expression appears to be indirect in this case.
Role of PDR18 expression in 2,4-D intracellular accumulation
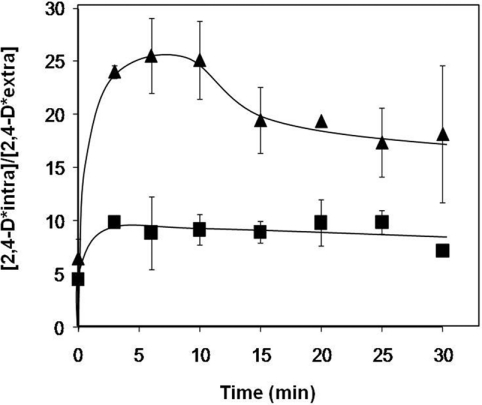
Given the presumed role of Pdr18 as a plasma membrane MDR transporter that confers resistance on yeast cells against 2,4-D-imposed stress, the effect of PDR18 expression on the intracellular accumulation of [14C]2,4-D was assessed. The accumulation of [14C]2,4-D in non-adapted yeast cells suddenly exposed to the presence of 0.3 mM 2,4-D (at pH 3.5), which induces a mild growth inhibition in both the parental strain and Δpdr18 cells (results not shown), was 2.5-fold higher in cells devoid of PDR18 than in parental cells (Figure 5). This result strongly suggests that Pdr18 activity increases yeast resistance towards 2,4-D by reducing the accumulation of the 2,4-D anion within yeast cells, presumably by catalysing the direct extrusion of the herbicide.
Figure 5. Accumulation of [14C]2,4-D.
Comparison of [14C]2,4-D accumulation in non-adapted cells of S. cerevisiae BY4741 (■) and the derived deletion mutant Δpdr18 (▲), during cultivation for 30 min in MM4 liquid medium (pH 3.5) supplemented with 0.3 mM non-radioactive 2,4-D (Sigma) and 0.5 μM [14C]2,4-D. The accumulation [2,4-D* intra/2,4-D* extra] values are the means±S.D. for at least three independent experiments.
PDR18 deletion causes changes in yeast plasma membrane sterol composition
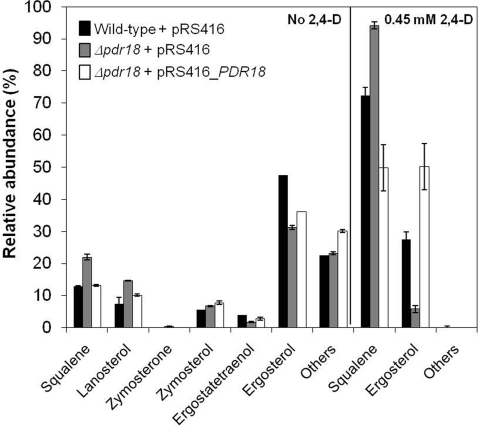
In the absence of 2,4-D supplementation, upon disruption of PDR18, a nearly 2-fold accumulation of squalene and lanosterol, the precursors of ergosterol biosynthetic pathway, and a 1.5-fold reduction of ergostatetraenol and ergosterol content, the end-products of the ergosterol biosynthetic pathways, were detected in the plasma membrane (Figure 6). Upon episomal complementation of PDR18, there was a partial complementation of the Δpdr18 plasma membrane sterol composition phenotype (Figure 6).
Figure 6. PDR18 expression affects plasma membrane sterol composition.
Comparison of the relative abundance of sterol content in yeast plasma membrane of the S. cerevisiae BY4741 strain and the derived deletion mutant Δpdr18 harbouring the PDR18 expression plasmid or the corresponding empty vector, both grown under control conditions or after 1 h of exposure to 0.45 mM 2,4-D, measured by GC-MS. Values are means±S.D. for at least three independent experiments.
