ABSTRACT
The protein YfeX from Escherichia coli has been proposed to be essential for the process of iron removal from heme by carrying out a dechelation of heme without cleavage of the porphyrin macrocycle. Since this proposed reaction is unique and would represent the first instance of the biological dechelation of heme, we undertook to characterize YfeX. Our data reveal that YfeX effectively decolorizes the dyes alizarin red and Cibacron blue F3GA and has peroxidase activity with pyrogallal but not guiacol. YfeX oxidizes protoporphyrinogen to protoporphyrin in vitro. However, we were unable to detect any dechelation of heme to free porphyrin with purified YfeX or in cellular extracts of E. coli overexpressing YfeX. Additionally, Vibrio fischeri, an organism that can utilize heme as an iron source when grown under iron limitation, is able to grow with heme as the sole source of iron when its YfeX homolog is absent. Plasmid-driven expression of YfeX in V. fischeri grown with heme did not result in accumulation of protoporphyrin. We propose that YfeX is a typical dye-decolorizing peroxidase (or DyP) and not a dechelatase. The protoporphyrin reported to accumulate when YfeX is overexpressed in E. coli likely arises from the intracellular oxidation of endogenously synthesized protoporphyrinogen and not from dechelation of exogenously supplied heme. Bioinformatic analysis of bacterial YfeX homologs does not identify any connection with iron acquisition but does suggest links to anaerobic-growth-related respiratory pathways. Additionally, some genes encoding homologs of YfeX have tight association with genes encoding a bacterial cytoplasmic encapsulating protein.
IMPORTANCE
Acquisition of iron from the host during infection is a limiting factor for growth and survival of pathogens. Host heme is the major source of iron in infections, and pathogenic bacteria have evolved complex mechanisms to acquire heme and abstract the iron from heme. Recently Létoffé et al. (Proc. Natl. Acad. Sci. U. S. A. 106:11719–11724, 2009) reported that the protein YfeX from E. coli is able to dechelate heme to remove iron and leave an intact tetrapyrrole. This is totally unlike any other described biological system for iron removal from heme and, thus, would represent a dramatically new feature with potentially profound implications for our understanding of bacterial pathogenesis. Given that this reaction has no precedent in biological systems, we characterized YfeX and a related protein. Our data clearly demonstrate that YfeX is not a dechelatase as reported but is a peroxidase that oxidizes endogenous porphyrinogens to porphyrins.
Introduction
Dye-decolorizing peroxidases, or DyP-type peroxidases, are a relatively recently recognized superfamily of heme-containing peroxidases that are found in fungi and bacteria (1, 2). They are structurally distinct from animal and plant peroxidases as well as classical fungal peroxidases, such as LmnP and Lip (3–5). DyPs are enzymatically unique in that they effectively oxidize high-redox-potential anthraquinone dyes, thereby either decolorizing or altering the color of the dye. Because of this property, DyPs are of industrial interest as biological decolorizers of synthetic dyes. While the fungal DyPs characterized to date are extracellular proteins (1), at least some bacterial DyPs are intracellular (6–8). The crystal structures for the bacterial DyPs TyrA of Shewanella oneidensis (8) and YcdB of Escherichia coli (6) have been solved and the axial ligands for the heme iron identified. However, the physiological role of these proteins in bacteria has not been determined. DyPs are members of the larger CDE structural superfamily that includes the hemoproteins chlorite dismutases (9) and HemQ (10).
Recently, Létoffé et al. (11) reported that the E. coli proteins YfeX and EfeB carry out the previously unobserved biological dechelation of heme to release free iron and an intact protoporphyrin molecule. Both of these proteins appear by sequence homology to be DyP-type peroxidases, although this was not tested. EfeB (YcdB) is one member of a tripartite iron transporter system (YcdNOB) and is a periplasmic hemoprotein (12). It is iron (ferric uptake regulator [Fur]) regulated and suggested to be a peroxidase involved in oxidation of ferrous iron during transport. There are no experimental data available that suggest an involvement of this system in heme uptake or utilization. The EfeB crystal structure was recently solved (6), and while the authors of that study repeat the suggestion that EfeB is involved in heme dechelation, they present no data in support of the statement and cite only Létoffé et al. Less is known about YfeX; however, Létoffé et al. proposed that YfeX, a cytoplasmic protein, and EfeB, a periplasmic protein, “are the sole proteins able to provide iron from exogenous heme sources to E. coli” (11).
The suggestion that YfeX can dechelate heme to yield free iron and an intact tetrapyrrole macrocycle would put YfeX in a unique class unlike all other enzymes that have been definitively shown to release iron from heme (13, 14). Although it has been claimed that mammalian ferrochelatase can catalyze a dechelation reaction (15), that reaction was reported to occur in vitro under non-physiologically relevant conditions at acidic pH and elevated temperature, conditions that are well outside the range where mammalian ferrochelatase is active or stable (16). Substantive evidence for metazoan and bacterial heme oxygenases exists in the literature, and in all cases the end product is a cleaved, linear tetrapyrrole, not an intact porphyrin.
