Abstract
O2 deprivation (hypoxia) and cellular proliferation engage opposite cellular pathways, yet often coexist during tumor growth. The ability of cells to grow during hypoxia results in part from crosstalk between hypoxia inducible factors (HIFs) and the proto-oncogene c-Myc. Acting alone, HIF and c-Myc partially regulate complex adaptations undertaken by tumor cells growing in low O2. However, acting in concert, these transcription factors reprogram metabolism, protein synthesis and cell cycle progression, to “fine tune” adaptive responses to hypoxic environments.
Tumor cell signaling pathways regulating energy production and macromolecular synthesis have garnered substantial interest in recent years. Proto-oncogenes such as c-Myc direct changes in metabolism and protein synthesis supporting enhanced proliferation rates. At the same time, hypoxia and other environmental stresses (e.g. growth factor or nutrient deprivation) redirect intermediate metabolites, to sustain bioenergetics and cell survival. Recent studies describe crosstalk between the c-Myc and HIF pathways, demonstrating an interplay between responses to oxygen (O2) deprivation and a key transcription factor regulating growth (Gordan et al., 2007; Koshiji et al., 2004; Zhang et al., 2007). In this review, we summarize the effects of c-Myc and HIFs on carbon metabolism, protein synthesis, and proliferation, highlighting their antagonist effects on carbon utilization and translation initiation. We will also describe direct effects of HIFs on c-Myc transcriptional activity.
In normal cells, c-Myc is induced upon growth factor stimulation, whereas c-Myc is constituitively high in transformed cells (Campisi et al., 1984). c-Myc overexpression is estimated to occur in 70% of human tumors, including lung and colon adencarcinoma and B cell lymphoma (Nilsson and Cleveland, 2003). c-Myc acts as both a transcriptional activator and repressor, promoting transcription (e.g. cyclin D2 and ornithine decarboxylase [ODC]) by binding E boxes (CACGTG) in a complex with Max, while inhibiting the expression of other genes (e.g. cyclin dependant kinase inhibitors [CKIs] p21 and p27) by binding their initiator elements in a complex with Max and either Miz1 or Sp1. A second group of transcription factors, including Mad1 and Mxi, also bind E box sequences in a complex with Max, but repress transcription (Adhikary and Eilers, 2005).
During rapid cellular proliferation, tumors outstrip their blood supply, limiting O2 and nutrient availability. HIF-α subunits are stabilized under hypoxia (typically less than 3–5% O2) due to decreased activity of prolyl hydroxylases marking them for recognition by the von Hippel-Lindau (VHL) tumor suppressor ubiquitin ligase complex and proteasomal degradation. HIF-α subunits then translocate to the nucleus, dimerize with the stable β-subunit ARNT and promote O2-regulated gene expression. HIF-1α and HIF-2α, the best characterized HIF-α subunits, are differentially expressed: HIF-1α is ubiquitously expressed and HIF-2α is restricted to endothelial, lung, renal and hepatic cells. While HIF-1α and HIF-2α have shared targets, such as vascular endothelial growth factor (VEGF) and adipose differentiation-related protein (ADRP), they also regulate unique gene targets, with HIF-1α regulating glycolytic enzymes (Hu et al., 2003), and HIF-2α activating the stem cell factor Oct4 (Covello et al, 2006). HIF has been recently reviewed (Kaelin, 2005; Semenza, 2003); we focus here on the metabolic outcomes of HIF stabilization. We will refer to effects downstream of both HIF-α subunits as being HIF-mediated, whereas those unique to HIF-1α vs. HIF-2α will be described separately.
Carbon Metabolism
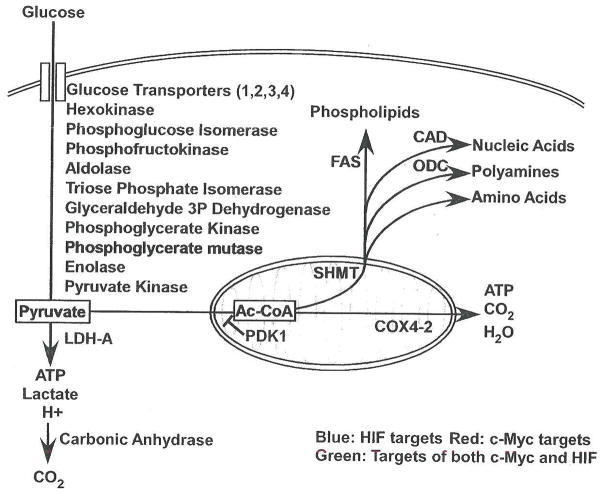
Growth factors induce coordinated transcriptional, translational and post-translational changes to support cell cycle progression, particularly by increasing nutrient uptake and glycolytic metabolism. The resulting elevation in glucose metabolism is striking, and occurs despite the presence of adequate O2 levels for mitochondrial oxidative phosphorylation, a more efficient form of ATP production (Thompson et al., 2005). Pyruvate is produced at a higher rate than it is metabolized by mitochondria, with excesses converted to lactate by lactate dehydrogenase (LDH-A). However, some pyruvate is converted to acetyl-CoA and then citrate in mitochondria, contributing to fatty acid synthesis (Bauer et al., 2004; Bauer et al., 2005). As discussed below, carbon units are modified to produce building blocks for other macromolecules, such as nucleotides and amino acids (Nikiforov et al., 2002).
