Abstract
Tuber formation in potato (Solanum tuberosum) is promoted by short photoperiods and is inhibited by gibberellins (GAs). Endogenous levels of GA1 were shown to decrease in stolons and leaves of potato plants induced to tuberize, which suggests that photoperiodic regulation of GA biosynthesis may play a role in tuber induction. We report the isolation of three potato cDNA clones (StGA20ox1–3) encoding GA 20-oxidase, a key regulatory enzyme in the GA-biosynthetic pathway. Using northern analysis, we detected a differential pattern of tissue-specific expression of the mRNAs corresponding to these clones. StGA20ox mRNAs were also very abundant in leaves of the potato ga1 mutant, which is blocked in the 13-hydroxylation step, and were strongly down-regulated by gibberellic acid, suggesting a feedback regulation of these genes. In plants grown in short-day (inductive) conditions, levels of the StGA20ox transcripts in leaves fluctuated during a 24-h period, with a peak of accumulation observed about 4 h after the lights were turned off. Interruption of the night with a 30-min “night break” of light (noninductive conditions) did not have a marked effect on the levels of accumulation of the three GA 20-oxidase mRNAs during the day, but it induced a second peak of expression of StGA20ox1 and StGA20ox3 transcripts late in the night. This observation, together with the finding that StGA20ox1 mRNA is expressed at high levels in leaves, suggests that night-break induction of this gene might play a role in the control of tuberization by regulating endogenous levels of GAs in response to daylength conditions.
GAs are cyclic, diterpenoid hormones with an essential role in plant growth and development. They control a variety of growth responses in higher plants, including stem elongation, fruit set, flower induction, seed germination, and mobilization of seed reserves (for review, see Hooley, 1994; Swain and Olszewski, 1996). In potato (Solanum tuberosum) exogenous application of GAs has a strong inhibitory effect on tuberization (Okazawa, 1960; Tizio, 1971).
Tuber formation in potato is promoted by short photoperiods, cool temperatures, and low rates of nitrogen fertilization (Ewing, 1990). GA activity was shown to decrease when leaves were exposed to short days (Pont-Lezica, 1970; Kumar and Wareing, 1974), with as few as two short days being sufficient to cause a decline in the GA activity of S. tuberosum subsp. andigena leaves (Railton and Wareing, 1973). Changes in GA activity were shown to occur not only in response to photoperiod, but also to the other environmental conditions that affect tuberization: (a) high temperatures increased GA activity in buds (Menzel, 1983); (b) a continuous supply of nitrate in the hydroponic solution increased GA activity in shoots and prevented tuberization (Krauss and Marschner, 1982); and (c) a decrease in GA1 level was observed when the stolon tips started to swell in in-vitro-cultured single-node cuttings grown in a high-Suc tuber-inducing medium (Xu et al., 1998). Furthermore, tuberization was improved by the application of inhibitors of GA synthesis such as paclobutrazol or ancymidol (Menzel, 1980; Hussey and Stacey, 1984; Ewing, 1995; Jackson and Prat, 1996).
Further evidence for the involvement of GAs in the control of tuberization has derived from the isolation of a mutant of the short-day plant S. tuberosum subsp. andigena, which appears to be blocked in the GA-biosynthetic pathway at the 13-hydroxylation step that catalyzes the conversion of GA12 into GA53. This mutant has a dwarf phenotype and can form tubers during long-day conditions (Bamberg and Hanneman, 1991), suggesting a correlation between decreasing levels of GA activity and tuber initiation.
GAs are synthesized from isopentenyl pyrophosphate via geranylgeranyl pyrophosphate (for review, see Graebe, 1987; Sponsel, 1995; Hedden and Kamiya, 1997; Lange, 1998). The first committed step of GA biosynthesis is the formation of ent-kaurene from geranylgeranyl pyrophosphate, with copalyl pyrophosphate as an intermediary. This reaction is catalyzed by the enzymes ent-copalyl diphosphate synthase and ent-kaurene synthase, which have been cloned from various plant species (for a recent review, see Sun and Kamiya, 1997). ent-Kaurene is metabolized to GAs by membrane-associated monooxygenases and soluble, 2-oxoglutarate-dependent dioxygenases (Graebe, 1987). The latter group includes the GA 20-oxidase enzymes responsible for successive oxidations of C-20, leading to its loss as CO2 and to the formation of C-19 GAs.
GA 20-oxidase activity is suggested to be one of the principal points of regulation in the GA-biosynthetic pathway. In spinach transfer from short- to long-day conditions is associated with an increase in GA 20-oxidase activity and higher levels of GA20 (Gilmour et al., 1986; Zeevaart et al., 1990). Also, a substantial increase in the levels of GA 20-oxidase mRNA has been detected in spinach shoot tips upon transfer to long-day conditions (Wu et al., 1996). Expression of the Arabidopsis GA5 gene encoding GA 20-oxidase is enhanced by transfer of plants from short to long days (Xu et al., 1995).
GA 20-oxidases have been cloned and expressed from a number of plant species. One cDNA clone was isolated from pumpkin, three from Arabidopsis, one from spinach, two from pea, one from rice, and three from French bean (Lange et al., 1994; Phillips et al., 1995; Xu et al., 1995; Martin et al., 1996; Wu et al., 1996; García-Martínez et al., 1997; Lester et al., 1997; Toyomasu et al., 1997). Comparison of these cDNAs has shown highly conserved domains in all GA 20-oxidase proteins. We used a PCR-based approach for the isolation of three potato cDNA clones encoding putative GA 20-oxidases. Northern analysis showed that the three cDNAs (StGA20ox) showed different patterns of tissue-specific expression. We have obtained evidence of a photoperiodic regulation of the expression of clone StGA20ox1, which is strongly expressed in leaves. Photoperiod also affected expression of clone StGA20ox3, although to a lower degree. These results suggest that regulated expression of clone StGA20ox1 may play a role in tuber formation under photoinductive conditions.
