Abstract
Four blaVIM-2 gene-harboring Pseudomonas aeruginosa strains were identified. These strains possessed a class 1 integron harboring ORF1, blaVIM-2, and aacA4 gene cassettes. The transposon-mediated horizontal spread of the blaVIM-2 gene among these strains was suggested, which increases the threat that the blaVIM-2 gene will disseminate among diverse genera of bacteria.
The emergence of metallo-β-lactamase (MBL)-producing bacilli that are resistant to carbapenems is becoming a severe therapeutic problem (9). Two types of MBLs, IMP and VIM, have been reported (13). IMP-1 was identified in Pseudomonas aeruginosa in Japan in 1991 (15). Strains producing IMP-type MBLs have also been reported in Hong Kong (3), Taiwan (17), and Italy (12). Strains producing VIM-type MBLs were originally reported in European countries. VIM-1 was identified in P. aeruginosa in Italy in 1999 (7), and VIM-2 was identified in France (10). Thereafter, VIM-3 was identified in Taiwan (16). However, there have been few reports describing VIM-type-MBL-producing bacteria in Japan. The genes of both IMP- and VIM-type MBLs (blaIMP and blaVIM, respectively) are often encoded on mobile gene cassettes inserted into class 1 integrons (1, 7, 10). The class 1 integrons are genetic elements capable of integrating gene cassettes by a site-specific recombination mechanism (4). Gene cassettes are mobile units composed of a gene, most often an antibiotic resistance gene, and a recombination site, the 59-base element (4). Integrons are sometimes found as a part of transposons (4), which is probably the reason that they are found in many different genetic locations. In this work, we report on the characterization of the blaVIM-2 gene cassette-harboring class 1 integron identified in the P. aeruginosa clinical strains isolated in one hospital in Akita Prefecture, Japan.
Clinical isolates were screened for MBL production by a disk diffusion test (2) modified for use with disks containing 3 mg of sodium mercaptoacetate. An increase of more than 5 mm in the diameter of the inhibition zone around the ceftazidime disk (30 μg) in the presence of the sodium mercaptoacetate disk indicated a screening test positive for MBL production. The blaIMP and blaVIM genes were detected by PCR by using the consensus primer pairs IMP S and IMP AS for the blaIMP genes and VIM S and VIM AS for the blaVIM genes (Table 1). The blaVIM gene was typed by direct sequencing by using the VIMseq S and VIMseq AS primers (Table 1). Four blaVIM-2 gene-positive P. aeruginosa strains, Mβ-2, Mβ-6, Mβ-7, and Mβ-9, were employed in this study. Two fragments, INT5CS-VIM AS and VIM S-INT3CS, were amplified from strain Mβ-7 by PCR with two primer sets, INT5/CSBH and VIM AS and VIM S and INT3/CSJY2ER, and sequenced by using the primers listed in Table 1, as described previously (18). The annealing sites of these primers are shown schematically in Fig. 1. Pulsed-field gel electrophoresis (PFGE) was performed as described by Speijer et al. (13) by using SpeI. The chromosomal DNA fragments were analyzed by Southern blot hybridization as described previously (18) by using the blaVIM-2 DNA probe, which was prepared by PCR by using the VIMseq S and VIMseq AS primers (Table 1). INT5CS-VIM AS and VIM S-INT3CS fragments amplified from strains Mβ-2, Mβ-6, and Mβ-9 were analyzed by Southern blot hybridization for the presence of the ORF1, blaVIM-2, and aacA4 genes. The ORF1 DNA probe and the aacA4 DNA probe were prepared by PCR by using the ORF1 S and ORF1 AS primers and the AACA4 S and AACA4 AS primers (Table 1), respectively.
TABLE 1.