In transformed cells, high levels of c-Myc promote energy production and biomolecule synthesis required for rapid proliferation, independent of growth factor stimulation. c-Myc potently enhances the glycolytic pathway, increasing target gene expression from glucose transporters through pyruvate kinase (listed in Figure 1), as well as LDH-A, which promotes pyruvate conversion to lactate, regenerates NAD+ from NADH and allows cells to shed glucose-derived carbon through lactate efflux, lowering extracellular pH (Osthus et al., 2000; Shim et al, 1997). Interestingly, LDH-A knockdown has been shown to inhibit Neu-transformed mouse mammary epithelial cell proliferation in vitro and in subcutaneous allografts, possibly by promoting mitochondrial respiration (Fantin et al., 2006). c-Myc activation of LDH-A diverts pyruvate away from mitochondria, but c-Myc also increases mitochondrial mass through gene targets such as mitochondrial transcription factor A (TFAM), and increased mitochondrial iron metabolism (Li et al., 2005; Wu et al., 1999). While c-Myc has been associated with enhanced reactive oxygen species (ROS) production (Vafa et al., 2002), it also increases levels of peroxiredoxin 3, a mitochondrial antioxidant protein whose expression is necessary for c-Myc induced transformation (Wonsey et al., 2002).
Figure 1. Schematic of HIF and c-Myc effects on carbon metabolism.
Both HIF and c-Myc act on multiple targets to regulate carbon utilization. HIF targets are shown in blue, c-Myc targets in red, and targets regulated by both HIF and c-Myc in green. Arrows are included to designate effects on flux through cellular pathways, with blue arrows showing pathways promoted by HIF, red arrows those promoted by c-Myc and green arrows by both.
Why does c-Myc promote both mitochondrial biogenesis and a metabolic shift towards glycolysis? c-Myc drives anabolic pathways, with targets including the nucleotide synthesis complex carbomyl phosphate synthetase aspartate transcarbomylase and dihydroorotase (CAD), serine hydroxymethyltransferase (SHMT), fatty acid synthase (FAS) and ODC, enzymes promoting nucleotide, amino acid, fatty acid and polyamine synthesis respectively (Coller et al., 2000; O’Connell et al., 2003). Each of these processes requires mitochondrial intermediates. The importance of mitochondrial biosynthesis in c-Myc driven proliferation and transformation has been confirmed via genetic manipulations: growth inhibition in c-Myc null fibroblasts is partially rescued by SHMT expression, producing single carbon units for purine synthesis and amino acids such as glycine and methionine (Nikiforov et al., 2002). Similarly, the polyamine synthetic enzyme ODC has been shown to be required for c-Myc mediated lymphomagenesis (Nilsson et al., 2005). Therefore, while the enhancement of glycolysis and diversion of pyruvate to lactate maintains ATP levels during cell growth, growth promotion by c-Myc also requires mitochondrial activity and the production of biosynthetic substrates.
HIF-1α/ARNT dimers also potently enhance glycolytic metabolism (Figure 1) with targets from glucose transporters through LDH-A (Hu et al., 2003; Iyer et al., 1998; Ryan et al., 1998). In contrast to c-Myc, HIF specifically blocks access of glycolytic end products to mitochondria. This effect is mediated by the HIF target Pyruvate Dehydrogenase Kinase 1 (PDK1), which inhibits conversion of pyruvate to acetyl-CoA by phosphorylating pyruvate dehydrogenase, leading to decreased respiration (Kim et al., 2006; Papandreou et al., 2006). At the same time, HIF mediates a shift in the components of cytochrome c oxidase (COX), substituting COX4-2 for COX4-1 via transcriptional upregulation of COX4-2 and the LON protease (which degrades COX4-1; (Fukuda et al., 2007). This results in enhanced electron transport chain (ETC) efficiency under hypoxia, with increased ATP production and decreased ROS generation from those substrates not excluded from mitochondria. HIF-2α/ARNT targets, (e.g. SOD2) also protect cellular and mitochondrial components in the presence of oxidative stress (Scortegagna et al., 2003), suggesting that ROS limitation is an important HIF metabolic adaptation to low O2. Finally, by indirect modulation of c-Myc transcriptional activity (see below), chronic HIF activation decreases overall mitochondrial mass (Hervouet et al., 2005; Zhang et al, 2007).
By blocking pyruvate conversion to acetyl-CoA, HIF decreases anabolic use of glycolytic end products. This has been shown to inhibit de novo fatty acid synthesis (Lum et al., 2007). Similarly, HIF promotes the packaging of extracellular lipid into triglyceride droplets through ADRP, limiting its use in biosynthetic pathways (Bostrom et al., 2006). This should cause a metabolic shift even when c-Myc is present, as the anabolic pathways c-Myc upregulates are substrate-limited (Newsholme et al., 1985). HIF can also attenuate glycolytic pH changes through the activation of carbonic anhydrase expression (Wykoff et al., 2000). Thus, HIF blocks the energetically costly effects of c-Myc and helps tumor cells survive, while leaving c-Myc-directed biosynthetic pathways intact for use after reoxygenation.