MATERIALS AND METHODS
Plant Material
We used autotetraploid photoperiodic potato (Solanum tuberosum subsp. andigena) plants and the ga1 dwarf mutant (Bamberg and Hanneman, 1991), which is blocked in the 13-hydroxylation step, in these studies. Plants were grown under 16-h light/8-h dark conditions at 22°C. Plant tissues were harvested from plants at the 14-leaf stage, frozen in liquid N2, and stored at −80°C until used. The apex samples corresponded to the apex plus the first expanding leaves that surround the apex. We harvested the first three expanded leaves as young leaves, and the fifth to ninth leaves as old leaves. Internodes were harvested from the stem. Solanum demissum plants provided the flower, fruit, and seed samples.
PCR Amplification
Potato genomic DNA was extracted as described by Dellaporta et al. (1983). Dr. Peter Hedden (IACR-Long Ashton Research Station, UK) provided the degenerate oligonucleotide primers N1 to N7, based on regions found to be highly conserved in the pumpkin and Arabidopsis GA 20-oxidase cDNA clones. Primer N1 was based on the conserved sequence KLPWKET, corresponding to the amino acid residues 147 to 153 in the Arabidopsis At-2301 clone. Primers N2 and N3 corresponded to sense and antisense oligonucleotides derived from the sequence TGPHCDP at residues 144 to 150 of At-2301. Primers N4 and N5 were sense and antisense primers, respectively, derived from the sequence 282-FVVNIGD-288; and primers N6 and N7 were antisense oligonucleotides complementary to the conserved residues 303-HRAVVNS-309 and 317-AFFLCPK-323, respectively. We carried out PCR amplification with these primers, using potato genomic DNA as the template. From the different combinations of primers only oligonucleotides N2 and N5, with nucleotide sequences 5′-ACX GGX CCX CA(CT) (TA)(CG)X GA(CT) CC-3′ and 5′-TC XCC (GAT)AT (AG)TT XAC XAC (CT)AA-3′ generated an amplification product of the expected size (180 bp). This PCR product was subcloned into pBluescript (Stratagene) and the nucleotide sequence was determined for 20 of the obtained subclones.
Library Screening
A cDNA library was constructed into λZAPII (Stratagene) from 2 μg of poly(A+) RNA prepared from leaves of the ga1 dwarf mutant of S. tuberosum subsp. andigena. Total RNA was extracted as described by Logemann et al. (1987). We enriched poly(A+) RNA using a magnetic resin with oligo(dT)25 according to the manufacturer's instructions (Dynal, Oslo, Norway). The original library contained 1.5 million independent plaques. 32P-labeled PCR fragments 7, 8, 10, and 13 were used to screen the cDNA library. Hybridization was carried out as described by Amasino (1986). Hybridization temperature was 37°C, and filters were washed with 3 × SSC containing 0.5% SDS at 55°C. We excised isolated positive plaques in vivo using the method described by Stratagene, and further characterized the plasmids containing the longest cDNA inserts .
DNA Sequencing
cDNA clone inserts in pBluescript SK+ were sequenced using an automated laser-fluorescent DNA-sequencer system (ALF, Pharmacia). We used the T3 and T7 primers and primers designed after the partial insert sequences for sequencing. Using programs from the Genetics Computer Group (Madison, WI) we performed the sequence processing and database searches.
DNA Gel-Blot Analysis
Potato genomic DNA was digested with restriction enzymes and fractionated on a 0.8% agarose gel before transfer to a nylon membrane (Hybond-N, Amersham). DNA probes corresponding to the 3′-end of the clones (HindII to XhoI fragments of each clone) were used for hybridization. We used a multiprime kit (Boehringer Mannheim) to label the probes radioactively. The hybridization temperature was 45°C. The filters were washed in 0.2 × SSC, 0.5% SDS at 70°C.
RNA Gel-Blot Analysis
We isolated the RNA according to the method of Logemann (1987), electrophoresed the samples (30 μg per lane) in 1.2% agarose/formaldehyde gels, and transferred them to nylon membranes. Hybridization and washing conditions were the same as for Southern analysis.
Treatment with GA3- and GA-Biosynthesis Inhibitors
Leaves and stems of 10-leaf plants were treated by spraying the whole plant to runoff with the GA3- or GA-inhibitor solutions. Stock solutions of GA3 and ancymidol (Sigma) of 0.1 m and 5 mg mL−1, respectively were dissolved in 95% ethanol and further diluted in water. GA3 was used at a 5 × 10−5 m final concentration and ancymidol as a 5 mg L−1 solution. Dr. W. Rademacher (BASF Agricultural Research Center, Limburgerhof, Germany) kindly provided the prohexadione-calcium (BAS125 10W, containing 10% prohexadione-calcium). It was directly diluted in water at a concentration of 50 mg L−1, and 50 mL of the resulting solution (250 ng prohexadione-calcium) was applied per plant.
Kinetic Studies
Plantlets were propagated in vitro, transferred to soil, and grown under greenhouse conditions until the 10-leaf stage. We then transferred the plants to growth cabinets under light-dark cycles of short days (8 h light/16 h dark) or short days plus a night break (8 h light/16 h dark with 30-min light interruption in the middle of the dark period). High-pressure sodium lamps (150–200 μmol m−2 s−1 PAR; SON-T AGRO 400, Philips, Eindhoven, The Netherlands) provided the lighting. Plants were adapted for 10 d to the new light regime, and leaf samples were then harvested approximately every 3 h (except during the night period, when samples were also harvested immediately before and after the night break). We used green safelights for manipulations during the dark period.