The levels of ergosterol in the plasma membrane of S. cerevisiae BY4741 exposed to 2,4-D were found to decrease 1.5-fold compared with unstressed conditions, whereas the levels of squalene increased 5.5-fold. This effect was exacerbated in the absence of the PDR18 gene. Indeed, in Δpdr18 cells 2,4-D exposure led to a 4.3-fold increase in squalene and a 5.2-fold decrease in ergosterol plasma membrane concentrations (Figure 6). Remarkably, under 2,4-D challenge, the relative abundance of squalene became higher than the relative abundance of ergosterol in both wild-type and Δpdr18 backgrounds, and the concentration of the other sterols detected in the plasma membrane of unstressed yeast cells became undetectable (Figure 6).
A lower plasma membrane potential is observed in Δpdr18 cells
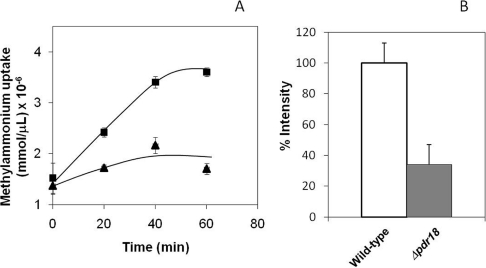
The role of PDR18 expression in the maintenance of yeast membrane potential was also analysed. Yeast plasma membrane potential was first estimated based on the uptake of methylammonium, a non-metabolizable ammonium analogue, whose influx is strongly dependent on the maintenance of the transmembrane potential [27]. The deletion of PDR18 was found to decrease to approximately 60% the level of methylammonium uptake in yeast cells (Figure 7A). Consistent with these results, the fluorescence intensity levels of cells loaded with the DiOC6(3) probe, whose accumulation inside yeast cells is dependent on the plasma membrane potential [18], was 3-fold higher in wild-type cells than in Δpdr18 cells (Figure 7B). Both methods indicate a strong depletion of the plasma membrane potential in the absence of PDR18.
Figure 7. PDR18 gene expression is important to maintain plasma membrane potential in yeast cells.
(A) Time-course accumulation of [14C]methylammonium was followed during incubation of S. cerevisiae BY4741 (■) and the derived deletion mutant Δpdr18 (▲), at 30°C, in growth MM4 liquid medium (pH 3.5), supplemented with the radiolabelled ammonium analogue. Relative levels of [14C]methylammonium, assessed as described in the Experimental section, are means±S.D. for at least three independent experiments. (B) Comparison of the average membrane potential using the fluorescent probe DiCO6(3). Values of membrane potential are set as the percentage of the value obtained for wild-type cells and are means±S.D. for at least three independent experiments.
DISCUSSION
The present study provides the first functional report on the uncharacterized yeast PDR transporter, Pdr18, encoded by ORF YNR070w. Although this gene was not previously characterized, a microarray analysis from our group showed that PDR18 is up-regulated in yeast cells exposed to inhibitory concentrations of the herbicide 2,4-D [13]. Guided by this preliminary result, the present study provides evidence showing that PDR18 is a determinant of yeast resistance to 2,4-D, to MCPA, another auxin like herbicide, and to several other unrelated chemical stresses, including barban, an herbicide of the carbanilate family, mancozeb, an agricultural fungicide, and the soil contaminant metals cadmium, copper, manganese and zinc. This study provides evidence showing that PDR18 gene expression decreases the intracellular accumulation of radiolabelled 2,4-D. The intracellular accumulation pattern observed for the Δpdr18 deletion mutant, compared with the wild-type strain, is similar to the one observed previously for the Δtpo1 deletion mutant [16]. Interestingly, the PDR18 homologue in the plant model Arabidopsis thaliana, AtPDR9, was seen to confer 2,4-D resistance in plants, also contributing to decreased 2,4-D accumulation in plant roots [28].