Primers used for detection of MBL genes and genes comprising the blaVIM-2 gene-containing integron and for sequencing of the blaVIM-2 gene containing integron
| Primer | Positionsa | Sequence (5′ to 3′) | Strand | Target gene | Accession no. | Amplicon size (bp) |
|---|---|---|---|---|---|---|
| VIM Sb | 1400-1419 | CCG ATG GTG TTT GGT CGC AT | + | |||
| VIM ASb | 1772-1790 | GAA TGC GCA GCA CCA GGA T | − | blaVIM | Y18050 | 391 |
| IMP S | 661-680 | AAA GAT ACT GAA AAG TTA GT | + | |||
| IMP AS | 1087-1106 | TCY CCA AYT TCA CTR TGA CT | − | blaIMP | S71932 | 446 |
| ORF1 S | 519-539 | ATG ATT ACC GGC ATC AAT CAC | + | |||
| ORF1 AS | 900-917 | TCA GCT CCA CAC CAG CCC | − | ORF1 | AY294333 | 399 |
| AACA4 S | 1975-1994 | ATG ACT GAG CAT GAC CTT GC | + | |||
| AACA4 AS | 2474-2493 | TTA GGC ATC ACT GCG TGT TC | − | aacA4 | AY294333 | 519 |
| INT5/CSBHc | 758-779 | AGC TAG ATC CTT CTA GAA AAC CGA GGA TGC | + | IntI1 | AF191564 | —e |
| INT3/CSJY2ERd | 2408-2431 | AGC TAA AAT TGC GAT GCC ATA ACC GAT TAT GAC | − | qacEΔ1 | AF191564 | — |
| VIMseq S | 1286-1305 | TGA CCG CGT CTG TCA TGG CT | + | blaVIM-2 | Y18050 | — |
| VIMseq AS | 1880-1899 | CAG ATC GGC ATC GGC CAC GT | − | blaVIM-2 | Y18050 | — |
| INT1 S | 321-338 | GAA CGC AGC GGT GGT AAC | + | Noncoding region | AY294333 | — |
| INT/5CS2 | 521-544 | GAT TAC CGG CAT CAA TCA CAT CAC | + | ORF1 | AY294333 | — |
| VIMS2 | 1665-1687 | CGT GGC CGA TGC CGA TCT GGC TG | + | blaVIM-2 | AY294333 | — |
Positions given in nucleotides.
Also employed for sequencing.
BamHI recognition site is added to the 5′ end.
EcoRI recognition site is added to the 5′ end.
—, sequence primer.
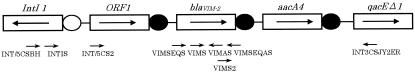
FIG. 1.
Schematic structure (not to scale) of the approximately 3-kb integron of P. aeruginosa strain Mβ-7, containing the blaVIM-2 gene cassette. The gene cassettes are boxed. Arrows indicate transcriptional orientation. The attI1 recombination site is represented by a white circle, and the 59-base elements are indicated by black circles. Primers used for sequencing are shown under the integron structure.
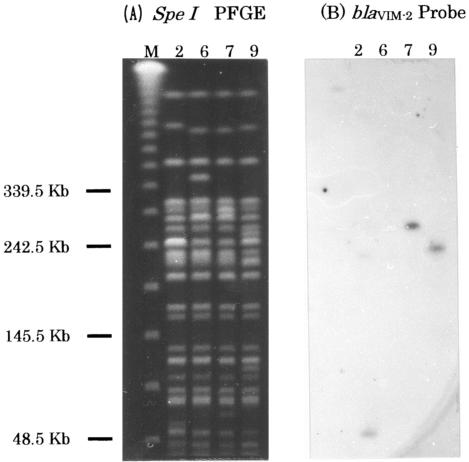
From September 2001 to October 2002, 16 isolates tested positive for MBL production by the disk diffusion screening test. Five strains, Mβ-3, Mβ-4, Mβ-5, Mβ-8, and Mβ-12, were positive for the blaIMP gene, and four P. aeruginosa isolates, Mβ-2, Mβ-6, Mβ-7, and Mβ-9, were positive for the blaVIM-2 gene. Sequence analysis of the integron from strain Mβ-7 revealed a class 1 integron structure. As shown in Fig. 1, this class 1 integron contained three gene cassettes. The first cassette contained a 399-bp open reading frame of unknown function, ORF1. The second cassette contained the blaVIM-2 gene, and the third cassette contained the aacA4 gene, which encodes aminoglycoside acetyltransferase. Southern blot hybridization analysis of the INT5CS-VIM AS and VIM S-INT3CS fragments amplified from strains Mβ-2, -6, and -9 revealed that all of these strains also contained the integron harboring the ORF1, blaVIM-2, and aacA4 genes (data not shown). SpeI PFGE patterns for the four isolates differed only within three bands, indicating that these isolates are closely related (Fig. 2A). The Southern blot analysis of the SpeI-digested chromosomal PFGE fragments revealed that the blaVIM-2 gene is located on an approximately 60-kb fragment in Mβ-2, a 280-kb fragment in Mβ-7, and a 240-kb fragment in Mβ-9 but on no fragment in Mβ-6 (Fig. 2B).