Replicas of the RNA blots were first probed with the StGA20ox-specific probes, and then hybridized to a constitutive probe corresponding to the potato S4 ribosomal protein (Braun et al., 1994). Intensities of the signals were quantified by densitometric scanning (Molecular Dynamics, Sunnyvale, CA) of the radiographic film, and we used the ratio of StGA20ox to S4mRNA to calculate the levels of StGA20ox mRNAs in the different samples. Graphics represent the levels of the transcripts estimated for each probe in reference to the sample showing the strongest hybridization signal, to which we assigned an arbitrary value of 100 (short days of 23.5 h for RNA blots hybridized with probe StGA20ox1). Values approximately reflect the relative levels of the different transcripts, as probes of similar sizes and specific activities were used for hybridization. We repeated the experiment four times for probe StGA20ox1 and twice for probes StGA20ox2 and StGA20ox3.
RESULTS
Cloning of the Potato GA 20-Oxidase cDNAs
In an attempt to isolate the potato cDNAs encoding GA 20-oxidases, we used degenerate oligonucleotide primers complementary to conserved regions in the pumpkin and Arabidopsis GA 20-oxidase clones. Combinations of three sense (N1, N2, and N4) and four antisense (N3, N5, N6, and N7) primers were used in PCRs with potato genomic DNA as the substrate. Only primers N2 and N5 generated a product of 180 bp, which was subcloned into pBluescript and subjected to DNA sequencing. Sequence analysis of 20 clones chosen at random identified a total of five different PCR products (7, 8, 10, 12, and 13), which coded for protein sequences sharing between 75% and 88% amino acid identity with the GA 20-oxidase clone At-2301 from Arabidopsis (Phillips et al., 1995).
Cross-hybridization studies of the different PCR-derived clones showed that fragments 8 and 12 hybridized to each other, and therefore only fragments 7, 8, 10, and 13 were used for further studies. Hybridization of PCR fragments 7, 8, and 10 to northern blots containing RNA from wild-type plants or the GA-deficient mutant of potato, ga1, identified transcripts of approximately 1.5, 1.4, and 1.3 kb, respectively. These transcripts were more abundant in the dwarf plants and their abundance was strongly reduced by treatment with GA3 (data not shown), which indicates that they are regulated by a mechanism of negative-feedback control, as Phillips et al. (1995) and Xu et al. (1995) have reported for other GA 20-oxidase genes.
We did not obtain any hybridization signal by probing the ga1 RNA blot or northern blots containing RNAs from different potato tissues with fragment 13.
Isolation of Full-Length cDNA Clones
Because the mRNAs corresponding to fragments 7, 8, and 10 were more abundant in RNA preparations of the ga1 mutant, we constructed a cDNA library in λZAP from poly(A+) RNA isolated from young leaves of this dwarf mutant. Screening of the library with PCR fragments 7, 8, and 10 yielded clones StGA20ox1, StGA20ox3, and StGA20ox2, respectively, which corresponded to those with the longest insert sizes. No hybridization signals were obtained by screening the ga1 cDNA library or a cDNA library prepared from potato tubers with PCR fragment 13.
Clones StGA20ox1 and StGA20ox2 are likely to contain full-length open reading frames, based on comparisons with other GA 20-oxidases (Phillips et al., 1995; Wu et al.; 1996; García-Martínez et al., 1997). The ATG start codons were located at positions 7 to 9 of clone 7 and 28 to 30 of clone 8. Both clones included a poly(A+) tail and allowed for proteins of 378 and 375 residues and 43.2 and 42.7 kD, respectively.
Clone StGA20ox3 corresponds to a partial copy of cDNA. The insert contains a poly(A+) tail, but approximately 300 bp are missing from the 5′ end. This clone corresponded to a transcript less abundant than clones 7 and 8, as fewer positive clones were identified from screening the library. In an attempt to isolate a full-length cDNA clone corresponding to this gene, the insert of clone StGA20ox3 was purified and used for rescreening the library. No longer inserts could be obtained.
The StGA20ox cDNA Clones Encode Potato GA 20-Oxidases
Compared with other GA 20-oxidases, potato proteins share 62% to 70% homology with the Arabidopsis, spinach, and pea clones (see Fig. 1). Homology to the pumpkin clone was lower (58%). This was not totally unexpected, because the pumpkin enzyme produces predominantly the inactive tricarboxylic acid GA25 from GA12 and yields GA9 only as a minor product (Lange et al., 1994). The potato GA20ox clones share significant homology with other 2-oxoglutarate-dependent dioxygenases, although they are more closely related to the GA 20-oxidase enzymes than to any other dioxygenase.
Figure 1.
Alignments of the deduced amino acid sequences of the potato GA 20-oxidase clones StGA20ox1, StGA20ox2, and StGA20ox3 with those of pea (accession no. PsU58830), spinach (accession no. SoU33330), pumpkin (Cm20ox; accession no. X73314), and Arabidopsis (At2301; accession no. X83379). Identical residues are boxed in black; similar residues are shaded in gray. The conserved LPWKET and NYYPXCQKP regions thought to be involved in binding the GA substrate and the 2-oxoglutarate cosubstrate are underlined. Conserved H and D residues involved in the binding of Fe2+ are also indicated (*). Alignments were made using the PileUp and PrettyBox programs of the Genetics Computer Group.
To confirm that the full-length StGA20ox1 and StGA20ox2 clones corresponded to GA20-oxidases, their coding regions were cloned in-frame in a pET Escherichia coli expression vector. The recombinant plasmids were transformed into the BL21 E. coli strain and protein expression was induced by the addition of 1 mm isopropylthio-β-galactoside. Soluble protein extracts from these cells were assayed for GA20-oxidase activity by incubation with [14C]GA12 and HPLC separation of the reaction products. Bacterial extracts expressing the StGA20ox1 and StGA20ox2 proteins were both capable of using GA12 as a substrate to yield GA24 and GA9 as major products, demonstrating that these two clones corresponded to active GA20-oxidases.
