Abstract
The peroxisome proliferator-activated receptor gamma (PPARγ or PPARG) belongs to the nuclear receptor superfamily, and is a potential drug target for a variety of diseases. In this work, we constructed a series of bacterial biosensors for the identification of functional PPARγ ligands. These sensors entail modified Escherichia coli cells carrying a four-domain fusion protein, comprised of the PPARγ ligand binding domain (LBD), an engineered mini-intein domain, the E. coli maltose binding protein (MBD), and a thymidylate synthase (TS) reporter enzyme. E. coli cells expressing this protein exhibit hormone ligand-dependent growth phenotypes. Unlike our published estrogen (ER) and thyroid receptor (TR) biosensors, the canonical PPARγ biosensor cells displayed pronounced growth in the absence of ligand. They were able to distinguish agonists and antagonists, however, even in the absence of agonist. To improve ligand sensitivity of this sensor, we attempted to engineer and optimize linker peptides flanking the PPARγ LBD insertion point. Truncation of the original linkers led to decreased basal growth and significantly enhanced ligand sensitivity of the PPARγ sensor, while substitution of the native linkers with optimized G4S (Gly-Gly-Gly-Gly-Ser) linkers further increased the sensitivity. Our studies demonstrate that the properties of linkers, especially the C-terminal linker, greatly influence the efficiency and fidelity of the allosteric signal induced by ligand binding. Our work also suggests an approach to increase allosteric behavior in this multidomain sensor protein, without modification of the functional LBD.
Keywords: PPARγ, biosensor, fusion protein, nuclear hormone receptor, hormone assay, engineered allostery
1. Introduction
Peroxisome proliferator-activated receptors (PPARs) are members of the nuclear receptor superfamily that are activated by fatty acids to regulate the expression of genes involved in lipid homeostasis (Krey et al. 1997). Three subtypes of PPARs have been identified, including PPARα, PPARγ and PPARδ/β (Committee 1999), each of which exhibit tissue-specific expression patterns (Kliewer et al. 2001). PPARγ is highly expressed in adipose tissue and is considered critical for fat cell development and differentiation (Chawla et al. 1994; Kliewer et al. 2001). PPARγ has been identified as the target for the antidiabetic action of glitazones (often called “insulin sensitizers”), which are derivatives of thiazolidinedione (TZD) (Kliewer et al. 2001). PPARγ has also been implicated in dyslipidemia, hypertension and inflammation, as well as cancer (Willson et al. 2000). Although PPARγ exhibits a similar domain structure to other members of the nuclear hormone receptor superfamily, the PPARγ ligand-binding domain (LBD) is distinctly different from the canonical LBD structure. In particular, the helix 12 molecular switch of this peroxisome proliferator-activated receptor is folded back toward the predicted ligand binding pocket in the apo-form (unliganded) of the LBD, resulting in a pre-active conformation that is similar to that of the holo-forms (ligand-bound) of other nuclear receptors (Uppenberg et al. 1998).
The identification of potential ligands for modulating PPARγ calls for highly sensitive, reliable and powerful analytical tools. Traditional binding assays, based on radioactivity (Lehmann et al. 1995) protease sensitivity (Camp et al. 2000), fluorescence displacement (Adamson and Palmer 2002), spectrophotometry (Adamson and Palmer 2002) and surface plasmon resonance (Yu et al. 2004), generally do not give detailed information regarding the biological function of tested ligands. Inspired by the mechanism of co-activator recruitment upon agonist ligand binding (Willson et al. 2000), transactivation assays have also been developed to monitor the biological function of ligands. Early transactivation assays relied on a cloned, full-length PPAR receptor and a PPAR response element driving the expression of a convenient reporter enzyme in a non-native host cell (Kliewer et al. 1992; McDonnell et al. 1993). To eliminate the effect of endogenous receptors in these host cells, chimeric receptors typically containing GAL-4 DNA binding domain (DBD) and PPAR LBD have also been constructed (Chen et al. 2004; Forman et al. 1995; Kliewer et al. 1995; Lehmann et al. 1995; T. Taniguchi 2002). Purified coactivators can also be used for in vitro identification of functional PPAR ligands (Krey et al. 1997), and the binding motif of the coactivator has been used to construct a fluorescent sensor in combination with the PPARγ LBD (Awais et al. 2007).
Recently, synthetic allosteric proteins have been engineered by several investigators via domain insertion (Ostermeier 2005) or sequence overlap (Sallee et al. 2007) to detect the presence of ligands and modulate the function of various reporter enzymes. In general, the functional domains of artificial chimeric proteins are connected using peptides of various lengths and sequences, called linkers or spacers (Nixon et al. 1997). Studies suggest that these linkers participate in the communication between the included functional domains (Gokhale and Khosla 2000), and that their length and sequence may affect the function and stability of the fusion protein (Gokhale and Khosla 2000; Raag and Whitlow 1995; Robinson and Sauer 1998; van Leeuwen et al. 1997). Using these principles, bacterial biosensors have been recently created in our laboratories, which involve an appropriate Escherichia coli (E. coli) strain expressing an allosteric four-domain fusion reporter protein (Skretas and Wood 2005a). In this system, LBDs from nuclear hormone receptors are inserted into an engineered intein splicing domain, which is in turn linked to a thymidylate synthase (TS) reporter enzyme. We have hypothesized that the intein domain serves as a general stabilizing scaffold, allowing the rapid development of new biosensors through simple LBD swapping with the intein domain. Several human biosensors have been generated using this scaffold, including estrogen receptors (ERα and ERβ) and thyroid hormone receptor β (TRβ) (Skretas and Wood 2005a, b). Importantly, these studies suggest that the segments connecting the intein domain to the LBD play a critical role in the sensor behavior (Skretas et al. 2007; Skretas and Wood 2005a, b).
In the present work, we created an allosteric PPARγ ligand biosensor protein based on the modular design of our previous ERβ sensor. In order to modulate the PPARγ biosensor ligand sensitivity, the spacer peptides connecting the intein splicing domain and inserted LBD were subjected to progressive truncation. Furthermore, artificial linkers comprised of glycine and serine repeats (Huston et al. 1988) were used to replace the original ERβ biosensor protein spacers in an attempt to enhance the performance of the sensor. The resulting biosensors were evaluated for growth phenotype in the presence of known PPARγ ligands at different incubation temperatures. Finally, the mechanism of our bacterial biosensor response to ligands is proposed.
