Abstract
A new strategy known as multiplex PCR amplimer conformation was developed for detection of mutation in the gyrA gene of 138 clinical isolates of Mycobacterium tuberculosis. The method generated a single-stranded and heteroduplex DNA banding pattern of multiplex PCR amplimers of the region of interest that was extremely sensitive to specific mutations, thus enabling much more sensitive and reliable mutation analysis compared to the standard single-stranded conformation polymorphism technique. The genetic profiles of the gyrA gene of the 138 isolates as detected by MPAC were confirmed by nucleotide sequencing and were found to correlate strongly with the in vitro susceptibilities of the mutant strains to six fluoroquinolones (ofloxacin, levofloxacin, sparfloxacin, moxifloxacin, gatifloxacin, and sitafloxacin). All 32 isolates that contained gyrA mutations exhibited cross-resistance to the six fluoroquinolones (ofloxacin MIC for 90% of strains > 16 mg/liter), although moxifloxacin, gatifloxacin, and sitafloxacin (MIC for 90% of strains ≤ 4 mg/liter) were apparently more active than ofloxacin, levofloxacin, and sparfloxacin (MIC for 90% of strains ≥ 16 mg/liter). All gyrA mutations were clustered in codons 90, 91, and 94, and aspartic acid 94 was most frequently mutated. Twenty-three isolates without gyrA mutations were also found to exhibit reduced susceptibility to ofloxacin (MIC for 90% of strains = 4 mg/liter), but largely remained susceptible to other drugs (MIC for 90% of strains ≤ 1 mg/liter). Another 83 isolates without mutations were fully susceptible to all six fluoroquinolones (ofloxacin MIC for 90% of strains = 1 mg/liter). In conclusion, high-level phenotypic resistance to fluoroquinolones among M. tuberculosis clinical isolates, which appears to be predominantly due to gyrA mutations, may be readily detected by genotyping techniques such as multiplex PCR amplimer conformation.
Fluoroquinolones have antimycobacterial activities that possibly contribute a pivotal role in the second-line drug regimens used in the treatment of multidrug-resistant tuberculosis (32). However, in some communities, the resistance rates of Mycobacterium tuberculosis to these agents are surging (6, 9). To date, the genetic basis of this phenomenon has been attributed predominantly to mutations in the quinolone resistance-determining region of the gyrA gene (1, 10, 28, 31).
It would be of great relevance not only to study the genetic mutations responsible for fluoroquinolone resistance, but also to establish, as far as possible, their correlation with the resistance phenotypes of clinical isolates of M. tuberculosis. Such information facilitates rapid determination of the fluoroquinolone susceptibility status of the isolates and helps improve the clinical utility of these drugs in the settings of multidrug-resistant tuberculosis, as in cases involving streptomycin- and rifampin-resistant strains of M. tuberculosis (19, 21). Hence, the major objective of this study was to identify the gyrA mutation(s), if any, in 138 clinical isolates of M. tuberculosis from Hong Kong that exhibited a range of in vitro susceptibility levels to ofloxacin as well as to newer members of the fluoroquinolone class. The relationships between the mutations and the phenotypic resistance profiles of the clinical isolates were analyzed, with the goal of establishing correlations that would permit the application of genotyping methods for rapid identification of fluoroquinolone resistance phenotypes among clinical isolates of M. tuberculosis.
To this end, we recently developed a new method for rapid and more reliable detection of mutations in DNA fragments compared to the traditional single-stranded conformation polymorphism technique. A second objective of this study was to evaluate this method, termed multiplex PCR amplimer conformation (MPAC) analysis, in detection of gyrA mutations that are highly correlated with phenotypic resistance in M. tuberculosis, and hence its potential in facilitating rapid identification of fluoroquinolone-resistant isolates of this slowly growing organism.
MATERIALS AND METHODS
Strains and isolates.
A total of 140 Mycobacterium tuberculosis strains and isolates were included in this study: two reference strains (American Type Culture Collection [ATCC] strains ATCC 27294 H37Rv and ATCC 25177 H37Ra), and a collection of 138 clinical isolates. Each of these isolates was collected from different patients over a period of 10 years (1991 to 2000) in Grantham Hospital and various other hospitals in Hong Kong. Among these 138 isolates were 92 multidrug-resistant strains which exhibited clinical resistance to two or more of the following drugs: ofloxacin, rifampin, ethambutol, isoniazid, and parazinamide. Among these 92 multidrug-resistant isolates, 55 were known to be resistant to ofloxacin (MIC ≥ 2 mg/liter). The other 37 ofloxacin-susceptible multidrug-resistant isolates and 46 randomly selected ofloxacin-susceptible clinical isolates (MIC < 2 mg/liter) were included as fluoroquinolone-susceptible controls. Preliminary susceptibility data showed that 53% and 56% of the 83 ofloxacin-susceptible isolates were resistant to rifampin and isoniazid, respectively, whereas all ofloxacin-resistant isolates were resistant to these two drugs. All the 138 strains were isolated from patients prior to any antibiotic treatment.
