Abstract
Bone morphogenetic proteins (BMPs) and Wnts are growth factors that provide essential patterning signals for cell proliferation and differentiation. Here, we describe a molecular mechanism by which the phosphorylation state of the Drosophila transcription factor Mad determines its ability to transduce either BMP or Wingless (Wg) signals. Previously, Mad was thought to function in gene transcription only when phosphorylated by BMP receptors. We found that the unphosphorylated form of Mad was required for canonical Wg signaling by interacting with the Pangolin-Armadillo transcriptional complex. Phosphorylation of the carboxyl terminus of Mad by BMP receptor directed Mad toward BMP signaling, thereby preventing Mad from functioning in the Wg pathway. The results show that Mad has distinct signal transduction roles in the BMP and Wnt pathways depending on its phosphorylation state.
INTRODUCTION
Wnts and bone morphogenetic proteins (BMPs) are crucial morphogens that instruct cells when to divide, differentiate, or die (1). Both signaling pathways use a distinct repertoire of molecules to carry out their specific intracellular functions. Binding of Wingless (Wg, the Drosophila homolog of Wnt) to its receptors causes the stabilization and nuclear accumulation of the protein Armadillo (called β-catenin in vertebrates), which forms a transcriptional complex with the DNA-binding HMG (high-mobility group) protein Pangolin [called T cell factor (Tcf) in vertebrates] (2). Decapentaplegic (Dpp, a BMP ligand in Drosophila) signals by binding to its serine-threonine kinase transmembrane receptors, causing the phosphorylation of two C-terminal serine residues in the transcription factor Mad (the Drosophila homolog of vertebrate Smad1). Mad then interacts with the co-Smad Medea (called Smad4 in vertebrates), accumulates in the nucleus, and activates target genes. Although both cascades can function independently of each other, an increasing number of interactions have been described between these two pathways. During development, the BMP and Wnt pathways can synergize positively (through separate binding sites in enhancer elements in the genome) (3, 4), or negatively by mutual antagonism at the level of growth factor transcription (5–7). In addition, we have previously described a positive node of integration between BMP and Wnt signals at the level of phosphorylation of Mad and Smad1 (8, 9).
Mad has three distinct structural domains: MH1 (Mad homology 1), which contains the DNA binding domain; MH2, which mediates protein-protein interactions; and the linker domain, which controls protein stability. Mad is phosphorylated by BMP receptors at the C terminus (Ser-Val-Ser) and by mitogen-activated protein kinase (MAPK) or cyclin-dependent kinases 8 and 9 (CDK8 and CDK9) in the linker region (10–13). These latter phosphorylation events prime for phosphorylation by glycogen synthase kinase 3 (GSK3), which triggers the polyubiquitinylation and degradation of Mad or Smad1, terminating the BMP signal (8, 9). Wnt regulates this step by sequestering GSK3 inside multivesicular bodies (MVBs) (14), preventing GSK3-mediated phosphorylation of Mad or Smad1 and therefore prolonging the BMP signal (15). Here, we unexpectedly found a function for Mad in Wg signaling that is independent of phosphorylation of the C terminus of Mad. Genetic and molecular experiments show that unphosphorylated Mad binds to the Wnt transcriptional complex to activate a Wnt reporter gene, independently of its well-known role in the BMP pathway. The choice between these two distinct functions is controlled by phosphorylation, so that Mad signals in the Wg Pangolin-Armadillo pathway only when not phosphorylated by BMP receptor and GSK3.
RESULTS
GSK3 phosphorylation of Mad inhibits both BMP and Wg signaling
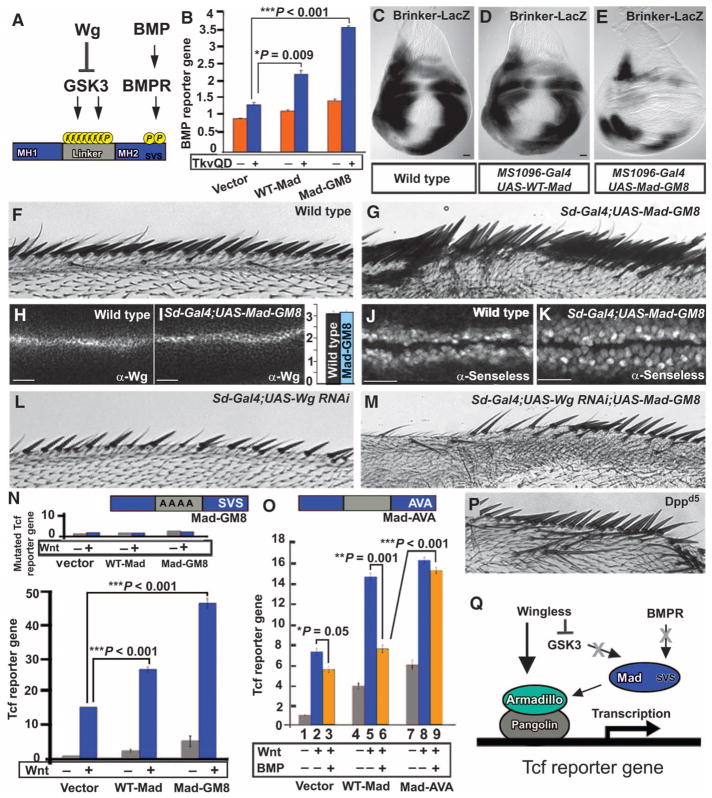
We noticed that the linker region of Mad contains more putative phosphorylation sites than previously reported (9), with at least 11 potential phosphorylation sites in its linker region (Fig. 1A and fig. S1A). Three are putative MAPK, CDK8, and CDK9 phosphorylation sites, which can serve as priming phosphates for a total of eight GSK3 phosphorylations (fig. S1A). Mad was stabilized by treating Drosophila S2R+ cells with Wg-conditioned medium (fig. S1, B and C). In addition, a form of Mad in which all eight GSK3 phosphorylation sites in the linker region were mutated into alanines (referred to as Mad-GM8) was no longer stabilized by Wg (fig. S1, B and C), indicating that the stabilization of Mad by Wg requires intact GSK3 phosphorylation sites in its linker region. As expected for a transcription factor involved in the BMP pathway (8, 9), the stabilized Mad mutant (Mad-GM8) increased the activity of a BMP reporter gene containing a BMP response element driving luciferase expression (Fig. 1B and fig. S1D), and inhibition of GSK3 by lithium chloride (LiCl) prolonged the duration of BMP signaling after a short BMP pulse (fig. S1E). In the wing imaginal disc, Brinker acts as a transcriptional repressor of genes activated by Dpp, and one of the functions of Dpp-activated Mad is to inhibit Brinker transcription (16). In vivo, expression of stabilized Mad (Mad-GM8) increased BMP signaling in wing imaginal discs as demonstrated by reduced expression of Brinker (Fig. 1, C to E). Mad-GM8 also induced ectopic wing vein formation, a phenotype typical of excess BMP signaling, to a greater extent than caused by the previously described form of Mad with mutations in two GSK3 sites (Mad-GM2) (9) (fig. S1, F to O). We conclude that GSK3 phosphorylation regulates the duration of the BMP signal through Mad (8, 9).
Fig. 1.