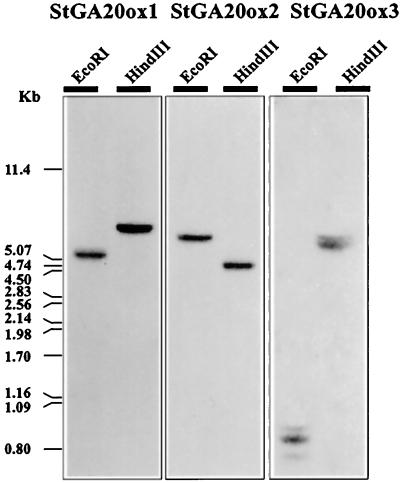
Southern Analysis
DNA-sequence comparisons of the StGA20 ox clones revealed a high degree of conservation in their coding regions but low nucleotide sequence homology in the 3′-noncoding regions. Therefore, we prepared subclones that included the 3′-noncoding regions and used them as specific probes in Southern and northern analyses. As shown in Figure 2, hybridization of the StGA20ox 3′-end probes to a Southern blot of potato genomic DNA revealed single EcoRI and HindIII fragments for the probes StGA20ox1 and StGA20ox2. Probe StGA20ox3 strongly hybridized to one EcoRI and HindIII fragment but identified more weakly two additional EcoRI and one HindIII fragments. This suggested the presence of an additional gene copy in the potato genome, which might correspond to PCR fragment 13. However, the fact that we did not detect hybridization with this probe either in RNA blots or by screening of the library suggests that it corresponded to a silent gene.
Figure 2.
Hybridization of clones StGA20ox1, StGA20ox2, and StGA20ox3 to a Southern blot of S. tuberosum subsp. andigena genomic DNA digested with EcoRI and HindIII. Ten micrograms of genomic DNA was loaded per lane. The HindII-XhoI fragments corresponding to the 3′-ends of the clones were used as probes. Molecular-mass markers are shown on the left (in kb).
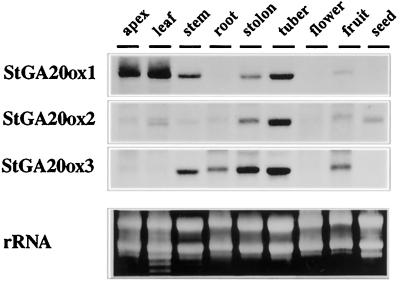
Each StGA20ox cDNA Clone Shows a Differential Pattern of Expression
Northern blots of RNA extracted from apex, leaves, stems, stolons, tubers, and roots of S. tuberosum subsp. andigena plants grown in short-day conditions were probed with the 32P-labeled StGA20ox 3′-end probes. Samples from flowers, fruits, and developing seeds were obtained from S. demissum plants grown under long-day conditions.
As shown in Figure 3, probe StGA20ox 1 identified a transcript of approximately 1.5 kb. This mRNA was abundant in shoot tips and leaves, and showed a moderate level of expression in stems, stolons, and tubers. Low levels of this mRNA were also detectable in fruits.
Figure 3.
Expression of the StGA20ox clones in different potato tissues. Northern analysis was carried out using 30 μg of total RNA from apex, young leaves, internodes (stem), roots, stolons, and tubers of S. tuberosum subsp. andigena plants grown under short days. Flower, fruit, and seed samples were obtained from S. demissum plants grown under long days. Ethidium-bromide staining of the gel is included to assess an equal loading of the lanes. RNA blots hybridized to probes StGA20ox1 and StGA20ox2 were exposed overnight, whereas the blot hybridized to probe StGA20ox3 was exposed for 4 d.
Probes StGA20ox 2 and StGA20ox 3 identified mRNA species of 1.3 and 1.4 kb, respectively (Fig. 3). StGA20ox 2 mRNA was relatively abundant in stolons and tubers. Moderate levels of expression of this mRNA were also found in fruits and developing seeds.
Clone StGA20ox 3 was expressed at significantly lowers levels than the two other clones. Whereas transcripts corresponding to clones StGA20ox 1 and StGA20ox2 were detectable after overnight exposure to the film, northern blots probed with clone StGA20ox3 required 4 to 5 d of exposition for signal detection. Low levels of this mRNA could be observed in the stems and roots as well as in the stolons and tubers. We also detected moderate levels of expression in fruits.
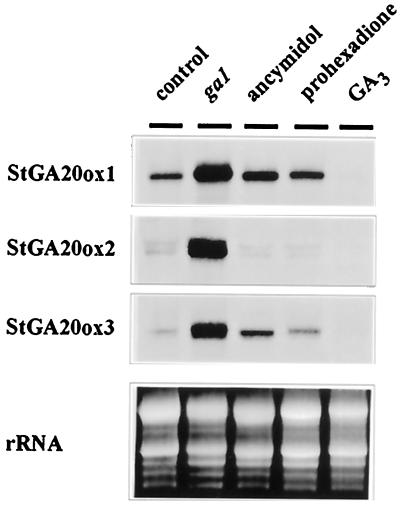
Feedback Regulation of StGA20ox Gene Expression
van den Berg (1995b) found that the ga1 dwarf mutant was deficient in the 13-hydroxylation step and therefore had low levels of active GAs. To investigate feedback inhibition of the StGA20ox genes, we analyzed the expression of the StGA20ox clones in the GA-deficient plants or in wild-type plants treated with inhibitors of GA biosynthesis such as ancymidol (which blocks the conversion of ent-kaurene into ent-kaurenoic) or prohexadione (which inhibits dioxygenases blocking GA biosynthesis at the 3β-hydroxylation step that converts GA20 into GA1) (Coolbaugh et al., 1978; Nakayama et al., 1990). GA-deficient plants were treated with GA3 to test for down-regulation of gene expression. As seen in Figure 4, levels of expression of all GA 20-oxidase mRNAs were very much increased in the ga1 dwarf mutant compared with wild-type plants. Treatment of wild-type plants with inhibitors of GA biosynthesis (ancymidol or prohexadione) increased the levels of expression of the StGA20ox1 and StGA20ox3 transcripts, but had little effect on the StGA20ox2 mRNA. In general, longer treatments or higher concentrations of the inhibitors were required to visualize induction of this transcript (data not shown). Treatment of ga1 plants with GA3 strongly reduced the abundance of mRNA corresponding to all clones (Fig. 4). These results indicate that, as shown for other plant species, expression of potato GA 20-oxidases is regulated by negative-feedback control by the biosynthetic end-product GA1.