Given the diversity of organisms that possess DyP-type proteins, the identification of this class of proteins as heme dechelatases would have profound physiological and environmental implications. Because of this and our interest in heme metabolism, we undertook to examine in more detail the protein YfeX. The data presented herein demonstrate that recombinant YfeX is a typical DyP-type peroxidase and does not possess the catalytic ability to dechelate iron from heme in vitro. In vivo experiments with YfeX in E. coli and its homolog in Vibrio fischeri revealed no evidence that YfeX either is involved in iron acquisition from heme or generates porphyrin from exogenously supplied heme.
RESULTS
The YfeX is a dye-decolorizing peroxidase that also oxidizes porphyrinogens.
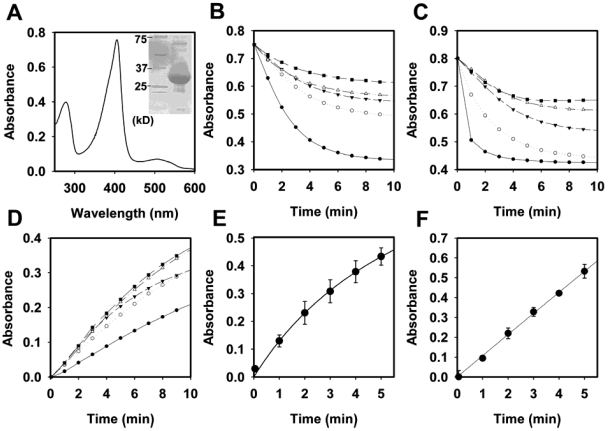
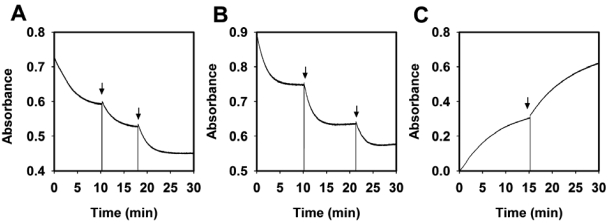
We first examined the properties of purified YfeX as a potential dye-decolorizing peroxidase, because the YfeX sequence is homologous to that of members of the DyP family of peroxidases. Purified overexpressed YfeX has little bound heme but is readily loaded with heme that binds with high affinity (Fig. 1A). Heme-loaded YfeX exhibits typical peroxidase activities. It efficiently decolorizes alizarin red and Cibacron blue F3GA (Fig. 1B and C) but not reactive red 120, reactive orange 14, reactive green, or reactive brown 5, and it utilizes pyrogallol (Fig. 1D) but not guiacol in a peroxidase assay. Heme-free, apo-YfeX had no peroxidase activity, and neither apo- nor holo-YfeX has catalase activity. The pH optima for dye decolorizations are acidic, as has been reported for extracellular fungal DyP-type peroxidases (1), but the optimum for pyrogallol oxidation is approximately 7 (Fig. 1D). Interestingly, holo-YfeX effectively oxidized both protoporphyrinogen IX and coproporphyrinogen III to their corresponding porphyrins (Fig. 1E and F), not unlike some intracellular plant peroxidases (17). It was noted that both dye decolorization and pyrogallol oxidation activity diminished with time even with an excess of substrates present. The average turnover of the protein before loss of activity varied considerably between dyes and pyrogallol. These numbers (total mol dye oxidized per mol enzyme) are 250 for alizarin red, 100 for Cibacron blue, and approximately 200,000 for pyrogallol (Fig. 2A to C). We could find no evidence that this loss of activity was accompanied by the appearance of free porphyrin.
FIG 1 .
Biochemical and enzymatic characterization of YfeX. (A) UV/visible scan of heme-loaded purified YfeX, as described in the text. The heme Soret peak has a maximum at 415 nm. The inset shows a sodium dodecyl sulfate-polyacrylamide gel that is overloaded with protein to demonstrate the purity of the YfeX protein employed in these studies. (B) Alizarin red dye decolorization by YfeX. Details are in Materials and Methods. (C) Cibacron blue dye decolorization by YfeX. (D) Pyrogallol oxidation by YfeX. Symbols (B, C, and D): solid circles, pH 5.5; open circles, pH 6.0; solid triangles, pH 6.5; open triangles, pH 7.0; and solid squares, pH 7.5. (E) Protoporphyrinogen IX oxidation to protoporphyrin IX by YfeX. (F) Coproporphyrinogen III oxidation to coproporphyrin III by YfeX.
FIG 2 .
Suicide loss of activity of YfeX during catalysis. (A) Decolorization of alizarin red by YfeX with hydrogen peroxide results in the eventual loss of peroxidase activity. Initially, 0.6 nmol of YfeX was added to start the reaction. At the indicated times (arrows), additional aliquots of 0.6 nmol YfeX were added to the reaction. (B and C) Decolorization of Cibacron blue by YfeX (B) and oxidation of pyrogallol by YfeX and hydrogen peroxide (C). Details are identical to those for panel A.
YfeX is not involved in biological iron removal from heme.