During the 2,4-D-induced lag-phase period preceding exponential growth resumption under herbicide stress, PDR18 transcript levels were shown to increase transiently. This fact, together with the reduction of the duration of the lag phase induced by 2,4-D due to PDR18 expression, indicates that the role of Pdr18 is preponderant during the period of adaptation to the herbicide, whereas the effect over the inhibition of specific growth exerted by the herbicide is not significant. Consistent with a broad role in stress defence, the transcriptional up-regulation of PDR18 was found to be partially reduced in mutants with either the PDR3 or YAP1 genes deleted, and completely abolished in a mutant devoid of NRG1. The partial effect of Pdr3 in PDR18 activation resembles the effect exerted by Pdr3 over the transcriptional up-regulation of TPO1 under 2,4-D stress described previously [15] and places PDR18 within the yeast PDR network. At the same time, the role of Yap1, the major regulator of S. cerevisiae oxidative stress response, in 2,4-D-induced PDR18 up-regulation may correlate with the observation that this herbicide exerts a pro-oxidant action in yeast [22]. Moreover, Yap1 also plays a role in the control of MDR, regulating the expression of at least ten other MDR proteins: the ABC drug efflux pumps Snq2, Ycf1 and Pdr5, and the drug:H+ antiporters Flr1, Tpo1, Tpo2, Tpo4, Azr1, Yhk8 and Qdr3 (http://www.yeastract.com) [5,23,24]. On the other hand, in the absence of NRG1 the 2,4-D-induced transcriptional up-regulation is completely abrogated. Consistent with the notion that Nrg1 acts as a transcriptional repressor, the action of Nrg1 in this case is proposed to be indirect, based on the fact that the abrogation of the Nrg1-binding site in the PDR18 promoter led only to a slight change in its herbicide-dependent transcriptional up-regulation. The action of Nrg1 is likely to occur through the regulation of other genes, possibly encoding other transcription factors. Interestingly, the transcript levels of the NRG1 gene suffer a 5-fold increase in yeast cells exposed for 15 min to 0.3 mM of 2,4-D, as described in a previous microarray analysis [13]. The same global analysis suggests that yeast cells challenged with toxic concentrations of 2,4-D experience a state of glucose and energy limitation, despite the saturating concentrations of this preferential carbon source in the surrounding medium, which could account for an Ngr1-mediated response [13].
Most significantly, in the present study the deletion of PDR18 in S. cerevisiae cells was found to lead to an accumulation of the precursors of the ergosterol biosynthetic pathway, squalene and lanosterol, and to a decrease in the content of ergostatetraenol and ergosterol, the end-products of the ergosterol biosynthetic pathway, in yeast plasma membrane. Under the same conditions, no changes in the phospholipid composition of the yeast plasma membrane were registered upon PDR18 deletion (results not shown). Although the exact role of Pdr18 in sterol homoeostasis requires clarification, Pdr18 is proposed to play a direct role in the incorporation of ergosterol in the plasma membrane as part of the non-vesicular ER (endoplasmic reticulum)-to-plasma membrane ergosterol transport mechanism [10]. Both in mammalian and yeast cells, newly synthesized cholesterol/ergosterol has been shown to be transported from the ER to the plasma membrane via two mechanisms: one dependent on vesicular transport and the other dependent on ATP, but independent of vesicular transport [29]. However, no specific transporter has so far been implicated in the mediation of this non-vesicular ergosterol movement. The results of the present study suggest that Pdr18 may contribute to this important physiological function. Given this proposed physiological role, the observed apparent inhibitory effect of PDR18 deletion on sterol biosynthesis could result from probing local sterol concentrations, thus influencing the activity of ergosterol-synthesizing enzymes, as suggested for Pdr16 [11].
The lipid composition of a cellular membrane has profound effects on its biophysical properties which may affect the fusibility of a membrane, including intrinsic curvature, thickness, stiffness and permeability [30–32]. Unlike intracellular membranes, the yeast plasma membrane is highly enriched in ergosterol. In various plant models, ergosterol induces changes in membrane potential [33,34] and modifications of H+ fluxes across the membranes [33,35,36], among other effects. A low level of ergosterol leads to disruption of the membrane lateral order [37], which results in membrane fluidization, compromising the physiological membrane potential. Consistent with the depletion of ergosterol in the plasma membrane of Δpdr18 cells, PDR18 expression was also found to be essential in the maintenance of yeast plasma membrane potential. Two probes were used to assess the differences between wild-type and Δpdr18 plasma membrane potential to rule out the hypothesis that the observed variation might result from the direct action of Pdr18 in the excretion of one of the selected probes.