Generation of PCR-amplified DNA.
Chromosomal DNA was extracted by the conventional method (29). For gyrA, a DNA fragment of 320 bp, corresponding to the quinolone resistance-determining region, was generated by PCR with the primer pair 5′-CAGCTACATCGACTATGAGA and 5′-GGGCTTCGGTGTACCTCAT. Amplification reactions consisted of a denaturation step of 3 min at 95°C, followed by 35 cycles of 1 min at 94°C, 2 min at 50°C, and 3 min at 72°C, and a final extension step of 5 min at 72°C (28). The amplification product was used as the template in direct nucleotide sequencing.
PCR-single-stranded conformation polymorphism, MPAC, and direct nucleotide sequencing.
The method of MPAC was developed with the rationale that single-stranded conformation differences between PCR products harboring sequence differences would be greatly enhanced if the PCR products were mixed with one or more reference DNA fragments prior to heat denaturation. Such reference fragments might be in the form of multiple PCR products, each covering part of the region of interest, thus further amplifying the single-stranded conformation differences if mutations were present therein.
In this study, the sensitivity of MPAC was evaluated by analyzing mixtures of different combinations of PCR products as well as the amplification products of a single multiplex PCR involving two or more of the following primer pairs, each covering part of the quinolone resistance-determining region of the gyrA gene: 1F, 5′-ATGACAGACACGACGTTG, 1R, 5′-TGCACGGGCTTGAGCCCGT; 2F, 5′-TCGGTTGCCGAGACCATG, 2R, 5′-TGGGTAGCACCGTCGGCTC; 3F, 5′-CAGCTACATCGACTATGCGA, and 3R, 5′-GGGCTTCGGTGTACCTCAT. Reaction conditions were the same as for the generation of the sequencing templates as described above.
The amplification products, ranging between 154 bp and 320 bp in size, were mixed with equal volumes of a denaturing solution (95% formamide, 0.05% xylene cyanol solution, 0.01% bromophenol blue) and heated at 95°C for 5 min. The mixture was immediately placed in ice-cold water. Five microliters of this mixture was loaded onto a 12% ExcelGel (Amersham Pharmacia Biotech, Uppsala, Sweden) and subjected to electrophoresis under 600 V constant voltage and a constant gel temperature of 15°C. The single-stranded and heteroduplex DNA conformation patterns of the PCR product mixture were visualized by silver staining. Samples that contained mutations were identified as those exhibiting different patterns from those of the reference strains. For comparison purposes, single-stranded conformation polymorphism analysis of each individual PCR product was also performed.
PCR products that harbored mutations were sequenced with the Silver Sequence sequencing system (Promega, Madison, Wis.). Sequencing templates were prepared by converting the double-stranded PCR products to single strands by heat denaturation in the sequencing reactions, the conditions of which were the same as that for template generation. Both forward and reverse primers used in PCRs were also used as sequencing primers in the corresponding sequencing reactions.
In vitro study of susceptibility to fluoroquinolones.
The ofloxacin susceptibility status of each of the 138 M. tuberculosis isolates was initially determined by the absolute concentration method with Löwenstein-Jensen medium; a MIC breakpoint of ≥2 mg/liter was used for defining ofloxacin resistance (2). The broth macrodilution method, as previously described (33), was then used to obtain the MICs of each of the six fluoroquinolones for the 138 isolates, including ofloxacin (Daiichi, Japan), levofloxacin (Daiichi, Japan), moxifloxacin (Bayer), gatifloxacin (Bristol-Myers Squibb), Sitafloxacin (Daiichi, Japan), and sparfloxacin (Rhone-Poulenc, France). This assessment also served to double-check the screening results of ofloxacin susceptibility.
RESULTS
One hundred thirty-eight M. tuberculosis clinical isolates and two ATCC control strains were studied for detection and identification of mutations in the gyrA gene by MPAC as well as DNA sequencing. The results of nucleotide sequencing showed that all 138 clinical isolates carried a mutation at codon 95 (AGC→ACC), which resulted in a serine 95-to-threonine substitution. This mutation was not detected in the two ATCC strains. Thirty-two of the 138 isolates were found to possess additional gyrA mutations apart from the Ser-95-Thr substitution that resulted in amino acid changes (Tables 1 and 2). These mutations, exhibiting a total of seven mutation types, were all clustered within nucleotides 2574 to 2586 of the gyrA gene (codons 90 to 94). Codon 94 was the most frequently substituted site. Twenty-three of the 32 isolates had mutations at this position, resulting in a total of five different types of amino acid changes (Asp→Ala, Asp→Gly, Asp→His, Asp→Tyr, and Asp→Asn). Five isolates contained a mutation at codon 90, with a resulting Ala→Val change. Four isolates had a mutation at codon 91, causing a Ser→Pro change at this site. No isolate was found to possess nucleotide changes at codons 92 and 93.