Figure 4.
Feedback regulation of StGA20ox gene expression in potato shoots. Total RNA was isolated from control and ga1 dwarf potato plants, ga1 plants treated for 2 d with GA3 (10 μm), and control plants treated with the GA-biosynthesis inhibitors ancymidol (5 mg L−1) and prohexadione (50 mg L−1). Thirty micrograms of total RNA was loaded per lane.
Expression of StGA20ox mRNAs Is Regulated by Light
A decrease in GA activity was shown to occur in potato plants exposed to short days and was associated with tuber induction (Pont-Lezica 1970; Kumar and Wareing, 1974). To determine whether changes in the levels of expression of the GA 20-oxidase transcripts could be responsible for this decline in GA activity, we investigated the expression of StGA20ox mRNAs in leaves of plants grown under short days (tuber-inducing conditions) and under short days with a night break (noninducing conditions).
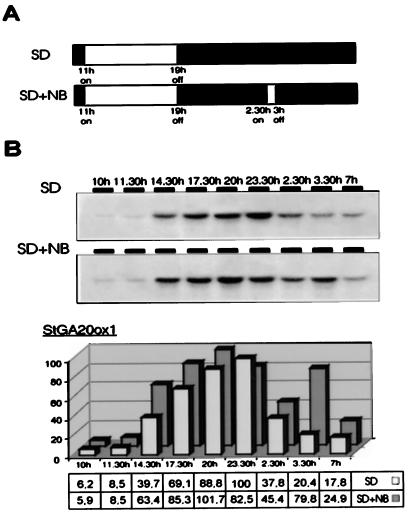
We detected fluctuating levels of mRNA for all three probes, as shown in Figures 5 and 6. Transcript StGA20ox1 accumulated in leaves to much higher levels than the other two transcripts, with peak levels approximately 5- and 20-fold those of transcripts StGA20ox2 and StGA20ox3, respectively. In plants grown in short-day conditions, StGA20ox1 mRNA started to accumulate with the beginning of the light period and increased throughout the day to become maximal at 23 h, 4 h after the lights were turned off. Afterward, the levels of transcript dropped, becoming minimal by the time the light period started again.
Figure 5.
Diurnal variation in the levels of accumulation of transcript StGA20ox1 in leaves. Plants were grown in short days (SD) or short days with a night break (SD + NB) for 3 weeks. Leaf samples were harvested 30 min after the lights were turned on and taken at intervals of approximately 3 h and immediately before and after the night break. A, Light regimes used to grow the plants. The light period and night break are indicated by white bars; the dark period by a black bar. B, RNA blots hybridized with the StGA20ox1 probe. After hybridization to the StGA20ox1 probe, blots were probed with a fragment corresponding to the ribosomal S4 protein (Braun et al., 1994). StGA20ox1 mRNA levels were quantified by densitometric scanning of the film and normalized to S4 mRNA levels. The graph represents the levels of StGA20ox1 mRNA estimated for each sample compared with the highest hybridization signal (short days, 23.5 h), to which we assigned an arbitrary value of 100. Thirty micrograms of total RNA was loaded per lane. The experiment was repeated four times.
Figure 6.
Diurnal variation of levels of StGA20ox2 and StGA20ox3 transcripts in leaves. Plants were grown in short days (SD) or short days with a night break (SD + NB) for 3 weeks. RNA blots were hybridized to probes StGA20ox2 and StGA20ox3, and subsequently to the ribosomal S4 protein probe for normalization. Densitometric scanning of the film quantified the levels of StGA20ox2 and StGA20ox3 mRNAs. The graphs represent the levels estimated for both transcripts compared with the strongest hybridization signal (short days, 23.5 h, in RNA blots hybridized with probe StGA200ox1), to which we assigned an arbitrary value of 100. Values correspond to approximately the levels of the respective mRNAs, as probes of about the same size and specific activities were used for hybridization. The experiment was repeated twice.
Interruption of the night period with 30 min of light did not have a strong effect on the levels of accumulation of the StGA20ox1 transcript, but did induce moderate changes in the cycling of expression. As seen in Figure 5, induction after lights-on was slightly hastened in these samples, with the peak of expression observed at 20 h instead of 23.5 h. A second peak of transcript accumulation was observed immediately after the night break, and resulted in higher levels of StGA20ox1 mRNA late in the night in plants grown under short days plus night break compared with plants grown under short days (Fig. 5).
Levels of StGA20ox2 transcripts also fluctuated during a 24-h period, with two peaks of expression observed in plants grown under short-day conditions (Fig. 6). Expression of StGA20ox2 mRNA was high at the end of the night period and increased further during the first hours of light. After this initial induction, levels of StGA20ox2 mRNA fell to basal levels and then increased again with the lights off, with maximal levels of expression observed shortly after the lights were turned off (20 h). Afterward, expression decreased again to reach low levels in the middle of the night period. Interruption of the night period did not have a marked effect on the levels of expression of this transcript; we observed only a slight increase in expression 7 h before the lights were turned on.
As shown in Figure 6, very low levels of the StGA20ox3 transcript were detected in leaves. The pattern of expression of this transcript was very similar to that observed for StGA20ox1, with maximal levels of transcript detected at 23.5 h in the dark period in plants kept under short-day conditions. Interruption of the night by a 30-min light application also induced a shift in the peak of expression to 20 h, with a second peak of mRNA observed after the night break. Night-break-induced up-regulation of this gene was, however, less pronounced than that of StGA20ox1.
Differences in GA 20-oxidase gene expression between plants grown under short days and those also given a night break were observed only for StGA20ox1 and StGA20ox3 transcripts. We found the levels of expression of the StGA20ox1 mRNA to be much higher (approximately 20- to 25-fold) than those of the StGA20ox3 mRNA. Therefore, of the three cDNA clones characterized, StGA20ox1 would be the best candidate for a role in the rise in leaf GA bioactivity observed in the plants grown in noninducing conditions.