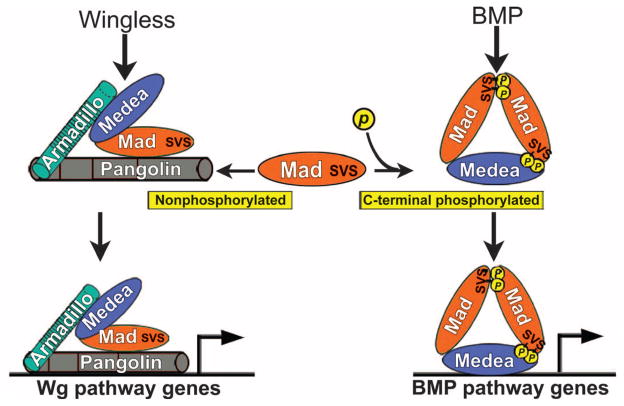
The phosphorylation state of Mad determines whether it signals through the BMP or the Wg pathway. (A) GSK3 and BMP receptor (BMPR) phosphorylation sites in Mad. (B) Wild-type Mad (WT-Mad) and Mad-GM8 increased the activity of the BMP reporter gene [*P = 0.009; ***P = 0.001, two-way analysis of variance (ANOVA) with Tukey’s post test]. The BMP pathway was stimulated with an activated BMP receptor, activated Thickveins (TkvQD), in Drosophila S2 cells. (C to E) Mad-GM8 increased BMP signaling as indicated by repressed Brinker expression in wing discs. n = 16 (C), n = 8 (D), and n = 7 (E). (F and G) WT (n = 31) and ectopic wing margin bristles induced by Mad-GM8 overexpression (n = 40). (H to K) Mad-GM8 did not alter endogenous Wg amounts (P > 0.66, n = 20 intensity measurements) but did increase the area of the Wnt target Senseless (P = 0.0086, n = 20 intensity measurements; Mann-Whitney Wilcoxon test). Numbers of wing discs examined were n = 40 (H), n = 10 (I), n = 16 (J), and n = 12 (K). (L and M) Sensory bristle induction by Mad-GM8 required endogenous Wg. n = 16 (L) and n = 17 (M). (N) WT-Mad and Mad-GM8 increased Tcf reporter gene activity in HEK293T cells, effects that were abolished by mutating the Tcf DNA binding sites (inset) (***P < 0.001, two-way ANOVA with Tukey’s post test; n = 6 experiments). (O) Tcf reporter gene induction by WT-Mad, but not by phosphorylation-resistant Mad-AVA, is repressed by treating cells with BMP4 (*P = 0.05; **P = 0.001; ***P < 0.001, two-way ANOVA with Tukey’s post test; n = 3 independent experiments). (P) Ectopic bristles in Dppd5 (24) mutant wings (n = 12, 7 wings with extra bristles). (Q) Proposed model in which Mad interacts with the Wg transcriptional complex. Scale bars, 20 μm.
In addition to the phenotypes noted above indicative of high BMP signaling, the stabilized form of Mad (Mad-GM8) caused induction of sensory bristles along the margin of the wing (Fig. 1, F and G). This phenotype is typically associated with increased Wg signaling (17), prompting us to investigate the molecular mechanism by which Mad functions in the Wg pathway. The induction of bristles by stabilized Mad took place without changes in Wg protein abundance (Fig. 1, H and I). However, the area of Wg targets such as Senseless (Fig. 1, J and K) and Distalless (fig. S2) was expanded by expression of Mad-GM8. This bristle induction required endogenous Wg (Fig. 1, G, L, and M, and fig. S3). Furthermore, Mad-GM8 overexpression induced phenotypes in eye imaginal discs reminiscent of those caused by gain-of-function mutations for Wg, such as transdetermination of adult eye cells into antenna-like tissue (fig. S4) (18, 19).
We next investigated whether Mad could activate a Tcf reporter gene (Super 7x TOPFLASH luciferase). Tcf reporter activity after Wnt3a treatment was increased by expression of wild-type Mad and to a greater extent by expression of stabilized Mad-GM8 (Fig. 1N). As a control, we used a mutated Tcf reporter gene (Super 7x FOPFLASH luciferase) that cannot bind Tcf, and, as expected, this reporter gene failed to respond to wild-type or stabilized Mad transfection (inset in Fig. 1N), demonstrating that Mad requires Tcf binding sites to activate Wnt signaling. Together, these data indicated that inhibition of the GSK3-mediated phosphorylations of Mad enhances cellular responsiveness not only to BMP but also to Wnt.
Mad activates Wg target genes independently of phosphorylation of its C terminus
To rule out the possibility that Mad caused ectopic margin bristles by increasing BMP signals, we carried out three experiments. First, Dpp misexpression in wing discs caused loss of margin bristles (fig. S1J). Second, addition of BMP4 inhibited stimulation of Wnt signaling in cells transfected with empty vector (Fig. 1O); inhibition by BMP4 was more marked in cells transfected with wild-type Mad (Fig. 1O). Third, a mutant with a partial Dpp loss of function (Dppd5) (20) developed ectopic anterior margin bristles (Fig. 1P), a phenotype indicative of increased Wg signaling. We conclude from these results that Dpp signaling normally inhibits canonical Wg signaling and that Mad may therefore induce Wg signaling through a Dpp-independent pathway.
To investigate this BMP-independent function further, we constructed a C-terminal phosphorylation-resistant mutant Mad by mutating the serine residues phosphorylated by BMP receptor (Ser-Val-Ser) into alanine residues (Ala-Val-Ala) (Mad-AVA) (Fig. 1O). This mutant form of Mad was inactive in the BMP pathway when compared to wild-type Mad (fig. S1D), but activated Tcf reporter gene assays (Fig. 1O, bar 8). In addition, the effect of Mad-AVA expression on Tcf reporter gene activity was no longer inhibited by BMP addition (Fig. 1O, bars 6 and 9, and fig. S5). These results suggested a molecular mechanism by which Mad interacts with the Wnt transcriptional complex in the absence of active BMP signaling (Fig. 1Q).
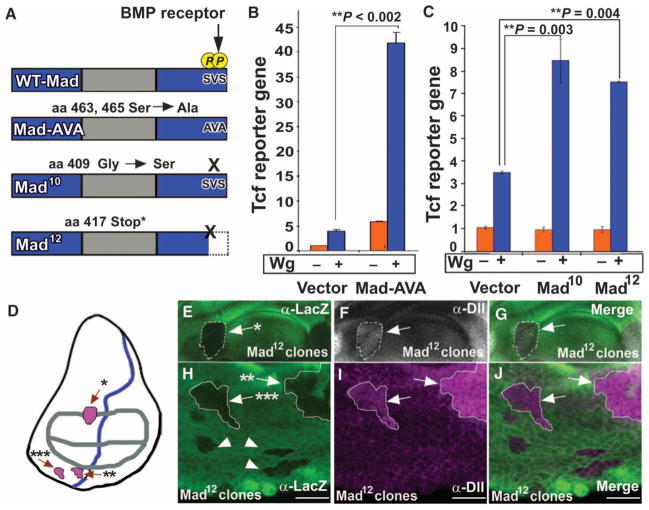
To demonstrate that nonphosphorylated Mad functions in Wg signaling in Drosophila, we tested two C-terminal mutant forms of Mad, Mad10 (21) and Mad12 (22), which lack C-terminal but retain linker phosphorylation sites (Fig. 2A) (9). Drosophila S2R+ cells transfected with phosphorylation-resistant Mad-AVA, Mad10, or Mad12 showed increased Tcf reporter gene activity in response to Wg (Fig. 2, B and C). In wing discs, Mad12 clones showed ectopic distribution of Distalless (arrows) close to sources of Wg protein production (Fig. 2, D to J). Distalless is a specific reporter for Wg signaling in the wing disc (23). Mad10 clones can also induce ectopic distribution of Distalless in wing imaginal discs (24). However, Mad12 clones distant from Wg sources failed to induce ectopic production of Distalless (Fig. 2H, arrowheads), suggesting that the induction of Wg signaling by Mad requires additional components, such as stabilized Armadillo, which is only found close to sources of Wg. These data demonstrate that the function of Mad in Wnt signaling occurs independently of BMP-induced phosphorylation of the C terminus of Mad. Increased abundance of the Wg target Distalless in Mad12 mutant clones (Fig. 2I) indicates that phosphorylation of the C terminus of Mad normally inhibits Wnt signaling at endogenous amounts of this transcription factor in vivo.