2. Experimental Procedures
2.1.Reagents
Rosiglitazone (ROSIG; PPARγ agonist) was obtained from Cayman Chemical Co. (Ann Arbor, MI), Pioglitazone (PIOG; PPARγ agonist) from AvaChem Scientific LLC (San Antonio, TX), 17β-estradiol (E2; ER agonist), GW9662 (PPARγ antagonist) and 15-deoxy-Δ12,14-Prostaglandin J2 (15D-PGJ2; PPARγ selective agonist) were purchased from Sigma-Aldrich (St. Louis, MO).
2.2.Construction of the initial PPARγ bacterial sensor protein
To generate insertional LBD fusions (Figure 1) with the Mycobacterium tuberculosis (Mtu) recA mini-intein (Derbyshire et al. 1997), gene splicing by overlap extension (SOE) was used with some modifications (Horton et al. 1989). All PCR reactions described in this work were performed using Phusion DNA polymerase (Finnzymes). The DNA sequence encoding residues 232–505 of the human PPARγ was amplified from a Mammalian Gene Collection (MGC) cDNA clone (Accession, BC006811; Clone ID, 3447380) using forward primer Delt_Int110_PPARG_for and reverse primer Delt_Int383_PPARG_rev (Supplementary Figure 1a). The sequences of these two primers and the ones mentioned below are shown in the Supplementary Table 1. At the same time, the Mtu intein DNA was amplified with the primers Int_Age_I_for / Int_110_rev, coding for N-terminal splicing domain of intein; and Int_383_for / Int_Xho_I_rev2, coding for C-terminal splicing domain, by PCR with overlap primers respectively. An assembly PCR was then performed using the amplified PPARγ DNA and two intein DNA segments using Int_Age_I_for and Int_Xho_I_rev2 as the outer primer, and the assembled DNA was used to replace the corresponding intein-LBD fusion section of pMIT∷ERβ* (Skretas et al. 2007; Skretas and Wood 2005b) by ligation into the Age I/Xho I digested vector backbone. The resulting construct, referred to as pMIT:110PPARγ383 (denoting the insertion of PPARγ between residues 110 and 383 of the full-length Mtu intein), was transformed into calcium competent E. coli DH5α for screening and amplification, and confirmed by sequencing. Confirmed clones were then transformed into the TS-deficient E. coli strain D1210ΔthyA∷KanR [F-Δ (gpt-proA)62 leuB6 supE44 ara-14 galK2 lacY1 Δ (mcrC-mrr) rpsL20 (Strr) xyl-5 mtl-1 recA13 lacIq] for the biosensor assay. For convenience, “pMIT:” has been omitted from the construct names throughout the text and figures.
Fig. 1.
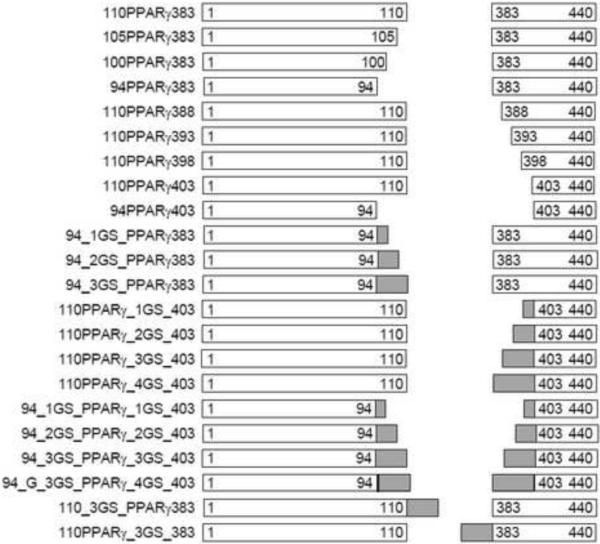
Schematic diagrams of the constructed intein-PPARγ LBD fusions with modified spacers. Open rectangles correspond to the N-terminal and C-terminal intein segments. The relative size of each intein segments is shown, with the included residues for each segment noted. Grey segments correspond to inserted GS linkers, with relative sizes shown. For the construct 94_G_3GS_PPARγ_4GS_403, the dark line at the end of the intein indicates the insertion of an additional Glycine residue before the GS linkers. The N-terminal MBP and C-terminal TS fusions are the same for all the constructs, and are therefore not shown in the diagrams.
2.3.Construction of sensors with truncated LBD-intein spacers
The PPARγ gene was amplified from 110PPARγ383 using PPARG_for / Int_383_rev, annealing to intein sequence as illustrated above) and Int_110_for / PPARG_rev (Supplementary Figure 1b). The resulting DNAs contain the full length of PPARγ LBD sequence with an overlap sequence annealing to the sequence coding the original N-terminal or C-terminal spacer respectively, acting as the template for assembly PCR together with intein segment DNAs. For C-terminal spacer truncation, the reverse primers including Int388_PPARG_rev, Int393_PPARGrev, Int398_PPARG_ rev, and Int403_ PPARG_rev were applied to construct 110PPARγ388, 110PPARγ393, 110PPARγ398, 110PPARγ403 respectively (Supplementary Figure 1b, left panel). For N-terminal spacer truncation, Int105_PPARG_for, Int100_PPARG_for, Int94_PPARG_for were used to create 105PPARγ383, 100PPARγ383 and 94PPARγ383 separately (Supplementary Figure 1b, right panel). For combined N-terminal and C-terminal truncations, one primer from each of the above groups was added. The resulting constructs were digested with Age I/Xho I and ligated into a similarly digested pMIT vector. Transformations were performed using the steps mentioned above. Schematic maps of the resulting plasmids together with those described below are shown in Figure 1. The nomenclature of the resulting clones indicates the last residue of N-terminal linker and the first residue of C-terminal linker.
2.4.Construction of sensors with GS linkers
The cloning processes were performed as described in the above section except that the primers, Int94_PPARG_for and Int403_PPARG_rev, contained the DNA sequences encoding different repeats of Gly-Gly-Gly-Gly-Ser between the PPARγ DNA sequence and the intein overlap DNA sequence (Supplementary Figure 1b). For convenience, “GS” was used in the context, figures and tables to denote “G4S” except for special comments. As a control, DNA fragments encoding GS linkers of different lengths were introduced into the primers Delt_Int110_PPARγ_for and Delt_Int383_PPARγ_rev respectively. As shown in Figure 1, the number ahead of “GS” for each construct denotes the number of “GS” unit repeats between the intein subunits and LBD at each junction.