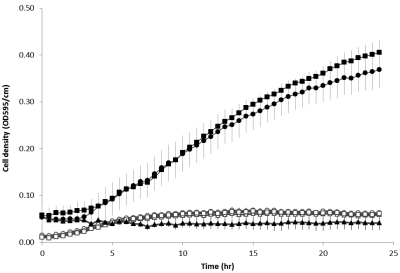
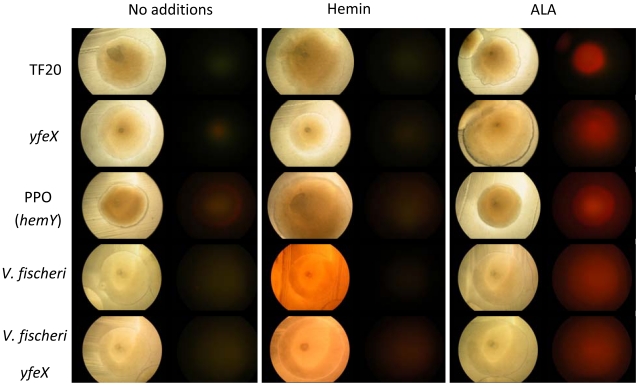
By following the published methods of Létoffé et al. (11), we were unable to reproduce the in vitro dechelation reaction with either meso- or protoheme as the substrate. We do not have an explanation for this, but we do note that YfeX is expressed at high levels in our system and is active as a peroxidase. As noted by Létoffé et al. (11), no dechelatase activity is measurable with purified enzyme. We also examined YfeX expression in V. fischeri, since it can utilize heme as its sole source of iron (18) and possesses a homolog of YfeX (VF_1898) but no homolog of EfeB. Additionally, as with E. coli, V. fischeri has no annotated heme oxygenase. Thus, if the proposal by Létoffé et al. is correct, V. fischeri VF_1898 should be essential for obtaining iron from heme. However, knockout of VF_1898 in V. fischeri had no impact on the ability of that organism to grow with heme as its sole iron source (Fig. 3), and wild-type cells overexpressing VF_1898 did not display red color or fluorescence characteristic of free porphyrin (Fig. 4). These data demonstrate that the homolog of YfeX, VF_1898, is not essential for iron removal from heme in V. fischeri and that when overexpressed in cells grown under iron-limiting conditions and supplemented with hemin, there is no evidence of a dechelation reaction. Thus, in neither E. coli nor V. fischeri were we able to obtain any data supporting the proposition that YfeX has dechelatase activity or that it participates in vivo in iron acquisition from heme.
FIG 3 .
Growth kinetics of V. fischeri strains with hemin as an iron source. Cultures of the wild type (circles), AKD910 (heme uptake mutant; triangles), and the ΔyfeX mutant (squares) were grown overnight in LBS medium and diluted 1:1,000 in mineral salts medium with 50 µM bipyridyl with (filled symbols) or without (open symbols) hemin (50 µg/ml) supplementation. Two hundred-microliter aliquots of the diluted cultures were incubated in a 96-well plate at 28°C, and cell density was measured at 595 nm every 30 min using a Bio-Tek Synergy 2 plate reader. Error bars show standard errors.
FIG 4 .
Impact of YfeX on porphyrin accumulation during growth of E. coli and V. fischeri. E. coli strains containing plasmid constructs as described in the Materials and Methods were grown on LB plates with or without 50 µg/ml hemin or 50 µg/ml ALA. V. fischeri was grown on LBS plates with 2 mM IPTG with or without 50 µg/ml hemin and 100 µM bipyridyl or 50 µg/ml ALA. Plates were incubated at 37°C (E. coli) and 28°C (V. fischeri) for 3 days and imaged using bright-field microscopy (F6.6, 1/60-s exposure) (left columns) and a red/green fluorescence filter (F6.6, 1/2-s exposure) (right columns).
Given that YfeX oxidizes porphyrinogens to porphyrins, the possibility exists that the fluorescence observed by Létoffé et al. (11) could have resulted from the in vivo intracellular oxidation by YfeX of endogenously produced porphyrinogens that would accumulate in the absence of available iron for ferrochelatase. To assess this possibility, we examined E. coli possessing plasmid-encoded human ferrochelatase (yfeX) or B. subtilis protoporphyrinogen oxidase (hemY). Since it has been suggested that E. coli K-12 strains are deficient in heme uptake, the heme transporter HasR was coexpressed with YfeX. Each of these was grown in LB medium with appropriate antibiotic alone or with hemin or 5-aminolevulinic acid (ALA) supplementation. The rationale for these choices was that (i) with overexpressed ferrochelatase, there should be free porphyrin only under conditions of endogenously synthesized excess porphyrin; (ii) if YfeX is a dechelatase, one should see fluorescence in the presence of hemin supplementation; and (iii) B. subtilis HemY has been shown to oxidize coproporphyrinogen and protoporphyrinogen to their corresponding porphyrins in vivo (16, 19), so when they are overexpressed one sees fluorescence from endogenously produced porphyrinogens but not exogenously supplied hemin. Our data showed that with overexpressed ferrochelatase, porphyrin fluorescence is seen only with ALA supplementation (Fig. 4). Cells overexpressing YfeX exhibit fluorescence on LB+ALA similar to what is seen with HemY expression, minimal fluorescence on LB alone, and no fluorescence when supplemented with hemin. Cells overexpressing HemY have some fluorescence on LB alone and high levels of fluorescence when supplemented with ALA. V. fischeri grown under similar conditions with and without the YfeX homolog expression vector had no fluorescence except when supplemented with ALA. These data, along with the observation that knockout of the YfeX homolog in V. fischeri resulted in cells with no detectable phenotype, strongly suggest that YfeX has no role in cellular iron acquisition from heme and does not function in vivo as a heme dechelatase.