TABLE 1.
Relative fluoroquinolone susceptibilities of 32 ofloxacin-resistant M. tuberculosis isolates exhibiting seven gyrA mutation typesa
| Strain | Amino acid change | MIC (mg/liter)
|
|||||
|---|---|---|---|---|---|---|---|
| Moxifloxacin | Gatifloxacin | Sitafloxacin | Ofloxacin | Levofloxacin | Sparfloxacin | ||
| ATCC 25177 | None | 0.25 | 0.25 | 0.25 | 0.25 | 0.25 | 0.06 |
| ATCC 27294 | 0.25 | 0.25 | 0.25 | 0.25 | 0.25 | 0.06 | |
| M5 | Ala90→Val | 1 | 1 | 0.5 | 8 | 4 | 2 |
| M22 | 1 | 1 | 0.5 | 8 | 4 | 2 | |
| M93 | 2 | 1 | 0.5 | 16 | 8 | 2 | |
| FCK | 2 | 2 | 1 | 4 | 2 | 0.5 | |
| G53211 | 1 | 1 | 0.5 | 4 | 2 | 1 | |
| SC1 | Ser91→Pro | 1 | 2 | 1 | 4 | 2 | 0.5 |
| SC14 | 1 | 2 | 1 | 2 | 1 | 0.25 | |
| 469 | 2 | 4 | >4 | 4 | 2 | 0.5 | |
| 484 | 4 | 4 | 4 | 4 | 2 | 0.5 | |
| M13 | Asp94→Gly | 2 | 2 | 1 | 16 | >8 | 4 |
| M29 | 4 | 2 | 1 | 16 | 8 | 4 | |
| M38 | 2 | 4 | 2 | 16 | >8 | >8 | |
| M70 | 2 | 4 | 2 | >16 | >8 | >8 | |
| M83 | 4 | 4 | 2 | >16 | >8 | 4 | |
| M88 | 2 | 4 | 2 | 16 | 8 | 4 | |
| M96 | 8 | 4 | 4 | >16 | >8 | >8 | |
| C855 | 1 | 2 | 1 | 4 | 2 | 0.5 | |
| SC15 | 2 | 2 | 1 | 8 | 2 | 1 | |
| Y777 | 4 | 4 | 2 | 8 | 4 | 1 | |
| TB72 | 1 | 4 | 1 | 8 | 4 | 0.5 | |
| M44 | Asp94→His | 2 | 4 | 2 | 16 | 8 | >8 |
| M46 | 2 | 4 | 2 | 16 | 8 | 4 | |
| M97 | 2 | 2 | 1 | 16 | 8 | 4 | |
| M3 | Asp94→Tyr | 2 | 2 | 4 | 16 | 8 | 2 |
| M28 | 2 | 2 | 1 | 16 | 8 | 2 | |
| M47 | 2 | 4 | 4 | >16 | >8 | >8 | |
| M69 | 1 | 2 | 1 | 16 | 8 | >8 | |
| M86 | 2 | 2 | 2 | 16 | 8 | 4 | |
| M59 | Asp94→Ala | 2 | 2 | 0.5 | 8 | 4 | 2 |
| M62 | 1 | 2 | 0.5 | 8 | 4 | 2 | |
| M68 | 1 | 1 | 0.5 | 8 | 4 | 2 | |
| TB392 | Asp94→Asn | 2 | 4 | 4 | 8 | 4 | 2 |
The Ser95→The substitution is not listed.
TABLE 2.
MICs relative to control strainsa
| Amino acid change | Relative MIC (fold)
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Moxifloxacin
|
Gatifloxacin
|
Sitafloxacin
|
Ofloxacin
|
Levofloxacin
|
Sparfloxacin
|
|||||||
| MIC50 | MIC90 | MIC50 | MIC90 | MIC50 | MIC90 | MIC50 | MIC90 | MIC50 | MIC90 | MIC50 | MIC90 | |
| Ala90→Val | 4 | 8 | 4 | 8 | 2 | 4 | 32 | 64 | 16 | 32 | 33 | 33 |
| Ser91→Pro | 4 | 16 | 8 | 16 | 4 | 16 | 16 | 16 | 8 | 8 | 8 | 8 |
| Asp94→Gly | 8 | 16 | 16 | 16 | 8 | 8 | 64 | 64 | 32 | 32 | 67 | 133 |
| Asp94→His | 8 | 8 | 16 | 16 | 8 | 8 | 64 | 64 | 32 | 32 | 67 | 133 |
| Asp94→Tyr | 8 | 8 | 8 | 16 | 8 | 16 | 64 | 64 | 32 | 32 | 67 | 133 |
| Asp94→Ala | 4 | 8 | 8 | 8 | 2 | 2 | 32 | 32 | 16 | 16 | 33 | 33 |
| Asp94→Asnb | 8 | 16 | 16 | 32 | 16 | 33 | ||||||
MICs are given relative to those for the two ATCC strains.