Protein Translation
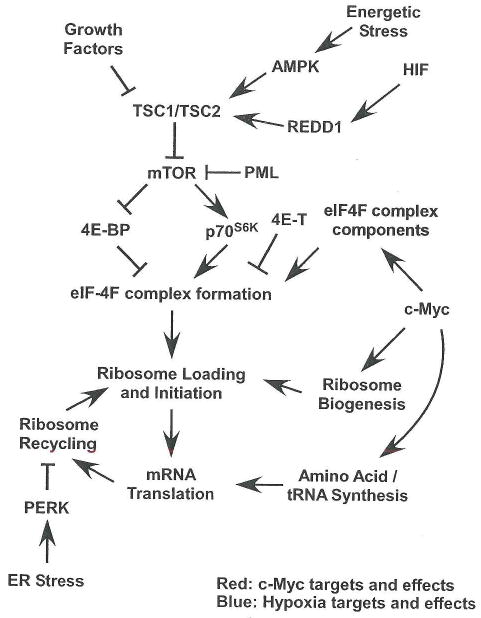
Cell division requires high levels of protein synthesis, effected by growth factor signaling pathway convergence on the tuberous sclerosis complex (TSC1 and TSC2), which regulates the mammalian target of rapamycin complex 1 (mTORC1). mTORC1 phosphorylates 4E-BP and p70 ribosomal protein S6 kinase (p70s6k), promoting assembly of the eIF4F complex (eIF4A, eIF4E and eIF4G), and initiation of cap-dependent translation. Supporting increased translation initiation (shown in Figure 2), c-Myc promotes ribosome and tRNA biogenesis through Pol I-dependent expression of the 45S pre-rRNA, and Pol III-dependent expression of tRNAs and the 5S rRNA. c-Myc enhances rRNA transcription through direct DNA binding, but must associate with the Pol III complex component TFIIIB to increase tRNA and 5S rRNA levels (Arabi et al., 2005; Gomez-Roman et al., 2003; Grandori et al., 2005). eIF4F complex components eIF4E and eIF4G, as well as eIF2α (described below), are also c-Myc transcriptional targets (Coller et al., 2000; Rosenwald et al., 1993).
Figure 2. Hypoxic, HIF and c-Myc effects on protein translation.
While c-Myc promotes ribosome biogenesis and expression of components of the translational machinery, hypoxia and HIFs modulate growth factor signaling pathways that normally upregulate translation. c-Myc targets and c-Myc promoted processes are highlighted in red, while direct HIF targets and hypoxia promoted processes are shown in blue.
Rather than regulating the expression of translation machinery components, O2 deprivation results in HIF-dependent and HIF-independent inhibition of translation initiation (outlined in Figure 2). Anoxia (0% O2) acutely induces eIF-2α phosphorylation (Koumenis et al., 2002), and causes eIF-4E sequestration in cytoplasmic P-bodies by the 4E transporter (4E-T) with more delayed kinetics (Koritzinsky et al., 2006). Even mild hypoxia (1.5% O2) triggers eIF-2α phosphorylation and 4E-BP and p70S6K hypophosphorylation (Arsham et al., 2003; Liu et al., 2006). eIF-2α phosphorylation, which blocks 43S pre-initiation complex regeneration, is mediated by the endoplasmic reticulum resident kinase PERK in a HIF-independent manner (Koumenis et al., 2002). 4E-BP hypophosphorylation is downstream of mTORC1 inhibition resulting from AMP-activated Kinase (AMPK) stimulation by energy depletion (Liu et al., 2006) and HIF induction of REDD1 (Brugarolas et al., 2004; Reiling and Hafen, 2004). REDD1 and AMPK both inhibit mTORC1 function via TSC2, although the mechanism by which REPD1 affects TSC2 is unclear. An additional HIF-independent effect on translation involves the PML tumor suppressor, where PML interacts directly with mTOR, disrupting its association with Rheb (Bernardi et al., 2006). In summary, hypoxia and HIF once again regulates substrate (in this case mRNA) access to biosynthetic machinery produced by c-Myc.
HIF effects on c-Myc and cell cycle control
c-Myc plays a central role in promoting the G1 to S phase cell cycle transition by regulating cyclins and CKIs (Adhikary and Eilers, 2005). The hypoxic induction of HIF-1 α suppresses cell proliferation: acute HIF-1α stabilization at moderate hypoxia (1% O2) results in cell cycle arrest by inhibiting c-Myc transcriptional activity (Koshiji et al., 2004). In contrast, HIF-2α induction promotes cell cycle progression by enhancing c-Myc function (Gordan et al., 2007). It should also be noted that the HIF-2α gene target transforming growth factor-α (TGF-α) promotes Cyclin D1 expression via autocrine signaling (Raval et al., 2005).