2.5.Bacterial growth phenotypes
A single colony from a freshly streaked plate consisting of Luria Bertani (LB) agar supplemented with 100 μg/mL ampicillin (Amp) and 50 μg/mL thymine was used to inoculate 5 mL of liquid LB medium containing the same amount of Amp and thymine. The culture was grown at 37°C for approximately 12 h to mid-log phase, represented by an optical density at 600 nm (OD600) of approximately 1.3. The culture was then diluted 1:200 into liquid thymine-free growth medium (-THY) (Belfort and Pedersen-Lane 1984), supplemented with 100 μg/mL ampicillin. The -THY medium used in growth phenotype tests is composed of 10 mL of 20% glucose, 10 mL of Thy Pool (2mg/mL each of L-Arg, L-His, L-Leu, L-Met, L-Pro, L-Thr), 10 mL of 10% casamino acids solution, 200 μL of 1% thiamine HCl, 1 mL of 0.1 M CaCl2, 200 mL of 5× Minimal Davis Broth (35 mg/mL dipotassium phosphate, 10 mg/mL monopotassium phosphate, 2.5 mg/mL sodium citrate, 0.5 mg/mL magnesium sulfate, 5 mg/mL ammonium sulfate), 4 mL of 25 mg/mL ampicillin, and q.s. to one liter using deionized water. The glucose, Thy Pool, casamino acids and ampicillin components were sterilized by microfiltration through a 0.2 μm membrane. All other stock solutions were sterilized by autoclaving at 121°C for 25 minutes, and the final medium was mixed after cooling all of the components to room temperature. All media and media stock solutions were prepared using deionized water.
To each well of a 96-well plate, 200 μl of the diluted cell suspension was dispensed and 2 μl of various ligands diluted into DMSO at different concentrations was applied. The resulting cell plates were shaken at different temperatures (34°C or 30°C), while the OD600 of each well was measured on a BioTek Synergy2 plate reader (Winooski, VT) at different time intervals. All assays were performed in triplicate, and repeated separately at least two times. The biosensor signal strength (ΔOD600) is represented by the difference between the OD600 of the test samples and the OD600 of the vehicle control (DMSO only). For determinations of the half-maximal effective concentration (EC50) of a given compound, the ΔOD600 values at a specific time where plotted as a function of the log of the ligand concentration, and fitted to a sigmoid dose-response curve using Origin software (OriginLab, Northampton, MA).
3. Results
3.1.Performance of the original PPARγ biosensor design
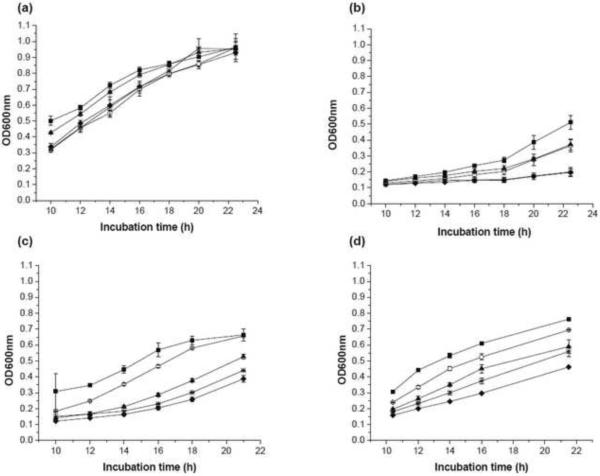
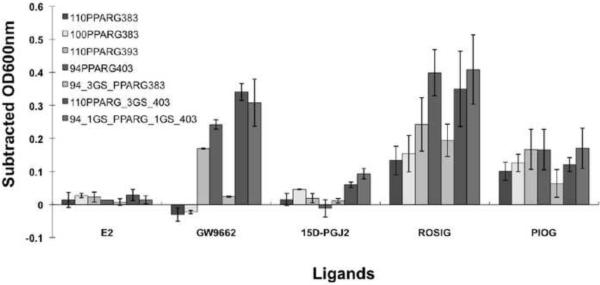
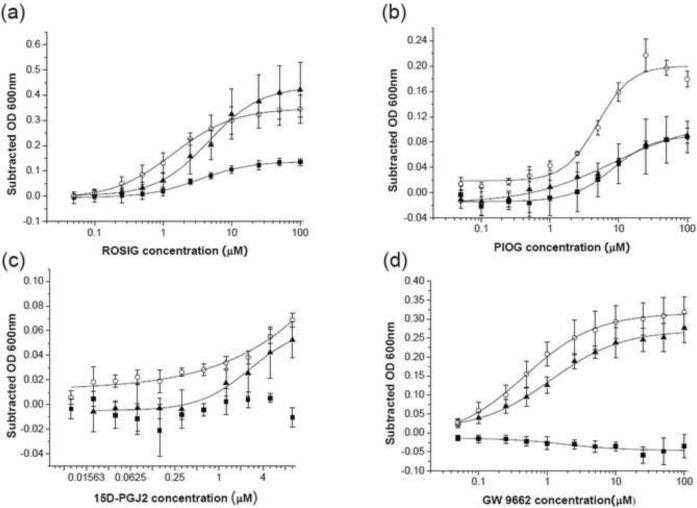
Our initial bacterial biosensor for PPARγ ligands was constructed by inserting the PPARγ LBD between residues 110 and 383 of the Mtu ΔI-SM mini-intein (Figure 1), effectively replacing the ERβ LBD in our previously reported pMIT∷ERβ* biosensor (Skretas et al. 2007; Skretas and Wood 2005b). The resulting construct is referred to here as 110PPARγ383, and is expressed from the same backbone plasmid as our previous biosensors. At 34°C, TS-deficient cells transformed with this construct exhibited a weak TS+ phenotype in the absence of ligand, as indicated by their ability to grow in –THY liquid medium (Figure 2a). The known agonists ROSIG and PIOG, however, significantly enhanced the 110PPARγ383 growth rate, while no significant effect was observed with the E2 control ligand (Figure 3). Comparatively, ROSIG caused stronger response than PIOG. A dose-response analysis of 110PPARγ383 (Figure 4a and b), with ROSIG and PIOG indicated an EC50 value of 3.4±0.2 μM for the former and 8.2±1.1 μM for the latter (Table 1). These results reflect previous reports that ROSIG is a stronger agonist than PIOG, and is consistent with their reported potencies (Chen et al. 2004; Henke et al. 1998). However, the weaker native PPARγ ligand, 15D-PGJ2 (Forman et al. 1995) didn't induce a significant cell growth signal (Figure 2 and figure 4c). Interestingly, the known antagonist GW9662 (Leesnitzer et al. 2002), inhibited cell growth (Figure 3 and Figure 4d). These observations suggest that our PPARγ biosensor is capable of detecting and assessing potent agonist ligands, and can also distinguish agonists from antagonists, as has been observed in our previous biosensors (Skretas et al. 2007; Skretas and Wood 2005a).