DISCUSSION
Sequence Homology to Other GA-20 Oxidases
The predicted StGA20ox proteins share high sequence homology with the GA 20-oxidase genes from other plant species. Homology to other oxoglutarate-dependent dioxygenases was also observed, but was always lower than with GA 20-oxidases. Highly conserved motifs such as the conserved consensus sequence NYYPXCQKP (postulated to be involved in binding the 2-oxoglutarate cofactor), the conserved H and D residues (involved in binding of Fe2+ at the active site of isopenicillin N synthase [Roach et al., 1995]), and the LPWKET motif (thought to be involved in binding the GA substrate [Xu et al., 1995]) were all present in the potato StGA20ox proteins (Fig. 1). This is consistent with StGA20ox gene products being GA 20-oxidases and also with in vitro assays in which we found that soluble extracts from E. coli expressing the full-length StGA20ox1 and StGA20ox2 proteins were capable of oxidation and subsequent loss of the C20 of GA12 to produce GA24 and GA9.
Feedback Regulation of StGA20ox Gene Expression
There is considerable evidence indicating that bioactive GAs may control their own synthesis through a negative-feedback mechanism of the GA 20-oxidase and 3β-hydroxylase genes (Chiang et al., 1995; Phillips et al., 1995; Xu et al., 1995; Martin et al., 1996). We found that the ga1 mutant of potato (thought to be blocked in the 13-hydroxylation step) accumulates high levels of all three GA 20-oxidase transcripts. Expression of the three GA 20-oxidase genes was, in addition, considerably reduced in the dwarf mutant after application of GA3. These results indicate that in potato, as in Arabidopsis and spinach, a negative-feedback mechanism of regulation controls the expression of the GA 20-oxidase genes.
Inhibition of GA biosynthesis by application of ancymidol or prohexadione resulted in a much stronger accumulation of StGA20ox1 and StGA20ox3 transcripts than of transcript StGA20ox2. Only when samples were taken in the dark period, after more prolonged treatments, or after application of a higher concentration of the inhibitors were we able to detect an accumulation of this latter transcript in the leaves (data not shown). This finding suggests that feedback regulation of the StGA20ox2 transcript might be stronger than that of the other two transcripts, and that lower levels of bioactive GAs would need to be attained to detect up-regulation of this gene.
Photoperiod-Regulated Expression of StGA20ox1
GA activity has been shown to decrease in potato when leaves are exposed to short days (Pont-Lezica 1970; Kumar and Wareing, 1974). The concentration of GA1 in apical cuttings of S. tuberosum subsp. andigena was also lower in plants grown under short- than under long-day photoperiods (van den Berg et al., 1995a; 1995b). These observations suggest that one of the steps in the potato GA-biosynthetic pathway may be subjected to regulation by daylength. In spinach and Arabidopsis, Wu et al. (1996) and Xu et al. (1995) found that the expression of GA 20-oxidase was regulated by photoperiod, with significantly higher levels of GA 20-oxidase mRNAs in long- compared with short-day conditions.
To investigate whether changes in GA 20-oxidase expression were also responsible for the decline in the GA content observed in potato plants grown in short days, we analyzed the levels of expression of the StGA20ox transcripts in leaves of S. tuberosum subsp. andigena plants grown in short days (tuber-inducing conditions) or in short days with a night break (noninducing conditions). In samples harvested during the day, we could not detect a higher level of the StGA20ox transcripts in plants grown in short days with a night break compared with short days alone (see Figs. 5 and 6). However, in time-course experiments we observed that an interruption of the dark period induced a second peak of accumulation of transcripts StGA20ox1 and StGA20ox3 late in the night. Interruption of the dark period with a 30-min night break induced a shift in the peak of expression from 23.5 to 20 h, and resulted in a rise in the levels of both transcripts immediately after the night break. As a result of this second peak of expression, the interval in which we found accumulation of both transcripts in leaves was prolonged (Figs. 5 and 6). The peak of expression was extended from 14.5 to 23.5 h in short-day conditions, whereas in short-day conditions with a night break, a rise in the levels of transcript could be observed until 3.5 h into the night period.
We obtained similar results in time-course studies of transcript StGA20ox1 in plants grown in long days (16 h light/8 h dark). As before, we did not detect a higher level of StGA20ox1 mRNA in the plants kept in long days, but we did observe that the peak of expression was extended, with up-regulated levels of transcript observed at all time points studied (data not shown). These results contrast with those obtained in spinach and Arabidopsis, in which higher levels of GA 20-oxidase mRNA were detected in long days, and suggest that GA 20-oxidase activity may be differently regulated in potato than in Arabidopsis or spinach. It will be interesting to discover whether this pattern of regulation is a unique feature of potato or if it reflects a general mechanism of regulation evolved by short-day plants (potato) compared with long-day plants (spinach and Arabidopsis).
At present it is unclear whether up-regulation of transcripts StGAox1 and StGA20ox3 caused by the night break is sufficient to account for the higher levels of GAs observed in the noninduced plants. Also, because StGA20ox1 transcripts were expressed at much higher levels in leaves than StGA20ox3 transcripts, it is likely that up-regulation of this transcript plays a more prevalent role in the rise of endogenous levels of GAs observed in plants kept in noninduced conditions. More experiments based on StGA20ox1 sense expression or antisense inhibition will be needed, however, to ascertain the possible function of this gene in the control of tuberization.