Fig. 2.
Mad transduces Wg signals independently of BMP activity. (A) Diagrams of WT-Mad and phosphorylation-resistant C-terminal Mad mutant proteins. (B and C) Mad C-terminal phosphorylation-resistant mutants increase Wg signaling, as measured by Tcf reporter gene activity in Drosophila S2R+ cells (**P = 0.002; **P = 0.004; **P = 0.003, two-way ANOVA with Tukey’s post test; n = 3 experiments). (D) Diagram of third instar wing imaginal disc with the location of clones shown indicated by arrows and asterisks; regions of Wg (gray) and Dpp (blue) sources are outlined. (E to J) Mitotic Mad12 clones in the wing imaginal disc activate the downstream Wg target Distalless near Wg sources. Clones distant to sources of Wg do not activate Distalless and are indicated by arrowheads. This induction of the Wg target occurred at endogenous amounts of Mad (n = 15 clones from seven wing discs with ectopic Distalless). Scale bars, 20 μm.
Mad and Medea are required for Wg signal transduction
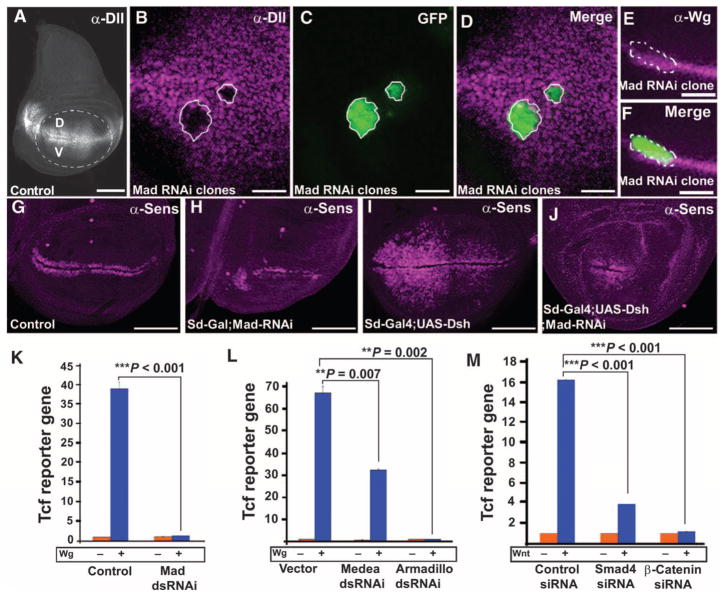
We next examined the requirement of full-length Mad in Wg signal transduction. Because deletion alleles of Mad are not available, we used Mad RNA interference (RNAi) clones (9). Wing discs with Mad RNAi clones lacked Distalless (Fig. 3, A to D), whereas endogenous Wg was unaffected (Fig. 3, E and F). The phenotype caused by this Mad RNAi was specific because it was rescued in the whole adult wing with a UAS-Smad1 transgene expressing the human homolog of Mad (fig. S6). Mad knockdown throughout the developing wing pouch reduced the abundance of Wnt target Senseless (Fig. 3, G and H) and prevented Dishevelled overexpression from inducing ectopic Senseless in the wing pouch (Fig. 3, compare I to J, and fig. S7, A to D). This epistatic experiment showed that Mad is required downstream of Dishevelled in the Wnt pathway. Similarly, formation of increased numbers of sensory bristles by Dishevelled overexpression was inhibited by Mad RNAi (fig. S7, E to L). Furthermore, Tcf reporter gene activity in S2R+ cells was reduced by Mad RNAi (Fig. 3K), an inhibitory effect that was rescued by overexpression of human Smad1 (fig. S7M).
Fig. 3.
Mad and Medea are required for Wg signal transduction. (A) Wild-type distribution of Distalless in a wing imaginal disc. Scale bar, 100 μm. (B to D) Decreased abundance of the Wnt target Distalless in Mad RNAi clones in the ventral (V) wing pouch. n = 10 clones in the region where Distalless is normally found, all showing decreased Distalless abundance. Scale bar, 20 μm. (E and F) Wg production was maintained in Mad RNAi clones (n = 14 clones; scale bar, 50 μm). (G to J) The area of Senseless in wing imaginal discs was reduced in clones expressing Mad RNAi or those expressing both UAS-Mad RNAi and UAS-Dishevelled. At least 12 discs per genotype were analyzed, with similar results. Scale bars, 100 μm. (K) Knockdown of Mad by dsRNA inhibited the activity of the Tcf reporter gene in Drosophila S2R+ cells (***P < 0.001, two-way ANOVA with Tukey’s post test, n = 6 experiments). (L) Knockdown of Medea or Armadillo by dsRNA blocked Wg signaling in S2R+ cells (**P = 0.007; **P = 0.002, two-way ANOVA with Tukey’s post test, n = 3 experiments). (M) siRNAs against Smad4 or β-catenin inhibited Wnt reporter responses in HEK293T cells (***P < 0.001, two-way ANOVA with Tukey’s post test, n = 3 experiments).
In addition, we found a requirement for Armadillo (25) and Medea in Wg signaling in Drosophila S2R+ cells and for Smad4 and β-catenin in human embryonic kidney (HEK) 293T cells (Fig. 3, L and M, and fig. S7, N to P). These observations are consistent with previous reports that Smad4 interacts with β-catenin in the activation of Xenopus homeobox genes (26, 27). We conclude that Mad and Medea are required for Wg signaling, both in Tcf reporter assays of Wg activity and in vivo in Drosophila.