FIG. 2.
Southern hybridization analysis of the SpeI-digested chromosomal DNA fragments from Mβ-2, -6, -7, and -9. PFGE patterns (A) and results of Southern hybridization using a blaVIM-2 DNA probe (B) are shown. Lanes: M, lambda molecular weight ladder; 2, Mβ-2; 6, Mβ-6; 7, Mβ-7; 9, Mβ-9.
We have shown in this study that P. aeruginosa strains harboring the blaVIM-2 gene have been disseminated in one hospital in Akita Prefecture, Japan, confirming that blaVIM-2 gene-harboring strains are now widespread in eastern Asian countries. Earlier studies reported that blaVIM-1 is located on the chromosome of P. aeruginosa strain VR-143/97 (7) but that blaVIM-2 is located on an approximately 45-kb plasmid (10) or on the fragments of XbaI-digested genomic DNA (8). In this study, the blaVIM-2 gene was found in various genetic locations, which suggests the horizontal spread of the blaVIM-2 gene among these four P. aeruginosa strains. The precise mechanism by which the blaVIM-2 gene achieved a horizontal spread among these four strains is unclear. Our results demonstrate that the four blaVIM-2 gene-positive P. aeruginosa isolates harbored integrons of the same size and containing three genes, ORF1, blaVIM-2, and aacA4, which suggests the horizontal spread of the integron itself, rather than of the blaVIM-2 gene-containing gene cassette among different integrons. Although integrons themselves are not mobile, several class 1 integrons have been found in Tn21 and Tn21-related transposons (4, 5, 14), which enables the integrons to be transposed. These findings raise the possibility that the class 1 integron described in this study is also part of a transposon. The structures of several blaVIM-2 gene-containing integrons (6, 8, 10, 11), including the integron identified in strain Mβ-7, are unique, indicating that the blaVIM-2 gene cassette has disseminated among various integrons worldwide. Moreover, our present results suggest the possibility that the blaVIM-2 gene cassette-harboring integron is associated with a transposon, which increases the threat that the blaVIM-2 gene will disseminate among diverse genera of bacteria.
Nucleotide sequence accession numbers.
Sequences for the blaVIM-2 genes from isolates Mβ-2, Mβ-6, Mβ-7, and Mβ-9 were submitted to GenBank under accession no. AY242981 to AY242984. The sequence for the integron from strain Mβ-7 was submitted under accession no. AY294333.
REFERENCES
- 1.Arakawa, Y., M. Murakami, K. Suzuki, H. Ito, R. Wacharotayankun, S. Ohsuka, N. Kato, and M. Ohta. 1995. A novel integron-like element carrying the metallo-β-lactamase gene blaIMP. Antimicrob. Agents Chemother. 39:1612-1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arakawa, Y., N. Shibata, K. Shibayama, H. Kurokawa, T. Yagi, H. Fujiwara, and M. Goto. 2000. Convenient test for screening metallo-β-lactamase-producing gram-negative bacteria by using thio compounds. J. Clin. Microbiol. 38:40-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chu, Y.-W., M. Afzal-Shah, E. T. S. Houang, M.-F. I. Palepou, D. J. Lyon, N. Woodford, and D. M. Livermore. 2001. IMP-4, a novel metallo-β-lactamase from nosocomial Acinetobacter spp. collected in Hong Kong between 1994 and 1998. Antimicrob. Agents Chemother. 45:710-714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fluit, A. C., and F. J. Schmitz. 1999. Class 1 integrons, gene cassettes, mobility, and epidemiology. Eur. J. Clin. Microbiol. Infect. Dis. 18:761-770. [DOI] [PubMed] [Google Scholar]
- 5.Heikkilä, E., M. Skurnik, L. Sundström, and P. Huovinen. 1993. A novel dihydrofolate reductase cassette inserted in an integron borne on a Tn21-like element. Antimicrob. Agents Chemother. 37:1297-1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jeong, S. H., K. Lee, Y. Chong, J. H. Yum, S. H. Lee, H. J. Choi, J. M. Kim, K. H. Park, B. H. Han, S. W. Lee, and T. S. Jeong. 2003. Characterization of a new integron containing VIM-2, a metallo-β-lactamase gene cassette, in a clinical isolate of Enterobacter cloacae. J. Antimicrob. Chemother. 51:397-400. [DOI] [PubMed] [Google Scholar]