Only one strain harboring the Asp-94-Asn change was detected.
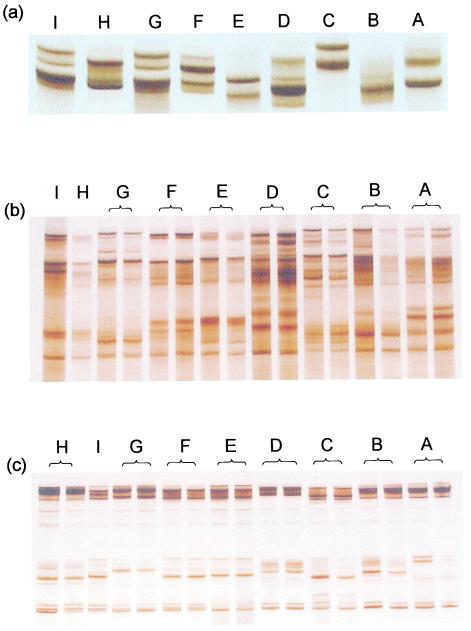
The patterns of MPAC analysis of all 138 isolates and the two ATCC strains correlated with mutations identified through nucleotide sequencing. Isolates that possessed a gyrA mutation exhibited a characteristic MPAC pattern readily distinguished by visual inspection from those of the wild-type sequence as well as from other types of mutation. Figure 1 shows the comparative MPAC and single-stranded conformation polymorphism patterns of each of the gyrA mutation types and the wild-type sequence. The MPAC patterns exhibited a much higher degree of complexity than those of single-stranded conformation polymorphism analysis. The sensitivity of MPAC was evaluated with different combinations of multiple and multiplex PCR amplimers covering the region of interest. Each treatment was found to give rise to a different MPAC pattern, yet still allow successful identification of the mutation types (Fig. 1). The temperature at which electrophoresis was performed was also found to affect the patterns to be obtained (results not shown).
FIG. 1.
Single-stranded conformation polymorphism and MPAC patterns of Mycobacterium tuberculosis strains harboring gyrA mutations. (a) Single-stranded conformation polymorphism patterns of PCR products generated with primers 3F/3R. (b) MPAC patterns of multiplex PCR products generated with primers 1F/1R, 2F/2R, and 3F/3R in a single reaction. (c) MPAC patterns of a mixture of two PCR products generated by primers 2F/2R and primers 3F/3R, respectively. A, Strains ATCC 27294 and ATCC 25177; B, no mutation in the region encoding amino acids 90 through 94; C, Ala90→Val; D, Ser91→Pro; E, Asp94→Gly; F, Asp94→His; G, Asp94→Tyr; H, Asp94→Ala; I, Asp94→Asn. Apart from the two ATCC strains, all isolates contained a Ser→Thr substitution at position 95; hence, patterns A and B are different.
Table 3 shows the fluoroquinolone susceptibility analysis of the clinical isolates and the ATCC strains. All 32 isolates that possessed a gyrA mutation exhibited high-level fluoroquinolone resistance, with a MIC for 90% of strains (MIC90) of ofloxacin exceeding 16 mg/liter, representing an increase of more than 64-fold compared to that of the two control strains. On the other hand, 23 isolates without discernible gyrA mutations, other than the Ser-95-Thr change, were also classified as resistant, with a breakpoint of ≥2 mg/liter for ofloxacin (2). It should be noted, however, that these 23 isolates exhibited reduced susceptibility, mainly to ofloxacin (MIC90 = 4 mg/liter, or 16-fold increase compared to that of the control strains) and to a lesser extent to other drugs (Table 3). In particular, these 23 isolates did not exhibit significant differences from 83 ofloxacin-susceptible isolates (ofloxacin MIC < 2 mg/liter) in terms of susceptibility to moxifloxacin, gatifloxacin, and sitafloxacin, as suggested by the distribution of MICs among isolates in these two categories (Table 3). For the 83 ofloxacin-susceptible isolates which also harbored the Ser-95-Thr substitution, all fluoroquinolones studied were found to have good in vitro activities (MIC90s 0.25 to 1 mg/liter), with sparfloxacin being the most active agent (83% of ofloxacin-susceptible isolates exhibiting an MIC of ≤0.125 mg/liter, Table 3).
TABLE 3.