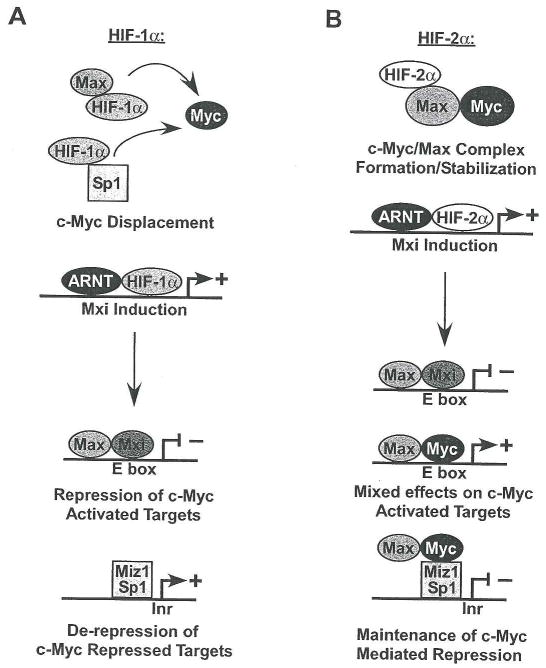
HIF-1α and HIF-2α exhibit opposing effects on c-Myc interaction with its transcription co-factors, disrupting or stabilizing c-Myc DNA binding complexes respectively (see Figure 3). HIF-1α binds directly to Sp1, resulting in c-Myc displacement from Sp1 complexes and decreased c-Myc promoter interaction. Surprisingly, this occurs at two c-Myc repressed targets, p21 and MutSα (Koshiji et al., 2004; Koshiji et al., 2005), where Sp1 is required, and one c-Myc activated target, Nbs1 (To et al., 2006). The Per/Arnt/Sim (PAS)-B domain of HIF-1α mediates its interaction with Sp1. While this domain is highly conserved in HIF-2α, the phosphorylation of a key threonine in the HIF-2α PAS-B domain blocks HIF-2α/Sp1 association (To et al., 2006). However, HIF-2α forms a complex with Max, causing a dose-dependent stabilization of c-Myc/Max association, and increased c-Myc effects on the cell cycle regulators Cyclin D2, E2F1, p21 and p27 (Gordan et al., 2007). These growth promoting effects of HIF-2α occur rapidly, and are detected within 1–2 hours at 0.5% O2. Furthermore, they are likely to be reversible. Of note, direct HIF-α effects on c-Myc transcriptional activity may be attenuated in c-Myc overexpressing cells due to altered c-Myc/Max stoichiometry (Dang CV, personal communication).
Figure 3. Acute and chronic HIF effects on c-Myc transcriptional activity.
When HIF-1α is induced (A), it acts immediately to disrupt c-Myc complexes. By inducing Mxi, it also causes transcriptional repression of some c-Myc target genes. Conversely (B), HIF-2α increases c-Myc transcriptional activity at specific targets, while inhibiting the expression of others via Mxi. By increasing c-Myc/Max interactions, HIF-2α promotes c-Myc mediated activation or repression of Cyclin D2, p21 and p27. However, Mxi induction inhibits expression of other c-Myc activated targets (e.g. CAD and ODC).
A more chronic adaptation results from HIF-mediated Mxi induction causing decreased levels of c-Myc targets ODC, CAD, and peroxisome proliferator-activated receptor gamma coactivator-1β (Zhang et al., 2007). This is correlated with decreased apoptosis under anoxic conditions (0.1% O2), and decreased mitochondrial biogenesis (Corn et al., 2005; Zhang et al., 2007). While this analysis focuses on HIF-1α, HIF-2α appears to also induce Mxi expression in VHL mutant renal cancer cell lines (Zhang et al., 2007). Mxi interacts with Max and binds E boxes to block transcription (Schreiber-Agus et al., 1995). Although Mxi interaction with Max inhibits c-Myc/Max complex formation, repression at E boxes is both necessary and sufficient for Mxi effects on transformation (Harper et al., 1996). Intriguingly, Mxi appears to act on only a subset of c-Myc targets, repressing ODC but not the DNA synthesis enzyme ribose-5-phosphate isomerase (O’Hagan et al., 2000). Similarly, Mxi induction by chronic HIF-α stabilization may reinforce HIF-1α effects. When induced by HIF-2α, Mxi represses some c-Myc targets even as HIF-2α enhances c-Myc transcriptional effects on others. This may promote tumor cell survival by limiting c-Myc influences on energy intensive processes and the production of toxic ROS, while causing increased proliferation rates.