Mad binds to Pangolin in the absence of phosphorylation of its C terminus
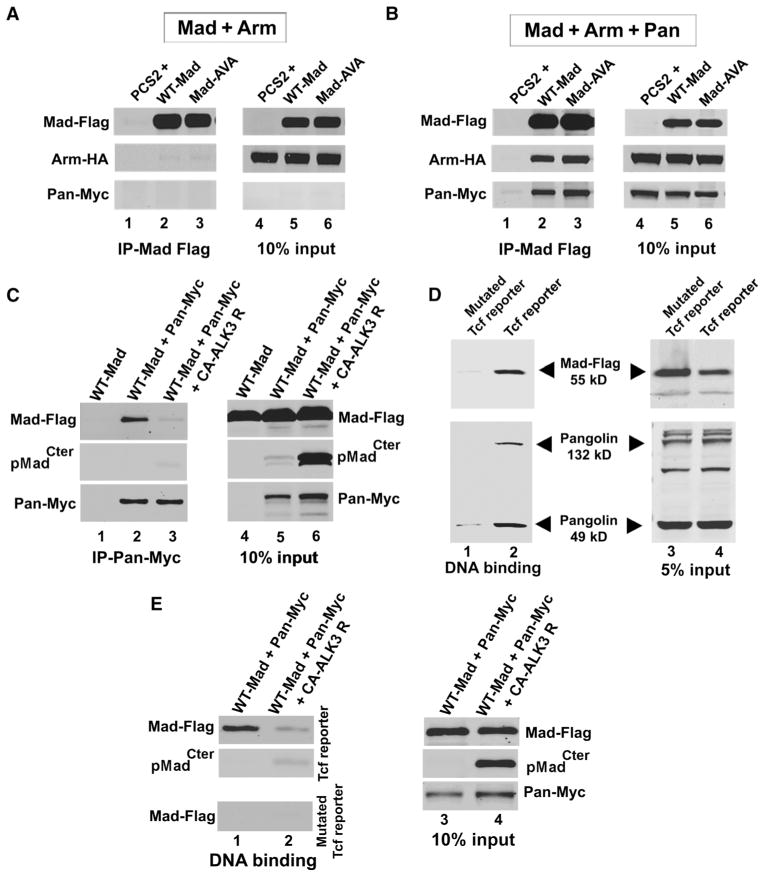
Next, we examined whether Mad could bind to the Pangolin-Armadillo complex. A previous report had described that the mammalian Mad homolog Smad1 interacted with β-catenin and Tcf to activate Myc transcription (28). Mad-Flag, Armadillo–HA (hemagglutinin), and Pangolin-Myc were transfected separately into HEK293T cells and cell lysates were prepared. To stabilize Mad and Armadillo, we treated all cultures with the GSK3 inhibitor 6-bromoindirubin-3′-oxime (BIO) (29). We later found that preventing GSK3 phosphorylation of Mad increased binding efficiency (fig. S8, A and B). Less Mad bound to Pangolin in lysates from transfected cells treated with a pharmacologically inactive form of BIO, 1-methyl BIO (29) (fig. S8C).
Mad and Armadillo failed to bind to each other (Fig. 4A). However, in the presence of Pangolin, Mad-Flag pulled down Pangolin and Armadillo (Fig. 4B). Thus, Pangolin mediates the binding of both Mad and Armadillo. Moreover, Mad-AVA bound Pangolin to a similar extent as wild-type Mad (Fig. 4B, compare lanes 2 and 3), suggesting that phosphorylation of the C terminus of Mad is not required for binding of Pangolin. Binding of Mad to Pangolin was previously reported, but it was suggested that the binding of Mad inhibited the interaction between Armadillo and Pangolin (24). The different findings might be due to our use of a GSK3 inhibitor, which stabilizes Mad and Armadillo. As described previously, Mad10 mitotic clones show increased abundance of the Wg target Distalless in wing discs (24). Therefore, we propose that because Mad10 or Mad12 cannot be phosphorylated by BMP receptors, more Mad protein is available to signal through the Wg pathway. We conclude from these biochemical experiments that Mad, Pangolin, and Armadillo can form protein complexes in cell extracts, independently of the phosphorylation of the C terminus of Mad.
Fig. 4.
Mad is a component of the Wnt transcriptional complex bound to DNA. (A) Mad-Flag did not immunoprecipitate with Armadillo in the absence of Pangolin (n = 4 experiments). (B) Mad-Flag bound Pangolin, which also coimmunoprecipitated with Armadillo (n = 3 independent experiments). (C) Pangolin-Myc immunoprecipitated unphosphorylated Mad-Flag (lane 2). Less Mad-Flag bound to Pangolin-Myc (lane 3) in cells expressing a constitutively activated BMP receptor (CA-ALK3 receptor) (which would be expected to trigger phosphorylation of sites in the C-terminal domain). Binding of Mad to Pangolin was significantly reduced (P = 0.0014, t test; n = 3 independent experiments). (D) Biotin-labeled PCR products containing Tcf binding sites bound both Pangolin and Mad-Flag proteins (lane 2) from S2R+ cell lysates in a DNA affinity precipitation assay. Pangolin was detected by immunoblotting. PCR products containing mutated Tcf binding sites failed to bind Pangolin or Mad proteins (lane 1) (n = 3 experiments). (E) Pangolin-Myc and Mad-Flag bound to PCR products containing Tcf binding sites (lane 1, top panel) but not to those containing mutated Tcf binding fragments (lane 1, bottom panel). C-terminally phosphorylated Mad-Flag extracts did not bind to the PCR products containing Tcf binding sites (lane 2, top panel). Inputs are shown in lanes 3 and 4.
Next, we asked whether binding of Mad to Pangolin was inhibited by phosphorylation of its C terminus. We cotransfected HEK293T cells with Mad-Flag and Pangolin-Myc, with or without a constitutively active Alk3 BMP receptor (CA-Alk3). As expected, expression of CA-Alk3 resulted in phosphorylation of the two C-terminal serine residues of Mad (Fig. 4C, compare lanes 5 and 6). Pangolin-Myc immunoprecipitated Mad-Flag only in cells lacking the constitutively active Alk3 receptor (Fig. 4C, compare lanes 2 and 3). Activation of BMP signaling with expression of the constitutively active Alk3 receptor significantly reduced binding of Mad to Pangolin. These experiments indicate that the phosphorylation of the C terminus of Mad has an inhibitory effect on the formation of complexes with Pangolin.
The Pangolin-Mad-Armadillo complex binds to Tcf DNA binding sites
Using a DNA affinity precipitation assay (30), we asked whether the protein complex of Pangolin and Mad could interact with Tcf DNA binding sites. A 5′ biotin-labeled and a 3′ unlabeled primer were used to amplify by polymerase chain reaction (PCR) the seven Tcf binding repeats (table S1) of the Tcf reporter gene (SuperTOPFLASH) or its mutated counterpart (SuperFOPFLASH), in which all seven specific binding sites for Tcf are mutated. Biotin-labeled PCR products were then bound to streptavidin-agarose beads and lysates from Wg-treated Drosophila S2R+ cells transfected with Mad-Flag were incubated with streptavidin-biotin-DNA beads. We found that Mad-Flag and two isoforms of endogenous Pangolin bound to Tcf reporter gene DNA (Fig. 4D, lane 2). Mad binding to the mutated Tcf reporter gene product was barely detectable (Fig. 4D, lane 1). In addition, extracts containing C-terminally phosphorylated Mad displayed decreased association with DNA containing Tcf binding sites (Fig. 4E). In conclusion, these biochemical experiments suggest that Mad preferentially binds to the Pangolin-Armadillo complex on Tcf binding sites in DNA in the absence of phosphorylation of the C terminus by BMP receptor. The results uncover a role for nonphosphorylated Mad in Wg signaling transduction.
