Abstract
The dynamics of mutations associated with resistance to antiretroviral drugs were analyzed after cessation of therapy. The results showed that the kinetics of the shift to wild-type amino acid residues were significantly faster for protease inhibitors, intermediate for nonnucleoside reverse transcriptase inhibitors, and slower for nucleoside reverse transcriptase inhibitors.
Structured treatment interruptions (STIs) have been proposed as a strategy for minimizing the cost and toxicity of long-term highly active antiretroviral therapy while also providing a mechanism for enhancing human immunodeficiency virus type 1 (HIV-1)-specific immunity (1, 9, 11, 12). This strategic treatment interruption is also proposed for individuals whose virus has become resistant to treatment, to induce reversion of resistance to the wild type and therefore to improve the success of subsequent salvage (6-8, 10). However, prospective randomized studies have reported conflicting results regarding the latter strategy. The CPCRA 064 study (J. Lawrence, D. Mayers, K. Huppler Hullsiek, G. Collins, D. Abrams, R. Reisler, L. Crane, B. Schmetter, T. Dionne, J. Saldanha, M. Jones, and J. Baxter, Abstr. 10th Conf. Retrovir. Opportunistic Infect., abstr. 67, 2003) showed no benefits of STI before changing therapy in patients with a multidrug-resistant HIV infection, while the ANRS GIGHAART study (C. Katlama, S. Dominguez, C. Duvivier, C. Delaugerre, G. Peytavin, M. Legrand, V. Calvez, K. Gourlain, and D. Costagliola, Abstr. 10th Conf. Retrovir. Opportunistic Infect., abstr. 68, 2003) described virologic and immunologic benefits after 8 weeks of treatment interruption and a subsequent salvage regimen including more than six drugs. Deeks et al. (4) also described a durable viral suppression with a subsequent therapy containing only one fully active agent. Therefore, further studies are required to specify the optimal conditions for conducting an STI strategy with success, especially as concerns the baseline level of resistance, the duration of the interruption, and the drugs included in the salvage therapy. Furthermore, the expected shift to wild-type amino acid residues which is associated with a virologic response is not systematically observed during the time of treatment discontinuation. Theoretically, the longer the duration of the interruption, the more reversion is observed (6, 7), but the dynamics of reversion seem to differ among the patients and among the resistance-associated mutations.
Devereux et al. (7) compared the proportions of primary and secondary mutations detectable on and off therapy. They reported a significant decline in primary and secondary mutations whether samples were tested a median of 6.4 and 12.9 weeks after therapy discontinuation, respectively. Birk et al. (2) described a significant difference between the outcome of primary and secondary protease inhibitor (PI) mutations and between primary PI versus primary reverse transcriptase mutations. Moreover, Deeks et al. (5) observed that phenotypic resistance seemed to wane simultaneously for nonnucleoside reverse transcriptase inhibitors (NNRTIs) and PIs but could be delayed for nucleoside reverse transcriptase inhibitors (NRTIs). So far, no study has clearly compared the dynamics of disappearance among the following three mutation groups: NRTIs, NNRTIs, and PIs. To know whether, during an STI, an antiretroviral class could recover a favorable genetic background faster than another, we compared these dynamics in patients harboring multiresistant viruses and in treatment interruption.
This study was conducted in 19 HIV-1-infected patients who were enrolled in a prospective STI study to obtain a reversion of resistance in plasma. Salvage therapy was resumed depending on clinical events, the patient's wish, or predetermined criteria of reversion to recover plasmatic viruses with no mutations for at least two classes of antiretrovirals. The patients were monitored at day 0 and weeks 2, 4, 8, 14, 20, and 26 with a clinical examination, biochemical tests, measurement of the HIV-1 RNA load in plasma (Roche Amplicor HIV-1 Monitor assay 1.5) and the CD4 cell count (flow cytometric analysis [Coulter]), and (except at weeks 2 and 4) genotypic HIV-1 drug resistance testing (automated, population-based full-sequence analysis [ABI system]).
In our study, the genotypes allowed a comparison of the dynamics of disappearance for the three mutation groups. For each antiretroviral class, we took into account the time point at which the genotype harbored a shift of all of the baseline resistance mutations reported in Table 1 to wild-type amino acid residues. These dynamics were compared by using the nonparametric log rank (Mantel-Cox) test.