Models for HIF/Myc interplay in tumors
In evaluating the multiple facets of c-Myc/HIFs interactions, it is useful to consider their distinct expression kinetics in solid tumors. Tumor O2 levels oscillate over the course of both hours and days, meaning that tumor cells experience periodic, fluctuating HIF expression (Dewhirst, 2007). While tumors harboring c-Myc amplification likely exhibit constitutively high c-Myc target gene expression, HIF-1α activation should transiently divert substrates away from anabolic synthesis and inhibit c-Myc transcriptional activity only when O2 levels are dangerously low (< 1% O2). However, any appreciable effect on mitochondrial mass or metabolic enzyme expression following short periods of HIF activation is unlikely. Similarly, hypoxia disrupts the eIF-4F complex, temporarily inhibiting translation without dismantling the translational machinery components. When O2 levels return to normal (2–9% for most cells), a tumor will then be able to return to rapid proliferation under the influence of c-Myc. HIF-2α likely has different effects in tumors. For example, HIF-2α appears to be stabilized at higher O2 levels (5%), and for longer time periods, than HIF-1α in neuroblastoma (Holmquist-Mengelbier et al., 2006). HIF-2α expression is also associated with worse prognosis than HIF-1α expression in some tumors (e.g. non-small cell lung and head and neck cancer; Semenza, 2003). HIF-2α does not promote glycolytic metabolism, and should not divert carbon away from mitochondria to the same extent as HIF-1α (Hu et al., 2003). This may allow it to promote angiogenesis while sparing c-Myc’s effect on cell cycle progression.
In VHL-deficient renal tumors, the situation becomes more complex, partly because some renal tumors express different HIF-α subunits (Mandriota et al., 2002; Raval et al., 2005). Those expressing HIF-2α exclusively exhibit enhanced c-Myc-dependent proliferation, while HIF effects on mitochondrial metabolism should decrease O2 consumption and ROS. When HIF-1α and HIF-2α are both present, they likely negate each others’ effect on c-Myc driven proliferation, causing a net decrease in protein translation and mitochondrial mass. In both cases, lipid accumulation is promoted, giving rise to the “clear cell” renal cancer phenotype. This is a very unusual metabolic status for a tumor. However, it is important to consider tumors in their full complexity. Studying such tumors will yield the identification of novel players in tumor metabolism and growth supporting proliferation independent of c-Myc.
References
- Adhikary S, Eilers M. Transcriptional regulation and transformation by Myc proteins. Nat Rev Mol Cell Biol. 2005;6:635–645. doi: 10.1038/nrm1703. [DOI] [PubMed] [Google Scholar]
- Arabi A, Wu S, Ridderstrale K, Bierhoff H, Shiue C, Fatyol K, Fahlen S, Hydbring P, Soderberg O, Grummt I, et al. c-Myc associates with ribosomal DNA and activates RNA polymerase I transcription. Nat Cell Biol. 2005;7:303–310. doi: 10.1038/ncb1225. [DOI] [PubMed] [Google Scholar]
- Arsham AM, Howell JJ, Simon MC. A novel hypoxia-inducible factor-independent hypoxic response regulating mammalian target of rapamycin and its targets. J Biol Chem. 2003;278:29655–29660. doi: 10.1074/jbc.M212770200. [DOI] [PubMed] [Google Scholar]
- Bauer DE, Harris MH, Plas DR, Lum JJ, Hammerman PS, Rathmell JC, Riley JL, Thompson CB. Cytokine stimulation of aerobic glycolysis in hematopoietic cells exceeds proliferative demand. Faseb J. 2004;18:1303–1305. doi: 10.1096/fj.03-1001fje. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer DE, Hatzivassiliou G, Zhao F, Andreadis C, Thompson CB. ATP citrate lyase is an important component of cell growth and transformation. Oncogene. 2005;24:6314–6322. doi: 10.1038/sj.onc.1208773. [DOI] [PubMed] [Google Scholar]
- Bernardi R, Guernah I, Jin D, Grisendi S, Alimonti A, Teruya-Feldstein J, Cordon-Cardo C, Simon MC, Rafii S, Pandolfi PP. PML inhibits HIF-1alpha translation and neoangiogenesis through repression of mTOR. Nature. 2006;442:779–785. doi: 10.1038/nature05029. [DOI] [PubMed] [Google Scholar]
- Bostrom P, Magnusson B, Svensson PA, Wiklund O, Boren J, Carlsson LM, Stahlman M, Olofsson SO, Hulten LM. Hypoxia converts human macrophages into triglyceride-loaded foam cells. Arterioscler Thromb Vase Biol. 2006;26:1871–1876. doi: 10.1161/01.ATV.0000229665.78997.0b. [DOI] [PubMed] [Google Scholar]