The action of Pdr18 in 2,4-D resistance can be explained in light of its contribution to sterol homoeostasis. It is interesting to see that exposure to the herbicide 2,4-D leads to several changes in membrane sterol composition similar to those caused by PDR18 deletion, including a decrease in ergosterol and an increase in squalene relative concentrations. These changes occurring under 2,4-D stress indicate a possible action of the herbicide as an inhibitor of ergosterol biosynthesis or transport into the plasma membrane, and are consistent with the requirement for PDR18 expression and the observed PDR18 up-regulation registered in the present study. Furthermore, in the absence of PDR18 the effect of 2,4-D in the plasma membrane sterol content is even more pronounced than in wild-type cells. Such a reduced ergosterol content in Δpdr18 cells is likely to increase the permeability of the plasma membrane towards 2,4-D and to affect the active export of 2,4-D to the outer medium, through dedicated transporters, eventually including Tpo1, Pdr5 [15] and Pdr18 itself, consistent with the observed increase accumulation of 2,4-D in yeast cells devoid of PDR18.
On the basis of the results presented in this paper, a physiological role for Pdr18 in the control of sterol homoeostasis specifically in maintaining ergosterol physiological levels in the plasma membrane is proposed. The role of Pdr18 as an MDR determinant is suggested to derive, at least partially, from its physiological role, which is expected to affect drug partition and transport across cell membranes. The results of the present study are expected to increase current knowledge on the action of this family of transporters with an effect on the design of strategies to deal with MDR. Given the particular role of Pdr18 in pesticide resistance, these results may also guide the design of new pesticide-resistant crops of agroeconomic interest.
AUTHOR CONTRIBUTION
Tânia Cabrito participated in the design and optimization of the experiments, and carried out most of the experimental work. Miguel Teixeira carried out the first preliminary experiments, contributing to the conception and co-ordination of the study. Ashutosh Singh performed the plasma membrane sterol composition assessment assays and contributed to the writing of this part of the paper, under the supervision of Rajendra Prasad. Tânia Cabrito, Miguel Teixeira and Isabel Sá-Correia participated in the writing of the paper. Isabel Sá-Correia co-ordinated the study and conceived and supervised all of the work. All authors read and approved the final paper.
ACKNOWLEDGEMENTS
We thank the Advanced Instrumentation Research Facility (AIRF), Jawaharlal Nehru University, New Delhi, India for providing the GC-MS equipment.
FUNDING
This work was supported by Fundação para a Ciência e a Tecnologia (FCT, Portugal) [contracts PTDC/AGR-AAM/102967/2008, PTDC/BIA-MIC/72577/2006 and SFRH/BD/40885/2007 (a Ph.D. fellowship awarded to T.R.C.)]. This work was also supported, in part, by the Department of Biotechnology, JNU, New Delhi, India [grant numbers BT/PR9100/Med/29/03/2007, BT/PR9563/BRB/10/567/2007, BT/PR11158/BRB/10/640/2008, BT/PR13641/MED/29/175/2010 (to R.P.)].