Fig. 2.
Growth phenotype of representative PPARγ biosensor strains in response to test ligands. Panels indicate growth rates for the following strains: (a) 110PPARγ383; (b) 110PPARγ393; (c) 110PPARγ_3GS_403; (d) 94_1GS_PPARγ_1GS_403. The strains 110PPARγ383, 110PPARγ393 and 110PPARγ_3GS_403 were incubated at 34°C, while 94_1GS_PPARγ_1GS_403 was incubated at 30°C. The final ligand concentration in each case was 10μM. Squares = ROSIG, triangles = PIOG, asterisks = 15D-PGJ2, open circles = GW 9662, and diamond = solvent only control (DMSO). The optical density at 600nm (OD600nm) is plotted as a function of incubation time. Error bars correspond to standard deviations calculated from the ODs of triplicate samples.
Fig. 3.
Comparison of signal strength of representative PPARγ biosensors in response to known agonist and antagonist ligands. The final concentration of each ligand is 10 μM. The strains 110PPARγ383, 100PPARγ383, 110PPARγ393, 94_3GS_PPARγ383, and 110PPARγ_3GS_403 were incubated at 34°C, while 94PPARγ403 and 94_1GS_PPARγ_1GS_403 were incubated at 30°C. Optical densities at 600nm (OD600nm) were measured every two hours after 10 hours of incubation, and the solvent control (1% DMSO) OD600nm measurements were subtracted from the OD600nm measurements for cultures with test ligands. The maximum subtracted OD600nm (Subtracted OD600nm) for E2 (non-binding control compound), GW9662 (known antagonist), 15D-PGJ2 (native ligand), ROSIG (strong agonist), and PIOG (strong agonist), within 24 hours is shown here. Error bars correspond to the standard deviations calculated from two or more average values based on repetitive assays with triplicates
Fig. 4.
Dose-dependence of ligand sensitivity of the wild type sensor and modified PPARγ sensors. Ligands shown are (a) ROSIG, (b) PIOG, (c) 15D-PGJ2, and (d) GW 9662. Solid squares = 110PPARγ383, solid triangles = 110PPARγ_3GS_403 and open circles = 94_1GS_PPARγ_1GS_403. As in Figure 3, after 10 hours of incubation, the optical density at 600nm (OD600nm) was measured every two hours, and the maximum Subtracted OD600nm within 24 hours total incubation was plotted against the concentration of each ligand (each curve shown was taken at a single time point). Dose-response curves were fitted to a sigmoid function for a determination of the EC50 value for each compound. In panels (a) and (d), dose response curves for 110PPARγ383 could not be fit due to weak ligand responses. Error bars correspond to the standard deviations calculated from triplicate samples in one assay. Average EC50 values based on two or more independent assays are summarized in Table 1.
Table 1.
Summary of EC50 (or IC50 for antagonism) for ligands shown in Figure 4.
| Ligands [μM] | Sensors | ||
|---|---|---|---|
| 110PPARγ383 | 110PPARγ_3GS_403 | 94_1GS_PPARγ_1GS_403 | |
| ROSIG | 3.4±0.2 | 4.6±0.4 | 1.4±0.1 |
| PIOG | 8.2±1.1 | 8.5±3.4 | 4.9±0.8 |
| GW9662 | 1.8±1.2 | 1.1±0.2 | 0.5±0.1 |
Note that the EC50 values shown in the table are calculated from two or more assays with triplicates, which may not be fully consistent with the observations in Fig. 4.
3.2.The effect of linker truncation between the intein and PPARγ LBD
We attempted to regulate the ligand sensitivity of the PPARγ biosensor by progressively truncating the peptide linkers connecting the intein segments to the PPARγ LBD. As summarized in Table 2, the first group of truncations focused on the N-terminal intein segment residues 95–110 (QPRRFDGFGDSAPIPA), while retaining the entire original C-terminal intein segment. In all cases, the PPARγ LBD was not modified. In the absence of any ligand, the basal growth of sensor strains in this group was lower at 34°C than that of the prototype. Notably, the 100PPARγ383 sensor strain, with a ten residue truncation of the N-terminal linker, displayed slightly increased sensitivity to ROSIG and PIOG at 34°C, and was able to generate a significant response to 15D-PGJ2 as well (Figure 3). The GW9662 antagonist exerted an inhibitory effect on the growth of the 100PPARγ383 strain at 34°C as well, which was very similar to that of the prototype. Interestingly, removal of the entire N-terminal linker (94PPARγ383) suppressed cell growth at 34°C regardless of ligand addition, but cell growth in the presence of ROSIG and PIOG was rescued at 30°C. These results indicate that truncated N-terminal intein segment linkers generally decrease background growth in the absence of ligand, while improving ligand sensitivity and retaining the ability to differentiate agonist from antagonist. Large truncations, however, significantly decrease the overall activity of the sensor, as well as its ability to detect weak ligands.
Table 2.
Summary of basal growth and ligand sensitivity of different PPARγ sensors.