TABLE 1.
Resistance-associated mutations at baseline and at the end of the STI period
| Patient and time point | Mutation(s) associated with resistance to:
|
||
|---|---|---|---|
| NRTIs | NNRTIs | PIs | |
| 1 | |||
| Baseline | M41L, D67N, T69D, MI84V, L210W, T215Y, K219N | K101E, Y181C, G190A | M46L, I54V, V82A |
| Wk 8 | M41L/M, D67N/D, L210W/L, T215Y/S | K101R | None |
| 2 | |||
| Baseline | M41L, D67N, V75S, MI84V, L210W, T215Y, K219N | V108I, Y181C, G190A | M46I, I54V, I84V, L90M |
| Wk 26 | M41L/M | None | None |
| 3 | |||
| Baseline | M41L, D67N, V75S, MI84V, T215F, K219W | K101E, V108I, Y181C, G190A | M46I, I54V, I84V, L90M |
| Wk 26 | T215F/S, K219Q | None | I84V/I, L90M/L |
| 4 | |||
| Baseline | M41L, D67N, L74V, MI84V, L210W, T215Y, K219N | K101E, V108I, Y181C, G190A | M46I, I54M, I84V, L90M |
| Wk 26 | M41L, D67N, L74V, MI84V, L210W, T215Y, K219N | K101E/K, V108I/V, Y181C, G190A | I84V, L90M |
| 5 | |||
| Baseline | D67N, K70R, MI84V, T215F, K219E | K101E, V108I, Y181C, G190A | G48M, I54M, V82A, L90M |
| Wk 8 | None | None | None |
| 6 | |||
| Baseline | M41L, D67N, T69N, K70R, T215F, K219Q | None | V82A, I84V, L90M |
| Wk 14 | M41L, T215F | None | None |
| 7 | |||
| Baseline | M41L, D67N, T69D, K70R, MI84V, T215F, K219Q | K103N, V108I, Y181C, G190A | M46I, I54S, I84V, L90M |
| Wk 26 | D67N, T69D, K70R, T215F, K219Q | None | None |
| 8 | |||
| Baseline | M41L, D67N, K70R, MI84V, T215Y, K219Q | None | M46I, I54V, V82A, L90M |
| Wk 26 | D67N/D, K70R/G, T215F/S, K219Q/K | None | None |
| 9 | |||
| Baseline | D67N, T69N, K70R, T215F, K219Q | K101E, K103R, G190A | I54M, I84V, L90M |
| Wk 8 | None | None | None |
| 10 | |||
| Baseline | M41L, L74V, Y115F, MI84V, L210W, T215Y, K219Q | Y188L | I50V, I54V, V82A, L90M |
| Wk 26 | M41L, L74V, MI84V, L210W/L, T215Y | Y188L | I50V, I54V, V82A, L90M |
| 11 | |||
| Baseline | M41L, D67N, L74V, V75T, MI84V, L210W, T215Y, K219N | Y181C, G190S | G48V, I54M, L90M |
| Wk 14 | M41L/M, T215Y/S | None | None |
| 12 | |||
| Baseline | M41L, D67N, MI84V, L210W, T215Y | Y188L | M46L, G48V, I50V, I54V, V82A |
| Wk 26 | None | None | None |
| 13 | |||
| Baseline | M41L, D67N, T69D, V75M, M184I, L210W, T215Y, K219R | K101E, Y188L | M46L, I54V, I84V, L90M |
| Wk 20 | M41L, D67N, T69D, V75M, M184I, L210W, T215Y, K219R | K101E, Y188L | M46L, I54V, I84V, L90M |
| 14 | |||
| Baseline | M41L, D67N, MI84V, L210W, T215Y | Y188L | M46I, I54M, V82A, I84V, L90M |
| Wk 8 | M41L, L210W, T215S | None | None |
| 15 | |||
| Baseline | M41L, D67N, T69D, Q151M, L210W, T215Y | Y181C, Y188L, G190A | M46I, I54V, V82A, L90M |
| Wk 26 | M41L/M, D67N/D, T69D, Q151M/L | None | I54M, V82A/V, L90M/L |
| 16 | |||
| Baseline | M41L, D67N, L74V, V75M, MI84V, L210W, T215Y, K219N | Y181C, G190C | I50V, I54V, V82A, L90M |
| Wk 20 | None | None | None |
| 17 | |||
| Baseline | M41L, D67N, T69N, K70R, V75T, MI84V, T215Y, K219Q | G190A | M46I, I54V, I84V, L90M |