- 7.Lauretti, L., M. L. Riccio, A. Mazzariol, G. Cornaglia, G. Amicosante, R. Fontana, and G. M. Rossolini. 1999. Cloning and characterization of blaVIM, a new integron-borne metallo-β-lactamase gene from a Pseudomonas aeruginosa clinical isolate. Antimicrob. Agents Chemother. 43:1584-1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee, K., J. B. Lim, J. H. Yum, D. Yong, Y. Chong, J. M. Kim, and D. M. Livermore. 2002. blaVIM-2 cassette-containing novel integrons in metallo-β-lactamase-producing Pseudomonas aeruginosa and Pseudomonas putida isolates disseminated in a Korean hospital. Antimicrob. Agents Chemother. 46:1053-1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Livermore, D. M., and N. Woodford. 2000. Carbapenemases: a problem in waiting? Curr. Opin. Microbiol. 3:489-495. [DOI] [PubMed] [Google Scholar]
- 10.Poirel, L., T. Naas, D. Nicolas, L. Collet, S. Bellais, J.-D. Cavallo, and P. Nordmann. 2000. Characterization of VIM-2, a carbapenem-hydrolyzing metallo-β-lactamase and its plasmid- and integron-borne gene from a Pseudomonas aeruginosa clinical isolate in France. Antimicrob. Agents Chemother. 44:891-897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Poirel, L., T. Lambert, S. Türkoglü, E. Ronco, J.-L. Gaillard, and P. Nordmann. 2001. Characterization of class 1 integrons from Pseudomonas aeruginosa that contain the blaVIM-2 carbapenem-hydrolyzing β-lactamase gene and of two novel aminoglycoside resistance gene cassettes. Antimicrob. Agents Chemother. 45:546-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Riccio, M. L., N. Franceschini, L. Boschi, B. Caravelli, G. Cornaglia, R. Fontana, G. Amicosante, and G. M. Rossolini. 2000. Characterization of the metallo-β-lactamase determinant of Acinetobacter baumannii AC-54/97 reveals the existence of blaIMP allelic variants carried by gene cassettes of different phylogeny. Antimicrob. Agents Chemother. 44:1229-1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Speijer, H., P. H. M. Savelkoul, M. J. Bonten, E. E. Stobberingh, and J. H. T. Tjhie. 1999. Application of different genotyping methods for Pseudomonas aeruginosa in a setting of endemicity in an intensive care unit. J. Clin. Microbiol. 37:3654-3661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sundström, L., G. Swedberg, and O. Sköld. 1993. Characterization of transposon Tn5086, carrying the site-specifically inserted gene dhfrVII mediating trimethoprim resistance. J. Bacteriol. 175:1796-1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Watanabe, M., S. Iyobe, M. Inoue, and S. Mitsuhashi. 1991. Transferable imipenem resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 35:147-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yan, J.-J., P.-R. Hsueh, W.-C. Ko, K.-T. Luh, S.-H. Tsai, H.-M. Wu, and J.-J. Wu. 2001. Metallo-β-lactamases in clinical Pseudomonas isolates in Taiwan and identification of VIM-3, a novel variant of the VIM-2 enzyme. Antimicrob. Agents Chemother. 45:2224-2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yan, J.-J., W.-C. Ko, S.-H. Tsai, H.-M. Wu, and J.-J. Wu. 2001. Outbreak of infection with multidrug-resistant Klebsiella pneumoniae carrying blaIMP-8 in a university medical center in Taiwan. J. Clin. Microbiol. 39:4433-4439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yatsuyanagi, J., S. Saito, Y. Miyajima, K. Amano, and K. Enomoto. 2003. Characterization of atypical enteropathogenic Escherichia coli strains harboring the astA gene that were associated with a waterborne outbreak of diarrhea in Japan. J. Clin. Microbiol. 41:2033-2039. [DOI] [PMC free article] [PubMed] [Google Scholar]