References
- 1.Roepe P. D., Wei L. Y., Hoffman M. M., Fritz F. Altered drug translocation mediated by the MDR protein: direct, indirect, or both? J. Bioenerg. Biomembr. 1996;28:541–555. doi: 10.1007/BF02110444. [DOI] [PubMed] [Google Scholar]
- 2.Sá-Correia I., Santos S., Teixeira M., Cabrito T., Mira N. Drug:H+ antiporters in chemical stress response in yeast. Trends Microbiol. 2009;17:22–31. doi: 10.1016/j.tim.2008.09.007. [DOI] [PubMed] [Google Scholar]
- 3.Jungwirth H., Kuchler K. Yeast ABC transporters: a tale of sex, stress, drugs and aging. FEBS Lett. 2006;580:1131–1138. doi: 10.1016/j.febslet.2005.12.050. [DOI] [PubMed] [Google Scholar]
- 4.Tomitori H., Kashiwagi K., Asakawa T., Kakinuma Y., Michael A. J., Igarashi K. Multiple polyamine transport systems on the vacuolar membrane in yeast. Biochem. J. 2001;353:681–688. doi: 10.1042/0264-6021:3530681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Teixeira M. C., Cabrito T. R., Hanif Z. M., Vargas R. C., Tenreiro S., Sá-Correia I. Yeast response and tolerance to polyamine toxicity involving the drug:H+ antiporter Qdr3 and the transcription factors Yap1 and Gcn4. Microbiology. 2011;157:945–956. doi: 10.1099/mic.0.043661-0. [DOI] [PubMed] [Google Scholar]
- 6.Vargas R. C., Garcia-Salcedo R., Tenreiro S., Teixeira M. C., Fernandes A. R., Ramos J., Sá-Correia I. Saccharomyces cerevisiae multidrug resistance transporter Qdr2 is implicated in potassium uptake, providing a physiological advantage to quinidine-stressed cells. Eukaryot. Cell. 2007;6:134–142. doi: 10.1128/EC.00290-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smriti, Krishnamurthy S., Dixit B. L., Gupta C. M., Milewski S., Prasad R. ABC transporters Cdr1p, Cdr2p and Cdr3p of a human pathogen Candida albicans are general phospholipid translocators. Yeast. 2002;19:303–318. doi: 10.1002/yea.818. [DOI] [PubMed] [Google Scholar]
- 8.Schuller C., Mamnun Y. M., Wolfger H., Rockwell N., Thorner J., Kuchler K. Membrane-active compounds activate the transcription factors Pdr1 and Pdr3 connecting pleiotropic drug resistance and membrane lipid homeostasis in Saccharomyces cerevisiae. Mol. Biol. Cell. 2007;18:4932–4944. doi: 10.1091/mbc.E07-06-0610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rockwell N. C., Wolfger H., Kuchler K., Thorner J. ABC transporter Pdr10 regulates the membrane microenvironment of Pdr12 in Saccharomyces cerevisiae. J. Membr. Biol. 2009;229:27–52. doi: 10.1007/s00232-009-9173-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li Y., Prinz W. A. ATP-binding cassette (ABC) transporters mediate nonvesicular, raft-modulated sterol movement from the plasma membrane to the endoplasmic reticulum. J. Biol. Chem. 2004;279:45226–45234. doi: 10.1074/jbc.M407600200. [DOI] [PubMed] [Google Scholar]
- 11.van den Hazel H. B., Pichler H., do Valle Matta M. A., Leitner E., Goffeau A., Daum G. PDR16 and PDR17, two homologous genes of Saccharomyces cerevisiae, affect lipid biosynthesis and resistance to multiple drugs. J. Biol. Chem. 1999;274:1934–1941. doi: 10.1074/jbc.274.4.1934. [DOI] [PubMed] [Google Scholar]
- 12.Hillenmeyer M. E., Fung E., Wildenhain J., Pierce S. E., Hoon S., Lee W., Proctor M., St Onge R. P., Tyers M., Koller D., et al. The chemical genomic portrait of yeast: uncovering a phenotype for all genes. Science. 2008;320:362–365. doi: 10.1126/science.1150021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Teixeira M. C., Fernandes A. R., Mira N. P., Becker J. D., Sá-Correia I. Early transcriptional response of Saccharomyces cerevisiae to stress imposed by the herbicide 2,4-dichlorophenoxyacetic acid. FEMS Yeast Res. 2006;6:230–248. doi: 10.1111/j.1567-1364.2006.00041.x. [DOI] [PubMed] [Google Scholar]