Previous studies performed with apices of potato plants exposed to 10- or 16-h photoperiods also did not show significant differences in [14C]GA12 metabolism in short-day plants (van den Berg, 1995b). Therefore, it is possible that the reduced levels of GA1 observed in S. tuberosum subsp. andigena plants in short days were mainly the result of photoperiodic control of a step in the GA-biosynthetic pathway other than GA 20-oxidase. Zeevaart and Gage (1993) have reported a photoperiodic effect on the biosynthesis of ent-kaurene in spinach and Agrostemma githago L. Isolation of the cDNAs encoding ent-copalyl diphosphate synthase and ent-kaurene synthase or the dioxygenase responsible for the 3β-hydroxylation step (which catalyzes conversion of GA20 to GA1) should elucidate whether these biosynthetic activities are subjected to daylength regulation and if they are therefore involved in the photoperiodic induction of tuber formation in potato.
ACKNOWLEDGMENTS
We are grateful to Drs. Peter Hedden and Andy Phillips for kindly providing the oligonucleotides used for PCR amplification. We thank Dr. W. Rademacher for the generous gift of prohexadione, Dr. John Bamberg of the Potato Introduction Station (NRSP–6, Sturgeon Bay, WI) for supplying the dwarf S. tuberosum subsp. andigena seeds, and Dr. J.L. García-Martínez for the GA 20-oxidase activity assay of the StGA20ox1 and StGA20ox2 proteins. We also thank Drs. Hedden, Phillips, and García-Martínez for helpful comments on the manuscript.
Footnotes
This work was supported by the Comision Interministerial de Ciencia y Tecnología Plan Nacional (grant no. BIO96-0532-C02-02).
LITERATURE CITED
- Amasino RM. Acceleration of nucleic acid hybridization rate by polyethylenglycol. Anal Biochem. 1986;152:304–307. doi: 10.1016/0003-2697(86)90413-6. [DOI] [PubMed] [Google Scholar]
- Bamberg JB, Hanneman RE. Characterization of a new gibberellin related dwarfism locus in potato (Solanum tuberosum L.) Am Potato J. 1991;68:45–52. [Google Scholar]
- Braun HP, Emmerman H, Mentzel H, Schmitz UK. Primary structure and expression of a gene encoding the cytosolic ribosomal protein S4 from potato. Biochim Biophys Acta. 1994;1218:435–438. doi: 10.1016/0167-4781(94)90200-3. [DOI] [PubMed] [Google Scholar]
- Chiang HH, Hwang I, Goodman HM. Isolation of the Arabidopsis GA4 locus. Plant Cell. 1995;7:195–201. doi: 10.1105/tpc.7.2.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coolbaugh RC, Hirano SS, West ChA Studies on the specificity and site of action of a-cyclopropyl-a-[p-methoxyphenyl]-5-pyrimidine methyl alcohol (ancymidol), a plant growth regulator. Plant Physiol. 1978;62:571–576. doi: 10.1104/pp.62.4.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellaporta SL, Wood J, Hicks JB. A plant DNA minipreparation: version II. Plant Mol Biol Rep. 1983;1:19–21. [Google Scholar]
- Ewing EE (1990) Induction of tuberization in potato. In ME Vayda, WD Park, eds, The Molecular and Cellular Biology of the Potato, CAB International, Wallinford, UK, pp 25–43
- Ewing EE. The role of hormones in potato (Solanum tuberosum L.) tuberization. In: Davies PJ, editor. Plant Hormones: Physiology, Biochemistry and Molecular Biology. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1995. pp. 698–724. [Google Scholar]
- García-Martínez JL, Lopez-Diaz I, Sánchez-Beltrán MJ, Phillips AL, Ward DA, Gaskin P, Hedden P. Isolation and transcript analysis of gibberellin 20-oxidase genes in pea and bean in relation to fruit development. Plant Mol Biol. 1997;33:1073–1084. doi: 10.1023/a:1005715722193. [DOI] [PubMed] [Google Scholar]
- Gilmour SJ, Zeevaart JAD, Schwenen L, Graebe JE. Gibberellin metabolism in cell-free extracts from spinach leaves in relation to photoperiod. Plant Physiol. 1986;82:190–195. doi: 10.1104/pp.82.1.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graebe JE. Gibberellin biosynthesis and control. Annu Rev Plant Physiol. 1987;38:419–465. [Google Scholar]
- Hedden P, Kamiya Y. Gibberellin biosynthesis: enzymes, genes and their regulation. Annu Rev Plant Physiol Plant Mol Biol. 1997;48:431–460. doi: 10.1146/annurev.arplant.48.1.431. [DOI] [PubMed] [Google Scholar]
- Hooley R. Gibberellins: perception, transduction and responses. Plant Mol Biol. 1994;26:1529–1555. doi: 10.1007/BF00016489. [DOI] [PubMed] [Google Scholar]
- Hussey G, Stacey NJ. ) Ann Bot. 1984;48:787–796. [Google Scholar]
- Jackson SD, Prat S. Control of tuberisation in potato by gibberellins and phytochrome B. Physiol Plant. 1996;98:407–412. [Google Scholar]
- Krauss A, Marschner H. Influence of nitrogen nutrition, daylength and temperature on contents of gibberellic and abscisic acid on tuberization of potato plants. Potato Res. 1982;25:13–21. [Google Scholar]
- Kumar D, Wareing PF. Studies on tuberization of Solanum andigena. New Phytol. 1974;73:833–840. [Google Scholar]
- Lange T, Hedden P, Graebe JE. Expression cloning of a gibberellin 20-oxidase, a multifunctional enzyme involved in gibberellin biosynthesis. Proc Natl Acad Sci USA. 1994;91:8552–8556. doi: 10.1073/pnas.91.18.8552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange T. Molecular biology of gibberellin synthesis. Planta. 1998;204:409–419. doi: 10.1007/s004250050274. [DOI] [PubMed] [Google Scholar]