DISCUSSION
We investigated how the phosphorylation state of the transcription factor Mad can determine its ability to transduce a BMP or a Wnt signal in the cell. The molecular mechanism depicted in Fig. 5 proposes a new layer of regulation in canonical Wg signaling. Mad is required for Wg signaling both in Tcf reporter gene assays and in vivo in Drosophila. This aspect of Mad activity is inhibited by phosphorylation by BMP receptor resulting from activation of the BMP pathway. Wnt signaling sequesters GSK3 from the cytosol into MVBs (14), causing stabilization of Armadillo and Mad, which then binds to Pangolin. This protein complex can bind to Tcf binding sites in DNA to control the transcription of Wnt target genes. The proteins encoded by the Drosophila Mad10 and Mad12 genetic mutants have lost the BMP receptor branch of their function, but retain their ability to engage in Wg signaling (Fig. 2). Interactions between Dpp and Wg signaling, both positive (3, 4) and negative (5–7), have been reported in Drosophila. We propose that the BMP pathway would have positive interactions with Wg only for genes with both BMP and Wg response elements in their promoters (3, 4). In most other cases, we suggest that the BMP and Wg pathways compete for the available pool of Mad (Fig. 5).
Fig. 5.
Proposed model in which Mad and Medea bind to the Wg Pangolin-Armadillo transcriptional complex in the absence of BMP signals. Previously, Mad was believed to be transcriptionally active only when phosphorylated at its C terminus by BMP receptor. In this model, Mad promotes activation of Wg pathway target genes when not phosphorylated. In the presence of BMP, Wg serves to prolong the signal from Mad phosphorylated in the C-terminal domain by inhibiting GSK3-mediated phosphorylation of the linker region of Mad (8, 9). In addition, BMP signaling also diverts Mad or Medea from the Wg pathway to promote activation of BMP target genes, thereby causing competition between the BMP and the Wingless pathway.
Our findings suggest that Dpp signaling can generate an inverse gradient of Wnt activity because high BMP activity competes for Mad and Medea, reducing their availability to signal in the Wg pathway. Unphosphorylated Mad can participate in Wg signaling when only Pangolin binding sites are present in an enhancer. When BMP receptor phosphorylates Mad, a trimer with Medea would be formed that would direct Mad to BMP responsive promoters (Fig. 5). It has been proposed that Smad4 is the limiting component in cells transducing BMP and transforming growth factor–β (TGF-β) signals (31). In a cell receiving both BMP and Wnt signals, Mad phosphorylated at the C terminus would compete with unphosphorylated Mad for limited amounts of Medea. Moreover, most cellular Smad1 has been reported to enter the nucleus in response to BMP2 treatment (32), suggesting that the competition between BMP and Wnt signaling could occur directly at the level of Mad as well.
This study advances our understanding of signaling crosstalk between the Wg and the BMP pathways in several ways. First, it shows that Mad, a transcription factor classically associated with the BMP pathway, is also an effector of canonical Wnt signaling. Second, BMP receptor–mediated phosphorylation of Mad is not required for activity in the Wnt pathway. Instead, mutations such as Mad12 that truncate the C terminus increase Wnt signaling at endogenous protein amounts. Third, phosphorylation of the linker region of Mad by GSK3 has an inhibitory effect on Wnt signaling. This previously unknown phosphorylation-independent function of Mad has implications for cell differentiation and cancer.
MATERIALS AND METHODS
Drosophila stocks
Drosophila strains used in this work were as follows: UAS-WT-Mad, UAS-Mad-GM8/Cyo, UAS-Mad-GM2, UAS-human-Smad1, UAS-Dishevelled (#9453), UAS-DN-GSK3, UAS-ArmS10 (#4782), hsflp;UAS-Mad RNAi;UAS-Mad-RNAi, UAS-Wg-RNAi (Vienna Drosophila RNAi Center, VDRC #13352). Gal4 drivers used (Bloomington stock number in parentheses) were as follows: MS1096-Gal4, Scalloped-Gal4 (#8609), Engrailed-Gal4 (#6356), Eyeless-Gal4, and yw;Act>y+>Gal4;UAS-GFP. Other strains used in this study were Brinker-LacZ, Dpp[d5]/Cyo (#2071), and Canton S.
Clonal analysis
For random heat shock “flip-out” clones, we crossed females of the genotype yw;Act>y+>Gal4;UAS-GFP to males of the genotype ywhsflp;Mad-RNAi/Mad-RNAi. Flies laid eggs for 6 to 8 hours, and eggs were incubated for a further 16 to 20 hours at 25°C. Larvae at first instar were administered single heat shocks (37°C) ranging from 5 to 15 min. An RNAi approach was used because no complete loss-of-function Mad mutants exist at present; the most precisely defined deletion of Mad Df(2L)C28 removes a number of other genes (33). The Mad RNAi construct used previously described Mad sequences (9) placed in a Gal4-inducible pWiz vector (34). Mitotic Mad12 clones were induced by crossing female ywhsflp;Arm-LacZM21FRT40/Cyo-GFP with ywhsflp;Mad12FRT40/Cyo males. Flies laid eggs for 8 hours, and eggs were incubated for a further 16 to 28 hours at 25°C. Larvae at the first instar were administered single heat shocks (37°C) ranging from 20 to 30 min. After heat shock, larvae were grown at 25°C for recovery and further development.
Mounting of adult wings
Wings were removed from adult flies and dehydrated in 100% ethanol for 5 min. The wings were placed on a slide with the dorsal side up, and the ethanol was allowed to evaporate. A small drop of Canada balsam was dropped onto the wing and a glass coverslip was placed on top. A 10-g weight was used to flatten the preparation.
Wing disc fixation and immunostaining
Wing discs were dissected from third instar larvae in cold 0.02% Triton X-100 phosphate-buffered saline (PBS) (PBST) solution. Discs were fixed in 4% formaldehyde for 20 min on ice and rinsed with PBST. Discs were then incubated in blocking solution (2.5% bovine serum albumin and 5% goat serum in PBS/0.02% Triton X-100) for 1 to 2 hours at room temperature. Primary antibodies—α-Senseless (1:10; gift of H. Bellen), α-Distalless (1:300; gift of I. Duncan) (35), α-LacZ (1:1000), or α-Wg (1:200; Developmental Hybridoma Bank)—were incubated in blocking solution overnight at 4°C and washed 10 times for 2 hours in PBST. Discs were incubated for 1 to 2 hours in blocking solution and incubated for 1 hour in anti-mouse Cy3-conjugated secondary antibody (1:1000, Jackson Laboratory) at room temperature. Wing discs were placed in DAPI (4′,6-diamidino-2-phenylindole)–containing Vectashield (Vector) overnight and mounted on glass slides.
Reporter gene assays in Drosophila S2 and HEK293T cells
To test whether stabilized Mad linker mutants could activate a BMP reporter gene (BRE-luciferase) (36) to a greater extent than wild-type Mad, we cotransfected pAC-Mad (0.1 μg), BMP reporter gene (0.1 μg), and thymidine kinase (TK)–Renilla (0.01 μg) constructs with an activated form of the Thickveins receptor (pAC-TKVQD, 0.1 μg) (37) into S2 cells. Luciferase readings were measured 48 hours after transfection with the Promega Dual-Luciferase Reporter Assay System. S2 cells produce the BMP ligands Dpp and Glass bottom boat (Gbb), and pMadCter is readily detected in nuclei. For this reason, we also used HEK293T cells, which have no detectable amounts of pSmad1Cter, to carry out Wnt reporter assays. HEK293 cells were cultured in 12-well plates and transiently transfected with Fugene (Roche). Renilla luciferase pRLCMV served as the internal control. Transfections contained 0.2 μg of SuperTOPFLASH Tcf reporter gene (38), 0.02 μg of pRL-CMV, and 0.5 μg of the various pCS2-Mad constructs. pCMV empty vector was used to reach a total of 0.72 μg of DNA per well. Wg-conditioned medium from permanently transfected S2 cells (Drosophila Genomics Resource Center, stock #165; originally from R. Nusse) was used to treat Drosophila S2R+ cells. For 293T cells, Wnt3a-conditioned medium from permanently transfected L cells (gift of R. Nusse) was used.