- 14.Teixeira M. C., Duque P., Sá-Correia I. Environmental genomics: mechanistic insights into toxicity of and resistance to the herbicide 2,4-D. Trends Biotechnol. 2007;25:363–370. doi: 10.1016/j.tibtech.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 15.Teixeira M. C., Sá-Correia I. Saccharomyces cerevisiae resistance to chlorinated phenoxyacetic acid herbicides involves Pdr1p-mediated transcriptional activation of TPO1 and PDR5 genes. Biochem. Biophys. Res. Commun. 2002;292:530–537. doi: 10.1006/bbrc.2002.6691. [DOI] [PubMed] [Google Scholar]
- 16.Cabrito T. R., Teixeira M. C., Duarte A. A., Duque P., Sá-Correia I. Heterologous expression of a Tpo1 homolog from Arabidopsis thaliana confers resistance to the herbicide 2,4-D and other chemical stresses in yeast. Appl. Microbiol. Biotechnol. 2009;84:927–936. doi: 10.1007/s00253-009-2025-5. [DOI] [PubMed] [Google Scholar]
- 17.Hoeberichts F. A., Perez-Valle J., Montesinos C., Mulet J. M., Planes M. D., Hueso G., Yenush L., Sharma S. C., Serrano R. The role of K+ and H+ transport systems during glucose- and H2O2-induced cell death in Saccharomyces cerevisiae. Yeast. 2010;27:713–725. doi: 10.1002/yea.1767. [DOI] [PubMed] [Google Scholar]
- 18.Madrid R., Gomez M. J., Ramos J., Rodriguez-Navarro A. Ectopic potassium uptake in trk1 trk2 mutants of Saccharomyces cerevisiae correlates with a highly hyperpolarized membrane potential. J. Biol. Chem. 1998;273:14838–14844. doi: 10.1074/jbc.273.24.14838. [DOI] [PubMed] [Google Scholar]
- 19.Monk B. C., Kurtz M. B., Marrinan J. A., Perlin D. S. Cloning and characterization of the plasma membrane H+-ATPase from Candida albicans. J. Bacteriol. 1991;173:6826–6836. doi: 10.1128/jb.173.21.6826-6836.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bligh E. G., Dyer W. J. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 21.Sharma M., Dhamgaye S., Singh A., Prasad R. Lipidome analysis reveals antifungal polyphenol curcumin affects membrane lipid homeostasis: targets Δ5,6 desaturase ERG3 of Candida albicans. Front. Biosci. 2011 doi: 10.2741/451. in the press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Teixeira M. C., Telo J. P., Duarte N. F., Sá-Correia I. The herbicide 2,4-dichlorophenoxyacetic acid induces the generation of free-radicals and associated oxidative stress responses in yeast. Biochem. Biophys. Res. Commun. 2004;324:1101–1107. doi: 10.1016/j.bbrc.2004.09.158. [DOI] [PubMed] [Google Scholar]
- 23.Abdulrehman D., Monteiro P. T., Teixeira M. C., Mira N. P., Lourenco A. B., dos Santos S. C., Cabrito T. R., Francisco A. P., Madeira S. C., Aires R. S., et al. YEASTRACT: providing a programmatic access to curated transcriptional regulatory associations in Saccharomyces cerevisiae through a web services interface. Nucleic Acids Res. 2011;39:D136–D140. doi: 10.1093/nar/gkq964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Teixeira M. C., Monteiro P., Jain P., Tenreiro S., Fernandes A. R., Mira N. P., Alenquer M., Freitas A. T., Oliveira A. L., Sá-Correia I. The YEASTRACT database: a tool for the analysis of transcription regulatory associations in Saccharomyces cerevisiae. Nucleic Acids Res. 2006;34:D446–D451. doi: 10.1093/nar/gkj013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harbison C. T., Gordon D. B., Lee T. I., Rinaldi N. J., Macisaac K. D., Danford T. W., Hannett N. M., Tagne J. B., Reynolds D. B., Yoo J., et al. Transcriptional regulatory code of a eukaryotic genome. Nature. 