| Wk 26 | None | G190A/G | None |
| 18 | |||
| Baseline | M41L, D67N, T69N, K70R, L74V, L210W, T215Y, K219Q | G190A | M46I, I54V, I84V, L90M |
| Wk 14 | D67N/D, K70R/K, L210F, K219Q/K | None | None |
| 19 | |||
| Baseline | M41L, D67N, V75M, MI84V, L210W, T215F, K219Q | K103N | M46I, I84V, L90M |
| Wk 20 | None | None | None |
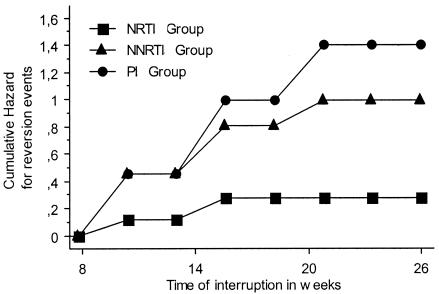
The median baseline HIV-1 RNA level was 5.06 log copies/ml (range, 4.51 to 5.69 log copies/ml), and the median CD4 cell count was 61.5/mm3 (range, 4 to 361/mm3). Genotypic HIV-1 drug resistance testing revealed a median of seven (range, five to eight) mutations associated with resistance to NRTIs, two (range, zero to four) mutations associated with resistance to NNRTIs, and four (range, three to five) major mutations associated with resistance to PIs (Table 1). The prior antiretroviral therapies experienced by the patients are summarized in Table 2. As illustrated in Fig. 1 (actuarial cumulative hazard curves), after therapy withdrawal, the shift of PI and, to a lesser extent, NNRTI mutations was on the increase during the period of treatment interruption while NRTI mutations were more stable. Finally, the data showed that the dynamics of the three mutation groups differed significantly according to the nonparametric log rank (Mantel-Cox) test (P < 0.05) and that the kinetics of the shift to wild-type amino acid residues was faster for the PIs, intermediate for the NNRTIs, and slower for the NRTIs.
TABLE 2.
Prior antiretroviral treatment of the patients in this study
| Patient | No. of prior antiretroviral regimens | No. of years on antiretroviral regimens | Antiretroviral compound(s) not received in prior regimens
|
||
|---|---|---|---|---|---|
| NRTIsa | NNRTIsb | PIsc | |||
| 1 | 8 | 10 | None | Efavirenz | Amprenavir |
| 2 | 17 | 6 | Zalcitadine | Nevirapine | None |
| 3 | 21 | 10 | None | None | None |
| 4 | 14 | 8 | None | None | Indinavir |
| 5 | 11 | 7 | None | None | Lopinavir |
| 6 | 13 | 7 | Zalcitadine | Nevirapine, efavirenz | Lopinavir |
| 7 | 14 | 11 | Abacavir | Efavirenz | Amprenavir |
| 8 | 9 | 7 | None | Nevirapine, efavirenz | Nelfinavir |
| 9 | 11 | 7 | Zalcitadine | None | Indinavir |
| 10 | 23 | 6 | None | None | None |
| 11 | 12 | 12 | None | None | Nelfinavir |
| 12 | 16 | 8 | Zalcitadine | None | None |
| 13 | 36 | 9 | None | None | None |
| 14 | 8 | 4 | Zalcitadine | Nevirapine | Saquinavir, nelfinavir |
| 15 | 10 | 8 | None | Efavirenz | Saquinavir, lopinavir |
| 16 | 4 | 7 | None | None | Indinavir, nelfinavir |
| 17 | 17 | 6 | Zalcitadine | None | None |
| 18 | 13 | 6 | Abacavir | None | Amprenavir |
| 19 | 19 | 8 | None | Nevirapine | None |
Zidovudine, didanosine, zalcitadine, lamivudine, stavudine, and abacavir.
Nevirapine and efavirenz.
Saquinavir, ritonavir, indinavir, nelfinavir, amprenavir, and lopinavir/ritonavir.