Bioinformatics data do not support a role for YfeX in dechelation of heme for iron acquisition.
For three bacteria that possess YfeX or a homolog, E. coli K-12 (YfeX), S. oneidensis MR-1 (SO_0740, TyrA), and P. aeruginosa PAO1 (PA2765), there are reasonable global expression profiling databases available. In all of these organisms, YfeX homologs appear to be expressed under a wide variety of conditions, albeit at different intensities. If YfeX is involved in iron acquisition, it would be reasonable to expect that it might be regulated by Fur. However, global iron-dependent gene regulation has been studied in detail in E. coli (20, 21), V. cholerae (22, 23), and P. aeruginosa (24), and notably, YfeX homologs in these organisms do not appear to be directly regulated by iron availability, nor do they contain sequences corresponding to Fur or IS boxes in their upstream regulatory regions (20, 21, 24). This appears contrary to what would be expected for a gene with the predicted physiological function of retrieving iron from heme (11).
DISCUSSION
Létoffé et al. (11) proposed that YfeX and EfeB are responsible for iron supply via dechelation of exogenously supplied heme in E. coli K-12 grown under iron starvation but in the presence of heme. Interestingly, their original identification of YfeX and EfeB was done by an expression plasmid library screen in wild-type cells grown on iron-replete rich medium without exogenous hemin. Under these conditions, they identified colonies that fluoresced red from free protoporphyrin. They suggested that this occurred from dechelation of endogenously produced heme, a proposition that would require the existence of an energetically futile cycle in these cells. The proposal that a bacterial heme dechelatase exists to provide iron while generating free protoporphyrin is unique and raises significant questions, the most important of which may be the fate of the highly reactive porphyrin that would be generated as a by-product. In organisms in which free protoporphyrin accumulates, such as the ΔhemH mutant of E. coli (25), plants treated with diphenyl ether herbicides (26), or animals with erythropoietic protoporphyria (27), one finds that the accumulation of free protoporphyrin in a cell has dire consequences. Indeed, in the field of cancer, photodynamic therapy is based upon the lethal effects of free cellular porphyrins under illumination (28). Given that there are no known biological mechanisms for the destruction of free porphyrins, the only possible fate would be either iron reincorporation via ferrochelatase, in a futile cycle, or excretion of porphyrin into the surrounding milieu. This last process has not been observed in bacteria and, if it does exist, it would be inadequate to eliminate intracellular protoporphyrin that accumulates when bacterial cultures are supplemented with ALA, the first committed intermediate in the biosynthesis of heme. In S. cerevisiae lacking the gene for ferrochelatase (hem15), protoporphyrin accumulates in the cytosol, and when the cells are illuminated, the porphyrin has been shown to be cytotoxic (29), supporting the observation that accumulation of protoporphyrin is undesirable. In mice that have a homozygous deletion of IRP2, the primary iron-regulatory protein in the erythron, lack of iron leads to accumulation of protoporphyrin (30, 31), further supporting the conclusion that in the absence of iron the demand for heme leads to protoporphyrin accumulation.
As a first step, we sought to biochemically characterize the dechelatase reaction, and in the present study, we focused on YfeX. However, we were unable to detect any dechelatase activity with either the purified enzyme or crude cell extracts of YfeX-overproducing cells, even though the Tac promoter-driven expression of YfeX was robust in the E. coli K-12 strain we employed, JM109. Additionally, we saw no fluorescence of cells expressing YfeX when they were grown with exogenously supplied heme. When YfeX-expressing JM109 cultures were grown with added ALA, there was visible fluorescing free porphyrin, but this was in an amount similar to what one finds when endogenously synthesized porphyrinogen is oxidized. The coexpression of YfeX with the heme transporter HasR gave results identical to those obtained with YfeX alone, showing that it was not a limitation of heme uptake that resulted in lack of observable dechelatase activity in vivo. Since V. fischeri possesses the ability to take up heme, coexpression of HasR and YfeX was not necessary.
Overall, our data demonstrate that YfeX is a typical DyP-type heme-containing peroxidase. As with other proteins of this general class, some heme destruction occurs via a currently undescribed suicide reaction, but this reaction does not generate any detectable free porphyrin. Significantly, we demonstrated that YfeX efficiently catalyzes the oxidation of both coproporphyrinogen III and protoporphyrinogen IX into the corresponding free porphyrins. Given that Létoffé et al. (11) never reported the production of porphyrin in cultures where iron was limited or provided exogenously via hemin, it seems likely that the fluorescence observed originated from YfeX-mediated oxidation of intracellular porphyrinogen that was diverted from the formation of heme rather than dechelation of newly synthesized heme.