Luciferase assays were performed with the Dual-Luciferase Reporter Assay System (Promega) according to the manufacturer’s instructions, and the results were standardized against internal Renilla controls. To assess the duration of BMP signals (fig. S1E), we transfected cells with BMP reporter gene (BRE-luciferase) (36) with wild-type Mad in a six-well plate. Twelve hours later, cells were starved in serum-free medium for 4 hours, and 5 nM BMP7 (R&D Systems) was added for 30 min. Cells were washed in serum-free medium and incubated with or without 30 mM LiCl to inhibit GSK3.
Gene silencing in Drosophila S2R+ cells and HEK293T cells
Double-stranded RNA was used to knock down Mad, Medea, and Armadillo in S2R+ cells. RNA design and treatment was based on the protocol by Clemens and co-workers (39). Primer sequences are found in table S1. The Ambion MEGAscript kit was used for double-stranded RNA (dsRNA) amplification. Cells were treated with Mad, Medea, or Armadillo dsRNA for 4 days. Cells were transfected on day 3 with Tcf reporter gene (0.4 μg) (38) and pTK-Renilla (0.04 μg) with Effectene (Qiagen). On day 4, cells were treated with Wg-conditioned medium or control medium for 8 hours.
To rescue Mad knockdown in S2R+ cells, we used a human Smad1 construct (in pAC 5.1 vector) transfected together with a copper-inducible pWiz-Mad RNAi construct (34). Transfection of pWiz-Mad RNAi (0.1 μg), metallothionein-Gal4 (0.1 μg), SuperTOPFLASH luciferase (0.1 μg), and pTK-Renilla (0.01 μg) was carried out with Qiagen Effectene. S2R+ cells were grown for a total of 4 days and then treated with either Wg-conditioned medium or control medium for 8 hours.
For small interfering RNAs (siRNAs) in HEK293T cells, siRNAs targeting human β-catenin and Smad4 were ON-TARGETplus SMARTpool from Thermo Scientific (#L-003482 and #L-003902, respectively). Control siRNA was BLOCK-iT Fluorescent Oligo (Invitrogen, #44-2926). siRNAs were transfected with Lipofectamine 2000 by means of the reverse transfection protocol (Invitrogen) and analyzed after 48 hours. Cells were first transfected with siRNA and then with DNA 24 hours later.
Coimmunoprecipitation assays
HEK293T cells were cultured in six-well plates and transiently transfected with DNA using Fugene (Roche). pCMV-Pangolin-Myc, pCS2-Mad-Flag, and pCMV Armadillo-HA plasmid DNAs were transfected separately into HEK293T cells and treated with 5 μM GSK3 inhibitor BIO for 4 hours (27). The use of BIO in this experiment was essential to ensure efficient binding, because GSK3 phosphorylations inhibit complex formation (fig. S8). The Pangolin and Armadillo constructs were gifts of E. Verheyen (24). Cells were lysed with a standard lysis buffer [50 mM tris (pH 7.4), 150 mM NaCl, 1 mM EDTA, and 1% Triton X-100]. Usually, 250 μl of cell lysate containing Mad proteins was incubated with 250 μl of lysates containing tagged proteins at 4°C with end-over-end rotation for 1 hour. For the binding of Mad-Flag, Armadillo-HA, and Pangolin-Myc, 166 μl of each lysate was added. Immunoprecipitation of Mad-Flag was carried out with anti-Flag beads (Sigma). Beads were centrifuged at 1000g for 1 min. The bound proteins were washed three times with 1 ml of tris-buffered saline (TBS) and then eluted with 200 μl of TBS containing FLAG peptide (100 μg/ml; Sigma) with end-over-end rotation for 30 min at 4°C. The results were analyzed by Western blot. The antibodies used were anti-Flag mouse (Sigma), 1:1000; anti–c-Myc mouse (Santa Cruz), 1:1000; anti-Smad1Cter, 1:1000 (40); and anti-HA (Sigma), 1:1000. Pangolin-Myc, Mad-Flag, and CA-ALK3 receptor were cotransfected into HEK293T cells for experiments testing whether C-terminally phosphorylated Mad bound Pangolin. Cells transfected with or without CA-ALK3 were treated with the GSK3 inhibitor BIO for 6 hours before lysate preparation. Pangolin-Myc immunoprecipitation was carried out with anti-Myc beads (Covance AFC-150P) under conditions in which Mad was phosphorylated or not. Elution was performed with Myc peptide (100 μg/ml; Sigma).
DNA affinity precipitation assay
The sequence encoding the seven Tcf binding sites present in the SuperTOPFLASH luciferase reporter (Tcf reporter gene) plasmid was amplified by PCR (38). The primers used were a forward biotin-labeled primer and a nonbiotinylated 3′ reverse primer (table S1). As a negative control, the same primers were used to amplify the sequence of the SuperFOPFLASH luciferase reporter (mutated Tcf reporter gene) (38). The biotinylated PCR products were incubated for 1 hour at 4°C with streptavidin-agarose beads (Sigma, S1638-5ML) (30). The beads were then washed three times with binding buffer [10% glycerol, 4 mM tris-Cl (pH 7.9), and 60 mM KCl] to remove unbound PCR products. DNA-biotin-avidin agarose beads were then added to S2R+ cell extracts overexpressing wild-type Mad-Flag. Transfected S2R+ cells were lysed with a modified lysis buffer: 10% glycerol, 4 mM tris-HCl (pH 7.9), 60 mM KCl, and 1% Triton X-100. Cell lysates and beads were incubated overnight at 4°C with end-over-end rotation to allow proteins to bind to DNA (30). The beads were then centrifuged at 1000g for 1 min. Bound proteins were washed three times with 1.5 ml of binding buffer [10% glycerol, 4 mM tris-HCl (pH 7.9), and 60 mM KCl] and then eluted by adding 12.5 μl of 5× SDS sample buffer and boiled for 5 min. Proteins were resolved on polyacrylamide gels followed by immunoblotting with mouse anti-Flag (Sigma; 1:1000) and rabbit anti-Pangolin (1:700).
For the custom Pangolin antibody, a synthetic peptide ([H]-CK-Acp-MPHTHTRHGSSGDDL-[NH2]) was used to immunize two rabbits (8) (Covance). The antiserum recognized three different isoforms of Pangolin in S2 cells (isoform J, 132 kD; isoforms B and H, 85 kD; isoform I, 49 kD), two of which specifically bound to Tcf binding sites.
Mad stability assay
To demonstrate stabilization of wild-type Mad by GSK3 inhibition, we transfected UAS–wild-type Mad-Flag or UAS–Mad-GM8 Flag with a metallothionein-Gal4 plasmid into S2R+ cells. Twelve hours after transfection, cells were treated with 100 mM CuSO4 for 24 hours. Cells were then grown for a further 24 hours in the absence of CuSO4, incubated with Wg medium for 12 hours, and lysed with radioimmunoprecipitation assay (RIPA) buffer. Western blots were performed using standard protocols. A rabbit α-Flag antibody (Sigma) was used to detect total amounts of wild-type Mad and Mad-GM8. A mouse α-Armadillo antibody was obtained from Developmental Studies Hybridoma Bank.