Comparative susceptibilities to six fluoroquinolones and distribution of MICs among isolatesa
| MIC (mg/liter) | % of strains or relative MICb
|
|||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Moxifloxacin
|
Gatifloxacin
|
Sitafloxacin
|
Ofloxacin
|
Levofloxacin
|
Sparfloxacin
|
|||||||||||||||||||
| C | S | R− | R+ | C | S | R− | R+ | C | S | R− | R+ | C | S | R− | R+ | C | S | R− | R+ | C | S | R− | R+ | |
| ≤0.125 | 16 | 17 | 24 | 13 | 33 | 39 | 16 | 100 | 83 | 26 | ||||||||||||||
| 0.25 | 100 | 61 | 43 | 100 | 61 | 65 | 100 | 55 | 26 | 100 | 31 | 100 | 60 | 4 | 13 | 61 | 3 | |||||||
| 0.5 | 16 | 30 | 12 | 13 | 10 | 26 | 22 | 40 | 22 | 52 | 4 | 9 | 19 | |||||||||||
| 1 | 6 | 9 | 31 | 1 | 4 | 16 | 1 | 9 | 34 | 29 | 2 | 35 | 3 | 9 | ||||||||||
| 2 | 1 | 53 | 4 | 44 | 25 | 83 | 3 | 9 | 22 | 4 | 28 | |||||||||||||
| 4 | 13 | 1 | 41 | 16 | 13 | 19 | 25 | 22 | ||||||||||||||||
| 8 | 3 | 3 | 4 | 28 | 31 | |||||||||||||||||||
| 16 | 38 | 19 | 19 | |||||||||||||||||||||
| >16 | 13 | |||||||||||||||||||||||
| MIC50 | 1 | 1 | 1 | 8 | 1 | 1 | 1 | 8 | 1 | 1 | 1 | 4 | 1 | 2 | 8 | 32 | 1 | 1 | 2 | 16 | 1 | 1 | 2 | 16 |
| MIC90 | 1 | 2 | 2 | 16 | 1 | 2 | 2 | 16 | 1 | 2 | 2 | 16 | 1 | 4 | 16 | >64 | 1 | 2 | 4 | 16 | 1 | 2 | 4 | 128 |
There were four categories of M tuberculosis strains: C, the two ATCC control strains; S, 83 ofloxacin-susceptible strains (ofloxacin MIC < 2 mg/liter); R−, 23 ofloxacin-resistant isolates without gyrA mutations (ofloxacin MIC ≥ 2mg/liter); and R+, 32 ofloxacin-resistant isolates with gyrA mutations (ofloxacin MIC ≥ 2mg/liter).
See Table 2, footnote a.
Variations in phenotypic resistance level and profile were observed among isolates with different gyrA mutations, with those carrying Asp-94-Gly, Asp-94-His, and Asp-94-Tyr being the most resistant (64- and 133-fold increase in MIC90 of ofloxacin and sparfloxacin, respectively, compared to the con-trol strains, Table 2). In contrast, the Ser-91-Pro change resulted in 16- and 8-fold increases in the MIC90 of ofloxacin and sparfloxacin, respectively. While cross-resistance existed among the fluoroquinolones tested, moxifloxacin, gatifloxacin, and sitafloxacin apparently exhibited better activities (2- to 16-fold increase in MIC90) than ofloxacin, levofloxacin and sparfloxacin (8- to 133-fold increase in MIC90) among the 32 ofloxacin-resistant isolates which had gyrA mutations (Table 2).
DISCUSSION
In this study, the MPAC technique was found to be a rapid and sensitive method for detection of gyrA mutations. The principle of this method is largely based on that of the traditional single-stranded conformation polymorphism technique (27) and a newly reported technique, reference strand-mediated conformation analysis (18). The involvement of multiple or multiplex PCR products in the analysis allowed much improved sensitivity and hence reliability in detecting mutations. The flexibility of comparing results of different PCR product combinations may further increase the sensitivity of mutation detection. In addition, analysis of mutation profile and comparison with those of known sequences may provide an alternative method for detecting genetic alterations in PCR products other than direct nucleotide sequencing, where it is not uncommon to miss or misinterpret mutations unless repeated analyses with both forward and reverse primers are performed. Importantly, the newly developed method is both economical and technically simple compared to nucleotide sequencing and real-time PCR. It can be readily performed even in suboptimally equipped laboratories such as some of those in the Third World countries confronting rampant drug-resistant tuberculosis.
A prerequisite for the use of molecular methods in delineating phenotypic resistance is a good correlation between the detected mutations and resistance phenotypes. Although it is commonly recognized that gyrA mutations are not the sole mechanism responsible for fluoroquinolone resistance in M. tuberculosis, such mutations, as exemplified in this study, were found to be consistently associated with clinically significant phenotypic resistance among isolates of M. tuberculosis (ofloxacin MIC90 > 16 mg/liter).