In addition to experimental approaches with purified YfeX and bacterial cell lines either lacking or overexpressing YfeX, we sought to determine if YfeX and homologs in other bacteria possessed a genetic or functional genomic context that might reveal the role this family of proteins plays in bacteria. It is noteworthy that proteins from a phylogenetic branch of the DyP superfamily closely related to yet distinct from YfeX, Enc_DyP, often occur in the same species as YfeX (e.g., Pseudomonas fluorescens, Burkholderia cenocepacia, Proteus mirabilis, etc). The Enc_DyP group of orthologous proteins is notable for its association with encapsulin, a protein observed to form icosahedral shells that function as a minimal bacterial intracellular compartment (32). This association is based on both genomic and functional observations. In over half of the organisms where members of the Enc_DyP subgroup occur, the corresponding gene is colocalized and likely forms an operon with the encapsulin-encoding gene (e.g., Rv0798c and Rv0799c in M. tuberculosis H37Rv). Furthermore, the encapsulation of an Enc_DyP protein in such nanocompartments has been demonstrated in Brevibacterium linens (32). We propose that YfeX and Enc_DyP perform distinct functions. This conjecture is supported by the fact that the two subfamilies may co-occur in the same microorganisms where functional redundancy is usually the exception rather than the rule, and by the observation that the Shewanella YfeX-like protein assembles into a dimer, while Enc_DyP from Bacteroides thetaiotaomicron is a hexamer (trimer of dimers) (33). Finally, the need for compartmentalization suggests that a substrate(s) or product(s) of the Enc_DyP branch is likely toxic or unstable, which apparently is not the case for the substrate(s) of the YfeX-type enzymes.
Based upon an analysis of the large collection of microarray data in the M3D database, in S. oneidensis and E. coli, the most significant induction of the YfeX orthologs is observed during logarithmic growth under anaerobic conditions. Virtually no signal is observed in stationary phase, and there is very low expression under aerobic conditions. Furthermore, in S. oneidensis, a specific increase in expression of SO_0740 is observed when fumarate is used as the terminal electron acceptor, compared to conditions of nitrate or trimethylamine-N-oxide (TMAO) anaerobic respiration. Interestingly, the list of the top ~50 genes correlated in expression with SO_0740 contains genes encoding a soluble periplasmic fumarate reductase FccA, various multiheme cytochromes, and components of the c-type cytochrome biogenesis machinery and quinone biosynthesis pathway, as well as several heme biosynthetic enzymes. Given that biosynthesis and assembly of these respiratory chain components in Shewanella in response to fumarate supplementation likely also requires heme and other cofactors (e.g., Fe-S centers, FAD for fumarate dehydrogenase, Mo for formate dehydrogenase, and Ni for hydrogenase), we suggest that the in vivo function of the YfeX ortholog in Shewanella could be associated with synthesis or assembly of some component(s) of the respiratory chain synthesized under anaerobic conditions. This association may involve the maturation of the c-type cytochromes (34, 35), given that the YfeX family is coexpressed with system I cytochrome c maturation components in both S. oneidensis (with the CcmH and CcmF subunits of cytochrome c heme lyase) and P. aeruginosa (with CcmD, as well as with multiple c-type cytochromes [PA5490, PA5300, and PA0541]), and that within the beta- and gammaproteobacteria, the YfeX-type DyP proteins never occur in organisms that lack genes associated with systems I and II of cytochrome c maturation (e.g., in Francisella tularensis, Bdellovibrio bacteriovorus, Coxiella burnetii, and all sequenced species of the genera Campylobacter, Helicobacter, “Candidatus Blochmannia,” Photorhabdus, and Buchnera) (see the SEED database subsystem “Biogenesis of c-type cytochromes” [http://theseed.uchicago.edu/FIG/seedviewer.cgi?page=Subsystems&subsystem=Biogenesis_of_c-type_cytochromes]).
Alternatively, YfeX could be required for the turnover or degradation of the respiratory complexes that are being replaced and disassembled during the switch from aerobic growth to anaerobiosis. Gene expression data available from P. aeruginosa PAO1 and E. coli do not contradict this proposal. The fact that in E. coli, YfeX has been reported to be a member of the sigma E (σE) regulon (36, 37) is in agreement with this hypothesis.
In the case of V. fischeri, recent work has shown that the VF_1220-to-VF_1228 gene cluster is required for using hemin as an iron source under iron-limiting conditions (18). This heme uptake/utilization gene cluster is regulated in response to iron levels in a Fur-dependent manner. If YfeX were involved in recovering iron from heme in V. fischeri, one might expect its regulation to be coordinated with the heme uptake and utilization gene cluster required for growth on hemin as an iron source. However, the yfeX gene in V. fischeri is the last gene in a putative three-gene operon with genes encoding hypothetical proteins, and a virtual footprint analysis (http://www.prodoric.de/vfp/) failed to identify any putative Fur binding sites upstream of the yfeX gene or upstream of the first gene in the putative operon (VF_1900). This analysis did reveal a putative binding site for the oxygen-sensitive FNR (fumarate/nitrate reduction) transcriptional regulator upstream of VF_1900. Interestingly, the yfeX containing putative operon (VF_1898– to VF_1900) is located near an operon encoding the components of the nitrate reductase complex (VF_1901 to VF_1907). These findings lend strength to our hypothesis that the function of YfeX in vivo may indeed be linked to maintenance of anaerobic respiratory chain components.