| Sensors | Group | Basal growth | Agonist | Antagonist | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 30°C | 34°C | 30°C | 34°C | 30°C | 34°C | ||||||
| ROSIG | PIOG | 15D-PGJ2 | ROSIG | PIOG | 15D-PGJ2 | ||||||
| 110PPARγ383 105PPARγ383 100PPARγ383 94PPARγ383 |
1 | ++++ ++ ++ ++ |
+++ ++ ++ +/− |
+/− +/− +/− + |
+/− + +/− + |
+ + +/− +/− |
+ + + +/− |
+ + + +/− |
+/− +/− + +/− |
+/− − +/− +/− |
--- --- -- +/− |
| 110PPARγ388 110PPARγ393 110PPARγ398 110PPARγ403 |
2 | +++ +++ ++ ++ |
++++ + +/− +/− |
+/− +/− + ++ |
+ + + + |
+/− +/− +/− + |
+ ++ +/− +/− |
+ ++ +/− +/− |
+/− +/− +/− +/− |
+/− +/− + + |
− ++ ++ +/− |
| 94PPARγ403 | 3 | + | +/− | ++++ | ++ | +/− | +/− | +/− | +/− | ++ | +/− |
| 94_1GS_PPARγ383 94_2GS_PPARγ383 94_3GS_PPARγ383 |
4 | ++ ++ ++ |
+ + + |
+ + +/− |
+ + +/− |
+/− +/− +/− |
++ ++ ++ |
+/− + + |
+/− +/− +/− |
+/− --- -- |
+/− +/− +/− |
| 110PPARγ_1GS_403 110PPARγ_2GS_403 110PPARγ_3GS_403 110PPARγ_4GS_403 |
5 | ++ ++ ++ ++ |
+/− +/− + + |
++ + + + |
+ + + + |
+/− + + +/− |
++++ +++++ +++ +++ |
+ +/− + + |
+/− +/− + + |
+ + + + |
++++ ++++ +++ +++ |
| 94_1GS_PPARγ_1GS_403 94_2GS_PPARγ_2GS_403 94_3GS_PPARγ_3GS_403 94_G3GS_PPARγ_4GS_403 |
6 | + + + + |
+/− +/− +/− +/− |
++++ +++ ++ + |
++ ++ + + |
++ ++ + + |
+/− + ++ ++++ |
+/− +/− +/− +/− |
+/− +/− +/− +/− |
+++ ++ + + |
+/− +/− + + |
| 110_3GS_PPARγ383 | 7 | ++ | ++ | +/− | +/− | +/− | + | + | +/− | +/− | +/− |
| 110PPARγ_3GS_383 | 8 | ++ | ++ | +/− | +/− | + | + | + | +/− | +/− | + |
For basal growth, the data corresponds to the change in OD600 within 12 h incubation time. Signal strengths for basal growth are defined as follows: “+/−” corresponds to an OD600 < 0.1; “+” is an OD600 ≥ 0.1”; “++” is an OD600 ≥ 0.2; “+++” is an OD600 ≥ 0.3; “++++” is an OD600 ≥ 0.4. For growth in the presence of PPARγ ligands, the data corresponds to the subtracted OD600 (ΔOD600 = test compound OD600 - vehicle control OD600) at an incubation time where the differences in growth were maximal. Signal strengths in table for ROSIG and PIOG are defined as follows: “+/−” corresponds to ΔOD600 ≤ 0.05; “+” is a ΔOD600 of 0.06–0.15; “++” is a ΔOD600 = 0.16–0.25; “+++” is a ΔOD600 = 0.26–0.35; “++++” is a ΔOD600 = 0.36–0.45; “+++++” is a ΔOD600 = 0.46–0.55; “++++++” is a ΔOD600 = 0.56–0.65. For 15D-PGJ2, signal strengths are defined as follows: “+” corresponds to ΔOD600 = 0.04–0.08; “++” is a ΔOD600 = 0.09–0.13. For GW 9662, signal strengths are defined as follows: “---” corresponds to ΔOD600 ≤ −0.03; “--” is a ΔOD600 ≤ −0.02; “−” is a ΔOD600 ≤−0.01; “+/−” is a ΔOD600 = (−0.01)–0.05; “+” is a ΔOD600 = 0.06–0.15; “++” is a ΔOD600 = 0.16–0.25; “+++” is a ΔOD600 = 0.26–0.35; “++++” is a ΔOD600 = 0.36–0.45.
Truncations from the C-terminal intein segment residues 383–402 (RVQALADALDDKFLHDMLAE) were also examined for their effects on strain sensitivity and ability to differentiate agonist from antagonist (Table 2, Group 2). The 110PPARγ388 strain, with a five-residue truncation of the C-terminal linker, displayed greater growth in the absence of ligand than the prototype at 34°C, and no improvement in ligand sensitivity was observed. Interestingly, the 110PPARγ393 sensor, with a ten residue truncation, exhibited much lower growth in the absence of ligand, while ROSIG and PIOG strongly facilitated its growth and 15D-PGJ2 had almost no effect (Table 2, Figures 2b and 3). Surprisingly, the 110PPARγ393 sensor exhibited an inverted antagonistic effect with GW9662, where the addition of GW9662 increased cell growth substantially (Figure 2b and 3). Further truncation of the C-terminal linker (110PPARγ398) produced responses to GW 9662 at 34°C and to ROSIG and PIOG at 30°C, while full deletion of C-terminal linker (110PPARγ403) abolished sensor growth at 34°C irrespective of any ligand, but could be rescued by all tested agonists and antagonist at 30°C (Table 2). Elimination of both linkers (94PPARγ403) produced strong positive responses to ROSIG, PIOG and GW9662 at 30°C (Table 2, Group 3 and Figure 3). Overall, these data suggest that a truncation of ten or more residues from the C-terminal intein-LBD linker inhibits basal growth, while providing improved sensitivity to strong agonists, but an inverted antagonist activity.
3.3.The effect of GS linker insertion between intein and PPARγ LBD
To further examine the effects of peptide linker composition on biosensor performance, we replaced the original intein linkers with different repeats of “Gly-Gly-Gly-Gly-Ser” (“GS”) flexible linkers (Freund et al. 1993). The first group of these sensors, included biosensor designs where the N-terminal linker was substituted with GS linkers of various lengths (94_1GS_PPARγ383, 94_2GS_PPARγ383 and 94_3GS_PPARγ383). These designs displayed much lower basal growth in the absence of ligands and higher sensitivity at 34°C to ROSIG compared to their respective prototypes of identical or similar linker length in Group 1 (Table 2, Group 4 and Figure 3). The antagonist GW9662 caused slight inhibitory cell growth at 30°C (Table 2 and data not shown), but exhibited no effect at 34°C (Table 2 and Figure 3).
Group five includes sensors with GS linkers replacing the original C-terminal intein linkers (Table 2). Note that the sensors of 110PPARγ_4GS_403, 110PPARγ_3GS_403, 110PPARγ_2GS_403 and 110PPARγ_1GS_403 have the same C-terminal linker length as that of 110PPARγ383, 110PPARγ388, 110PPARγ393 and 110PPARγ398, respectively, but they exhibited much lower background growth and higher ligand sensitivity at 34°C (Table 2). Among the designs in this group, 110PPARγ_3GS_403 (Figure 2c and Figure 3) and 110PPARγ_4GS_403 also produced promising responses to PIOG and 15D-PGJ2. Surprisingly, GW 9662 acted as a strong agonist in these sensor proteins, exhibiting a signal strength almost ten times stronger than that of the prototype. To assess the ligand potency for the 110PPARγ_3GS_403 sensor, dose-responses were also investigated and compared to the prototype sensor (Figure 4 and Table 1). Interestingly, while the 110PPARγ_3GS_403 design produced a stronger signal than 110PPARγ383 with ROSIG and PIOG, the EC50 values were similar for both (Table 1). It should be noted that EC50 for 15D-PGJ2 could not be calculated due to its weak potency with these sensors. A surprising finding is that the agonist EC50 for GW9662 acting on the 110PPARγ_3GS_403 sensor is close to the antagonist IC50 for this ligand on the prototype 110PPARγ383 sensor, despite the fact that the signals in these cases are distinctly different. These observations suggest that the C-terminal GS linker substitution for the original linker elevates the ligand sensitivity of the PPARγ sensors, but inverts the antagonist behavior of GW9662 without significantly changing its inherent binding properties.