- Brugarolas J, Lei K, Hurley RL, Manning BD, Reiling JH, Hafen E, Witters LA, Ellisen LW, Kaelin WG., Jr Regulation of mTOR function in response to hypoxia by REDD1 and the TSC1/TSC2 tumor suppressor complex. Genes Dev. 2004;18:2893–2904. doi: 10.1101/gad.1256804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campisi J, Gray HE, Pardee AB, Dean M, Sonenshein GE. Cell-cycle control of c-myc but not c-ras expression is lost following chemical transformation. Cell. 1984;36:241–247. doi: 10.1016/0092-8674(84)90217-4. [DOI] [PubMed] [Google Scholar]
- Coller HA, Grandori C, Tamayo P, Colbert T, Lander ES, Eisenman RN, Golub TR. Expression analysis with oligonucleotide microarrays reveals that MYC regulates genes involved in growth, cell cycle, signaling, and adhesion. Proc Natl Acad Sci U S A. 2000;97:3260–3265. doi: 10.1073/pnas.97.7.3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corn PG, Ricci MS, Scata KA, Arsham AM, Simon MC, Dicker DT, El-Deiry WS. Mxi1 is induced by hypoxia in a HIF-1-dependent manner and protects cells from c-Myc-induced apoptosis. Cancer Biol Ther. 2005;4:1285–1294. doi: 10.4161/cbt.4.11.2299. [DOI] [PubMed] [Google Scholar]
- Covello KL, Kehler J, Yu H, Gordan JD, Arsham AM, Hu CJ, Labosky PA, Simon MC, Keith B. HIF-2alpha regulates Oct-4: effects of hypoxia on stem cell function, embryonic development, and tumor growth. Genes Dev. 2006;20:557–570. doi: 10.1101/gad.1399906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewhirst MW. Intermittent hypoxia furthers the rationale for hypoxia-inducible factor-1 targeting. Cancer Res. 2007;67:854–855. doi: 10.1158/0008-5472.CAN-06-4744. [DOI] [PubMed] [Google Scholar]
- Fantin VR, St-Pierre J, Leder P. Attenuation of LDH-A expression uncovers a link between glycolysis, mitochondrial physiology, and tumor maintenance. Cancer Cell. 2006;9:425–434. doi: 10.1016/j.ccr.2006.04.023. [DOI] [PubMed] [Google Scholar]
- Fukuda R, Zhang H, Kim JW, Shimoda L, Dang CV, Semenza GL. HIF-1 regulates cytochrome oxidase subunits to optimize efficiency of respiration in hypoxic cells. Cell. 2007;129:111–122. doi: 10.1016/j.cell.2007.01.047. [DOI] [PubMed] [Google Scholar]
- Gomez-Roman N, Grandori C, Eisenman RN, White RJ. Direct activation of RNA polymerase III transcription by c-Myc. Nature. 2003;421:290–294. doi: 10.1038/nature01327. [DOI] [PubMed] [Google Scholar]
- Gordan JD, Bertout JA, Hu CJ, Diehl JA, Simon MC. HIF-2alpha Promotes Hypoxic Cell Proliferation by Enhancing c-Myc Transcriptional Activity. Cancer Cell. 2007;11:335–347. doi: 10.1016/j.ccr.2007.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandori C, Gomez-Roman N, Felton-Edkins ZA, Ngouenet C, Galloway DA, Eisenman RN, White RJ. c-Myc binds to human ribosomal DNA and stimulates transcription of rRNA genes by RNA polymerase I. Nat Cell Biol. 2005;7:311–318. doi: 10.1038/ncb1224. [DOI] [PubMed] [Google Scholar]
- Harper SE, Qiu Y, Sharp PA. Sin3 corepressor function in Myc-induced transcription and transformation. Proc Natl Acad Sci U S A. 1996;93:8536–8540. doi: 10.1073/pnas.93.16.8536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hervouet E, Demont J, Pecina P, Vojtiskova A, Houstek J, Simonnet H, Godinot C. A new role for the von Hippel-Lindau tumor suppressor protein: stimulation of mitochondrial oxidative phosphorylation complex biogenesis. Carcinogenesis. 2005;26:531–539. doi: 10.1093/carcin/bgi001. [DOI] [PubMed] [Google Scholar]
- Holmquist-Mengelbier L, Fredlund E, Lofstedt T, Noguera R, Navarro S, Nilsson H, Pietras A, Vallon-Christersson J, Borg A, Gradin K, et al. Recruitment of HIF-1alpha and HIF-2alpha to common target genes is differentially regulated in neuroblastoma: HTP-2alpha promotes an aggressive phenotype. Cancer Cell. 2006;10:413–423. doi: 10.1016/j.ccr.2006.08.026. [DOI] [PubMed] [Google Scholar]
- Hu CJ, Wang LY, Chodosh LA, Keith B, Simon MC. Differential roles of hypoxia-inducible factor 1alpha (HIF-1alpha) and HIF-2alpha in hypoxic gene regulation. Mol Cell Biol. 2003;23:9361–9374. doi: 10.1128/MCB.23.24.9361-9374.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer NV, Kotch LE, Agani F, Leung SW, Laughner E, Wenger RH, Gassmann M, Gearhart JD, Lawler AM, Yu AY, Semenza GL. Cellular and developmental control of O2 homeostasis by hypoxia-inducible factor 1 alpha. Genes Dev. 1998;12:149–162. doi: 10.1101/gad.12.2.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaelin WG. Proline hydroxylation and gene expression. Annu Rev Biochem. 2005;74:115–128. doi: 10.1146/annurev.biochem.74.082803.133142. [DOI] [PubMed] [Google Scholar]