In conclusion, we propose that YfeX is a heme-containing peroxidase typical of the DyP type. Its intracellular role remains to be identified, but the earlier suggestion that it possesses heme dechelatase activity is not supported by either in vitro or in vivo data, nor does it appear to be consistent with our bioinformatic analyses. Finally, it should be noted that the ability of the V. fischeri yfeX mutant to grow on heme as an iron source strongly suggests that another protein(s) exists to catalyze the removal of iron from exogenously supplied heme and that the proposal of Létoffé et al. that YfeX and EfeB “are the sole proteins able to provide iron from exogenous heme sources” is not justified (11). This point gathers additional support from the fact that V. fischeri and E. coli possess homologs of the hutWXZ operon, which has been shown to complement heme oxygenase mutants of Corynebacterium diphtheriae (38). We further propose that the accumulation of protoporphyrin that was reported to occur in E. coli overproducing YfeX resulted not from dechelation of exogenously supplied heme but from YfeX-mediated oxidation of endogenously synthesized porphyrinogens. This proposal is consistent with the observation that overexpression of B. subtilis HemY, which catalyzes the oxidation of both coproporphyrinogen and protoporphyrinogen, in E. coli results in the accumulation of porphyrin from porphyrinogens. Additionally, it is known that under conditions in which intracellular porphyrinogen concentrations are increased in plants, cytoplasmic peroxidases oxidize porphyrinogens to porphyrins (17). We propose that a similar event occurs in E. coli when the peroxidase YfeX is as abundant as it is in overexpressing cells.
MATERIALS AND METHODS
YfeX purification.
The gene encoding YfeX was obtained by PCR amplification of genomic DNA from E. coli JM109. The open reading frame was cloned into pTrcHisA (Invitrogen) to yield a six-histidine amino-terminal tag fusion. The sequence of the yfeX gene in the plasmid was confirmed by the Georgia Genomics Facility at the University of Georgia. Expression, isolation, and purification of YfeX using HisPur cobalt resin (Thermo Scientific) was as described previously for ferrochelatase (39). One additional step was column chromatography employing an AKTA Prime FPLC system with a HiPrep 16/60 Sephacryl S-300 column (GE Healthcare), where the buffer was 100 mM Tris-HCl (pH 7.5), 100 mM KCl. The fractions containing YfeX were combined and concentrated by centrifugation with an Amicon Ultra centrifugal filter (Millipore). The protein concentration was determined spectrophotometrically (ЄmM = 327 at 280 nm).
Strain and plasmid construction.
The Serratia marcescens hasR (heme transporter) gene was amplified by PCR of genomic DNA and cloned into the NheI/HindIII sites of pTrcHisA (Invitrogen), which had been modified to contain the kanamycin resistance gene. The resulting plasmid was transformed into E. coli JM109 along with the previously described yfeX expression plasmid and grown on plates containing LB plus ampicillin (100 µg/ml) and kanamycin (30 µg/ml), LB plus ampicillin (100 µg/ml), kanamycin (30 µg/ml), and heme (50 µg/ml), and LB plus ampicillin (100 µg/ml), kanamycin (30 µg/ml), and ALA (50 µg/ml).
A V. fischeri mutant (strain AKD915) with an in-frame deletion of the putative yfeX gene (VF_1898) was constructed in the wild-type parent V. fischeri ES114 using allelic exchange (40). Briefly, approximately 1.6 kb of DNA upstream of VF_1898 was amplified by PCR and fused to an approximately 1.6-kb DNA fragment downstream of VF_1898 using an engineered ClaI 6-bp restriction site added to the PCR primers. Additional codons were included in order to design specific primers with reasonable G+C content, resulting in inclusion of the first three codons of VF_1898.
To construct plasmid pAS101, which has yfeX controlled by an IPTG (isopropyl-β-d-thiogalactopyranoside)-inducible promoter, the yfeX homolog (VF_1898) was PCR amplified and directionally cloned into the KpnI and NheI sites of the IPTG-inducible expression vector pAKD601B (41), which is derived from stable shuttle vectors based on native V. fischeri plasmid pES213 (32, 42). The sequence of yfeX in the vector was confirmed at the University of Michigan DNA Sequencing Core facility.
Assays.
For all dye-decolorizing assays, stock solutions of each dye were prepared in buffers employed in the assays and diluted to give an optical density (OD) reading of approximately 1.0 at the wavelength of maximum absorbance, which was determined experimentally for each dye and buffer system. Dye decolorization assays were run in 50 mM HEPES buffers prepared at pHs of 5.5, 6.0, 6.5, 7.0, 7.5, and 8.0. Initial assays demonstrated that YfeX could decolorize alizarin red and Cibacron blue but not reactive brown 5, reactive red 120, reactive orange 14, or reactive green 10 (Procion green). The final assay mixture contained 0.03% H2O2, 0.2 mM Cibacron blue, or 0.45 mM alizarin red as well as 0.6 µM YfeX in 50 mM HEPES. Repetitive scans were run between 400 and 700 nM at room temperature. For pH optimum determinations for pyrogallol, 50 mM sodium phosphate buffers were prepared at pHs of 6.0, 6.5, 7.0, and 7.5. Each assay mixture contained 1% pyrogallol, 0.06% H2O2, 0.1 µM YfeX, and 50 mM phosphate buffer. All assays were repeated using 0.3% H2O2 (final concentration) and yielded similar results.