Our data also suggested that low-level phenotypic resistance (ofloxacin MIC around 2 mg/liter) could occur without gyrA mutations. Low-level ofloxacin resistance in the 23 isolates without gyrA mutations (Table 3) might be related to mutations outside the quinolone resistance-determining region of gyrA or genetic alterations leading to impeded drug accumulation, perhaps through changes in outer membrane permeability or enhanced expression of an efflux pump that exports fluoroquinolones (1, 14). Such resistance mechanisms may have limited clinical relevance by themselves as their effects appeared to be limited to ofloxacin only (Table 3), yet their possible roles in the development of high-level fluoroquinolone resistance, especially in combinations with target site mutations, should be further studied. The polymorphism at codon 95, encoding either serine or threonine, was also shown in this study not to have a significant impact on fluoroquinolone susceptibility.
In this study, gyrA mutations were predominantly found to occur in codons 90, 91, and 94, largely corroborating the findings of some other studies (1, 14, 28, 31). Previously reported mutations involving codon 88 (22) were not found in our strains. Double mutations in gyrA, while not uncommon in the isolates obtained through antimicrobial selection in the laboratory (14, 35), were not found among our present collection of clinical isolates, corroborating the findings of another group of investigators (31). Previous data suggested a 4- to 16-fold increase in MICs among laboratory mutants with a single gyrA mutation; for mutants with double mutations, the MICs were increased 32-fold or more compared to those for the parent (14). In this study, it was found that a single amino acid substitution may be associated with 2- to 133-fold increases in MIC, depending on the type of substitution as well as the test drug (Table 1).
It is important to note that cross-resistance to all study fluoroquinolones existed among the clinical isolates; sitafloxacin, moxifloxacin, and gatifloxacin were found to exhibit better activities than the other drugs when tested against the resistant isolates except for those harboring the Ser-91-Pro change (Table 1). The effective antimycobacterial activity of sparfloxacin has been reported previously by other investigators (1, 14), yet our results showed that this drug was not particularly active against the resistant isolates that harbored gyrA mutations. For moxifloxacin, gatifloxacin, and sitafloxacin, their superior activities have been attributed to the special structure-activity relationship of the C-8-methoxy and C-8-halogen substitution in the chemical structure of the prototype fluoroquinolones (3, 13, 16, 17, 24, 34). These new members with this substitutions can have stronger bacteriostatic and lethal activities against M. tuberculosis, as shown by their lower MICs even among the gyrA mutants (7, 8, 12, 20, 25, 30). In addition, acquired resistance to these new drugs would be expected to develop less readily, as reflected by their lower mutant prevention concentrations (4, 5), a new pharmacodynamic parameter that correlates well with the MICs of these new members.
All six fluoroquinolones studied were found to have good in vitro activities against the ofloxacin-susceptible isolates (MIC90s 0.25 to 0.5 mg/liter). Sparfloxacin was found to be particularly effective (Table 3). These observations concur with the results of other reports (7, 8, 11, 12, 15, 20, 23, 25, 26, 30).
Finally, we believe that expansion of the present collection of clinical isolates and continuing molecular studies may provide a gene library of DNA gyrase gene mutations which, together with reliable and convenient molecular mutation detection methods such as MPAC, would allow microbiologists and clinicians to predict the likely profile of phenotypic resistance to fluoroquinolones of a strain isolated in a setting in which multidrug-resistant tuberculosis might be expected.
Acknowledgments
This work was supported by a research grant from the Research Grants Council (CUHK4098/00 M).
REFERENCES
- 1.Alangaden, G. J., E. K. Manavathu, S. B. Vakulenko, N. M. Zvonok, and S. A. Lerner. 1995. Characterization of fluoroquinolone-resistant mutant strains of Mycobacterium tuberculosis selected in the laboratory and isolated from patients. Antimicrob. Agents Chemother. 39:1700-1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Canetti, G., F. Wallace, A. Khomenko, H. Mahler, N. Menon, D. Mitchison, N. Rist, and N. Smelev. 1969. Advances in techniques of testing mycobacterial drug sensitivity and the use of sensitivity tests in tuberculosis control programmes. Bull. W.H.O. 41:21-43. [PMC free article] [PubMed] [Google Scholar]