The sensors included in Group 6 have GS linkers replacing both of the original N-terminal and C-terminal intein linkers. In this case, basal growth in the absence of ligand was entirely blocked at 34°C, and was significantly inhibited at 30°C (Table 2). ROSIG was able to enhance cell growth at 34°C, and the growth rate enhancement increased with GS linker length. Interestingly, PIOG and PGJ2 failed to have any effect with any of these linkers at 34°C. As with the Group five sensors, GW9662 also generated an agonist signal. At 30°C the sensors exhibited some response to all the ligands, but unlike the behavior at 34°C, the signal strength increased as the length of the GS linkers was reduced. In particular, the 94_1GS_PPARγ_1GS_403 design exhibited the strongest observed response at 30°C of any of our sensors to the weak native ligand, 15D-PGJ2 (Figure 3). As with the previous sensors, growth-response curves were generated. In this case, the 94_1GS_PPARγ_1GS_403 design produced a higher signal at 34°C than that of both the prototype and the modified designs with only C-terminal GS linkers (Figure 4). The derived EC50 values for PIOG acting on 94_1GS_PPARγ_1GS_403 are slightly lower than that of the prototype, while the EC50s for ROSIG and GW9662 are much lower than that of the prototype (Table 1). These results indicate that GS linker substitutions at both LBD termini are able to not only improve the response of PPARγ sensor to various ligands at 30°C, but in some cases can also lead to increased apparent ligand potency.
Finally, we constructed two additional control sensors, with 3GS linkers inserted into the prototype design at either the N-terminal fusion junction (110_3GS_PPARγ383, group seven), or C-terminal fusion junction (110PPARγ_3GS_383, group eight). While the former sensor exhibited behavior very much like the original prototype, the latter produced a weak positive response to GW9662 (Table 2). These data imply that in the presence of original linker, the addition of a flexible GS linker cannot efficiently improve ligand sensitivity, but can invert the signal associated with a known antagonist.
4. Discussion
In view of the clinical significance of the PPARγ NHR, we have created reliable and economical allosteric bacterial biosensors for the identification of functional PPARγ ligands. These sensors are not affected by endogenous eukaryotic cell factors, as with most transactivation assays, and do not require purified NHR proteins. Further, the ligand response of the sensor can be quantified by simply measuring cell growth via a common laboratory spectrophotometer. The initially designed PPARγ sensor exhibits a constitutive positive TS phenotype, which is distinctly different from that of our previously published ER and TR sensors (Skretas et al. 2007; Skretas and Wood 2005a). We hypothesize that this is due to the pre-active configuration of the unbound PPARγ LBD switch domain (apo form), which is similar to that of agonist-bound ER and TR LBDs (Uppenberg et al. 1998). This observation suggests that our sensor is able to faithfully reflect the conformational status of the apo-form PPARγ LBD, which has not been observed in previously reported in vitro coactivator recruitment assays. Moreover, our sensors are able to not only assess agonist potency, but can detect intrinsic antagonists. Previous to this work, approaches to observe antagonism generally entail assays based on the competition between antagonists and agonists (Awais et al. 2007; Chen et al. 2004; Skretas et al. 2007). In this case, the signal originates from the replacement of the antagonists. In our assay, however, GW9662 alone decreased cell growth in the absence of agonist, verifying its intrinsic antagonism. This feature can be very useful for the identification of noncompeting antagonists. Our observations also demonstrate that modifications of the linkers connecting intein and LBD, through either truncation or GS linker substitution, are able to modulate basal growth rates of the PPARγ biosensor strains, as well as their responses to ligands.
Previously, we proposed that the ability of our allosteric bacterial biosensor protein to correctly reflect the binding and activity of agonist and antagonist compounds is based on the transmission a conformational change signal upon ligand binding through the intein, to the TS reporter enzyme (Skretas and Wood 2005b). However, the sensor signal transmission process remained unclear. Based on the present observations, a hypothesis is proposed below to shed light on the mechanism of sensor signal generation, which may potentially assist the design of novel biosensors.
We hypothesize that the four domain fusion protein containing MBP, TS, the LBD and the intein splicing domain may create a crowded local microenvironment surrounding TS (Van Roey et al. 2007) (Supplementary figure 2), and thus sterically block TS dimerization and activation (Wood et al. 1999). The proper intein fold putatively requires the approximation of N-terminal and C-terminal intein linkers to maintain the overall intein structure (Supplementary figure 2). The conformational changes of the LBD inserted into the intein via these two linkers are likely to alter the orientation and distance of the intein linkers, and hence influence the intein fold. As we have previously postulated (Skretas and Wood 2005b), TS dimerization possibly involves an intein structure perturbation. In the absence of ligand, helix 12 of a canonical nuclear hormone receptor LBD (e.g., ERα LBD) is rather mobile (Nagy and Schwabe 2004), which would allow the intein to remain highly folded, interfere TS dimerization and cause a negative growth phenotype. Upon agonist binding, helix 12 folds over the binding site and is moved away from the LBD N-terminus (Paulmurugan and Gambhir 2006), which would pull the C-terminal intein linker away from N-terminal linker, thus destabilizing the intein and allowing greater dimerization of TS (Supplementary figure 3a). In the absence of ligand, the PPARγ sensor has a similar intein fold to that of agonist-bound ER sensor, and correspondingly exhibits significant basal growth (Supplementary figure 3b). In this case, agonist (e.g., 15D-PGJ2) binding only slightly increases growth, primarily by stabilizing the active form of the PPARγ LBD (Kallenberger et al. 2003; Nettles and Greene 2005), and this effect depends somewhat on ligand potency. In contrast, the PPARγ antagonist moves helix 12 closer to the N-terminus, thus inhibiting TS activity and cell growth.
As helix 12 in the unbound form of the PPARγ LBD exhibits somewhat more mobility compared to that in the bound form (Kallenberger et al. 2003), the length of the LBD-intein peptide linkers may exert significant effects on the intein fold (groups 1, 2 and 3 of Table 2). A short linker (110PPARγ393) will inhibit helix 12 repositioning to the active site, which allows the intein to recover a native-like fold, and thus leads to decreased basal growth (Supplementary figure 3c). Agonist binding is proposed to stabilize the active conformation of the PPARγ LBD, thus changing the intein conformation and facilitating TS dimerization. As expected, increasingly potent ligands generate stronger signals. On the other hand, antagonist binding to the PPARγ sensor with a truncated C-terminal linker will push helix 12 to a site that is away from LBD N-terminus, although not as far as the agonist site. This will be sufficient to unfold the intein and activate TS, leading to apparent agonistic behavior. In the case of a truncated N-terminal linker (group 1), helix 12 movement is probably less restricted, while the steric interference between LBD and intein may be augmented, which will also influence TS dimerization. In this case, however, the basic features associated with agonist or antagonist binding will remain.