- Kim JW, Tchernyshyov I, Semenza GL, Dang CV. HIF-1 -mediated expression of pyruvate dehydrogenase kinase: a metabolic switch required for cellular adaptation to hypoxia. Cell Metab. 2006;3:177–185. doi: 10.1016/j.cmet.2006.02.002. [DOI] [PubMed] [Google Scholar]
- Koritzinsky M, Magagnin MG, van den Beucken T, Seigneuric R, Savelkouls K, Dostie J, Pyronnet S, Kaufman RJ, Weppler SA, Voncken JW, et al. Gene expression during acute and prolonged hypoxia is regulated by distinct mechanisms of translational control. Embo J. 2006;25:1114–1125. doi: 10.1038/sj.emboj.7600998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koshiji M, Kageyama Y, Pete EA, Horikawa I, Barrett JC, Huang LE. HIF-1alpha induces cell cycle arrest by functionally counteracting Myc. Embo J. 2004;23:1949–1956. doi: 10.1038/sj.emboj.7600196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koshiji M, To KK, Hammer S, Kumamoto K, Harris AL, Modrich P, Huang LE. HIF-1alpha induces genetic instability by transcriptionally downregulating MutSalpha expression. Mol Cell. 2005;77:793–803. doi: 10.1016/j.molcel.2005.02.015. [DOI] [PubMed] [Google Scholar]
- Koumenis C, Naczki C, Koritzinsky M, Rastani S, Diehl A, Sonenberg N, Koromilas A, Wouters BG. Regulation of protein synthesis by hypoxia via activation of the endoplasmic reticulum kinase PERK and phosphorylation of the translation initiation factor eIF2alpha. Mol Cell Biol. 2002;22:7405–7416. doi: 10.1128/MCB.22.21.7405-7416.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Wang Y, Zeller KI, Potter JJ, Wonsey DR, O’Donnell KA, Kim JW, Yustein JT, Lee LA, Dang CV. Myc stimulates nuclearly encoded mitochondrial genes and mitochondrial biogenesis. Mol Cell Biol. 2005;25:6225–6234. doi: 10.1128/MCB.25.14.6225-6234.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Cash TP, Jones RG, Keith B, Thompson CB, Simon MC. Hypoxia-induced energy stress regulates mRNA translation and cell growth. Mol Cell. 2006;21:521–531. doi: 10.1016/j.molcel.2006.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lum JJ, Bui T, Gruber M, Gordan JD, Deberardinis RJ, Covello KL„, Simon MC, Thompson CB. The transcription factor HIF-1{alpha} plays a critical role in the growth factor-dependent regulation of both aerobic and anaerobic glycolysis. Genes Dev. 2007 doi: 10.1101/gad.1529107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandriota SJ, Turner KJ, Davies D, R„Murray PG, Morgan NV, Sowter HM, Wykoff CC, Maher ER, Harris AL, Ratcliffe PJ, Maxwell PH. HIF activation identifies early lesions in VHL kidneys: evidence for site-specific tumor suppressor function in the nephron. Cancer Cell. 2002;1:459–468. doi: 10.1016/s1535-6108(02)00071-5. [DOI] [PubMed] [Google Scholar]
- Newsholme EA, Crabtree B, Ardawi MS. The role of high rates of glycolysis and glutamine utilization in rapidly dividing cells. Biosci Rep. 1985;5:393–400. doi: 10.1007/BF01116556. [DOI] [PubMed] [Google Scholar]
- Nikiforov MA, Chandriani S, O’Connell B, Petrenko O, Kotenko I, Beavis A, Sedivy JM, Cole MD. A functional screen for Myc-responsive genes reveals serine hydroxymethyltransferase, a major source of the one-carbon unit for cell metabolism. Mol Cell Biol. 2002;22:5793–5800. doi: 10.1128/MCB.22.16.5793-5800.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson JA, Cleveland JL. Myc pathways provoking cell suicide and cancer. Oncogene. 2003;22:9007–9021. doi: 10.1038/sj.onc.1207261. [DOI] [PubMed] [Google Scholar]
- Nilsson JA, Keller UB, Baudino TA, Yang C, Norton S, Old JA, Nilsson LM, Neale G, Kramer DL, Porter CW, Cleveland JL. Targeting ornithine decarboxylase in Myc-induced lymphomagenesis prevents tumor formation. Cancer Cell. 2005;7:433–444. doi: 10.1016/j.ccr.2005.03.036. [DOI] [PubMed] [Google Scholar]
- O’Connell BC, Cheung AF, Simkevich CP, Tam W, Ren X, Mateyak MK, Sedivy JM. A large scale genetic analysis of c-Myc-regulated gene expression patterns. J Biol Chem. 2003;278:12563–12573. doi: 10.1074/jbc.M210462200. [DOI] [PubMed] [Google Scholar]
- O’Hagan RC, Schreiber-Agus N, Chen K, David G, Engelman JA, Schwab R, Alland L, Thomson C, Ronning DR, Sacchettini JC, et al. Gene-target recognition among members of the myc superfamily and implications for oncogenesis. Nat Genet. 2000;24:113–119. doi: 10.1038/72761. [DOI] [PubMed] [Google Scholar]