FIG. 1.
Actuarial cumulative hazard curves of the three mutation groups for time spent in treatment interruption (P < 0.05).
These results describe the kinetics of antiretroviral resistance mutations in patients with extensive treatment experience who harbored virologic failures and stopped their treatment. In our study, we observed that the mutations associated with resistance to PIs and NNRTIs disappeared faster than those selected by NRTIs. The actuarial curves presented in Fig. 1 show that if the goal is to obtain maximum disappearance of resistance-associated mutations, it could be interesting to increase the duration of treatment interruption for PIs and NNRTIs. However, for NRTIs, some mutations seem to be very stable (M41L, T215Y/F, K219Q/E/N) and increasing the duration of the treatment interruption does not rapidly enhance the probability of their disappearance. This is probably linked to the fact that these mutations do not impact the fitness of the viruses. Further studies can be useful to confirm whether these kinetic results could be observed with other resistance profiles selected by different therapeutic histories.
The higher number of NRTI mutations cannot explain these results. For example, the median number of NNRTI mutations was only two, but these mutations are not the first ones to disappear over the time. In all likelihood, as illustrated by four examples in Table 3, most of the resistance mutations are linked in different ways and do not disappear simply one after the other. The difference in kinetics observed between PI and NRTI resistance mutations could probably be explained by the large impairment of fitness described as associated with PI mutations (3).
TABLE 3.
Examples of genotypic reversions during the STI
| Patient and time point | Viral load (no. of copies/ml) | Mutation(s) associated with resistance to:
|
||
|---|---|---|---|---|
| NRTIs | NNRTIs | PIs | ||
| 2 | ||||
| Baseline | 111,000 | M41L, D67N, V75S, MI84V, L210W, T215Y, K219N | V108I, Y181C, G190A | M46I, I54V, I84V, L90M |
| Wk 8 | 110,000 | M41L, D67N, V75S, MI84V, L210W, T215Y, K219N | V108I, Y181C, G190A | M46I, I54V, I84V, L90M |
| Wk 14 | 128,000 | M41L, D67N, V75S, MI84V, L210W, T215Y, K219N | V108I, Y181C, G190A | None |
| Wk 20 | 1,290,000 | M41L, T215Y | None | None |
| Wk 26 | 722,000 | M41L/M | None | None |
| 3 | ||||
| Baseline | 32,100 | M41L, D67N, V75S, MI84V, T215F, K219W | K101E, V108I, Y181C, G190A | M46I, I54V, I84V, L90M |
| Wk 8 | 171,000 | T215F/S, K219Q | None | I84V/I, L90M |
| Wk 14 | 108,000 | T215F/S, K219Q | None | I84V/I, L90M |
| Wk 20 | 303,000 | T215F/S, K219Q | None | I84V/I, L90M |
| Wk 26 | 583,000 | T215F/S, K219Q | None | I84V/I, L90M/L |
| 5 | ||||
| Baseline | 143,000 | D67N, K70R, MI84V, T215F, K219E | K101E, V108I, Y181C, G190A | G48M, I54M, V82A, L90M |
| Wk 8 | 836,000 | None | None | None |
| 11 | ||||
| Baseline | 144,000 | M41L, D67N, L74V, V75T, MI84V, L210W, T215Y, K219N | Y181C, G190S | G48V, I54M, L90M |
| Wk 8 | 700,000 | M41L, D67N/D, L74V, V75T, L210W, T215Y | None | None |
| Wk 14 | 1,700,000 | M41L/M, T215Y/S | None | None |
These data are of some importance for strategies based on resistance reversions, such as those observed during the ANRS GIGHAART trial, and it would be interesting to use multiple PIs in a salvage regimen if it is confirmed that the mutations associated with resistance to this antiretroviral class disappeared significantly faster than those associated with resistance to NRTIs. Other results are that it is difficult to achieve reversion of some NRTI resistance-associated mutations (M41L, T215Y/F, K219Q/E/N) and that the time needed to do so is probably longer. Thus, the benefit/risk ratio for this class has to be discussed carefully for trials based on resistance mutation reversion during treatment interruption.