Supplementary Material
Fig. S1. Increased BMP signals generated by a stabilized Mad protein.
Fig. S2. Mad-GM8 expression increases the area of Distalless, a downstream target of Wg.
Fig. S3. Inducible RNAi directed against Wg depletes Wg protein and its downstream target Senseless.
Fig. S4. Mad-GM8 expression in the eye imaginal disc produces phenotypes suggestive of high Wg signaling.
Fig. S5. C-terminal phosphorylation of Mad enables BMP4 to repress the Mad-induced increase in Tcf reporter gene activity.
Fig. S6. The Mad RNAi phenotype is rescued by coexpression of a human Smad1 transgene.
Fig. S7. Mad is required for Wg signal transduction during wing margin development.
Fig. S8. Inhibition of GSK3 activity by BIO enhances the binding of Mad to Pangolin.
Table S1. Primer sequences.
Acknowledgments
We thank E. Verheyen, I. Duncan, H. Bellen, M. Affolter, S. Newfeld, M. OConnor, N. Baker, C. Heldin, R. Nusse, and R. Moon, Developmental Studies’ Hybridoma Bank, Bloomington Stock Center, and the Vienna Drosophila RNAi Center for reagents; R. Dobrowolski and D. Ploper for comments on the manuscript; and K. Ngo for help with statistical analysis. We thank J. Olsen and U. Banerjee for the use of their scanning electron microscope.
Funding: This work was supported by the NIH (HD21502-25) and the Howard Hughes Medical Institute, of which E.M.D.R. is an investigator.
Footnotes
Author contributions: Experiments were performed by E.E. and H.D. (Figs. 1 to 4) with assistance from R.H.C. (parts of Fig. 1). E.E., H.D., and E.M.D.R. contributed to the experimental design and writing of the manuscript.
Competing interests: The authors declare that they have no competing interests.
REFERENCES AND NOTES
- 1.Schwank G, Basler K. Regulation of organ growth by morphogen gradients. Cold Spring Harb Perspect Biol. 2010;2:a001669. doi: 10.1101/cshperspect.a001669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Logan CY, Nusse R. The Wnt signaling pathway in development and disease. Annu Rev Cell Dev Biol. 2004;20:781–810. doi: 10.1146/annurev.cellbio.20.010403.113126. [DOI] [PubMed] [Google Scholar]
- 3.Takaesu NT, Bulanin DS, Johnson AN, Orenic TV, Newfeld SJ. A combinatorial enhancer recognized by Mad, TCF and Brinker first activates then represses dpp expression in the posterior spiracles of Drosophila. Dev Biol. 2008;313:829–843. doi: 10.1016/j.ydbio.2007.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Estella C, McKay DJ, Mann RS. Molecular integration of wingless, decapentaplegic, and autoregulatory inputs into Distalless during Drosophila leg development. Dev Cell. 2008;14:86–96. doi: 10.1016/j.devcel.2007.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Theisen H, Haerry TE, O’Connor MB, Marsh JL. Developmental territories created by mutual antagonism between Wingless and Decapentaplegic. Development. 1996;122:3939–3948. doi: 10.1242/dev.122.12.3939. [DOI] [PubMed] [Google Scholar]
- 6.Morimura S, Maves L, Chen Y, Hoffmann FM. decapentaplegic overexpression affects Drosophila wing and leg imaginal disc development and wingless expression. Dev Biol. 1996;177:136–151. doi: 10.1006/dbio.1996.0151. [DOI] [PubMed] [Google Scholar]
- 7.Domínguez M, Hafen E. Hedgehog directly controls initiation and propagation of retinal differentiation in the Drosophila eye. Genes Dev. 1997;11:3254–3264. doi: 10.1101/gad.11.23.3254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fuentealba LC, Eivers E, Ikeda A, Hurtado C, Kuroda H, Pera EM, De Robertis EM. Integrating patterning signals: Wnt/GSK3 regulates the duration of the BMP/Smad1 signal. Cell. 2007;131:980–993. doi: 10.1016/j.cell.2007.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eivers E, Fuentealba LC, Sander V, Clemens JC, Hartnett L, De Robertis EM. Mad is required for Wingless signaling in wing development and segment patterning in Drosophila. PLoS One. 2009;4:e6543. doi: 10.1371/journal.pone.0006543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kretzschmar M, Doody J, Massagué J. Opposing BMP and EGF signalling pathways converge on the TGF-β family mediator Smad1. Nature. 1997;389:618–622. doi: 10.1038/39348. [DOI] [PubMed] [Google Scholar]
- 11.Pera EM, Ikeda A, Eivers E, De Robertis EM. Integration of IGF, FGF, and anti-BMP signals via Smad1 phosphorylation in neural induction. Genes Dev. 2003;17:3023–3028. doi: 10.1101/gad.1153603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alarcón C, Zaromytidou AI, Xi Q, Gao S, Yu J, Fujisawa S, Barlas A, Miller AN, Manova-Todorova K, Macias MJ, Sapkota G, Pan D, Massagué J. Nuclear CDKs drive Smad transcriptional activation and turnover in BMP and TGF-β pathways. Cell. 2009;139:757–769. doi: 10.1016/j.cell.2009.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aragón E, Goerner N, Zaromytidou AI, Xi Q, Escobedo A, Massagué J, Macias MJ. A Smad action turnover switch operated by WW domain readers of a phosphoserine code. Genes Dev. 2011;25:1275–1288. doi: 10.1101/gad.2060811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Taelman VF, Dobrowolski R, Plouhinec JL, Fuentealba LC, Vorwald PP, Gumper I, Sabatini DD, De Robertis EM. Wnt signaling requires sequestration of glycogen synthase kinase 3 inside multivesicular endosomes. Cell. 2010;143:1136–1148. doi: 10.1016/j.cell.2010.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eivers E, Fuentealba LC, De Robertis EM. Integrating positional information at the level of Smad1/5/8. Curr Opin Genet Dev. 2008;18:304–310. doi: 10.1016/j.gde.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Müller B, Hartmann B, Pyrowolakis G, Affolter M, Basler K. Conversion of an extracellular Dpp/BMP morphogen gradient into an inverse transcriptional gradient. Cell. 2003;113:221–233. doi: 10.1016/s0092-8674(03)00241-1. [DOI] [PubMed] [Google Scholar]
- 17.Couso JP, Bishop SA, Martinez Arias A. The wingless signalling pathway and the patterning of the wing margin in Drosophila. Development. 1994;120:621–636. doi: 10.1242/dev.120.3.621. [DOI] [PubMed] [Google Scholar]
- 18.Duong HA, Wang CW, Sun YH, Courey AJ. Transformation of eye to antenna by misexpression of a single gene. Mech Dev. 2008;125:130–141. doi: 10.1016/j.mod.2007.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schubiger M, Sustar A, Schubiger G. Regeneration and transdetermination: The role of Wingless and its regulation. Dev Biol. 2010;347:315–324. doi: 10.1016/j.ydbio.2010.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bryant PJ. Localized cell death caused by mutations in a Drosophila gene coding for a transforming growth factor-β homolog. Dev Biol. 1988;128:386–395. doi: 10.1016/0012-1606(88)90300-4. [DOI] [PubMed] [Google Scholar]