- Osthus RC, Shim H, Kim S, Li Q, Reddy R, Mukherjee M, Xu Y, Wonsey D, Lee LA, Dang CV. Deregulation of glucose transporter 1 and glycolytic gene expression by c-Myc. J Biol Chem. 2000;275:21797–21800. doi: 10.1074/jbc.C000023200. [DOI] [PubMed] [Google Scholar]
- Papandreou I, Cairns RA, Fontana L, Lim AL, Denko NC. HIF-1 mediates adaptation to hypoxia by actively downregulating mitochondrial oxygen consumption. Cell Metab. 2006;3:187–197. doi: 10.1016/j.cmet.2006.01.012. [DOI] [PubMed] [Google Scholar]
- Raval RR, Lau KW, Tran MG, Sowter HM, Mandriota SJ, Li JL, Pugh CW, Maxwell PH, Harris AL, Ratcliffe PJ. Contrasting properties of hypoxia-inducible factor 1 (HIF-1) and HIF-2 in von Hippel-Lindau-associated renal cell carcinoma. Mol Cell Biol. 2005;25:5675–5686. doi: 10.1128/MCB.25.13.5675-5686.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiling JH, Hafen E. The hypoxia-induced paralogs Scylla and Charybdis inhibit growth by down-regulating S6K activity upstream of TSC in Drosophila. Genes Dev. 2004;18:2879–2892. doi: 10.1101/gad.322704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenwald IB, Rhoads DB, Callanan LD, Isselbacher KJ, Schmidt EV. Increased expression of eukaryotic translation initiation factors eIF-4E and eIF-2 alpha in response to growth induction by c-myc. Proc Natl Acad Sci U S A. 1993;90:6175–6178. doi: 10.1073/pnas.90.13.6175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan HE, Lo J, Johnson RS. HIF-1 alpha is required for solid tumor formation and embryonic vascularization. Embo J. 1998;17:3005–3015. doi: 10.1093/emboj/17.11.3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber-Agus N, Chin L, Chen K, Torres R, Rao G, Guida P, Skoultchi AI, DePinho RA. An ammo-terminal domain of Mxi1 mediates anti-Myc oncogenic activity and interacts with a homolog of the yeast transcriptional repressor SIN3. Cell. 1995;80:777–786. doi: 10.1016/0092-8674(95)90356-9. [DOI] [PubMed] [Google Scholar]
- Scortegagna M, Ding K, Oktay Y, Gaur A, Thurmond F, Yan LJ, Marck BT, Matsumoto AM, Shelton JM, Richardson JA, et al. Multiple organ pathology, metabolic abnormalities and impaired homeostasis of reactive oxygen species in Epas1−/− mice. Nat Genet. 2003;35:331–340. doi: 10.1038/ng1266. [DOI] [PubMed] [Google Scholar]
- Semenza GL. Targeting HIF-1 for cancer therapy. Nat Rev Cancer. 2003;3:721–732. doi: 10.1038/nrc1187. [DOI] [PubMed] [Google Scholar]
- Shim H, Dolde C, Lewis BC, Wu CS, Dang G, Jungmann RA, Dalla-Favera R, Dang CV. c-Myc transactivation of LDH-A: implications for tumor metabolism and growth. Proc Natl Acad Sci U S A. 1997;94:6658–6663. doi: 10.1073/pnas.94.13.6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson CB, Bauer DE, Lum JJ, Hatzivassiliou G, Zong WX, Zhao F, Ditsworth D, Buzzai M, Lindsten T. How do cancer cells acquire the fuel needed to support cell growth? Cold Spring Harb Symp Quant Biol. 2005;70:357–362. doi: 10.1101/sqb.2005.70.011. [DOI] [PubMed] [Google Scholar]
- To KK, Sedelnikova OA, Samons M, Bonner WM, Huang LE. The phosphorylation status of PAS-B distinguishes HIF-1 alpha from HIF-2alpha in NBS1 repression. Embo J. 2006;25:4784–4794. doi: 10.1038/sj.emboj.7601369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vafa O, Wade M, Kern S, Beeche M, Pandita TK, Hampton GM, Wahl GM. c-Myc can induce DNA damage, increase reactive oxygen species, and mitigate p53 function: a mechanism for oncogene-induced genetic instability. Mol Cell. 2002;9:1031–1044. doi: 10.1016/s1097-2765(02)00520-8. [DOI] [PubMed] [Google Scholar]
- Wonsey DR, Zeller KI, Dang CV. The c-Myc target gene PRDX3 is required for mitochondrial homeostasis and neoplastic transformation. Proc Natl Acad Sci U S A. 2002;99:6649–6654. doi: 10.1073/pnas.102523299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu KJ, Polack A, Dalla-Favera R. Coordinated regulation of iron-controlling genes, H-ferritin and IRP2, by c-MYC. Science. 1999;283:676–679. doi: 10.1126/science.283.5402.676. [DOI] [PubMed] [Google Scholar]
- Wykoff CC, Beasley NJ, Watson PH, Turner KJ, Pastorek J, Sibtain A, Wilson GD, Turley H, Talks KL, Maxwell PH, et al. Hypoxia-inducible expression of tumor-associated carbonic anhydrases. Cancer Res. 2000;60:7075–7083. [PubMed] [Google Scholar]
- Zhang H, Gao’ P, Fukuda R, Kumar G, Krishnamachary B, Zeller KI, Dang CV, Semenza GL. HIF-1 Inhibits Mitochondrial Biogenesis and Cellular Respiration in VHL-Deficient Renal Cell Carcinoma by Repression of C-MYC Activity. Cancer Cell. 2007;11:407–420. doi: 10.1016/j.ccr.2007.04.001. [DOI] [PubMed] [Google Scholar]