Our results also demonstrate that the physical property of the linker may also influence the intein fold (Table 2, groups 4, 5 and 6). Flexible GS linkers were assumed to alleviate the energy transferred to the intein by PPARγ pre-active helix 12 configuration, allowing the intein to retain its fold and minimizing basal biosensor growth (Supplementary figure 3d). Benefitting from the low basal growth, the ligand-induced growth phenotype signal generated by agonist binding will stabilize helix 12 and provide extra energy to change the intein fold and activate TS. Since the C-terminal GS linker is directly connected to the position of helix 12, it is reasonable that C-terminal GS linkers (Group 5, Table 2) are more favorable for the transmission of this generated energy than N-terminal (Group 4, Table 2) or two-terminal GS linkers (Group 6, Table 2), resulting in a higher ligand-response. The effect of GS linker length and temperature on ligand sensitivity indicates that GS linker is more flexible with increased length and temperature. It was also observed that a C-terminal GS linker exerted agonism upon antagonist binding, implying that antagonist binding to the PPARγ LBD may also stabilize helix 12 and provide energy to drag the C-terminal GS linker away from the N-terminal linker to unfold the intein. In contrast, N-terminal GS linker retained antagonism of antagonist at 30°C. Taken together, these observations suggest that the linkers significantly affect the intein fold, which dictates TS activity. Further, the C-terminal linker plays a critical role in distinguishing agonist and antagonists.
Table 2 also summarizes the effect of temperature on ligand sensitivity. In general, higher temperatures offer more stringent conditions for sensor cell growth, and require greater TS activity to generate a positive TS phenotype in -THY medium (Skretas and Wood 2005a). We observed that sensor cells grew much slower at 37°C than at 34°C in -THY medium, and ligand sensitivity at 37°C decreased due to a complete loss of growth at this temperature (data not shown). At room temperature, the stringency for TS activity is much lower, and the observed ligand responses became much weaker due to excessive background growth in the absence of ligand (data not shown). Therefore, we adopted either 34°C or 30°C for the sensor signal detection. It is apparent that all of the sensors grow faster at 30°C than at 34°C, and many of them exhibit improved ligand sensitivity at 30°C. The effect of temperature on sensor behavior may be due to higher stability of the LBD at low temperature. The higher potency of ligands for 94_1GS_PPARγ_1GS_403 at lower temperature (30°C) than that of 110PPARγ_3GS_403 and 110PPARγ383 at 34°C (Table 1) also implies that there might be an equilibrium shift of different conformation ensemble (Gunasekaran et al. 2004; Ma et al. 1999; Vallee-Belisle et al. 2009) along with temperature change. In this case, the lower temperature would allow a more active conformation ensemble within the cells.
5. Conclusion
The modular allosteric PPARγ bacterial biosensor design was able to detect the constitutive conformation of the PPARγ LBD, as well as assess agonist potency and report the intrinsic antagonism of antagonists. Modifications of our PPARγ allosteric bacterial biosensor by spacer truncation and substitution, as well as temperature optimization, produced a series of reliable tools to regulate sensor signal. This strategy would be potentially applicable to other types of biosensors based on allosteric proteins and even for the creation of artificial allosteric proteins.
Supplementary Material
Acknowledgement
This work was supported by NIH GRANT 1R21ES16630 and the Nancy Lurie Marks Family Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adamson DJ, Palmer CN. Methods Enzymol. 2002;357:188–197. doi: 10.1016/s0076-6879(02)57678-3. [DOI] [PubMed] [Google Scholar]
- Awais M, Sato M, Umezawa Y. Biosens Bioelectron. 2007;22:2564–2569. doi: 10.1016/j.bios.2006.10.013. [DOI] [PubMed] [Google Scholar]
- Belfort M, Pedersen-Lane J. J Bacteriol. 1984;160:371–378. doi: 10.1128/jb.160.1.371-378.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camp HS, Li O, Wise SC, Hong YH, Frankowski CL, Shen X, Vanbogelen R, Leff T. Diabetes. 2000;49:539–547. doi: 10.2337/diabetes.49.4.539. [DOI] [PubMed] [Google Scholar]
- Chawla A, Schwarz EJ, Dimaculangan DD, Lazar MA. Endocrinology. 1994;135:798–800. doi: 10.1210/endo.135.2.8033830. [DOI] [PubMed] [Google Scholar]
- Chen Q, Chen J, Sun T, Shen J, Shen X, Jiang H. Anal Biochem. 2004;335:253–259. doi: 10.1016/j.ab.2004.09.004. [DOI] [PubMed] [Google Scholar]
- Committee NRN. Cell. 1999;97:161–163. [Google Scholar]
- Derbyshire V, Wood DW, Wu W, Dansereau JT, Dalgaard JZ, Belfort M. Proc Natl Acad Sci U S A. 1997;94:11466–11471. doi: 10.1073/pnas.94.21.11466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forman BM, Tontonoz P, Chen J, Brun RP, Spiegelman BM, Evans RM. Cell. 1995;83:803–812. doi: 10.1016/0092-8674(95)90193-0. [DOI] [PubMed] [Google Scholar]
- Freund C, Ross A, Guth B, Pluckthun A, Holak TA. FEBS Lett. 1993;320:97–100. doi: 10.1016/0014-5793(93)80070-b. [DOI] [PubMed] [Google Scholar]