- 3.Dong, Y., C. Xu, X. Zhao, J. Domagala, and K. Drlica. 1998. Fluoroquinolone action against mycobacteria: effects of C-8 substituents on growth, survival, and resistance. Antimicrob. Agents Chemother. 42:2978-2984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dong, Y., X. Zhao, J. Domagala, and K. Drlica. 1999. Effect of fluoroquinolone concentration on selection of resistant mutants of Mycobacterium bovis BCG and Staphylococcus aureus. Antimicrob. Agents Chemother. 43:1756-1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dong, Y., X. Zhao, B. N. Kreiswirth, and K. Drlica. 2000. Mutant prevention concentration as a measure of antibiotic potency: studies with clinical isolates of Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 44:2581-2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fattorini, L., E. Iona, M. L. Ricci, O. F. Thoresen, G. Orru, M. R. Oggioni, E. Tortoli, C. Piersimoni, P. Chiaradonna, M. Tronci, G. Pozzi, and G. Orefici. 1999. Activity of 16 antimicrobial agents against drug-resistant strains of Mycobacterium tuberculosis. Microb. Drug Resist. 5:265-270. [DOI] [PubMed] [Google Scholar]
- 7.Fung-Tomc, J., B. Minassian, B. Kolek, T. Washo, E. Huczko, and D. Bonner. 2000. In vitro antibacterial spectrum of a new broad-spectrum 8-methoxy fluoroquinolone, gatifloxacin. J. Antimicrob. Chemother. 45:437-446. [DOI] [PubMed] [Google Scholar]
- 8.Gillespie, S. H., and O. Billington. 1999. Activity of moxifloxacin against mycobacteria. J. Antimicrob. Chemother. 44:393-395. [DOI] [PubMed] [Google Scholar]
- 9.Grimaldo, E. R., T. E. Tupasi, A. B. Rivera, M. I. Quelapio, R. C. Cardano, J. O. Derilo, and V. A. Belen. 2001. Increased resistance to ciprofloxacin and ofloxacin in multidrug-resistant Mycobacterium tuberculosis isolates from patients seen at a tertiary hospital in the Philippines. Int. J. Tuberc. Lung Dis. 5:546-550. [PubMed] [Google Scholar]
- 10.Guillemin, I., V. Jarlier, and E. Cambau. 1998. Correlation between quinolone susceptibililty patterns and sequences in the A and B subunits of DNA gyrase in mycobacteria. Antimicrob. Agents Chemother. 42:2084-2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jesus Ruiz-Serrano, M., L. Alcala, L. Martinez, M. Diaz, M. Marin, M. Jose Gonzalez-Abad, and E. Bouza. 2000. In vitro activities of six fluoroquinolones against 250 clinical isolates of Mycobacterium tuberculosis susceptible or resistant to first-line antituberculosis drugs. Antimicrob. Agents Chemother. 44:2567-2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ji, B., N. Lounis, C. Maslo, C. Truffot-Pernot, P. Bonnafous, and J. Grosset. 1998. In vitro and in vivo activities of moxifloxacin and clinafloxacin against Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 42:2066-2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kitamura, A., K. Hoshino, Y. Kimura, I. Hayakawa, and K. Sato. 1995. Contribution of the C-8 substituent of DU-6859a, a new potent fluoroquinolone, to its activity against DNA gyrase mutants of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 39:1467-1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kocagoz, T., C. J. Hackbarth, I. Unsal, E. Y. Rosenberg, H. Nikaido, and H. F. Chambers. 1996. Gyrase mutations in laboratory-selected, fluoroquinolone-resistant mutants of Mycobacterium tuberculosis H37Ra. Antimicrob. Agents Chemother. 40:1768-1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lalande, V., C. Truffot-Pernot, A. Paccaly-Moulin, J. Grosset, and B. Ji. 1993. Powerful bactericidal activity of sparfloxacin (AT-4140) against Mycobacterium tuberculosis in mice. Antimicrob. Agents Chemother. 37:407-413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu, T., X. Zhao, and K. Drlica. 1999. Gatifloxacin activity against quinolone-resistant gyrase: allele-specific enhancement of bacteriostatic and bactericidal activities by the C-8-methoxy group. Antimicrob. Agents Chemother. 43:2969-2974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu, T., X. Zhao, X. Li, A. Drlica-Wagner, J. Y. Wang, J. Domagala, and K. Drlica. 2001. Enhancement of fluoroquinolone activity by C-8 halogen and methoxy moieties: action against a gyrase resistance mutant of Mycobacterium smegmatis and a gyrase-topoisomerase IV double mutant of Staphylococcus aureus. Antimicrob. Agents Chemother. 45:2703-2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McIlhatton, B. P., C. Keating, M. D. Curran, M. F. McMullin, J. G. Barr, J. A. Madrigal, and D. Middleton. 2002. Identification of medically important pathogenic fungi by reference strand-mediated conformational analysis (RSCA). J. Med. Microbiol. 51:468-478. [DOI] [PubMed] [Google Scholar]