2004;431:99–104. doi: 10.1038/nature02800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee T. I., Rinaldi N. J., Robert F., Odom D. T., Bar-Joseph Z., Gerber G. K., Hannett N. M., Harbison C. T., Thompson C. M., Simon I., et al. Transcriptional regulatory networks in Saccharomyces cerevisiae. Science. 2002;298:799–804. doi: 10.1126/science.1075090. [DOI] [PubMed] [Google Scholar]
- 27.van de Mortel J. B., Mulders D., Korthout H., Theuvenet A. P., Borst-Pauwels G. W. Transient hyperpolarization of yeast by glucose and ethanol. Biochim. Biophys. Acta. 1988;936:421–428. doi: 10.1016/0005-2728(88)90019-9. [DOI] [PubMed] [Google Scholar]
- 28.Ito H., Gray W. M. A gain-of-function mutation in the Arabidopsis pleiotropic drug resistance transporter PDR9 confers resistance to auxinic herbicides. Plant Physiol. 2006;142:63–74. doi: 10.1104/pp.106.084533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sullivan D. P., Ohvo-Rekila H., Baumann N. A., Beh C. T., Menon A. K. Sterol trafficking between the endoplasmic reticulum and plasma membrane in yeast. Biochem. Soc. Trans. 2006;34:356–358. doi: 10.1042/BST0340356. [DOI] [PubMed] [Google Scholar]
- 30.Hancock J. F. Lipid rafts: contentious only from simplistic standpoints. Nat. Rev. Mol. Cell Biol. 2006;7:456–462. doi: 10.1038/nrm1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mukherjee S., Maxfield F. R. Membrane domains. Annu. Rev. Cell Dev. Biol. 2004;20:839–866. doi: 10.1146/annurev.cellbio.20.010403.095451. [DOI] [PubMed] [Google Scholar]
- 32.Dupont S., Beney L., Ferreira T., Gervais P. Nature of sterols affects plasma membrane behavior and yeast survival during dehydration. Biochim. Biophys. Acta. 2011;1808:1520–1528. doi: 10.1016/j.bbamem.2010.11.012. [DOI] [PubMed] [Google Scholar]
- 33.Amborabe B. E., Rossard S., Perault J. M., Roblin G. Specific perception of ergosterol by plant cells. C R Biol. 2003;326:363–370. doi: 10.1016/s1631-0691(03)00118-5. [DOI] [PubMed] [Google Scholar]
- 34.Rossard S., Luini E., Perault J. M., Bonmort J., Roblin G. Early changes in membrane permeability, production of oxidative burst and modification of PAL activity induced by ergosterol in cotyledons of Mimosa pudica. J. Exp. Bot. 2006;57:1245–1252. doi: 10.1093/jxb/erj090. [DOI] [PubMed] [Google Scholar]
- 35.Granado J., Felix G., Boller T. Perception of fungal sterols in plants (subnanomolar concentrations of ergosterol elicit extracellular alkalinization in tomato cells) Plant Physiol. 1995;107:485–490. doi: 10.1104/pp.107.2.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kasparovsky T., Blein J. P., Mikes V. Ergosterol elicits oxidative burst in tobacco cells via phospholipase A2 and protein kinase C signal pathway. Plant Physiol. Biochem. 2004;42:429–435. doi: 10.1016/j.plaphy.2004.04.003. [DOI] [PubMed] [Google Scholar]
- 37.Tierney K. J., Block D. E., Longo M. L. Elasticity and phase behavior of DPPC membrane modulated by cholesterol, ergosterol, and ethanol. Biophys. J. 2005;89:2481–2493. doi: 10.1529/biophysj.104.057943. [DOI] [PMC free article] [PubMed] [Google Scholar]