- Gokhale RS, Khosla C. Curr Opin Chem Biol. 2000;4:22–27. doi: 10.1016/s1367-5931(99)00046-0. [DOI] [PubMed] [Google Scholar]
- Gunasekaran K, Ma B, Nussinov R. Proteins. 2004;57:433–443. doi: 10.1002/prot.20232. [DOI] [PubMed] [Google Scholar]
- Henke BR, Blanchard SG, Brackeen MF, Brown KK, Cobb JE, Collins JL, Harrington WW, Jr., Hashim MA, Hull-Ryde EA, Kaldor I, Kliewer SA, Lake DH, Leesnitzer LM, Lehmann JM, Lenhard JM, Orband-Miller LA, Miller JF, Mook RA, Jr., Noble SA, Oliver W, Jr., Parks DJ, Plunket KD, Szewczyk JR, Willson TM. J Med Chem. 1998;41:5020–5036. doi: 10.1021/jm9804127. [DOI] [PubMed] [Google Scholar]
- Horton RM, Hunt HD, Ho SN, Pullen JK, Pease LR. Gene. 1989;77:61–68. doi: 10.1016/0378-1119(89)90359-4. [DOI] [PubMed] [Google Scholar]
- Huston JS, Levinson D, Mudgett-Hunter M, Tai MS, Novotny J, Margolies MN, Ridge RJ, Bruccoleri RE, Haber E, Crea R, et al. Proc Natl Acad Sci U S A. 1988;85:5879–5883. doi: 10.1073/pnas.85.16.5879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallenberger BC, Love JD, Chatterjee VK, Schwabe JW. Nat Struct Biol. 2003;10:136–140. doi: 10.1038/nsb892. [DOI] [PubMed] [Google Scholar]
- Kliewer SA, Lenhard JM, Willson TM, Patel I, Morris DC, Lehmann JM. Cell. 1995;83:813–819. doi: 10.1016/0092-8674(95)90194-9. [DOI] [PubMed] [Google Scholar]
- Kliewer SA, Umesono K, Mangelsdorf DJ, Evans RM. Nature. 1992;355:446–449. doi: 10.1038/355446a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliewer SA, Xu HE, Lambert MH, Willson TM. Recent Prog Horm Res. 2001;56:239–263. doi: 10.1210/rp.56.1.239. [DOI] [PubMed] [Google Scholar]
- Krey G, Braissant O, L'Horset F, Kalkhoven E, Perroud M, Parker MG, Wahli W. Mol Endocrinol. 1997;11:779–791. doi: 10.1210/mend.11.6.0007. [DOI] [PubMed] [Google Scholar]
- Leesnitzer LM, Parks DJ, Bledsoe RK, Cobb JE, Collins JL, Consler TG, Davis RG, Hull-Ryde EA, Lenhard JM, Patel L, Plunket KD, Shenk JL, Stimmel JB, Therapontos C, Willson TM, Blanchard SG. Biochemistry. 2002;41:6640–6650. doi: 10.1021/bi0159581. [DOI] [PubMed] [Google Scholar]
- Lehmann JM, Moore LB, Smith-Oliver TA, Wilkison WO, Willson TM, Kliewer SA. J Biol Chem. 1995;270:12953–12956. doi: 10.1074/jbc.270.22.12953. [DOI] [PubMed] [Google Scholar]
- Ma B, Kumar S, Tsai CJ, Nussinov R. Protein Eng. 1999;12:713–720. doi: 10.1093/protein/12.9.713. [DOI] [PubMed] [Google Scholar]
- McDonnell DP, Vegeto E, Gleeson MA. Biotechnology (N Y) 1993;11:1256–1261. doi: 10.1038/nbt1193-1256. [DOI] [PubMed] [Google Scholar]
- Nagy L, Schwabe JW. Trends Biochem Sci. 2004;29:317–324. doi: 10.1016/j.tibs.2004.04.006. [DOI] [PubMed] [Google Scholar]
- Nettles KW, Greene GL. Annu Rev Physiol. 2005;67:309–333. doi: 10.1146/annurev.physiol.66.032802.154710. [DOI] [PubMed] [Google Scholar]
- Nixon AE, Warren MS, Benkovic SJ. Proc Natl Acad Sci U S A. 1997;94:1069–1073. doi: 10.1073/pnas.94.4.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostermeier M. Protein Eng Des Sel. 2005;18:359–364. doi: 10.1093/protein/gzi048. [DOI] [PubMed] [Google Scholar]
- Paulmurugan R, Gambhir SS. Proc Natl Acad Sci U S A. 2006;103:15883–15888. doi: 10.1073/pnas.0607385103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raag R, Whitlow M. Faseb J. 1995;9:73–80. doi: 10.1096/fasebj.9.1.7821762. [DOI] [PubMed] [Google Scholar]
- Robinson CR, Sauer RT. Proc Natl Acad Sci U S A. 1998;95:5929–5934. doi: 10.1073/pnas.95.11.5929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sallee NA, Yeh BJ, Lim WA. J Am Chem Soc. 2007;129:4606–4611. doi: 10.1021/ja0672728. [DOI] [PubMed] [Google Scholar]
- Skretas G, Meligova AK, Villalonga-Barber C, Mitsiou DJ, Alexis MN, Micha-Screttas M, Steele BR, Screttas CG, Wood DW. J Am Chem Soc. 2007;129:8443–8457. doi: 10.1021/ja067754j. [DOI] [PubMed] [Google Scholar]
- Skretas G, Wood DW. J Mol Biol. 2005a;349:464–474. doi: 10.1016/j.jmb.2005.04.009. [DOI] [PubMed] [Google Scholar]
- Skretas G, Wood DW. Appl Environ Microbiol. 2005b;71:8995–8997. doi: 10.1128/AEM.71.12.8995-8997.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi T, Mizukami J. U.S. Patent 6,365,361. 2002
- Uppenberg J, Svensson C, Jaki M, Bertilsson G, Jendeberg L, Berkenstam A. J Biol Chem. 1998;273:31108–31112. doi: 10.1074/jbc.273.47.31108. [DOI] [PubMed] [Google Scholar]
- Vallee-Belisle A, Ricci F, Plaxco KW. Proc Natl Acad Sci U S A. 2009;106:13802–13807. doi: 10.1073/pnas.0904005106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Leeuwen HC, Strating MJ, Rensen M, de Laat W, van der Vliet PC. Embo J. 1997;16:2043–2053. doi: 10.1093/emboj/16.8.2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Roey P, Pereira B, Li Z, Hiraga K, Belfort M, Derbyshire V. J Mol Biol. 2007;367:162–173. doi: 10.1016/j.jmb.2006.12.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willson TM, Brown PJ, Sternbach DD, Henke BR. J Med Chem. 2000;43:527–550. doi: 10.1021/jm990554g. [DOI] [PubMed] [Google Scholar]
- Wood DW, Wu W, Belfort G, Derbyshire V, Belfort M. Nat Biotechnol. 1999;17:889–892. doi: 10.1038/12879. [DOI] [PubMed] [Google Scholar]
- Yu C, Chen L, Luo H, Chen J, Cheng F, Gui C, Zhang R, Shen J, Chen K, Jiang H, Shen X. Eur J Biochem. 2004;271:386–397. doi: 10.1046/j.1432-1033.2003.03937.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.