- Lester DR, Ross JJ, Davies PJ, Reid JB. Mendel's stem length gene (Le) encodes a gibberellin 3 beta-hydroxylase. Plant Cell. 1997;9:1435–1443. doi: 10.1105/tpc.9.8.1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logemann J, Schell J, Willmitzer L. Improved method for the isolation of RNA from plant tissue. Anal Biochem. 1987;163:16–20. doi: 10.1016/0003-2697(87)90086-8. [DOI] [PubMed] [Google Scholar]
- Martin DN, Proebsting WM, Parks TD, Dougherty WG, Lange T, Lewis MJ, Gaskin P, Hedden P. Feed-back regulation of gibberellin biosynthesis and gene expression in Pisum sativum L. Planta. 1996;200:159–166. doi: 10.1007/BF00208304. [DOI] [PubMed] [Google Scholar]
- Menzel BM. Tuberization in potato Solanum tuberosum cultivar Sebago at high temperatures: responses to gibberellin and growth inhibitors. Ann Bot. 1980;46:259–266. [Google Scholar]
- Menzel BM. Tuberization in potato (Solanum tuberosum cultivar Sebago) at high temperatures: gibberellin content and transport from buds. Ann Bot. 1983;52:697–702. [Google Scholar]
- Nakayama I, Miyazawa T, Kobayashi M, Kamiya Y, Abe H, Sakurai A. Effects of a new plant growth regulator prohexadione calcium (BX-112) on shoot elongation caused by exogenously applied gibberellins in rice (Oryza sativa L.) seedlings. Plant Cell Physiol. 1990;31:195–200. [Google Scholar]
- Okazawa Y. Studies on the relation between the tuber formation of potato plant and its natural gibberellin content. Proc Crop Sci Soc Jpn. 1960;29:121–124. [Google Scholar]
- Phillips AL, Ward DA, Uknes S, Appleford NEJ, Lange T, Huttly AK, Gaskin P, Graebe JE, Hedden P. Isolation and expression of three gibberellin 20-oxidase cDNA clones from Arabidopsis. Plant Physiol. 1995;108:1049–1057. doi: 10.1104/pp.108.3.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pont-Lezica RF. Evolution des substances de type gibberellines chez la pomme de terre pendant la tuberisation, en relation avec la longueur du jour et la temperature. Potato Res. 1970;13:323–331. [Google Scholar]
- Railton ID, Wareing PF. Effects of daylength on endogenous gibberellins in leaves of Solanum andigena. Changes in levels of free acidic gibberellin-like substances. Physiol Plant. 1973;28:88–94. doi: 10.1007/BF00386032. [DOI] [PubMed] [Google Scholar]
- Roach PL, Clifton IJ, Fülöp V, Harlos K, Barton GJ, Hadju J, Andersson I, Schofield CJ, Baldwin JE. Crystal structure of isopenicillin N synthase is the first from a new structural family of enzymes. Nature. 1995;375:700–704. doi: 10.1038/375700a0. [DOI] [PubMed] [Google Scholar]
- Sponsel VM. The biosynthesis and metabolism of gibberellins in higher plants. In: Davies PJ, editor. Plant Hormones: Physiology, Biochemistry and Molecular Biology. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1995. pp. 66–97. [Google Scholar]
- Sun T-P, Kamiya Y. Regulation and cellular localization of ent-kaurene synthesis. Physiol Plant. 1997;101:701–708. [Google Scholar]
- Swain SM, Olszewski NE. Genetic analysis of gibberellin signal transduction. Plant Physiol. 1996;112:11–17. doi: 10.1104/pp.112.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talon M, Zeevaart JAD, Gage DA. Identification of gibberellins in spinach and effects of light and darkness on their levels. Plant Physiol. 1991;97:1521–1526. doi: 10.1104/pp.97.4.1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tizio R. Action et role probable de certaines gibberellines (A1, A3, A4, A5, A7, A9 et A13) sur la croissance des stolons et la tuberisation de la pomme de terre (Solanum tuberosum L.) Potato Res. 1971;14:193–204. [Google Scholar]
- Toyomasu T, Kawaide H, Sekimoto C, von Numers C, Phillips AL, Hedden P, Kamiya Y. Cloning and characterization of a cDNA encoding gibberellin 20-oxidase from rice (Oryza sativa L.) seedlings. Physiol Plant. 1997;99:111–118. [Google Scholar]
- van den Berg JH, Davies PJ, Ewing EE, Halinska A. Metabolism of gibberellin A12 and A12-aldehyde and the identification of endogenous gibberellins in potato (Solanum tuberosum ssp. andigena) shoots. J Plant Physiol. 1995a;146:459–466. [Google Scholar]
- van den Berg JH, Simko I, Davies PJ, Elmer EE, Halinska A. Morphology and [14C]gibberellin A12 metabolism in wild-type and dwarf Solanum tuberosum ssp. andigena grown under long and short photoperiods. J Plant Physiol. 1995b;146:467–473. [Google Scholar]
- Wu K, Li L, Gage DA, Zeevaart JAD. Molecular cloning and photoperiod-regulated expression of gibberellin 20-oxidase from the long-day plant spinach. Plant Physiol. 1996;110:547–554. doi: 10.1104/pp.110.2.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, van Lammeren AA, Vermeer E, Vreugdenhil D. The role of gibberellin, abscisic acid, and sucrose in the regulation of potato tuber formation in vitro. Plant Physiol. 1998;117:575–584. doi: 10.1104/pp.117.2.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y-L, Li L, Wu K, Peeters AJM, Gage DA, Zeevaart JAD. The GA5 locus of Arabidopsis thaliana encodes a multifunctional gibberellin 20-oxidase: molecular cloning and functional expression. Proc Natl Acad Sci USA. 1995;92:6640–6644. doi: 10.1073/pnas.92.14.6640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeevaart JAD, Gage DA. ent-kaurene biosynthesis is enhanced by long photoperiods in the long-day plants Spinacia oleracea L. and Agrostemma githago L. Plant Physiol. 1993;101:25–29. doi: 10.1104/pp.101.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeevaart JAD, Talon M, Wilson TM. Stem growth and gibberellin metabolism in spinach in relation to photoperiod. In: Takahashi N, Phinney BO, Macmillan J, editors. Gibberellins. Berlin: Springer-Verlag; 1990. pp. 2273–2279. [Google Scholar]