REFERENCES
- 1.Autran, B., and G. Carcelain. 2000. AIDS. Boosting immunity to HIV—can the virus help? Science 290:946-949. [DOI] [PubMed] [Google Scholar]
- 2.Birk, M., V. Svedhem, and A. Sonnerborg. 2001. Kinetics of HIV-1 RNA and resistance-associated mutations after cessation of antiretroviral combination therapy. AIDS 15:1359-1368. [DOI] [PubMed] [Google Scholar]
- 3.Croteau, G., L. Doyon, D. Thibeault, G. McKercher, L. Pilote, and D. Lamarre. 1997. Impaired fitness of human immunodeficiency virus type 1 variants with high-level resistance to protease inhibitors. J. Virol. 71:1089-1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Deeks, S. G., R. M. Grant, T. Wrin, E. E. Paxinos, T. Liegler, R. Hoh, J. N. Martin, and C. J. Petropoulos. 2003. Persistence of drug-resistant HIV-1 after a structured treatment interruption and its impact on treatment response. AIDS 17:361-370. [DOI] [PubMed] [Google Scholar]
- 5.Deeks, S. G., T. Wrin, T. Liegler, R. Hoh, M. Hayden, J. D. Barbour, N. S. Hellmann, C. J. Petropoulos, J. M. McCune, M. K. Hellerstein, and R. M. Grant. 2001. Virologic and immunologic consequences of discontinuing combination antiretroviral-drug therapy in HIV-infected patients with detectable viremia. N. Engl. J. Med. 344:472-480. [DOI] [PubMed] [Google Scholar]
- 6.Delaugerre, C., M. A. Valantin, M. Mouroux, M. Bonmarchand, G. Carcelain, C. Duvivier, R. Tubiana, A. Simon, F. Bricaire, H. Agut, B. Autran, C. Katlama, and V. Calvez. 2001. Re-occurrence of HIV-1 drug mutations after treatment re-initiation following interruption in patients with multiple treatment failure. AIDS 15:2189-2191. [DOI] [PubMed] [Google Scholar]
- 7.Devereux, H. L., M. Youle, M. A. Johnson, and C. Loveday. 1999. Rapid decline in detectability of HIV-1 drug resistance mutations after stopping therapy. AIDS 13:F123-F127. [PubMed] [Google Scholar]
- 8.Izopet, J., P. Massip, C. Souyris, K. Sandres, B. Puissant, M. Obadia, C. Pasquier, E. Bonnet, B. Marchou, and J. Puel. 2000. Shift in HIV resistance genotype after treatment interruption and short-term antiviral effect following a new salvage regimen. AIDS 14:2247-2255. [DOI] [PubMed] [Google Scholar]
- 9.Lisziewicz, J., E. Rosenberg, J. Lieberman, H. Jessen, L. Lopalco, R. Siliciano, B. Walker, and F. Lori. 1999. Control of HIV despite the discontinuation of antiretroviral therapy. N. Engl. J. Med. 340:1683-1684. [DOI] [PubMed] [Google Scholar]
- 10.Miller, V., C. Sabin, K. Hertogs, S. Bloor, J. Martinez-Picado, R. D'Aquila, B. Larder, T. Lutz, P. Gute, E. Weidmann, H. Rabenau, A. Phillips, and S. Staszewski. 2000. Virological and immunological effects of treatment interruptions in HIV-1 infected patients with treatment failure. AIDS 14:2857-2867. [DOI] [PubMed] [Google Scholar]
- 11.Ortiz, G. M., D. F. Nixon, A. Trkola, J. Binley, X. Jin, S. Bonhoeffer, P. J. Kuebler, S. M. Donahoe, M. A. Demoitie, W. M. Kakimoto, T. Ketas, B. Clas, J. J. Heymann, L. Zhang, Y. Cao, A. Hurley, J. P. Moore, D. D. Ho, and M. Markowitz. 1999. HIV-1-specific immune responses in subjects who temporarily contain virus replication after discontinuation of highly active antiretroviral therapy. J. Clin. Investig. 104:R13-R18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rosenberg, E. S., M. Altfeld, S. H. Poon, M. N. Phillips, B. M. Wilkes, R. L. Eldridge, G. K. Robbins, R. T. D'Aquila, P. J. Goulder, and B. D. Walker. 2000. Immune control of HIV-1 after early treatment of acute infection. Nature 407:523-526. [DOI] [PubMed] [Google Scholar]