For porphyrinogen oxidation assays, coproporphyrinogen III and protoporphyrinogen IX were freshly prepared with sodium amalgam as previously described (43). Assays were as described for the dyes, except that coproporphyrinogen oxidation was monitored at 370 nm and protoporphyrinogen oxidation was monitored at 357 nm. In assays where Bacillus subtilis HemY (44) was substituted for YfeX, the concentration of HemY was approximately 1 nM and no peroxide was present.
To assay for heme dechelatase activity in crude extracts of YfeX-expressing cells, the basic assay described previously was employed (11). Three 1.5-ml samples were added to microcentrifuge tubes. To one tube no heme was added, to a second 50 µM heme (1 mM stock in dimethyl sulfoxide [DMSO]) was added, and to a third 50 µM mesoheme (1 mM stock in DMSO) was added. At time points of 0, 10, 20, 30, and 60 min, 250 µl was collected, acidified, and solvent extracted with ethyl acetate-acetic acid, 4:1 (vol/vol). Solvent-extracted samples were dried and porphyrin profiles analyzed as previously described (45).
To assay the ability of V. fischeri strains to grow on hemin as a sole iron source, cultures of the wild type and the ΔyfeX mutant were grown in LBS medium (46) overnight. These cultures were diluted 1:1,000 into mineral salts medium (18) without an iron source and supplemented with 50 µM 2,2′-bipyridyl chelator (Sigma, St. Louis, MO), with or without 50 µg ml−1 hemin (Sigma). The minimal medium cultures were grown in 200-µl volumes in a 96-well Falcon polystyrene plate (Becton Dickinson, Franklin Lakes, NJ) at 28°C for 24 to 30 h. Cell density was determined by measuring the absorbance at 595 nm with a Bio-Tek Synergy 2 plate reader every 30 min for the duration of the experiment. The assay was performed three times with biological duplicates, and results from a representative experiment with optical density values normalized to a 1-cm path length are shown.
To observe porphyrin accumulation in E. coli JM109, cells expressing yfeX, hasR plus yfeX, B. subtilis hemY, or human ferrochelatase were grown on LB alone or with either 50 µg ml−1 hemin or 50 µg ml−1 5-aminolevulinic acid (ALA) (Sigma). To observe porphyrin accumulation in V. fischeri, wild-type cells with or without the yfeX expression vector (pAS101) were grown on LBS plates with 2 mM isopropyl β-d-1-thiogalactopyranoside (IPTG), (Sigma) supplemented with either 100 µM 2,2′-bipyridyl chelator and 50 µg ml−1 hemin or 50 µg ml−1 ALA. Plates were incubated at 28°C for 3 days, and individual colonies were imaged for porphyrin accumulation using a Nikon (Melville, NY) Eclipse E600 epifluorescence microscope with a Nikon red/green dual color filter cube (51004v2; fluorescein isothiocyanate/tetramethyl rhodamine isothiocyanate FITC/TRITC) and a Nikon Coolpix 5000 camera. All images were taken using a 10× objective and a 1/60-s exposure for light images and 1/2-s exposure for fluorescent images.
Bioinformatics.
The SEED database and its tools were used as the primary comparative genomics platform in this study (47). Functional genomics data, namely, predicted or experimentally determined regulation patterns of YfeX orthologs in several model microorganisms, were explored to determine the functional expression context. For this, the following global expression profiling databases were employed: (i) MicrobesOnline at http://www.microbesonline.org/, (ii) M3D at http://m3d.bu.edu/, and (iii) SEED at http://www.theseed.org. Additional online resources on gene regulation employed were RegulonDB (http://regulondb.ccg.unam.mx/index.jsp) and RegPrecise (http://regprecise.lbl.gov/). Sufficient data on the regulation (predicted or perceived) of YfeX homologs are available for: Escherichia coli K-12 (YfeX), Shewanella oneidensis MR-1 (SO_0740), and Pseudomonas aeruginosa PAO1 (PA2765). Data available online for Vibrio fischeri ES114 were insufficient to resolve detailed expression correlations of the YfeX homolog VF_1898 with other genes.
The DyP family in the Pfam database (48) currently includes 1,094 sequences in 760 bacterial and fungal species. These constitute a divergent superfamily that can be partitioned into distinct branches of a phylogenetic tree. A neighbor-joining tree analysis (49) in the SEED database of the YfeX branch of the DyP superfamily includes 192 proteins that occur in 185 bacterial species of beta- and gammaproteobacteria out of 916 bacterial and archaeal organisms examined. These share end-to-end homology, with an E value of e−40 or better, with the E. coli YfeX protein and are likely isofunctional [see the encoded subsystem “Predicted dye-decolorizing peroxidases (DyP) of YfeX-like subgroup” http://theseed.uchicago.edu/FIG/seedviewer.cgi?page=Subsystems&subsystem=Predicted_dye-decolorizing_peroxidases_(DyP)_of_YfeX-like_Subgroup].