- 21.Hoodless P, Haerry T, Abdollah S, Stapleton M, O’Connor MB, Attisano L, Wrana JL. MADR1, a MAD-related protein that functions in BMP2 signaling pathways. Cell. 1996;85:489–500. doi: 10.1016/s0092-8674(00)81250-7. [DOI] [PubMed] [Google Scholar]
- 22.Sekelsky JJ, Newfeld SJ, Raftery LA, Chartoff EH, Gelbart WM. Genetic characterization and cloning of Mothers against dpp, a gene required for decapentaplegic function in Drosophila melanogaster. Genetics. 1995;139:1347–1358. doi: 10.1093/genetics/139.3.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Neumann CJ, Cohen SM. Long-range action of Wingless organizes the dorsal-ventral axis of the Drosophila wing. Development. 1997;124:871–880. doi: 10.1242/dev.124.4.871. [DOI] [PubMed] [Google Scholar]
- 24.Zeng YA, Rahnama M, Wang S, Lee W, Verheyen EM. Inhibition of Drosophila Wg signaling involves competition between Mad and Armadillo/β-catenin for dTcf binding. PLoS One. 2008;3:e3893. doi: 10.1371/journal.pone.0003893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Noordermeer J, Klingensmith J, Perrimon N, Nusse R. dishevelled and armadillo act in the Wingless signalling pathway in Drosophila. Nature. 1994;367:80–83. doi: 10.1038/367080a0. [DOI] [PubMed] [Google Scholar]
- 26.Nishita M, Hashimoto MK, Ogata S, Laurent MN, Ueno N, Shibuya H, Cho KW. Interaction between Wnt and TGF-β signalling pathways during formation of Spemann’s organizer. Nature. 2000;403:781–785. doi: 10.1038/35001602. [DOI] [PubMed] [Google Scholar]
- 27.Labbé E, Letamendia A, Attisano L. Association of Smads with lymphoid enhancer binding factor 1/T cell-specific factor mediates cooperative signaling by the transforming growth factor-β and Wnt pathways. Proc Natl Acad Sci USA. 2000;97:8358–8363. doi: 10.1073/pnas.150152697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hu MC, Rosenblum ND. Smad1, β-catenin and Tcf4 associate in a molecular complex with the Myc promoter in dysplastic renal tissue and cooperate to control Myc transcription. Development. 2005;132:215–225. doi: 10.1242/dev.01573. [DOI] [PubMed] [Google Scholar]
- 29.Meijer L, Skaltsounis AL, Magiatis P, Polychronopoulos P, Knockaert M, Leost M, Ryan XP, Vonica CA, Brivanlou A, Dajani R, Crovace C, Tarricone C, Musacchio A, Roe SM, Pearl L, Greengard P. GSK-3-selective inhibitors derived from Tyrian purple indirubins. Chem Biol. 2003;10:1255–1266. doi: 10.1016/j.chembiol.2003.11.010. [DOI] [PubMed] [Google Scholar]
- 30.Lei S, Dubeykovskiy A, Chakladar A, Wojtukiewicz L, Wang TC. The murine gastrin promoter is synergistically activated by transforming growth factor-β/Smad and Wnt signaling pathways. J Biol Chem. 2004;279:42492–42502. doi: 10.1074/jbc.M404025200. [DOI] [PubMed] [Google Scholar]
- 31.Candia AF, Watabe T, Hawley SH, Onichtchouk D, Zhang Y, Derynck R, Niehrs C, Cho KW. Cellular interpretation of multiple TGF-β signals: Intracellular antagonism between activin/BVg1 and BMP-2/4 signaling mediated by Smads. Development. 1997;124:4467–4480. doi: 10.1242/dev.124.22.4467. [DOI] [PubMed] [Google Scholar]
- 32.Schwappacher R, Weiske J, Heining E, Ezerski V, Marom B, Henis YI, Huber O, Knaus P. Novel crosstalk to BMP signalling: cGMP-dependent kinase I modulates BMP receptor and Smad activity. EMBO J. 2009;28:1537–1550. doi: 10.1038/emboj.2009.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wisotzkey RG, Johnson AN, Takaesu NT, Newfeld SJ. α/β Hydrolase2, a predicted gene adjacent to Mad in Drosophila melanogaster, belongs to a new global multigene family and is associated with obesity. J Mol Evol. 2003;56:351–361. doi: 10.1007/s00239-002-2411-4. [DOI] [PubMed] [Google Scholar]
- 34.Lee YS, Carthew RW. Making a better RNAi vector for Drosophila: Use of intron spacers. Methods. 2003;30:322–329. doi: 10.1016/s1046-2023(03)00051-3. [DOI] [PubMed] [Google Scholar]
- 35.Duncan DM, Burgess EA, Duncan I. Control of distal antennal identity and tarsal development in Drosophila by spineless–aristapedia, a homolog of the mammalian dioxin receptor. Genes Dev. 1998;12:1290–1303. doi: 10.1101/gad.12.9.1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Korchynskyi O, ten Dijke P. Identification and functional characterization of distinct critically important bone morphogenetic protein-specific response elements in the Id1 promoter. J Biol Chem. 2002;277:4883–4891. doi: 10.1074/jbc.M111023200. [DOI] [PubMed] [Google Scholar]
- 37.Inoue H, Imamura T, Ishidou Y, Takase M, Udagawa Y, Oka Y, Tsuneizumi K, Tabata T, Miyazono K, Kawabata M. Interplay of signal mediators of decapentaplegic (Dpp): Molecular characterization of mothers against dpp, Medea, and daughters against dpp. Mol Biol Cell. 1998;9:2145–2156. doi: 10.1091/mbc.9.8.2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Veeman MT, Slusarski DC, Kaykas A, Louie SH, Moon RT. Zebrafish prickle, a modulator of noncanonical Wnt/Fz signaling, regulates gastrulation movements. Curr Biol. 2003;13:680–685. doi: 10.1016/s0960-9822(03)00240-9. [DOI] [PubMed] [Google Scholar]
- 39.Clemens JC, Worby CA, Simonson-Leff N, Muda M, Maehama T, Hemmings BA, Dixon JE. Use of double-stranded RNA interference in Drosophila cell lines to dissect signal transduction pathways. Proc Natl Acad Sci USA. 2000;97:6499–6503. doi: 10.1073/pnas.110149597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Persson U, Izumi H, Souchelnytskyi S, Itoh S, Grimsby S, Engström U, Heldin CH, Funa K, ten Dijke P. The L45 loop in type I receptors for TGF-β family members is a critical determinant in specifying Smad isoform activation. FEBS Lett. 1998;434:83–87. doi: 10.1016/s0014-5793(98)00954-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Increased BMP signals generated by a stabilized Mad protein.
Fig. S2. Mad-GM8 expression increases the area of Distalless, a downstream target of Wg.
Fig. S3. Inducible RNAi directed against Wg depletes Wg protein and its downstream target Senseless.
Fig. S4. Mad-GM8 expression in the eye imaginal disc produces phenotypes suggestive of high Wg signaling.
Fig. S5. C-terminal phosphorylation of Mad enables BMP4 to repress the Mad-induced increase in Tcf reporter gene activity.
Fig. S6. The Mad RNAi phenotype is rescued by coexpression of a human Smad1 transgene.
Fig. S7. Mad is required for Wg signal transduction during wing margin development.
Fig. S8. Inhibition of GSK3 activity by BIO enhances the binding of Mad to Pangolin.
Table S1. Primer sequences.