- 19.Meier, A., P. Sander, K. J. Schaper, M. Scholz, and E. C. Bottger. 1996. Correlation of molecular resistance mechanisms and phenotypic resistance levels in streptomycin-resistant Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 40:2452-2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miyazaki, E., M. Miyazaki, J. M. Chen, R. E. Chaisson, and W. R. Bishai. 1999. Moxifloxacin (BAY 12-8039), a new 8-methoxyquinolone, is active in a mouse model of tuberculosis. Antimicrob. Agents Chemother. 43:85-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moghazeh, S. L., X. Pan, T. Arain, C. K. Stover, J. M. Musser, and B. N. Kreiswirth. 1996. Comparative antimicrobial activities of rifampin, rifapentine, and KRM-1648 against a collection of rifampin-resistant Mycobacterium tuberculosis isolates with known rpoB Mutations. Antimicrob. Agents Chemother. 40:2655-2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Perlman, D. C., W. M. El Sadr, L. B. Heifets, E. T. Nelson, J. P. Matts, K. Chirgwin, N. Salomon, E. E. Telzak, O. Klein, B. N. Kreiswirth, J. M. Musser, and R. Hafner. 1997. Susceptibility to levofloxacin of Mycobacterium tuberculosis isolates from patients with HIV-related tuberculosis and characterization of a strain with levofloxacin monoresistance. AIDS 11:1473-1478. [DOI] [PubMed] [Google Scholar]
- 23.Rastogi, N., V. Labrousse, and K. S. Goh. 1996. In vitro activities of fourteen antimicrobial agents against drug susceptible and resistant clinical isolates of Mycobacterium tuberculosis and comparative intracellular activities against the virulent H37Rv strain in human macrophages. Curr. Microbiol. 33:167-175. [DOI] [PubMed] [Google Scholar]
- 24.Renau, T. E., J. W. Gage, J. A. Dever, G. E. Roland, E. T. Joannides, M. A. Shapiro, J. P. Sanchez, S. J. Gracheck, J. M. Domagala, M. R. Jacobs, and R. C. Reynolds. 1996. Structure-activity relationships of quinolone agents against mycobacteria: effect of structural modifications at the 8 position. Antimicrob. Agents Chemother. 40:2363-2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saito, H., H. Tomioka, K. Sato, and S. Dekio. 1994. In vitro and in vivo antimycobacterial activities of a new quinolone DU-6859a. Antimicrob. Agents Chemother. 38:2877-2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saito, H., K. Sato, H. Tomioka, and S. Dekio. 1995. In vitro antimycobacterial activity of a new quinolone, levofloxacin (DR-3355). Tuberc. Lung Dis. 76:377-380. [DOI] [PubMed] [Google Scholar]
- 27.Sougakoff, W., N. Lemaitre, E. Cambau, M. Szpytma, V. Revel, and V. Jarlier. 1997. Nonradioactive single-strand conformation polymorphism analysis for detection of fluoroquinolone resistance in mycobacteria. Eur. J. Clin. Microbiol. Infect. Dis. 16:395-398. [DOI] [PubMed] [Google Scholar]
- 28.Takiff, H., L. Salazar, C. Guerrero, W. Philipp, W. M. Huang, B. Kreiswirth, S. T. Cole, W. R. Jacobs Jr., and A. Telenti. 1994. Cloning and nucleotide sequence of the Mycobacterium tuberculosis gyrA and gyrB genes, and detection of quinolone resistance mutations. Antimicrob. Agents Chemother. 38:773-780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Telenti, A., P. Imboden, F. Marchesi, T. Schmidheini, and T. Bodmer. 1993. Direct automated detection of rifampin-resistant mutants of Mycobacterium tuberculosis by PCR and single-strand conformation polymorphism. Antimicrob. Agents Chemother. 37:2054-2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Woodcock, J. M., J. M. Andrews, F. J. Boswell, N. P. Brenwald, and R. Wise. 1997. In vitro activity of BAY 12-8039, a new fluoroquinolone. Antimicrob. Agents Chemother. 41:101-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu, C., B. N. Kreiswirth, S. Sreeratsan, J. M. Musser, and K. Drlica. 1996. Fluoroquinolone resistance associated with specific gyrase mutations in clinical isolates of multidrug-resistant Mycobacterium tuberculosis. J. Infect. Dis. 174:1127-1130. [DOI] [PubMed] [Google Scholar]
- 32.Yew, W. W., C. K. Chan, C. H. Chau, C. M. Tam, C. C. Leung, P. C. Wong, and J. Lee. 2000. Outcomes of patients with multidrug-resistant pulmonary tuberculosis treated with ofloxacin / levofloxacin-containing regimens. Chest 117:744-751. [DOI] [PubMed] [Google Scholar]
- 33.Yew, W. W., L. J. Piddock, M. S. Li, D. Lyon, C. Y. Chan, and A. F. Cheng. 1994. In-vitro activity of quinolones and macrolides against mycobacteria. J. Antimicrob. Chemother. 34:343-351. [DOI] [PubMed] [Google Scholar]
- 34.Zhao, B. Y., R. Pine, J. Domagala, and K. Drlica. 1999. Fluoroquinolone action against clinical isolates of Mycobacterium tuberculosis: effects of a C-8 methoxyl group on survival in liquid media and in human macrophages. Antimicrob. Agents Chemother. 43:661-666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou, J., Y. Dong, X. Zhao, S. Lee, A. Amin, S. Ramaswamy, J. Domagala, J. M. Musser, and K. Drlica. 2000. Selection of antibiotic-resistant bacterial mutants: allelic diversity among fluoroquinolone-resistant mutations. J. Infect. Dis. 182:517-525. [DOI] [PubMed] [Google Scholar]