ACKNOWLEDGMENTS
This work was supported by grant DK32302 (H.A.D.), NIAID contract no. HHSN266200400042C (S.G.), National Science Foundation grants MCB-0347317 and OCE-0929081 (E.V.S.), and grant DK02503 (J.D.P.). A.N.S. was supported by funds awarded by DoD, Air Force Office of Scientific Research, National Defense Science and Engineering Graduate (NDSEG) Fellowship, 32 CFR 168a.
Footnotes
Citation Dailey HA, et al. 2011. The Escherichia coli protein YfeX functions as a porphyrinogen oxidase, not a heme dechelatase. mBio 2(6):e00248-11. doi:10.1128/mBio.00248-11.
REFERENCES
- 1. Sugano Y. 2009. DyP-type peroxidases comprise a novel heme peroxidase family. Cell. Mol. Life Sci. 66:1387–1403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sugano Y, Muramatsu R, Ichiyanagi A, Sato T, Shoda M. 2007. DyP, a unique dye-decolorizing peroxidase, represents a novel heme peroxidase family: ASP171 replaces the distal histidine of classical peroxidases. J. Biol. Chem. 282:36652–36658 [DOI] [PubMed] [Google Scholar]
- 3. Glenn JK, Gold MH. 1985. Purification and characterization of an extracellular Mn(II)-dependent peroxidase from the lignin-degrading basidiomycete, Phanerochaete chrysosporium. Arch. Biochem. Biophys. 242:329–341 [DOI] [PubMed] [Google Scholar]
- 4. Gold MH, Kuwahara M, Chiu AA, Glenn JK. 1984. Purification and characterization of an extracellular H2O2-requiring diarylpropane oxygenase from the white rot basidiomycete, Phanerochaete chrysosporium. Arch. Biochem. Biophys. 234:353–362 [DOI] [PubMed] [Google Scholar]
- 5. Welinder KG. 1992. Superfamily of plant, fungal and bacterial peroxidases. Curr. Opin. Struct. Biol. 2:388–393 [Google Scholar]
- 6. Liu X, et al. 2011. Crystal structure and biochemical features of EfeB/YcdB from Escherichia coli O157: ASP235 plays divergent roles in different enzyme-catalyzed processes. J. Biol. Chem. 286:14922–14931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ogola HJ, et al. 2009. Molecular characterization of a novel peroxidase from the cyanobacterium Anabaena sp. strain PCC 7120. Appl. Environ. Microbiol. 75:7509–7518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zubieta C, et al. 2007. Identification and structural characterization of heme binding in a novel dye-decolorizing peroxidase, TyrA. Proteins 69:234–243 [DOI] [PubMed] [Google Scholar]
- 9. Goblirsch B, Kurker RC, Streit BR, Wilmot CM, DuBois JL. 2011. Chlorite dismutases, DyPs, and EfeB: 3 microbial heme enzyme families comprise the CDE structural superfamily. J. Mol. Biol. 408:379–398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dailey TA, et al. 2010. Discovery and characterization of HemQ: an essential heme biosynthetic pathway component. J. Biol. Chem. 285:25978–25986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Létoffé S, Heuck G, Delepelaire P, Lange N, Wandersman C. 2009. Bacteria capture iron from heme by keeping tetrapyrrol skeleton intact. Proc. Natl. Acad. Sci. U. S. A. 106:11719–11724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cao J, Woodhall MR, Alvarez J, Cartron ML, Andrews SC. 2007. EfeUOB (YcdNOB) is a tripartite, acid-induced and CpxAR-regulated, low-pH Fe2+ transporter that is cryptic in Escherichia coli K-12 but functional in E. coli O157:H7. Mol. Microbiol. 65:857–875 [DOI] [PubMed] [Google Scholar]
- 13. Reniere ML, et al. 2010. The IsdG-family of haem oxygenases degrades haem to a novel chromophore. Mol. Microbiol. 75:1529–1538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Schuller DJ, Zhu W, Stojiljkovic I, Wilks A, Poulos TL. 2001. Crystal structure of heme oxygenase from the gram-negative pathogen Neisseria meningitidis and a comparison with mammalian heme oxygenase-1. Biochemistry 40:11552–11558 [DOI] [PubMed] [Google Scholar]
- 15. Chau TT, Ishigaki M, Kataoka T, Taketani S. 2010. Porcine ferrochelatase: the relationship between iron-removal reaction and the conversion of heme to Zn-protoporphyrin. Biosci. Biotechnol. Biochem. 74:1415–1420 [DOI] [PubMed] [Google Scholar]
- 16. Dailey HA, Sellers VM, Dailey TA. 1994. Mammalian ferrochelatase. Expression and characterization of normal and two human protoporphyric ferrochelatases. J. Biol. Chem. 269:390–395 [PubMed] [Google Scholar]
- 17. Jacobs JM, Jacobs NJ, Sherman TD, Duke SO. 1991. Effect of diphenyl ether herbicides on oxidation of protoporphyrinogen to protoporphyrin in organellar and plasma membrane enriched fractions of barley. Plant Physiol. 97:197–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Septer AN, Wang Y, Ruby EG, Stabb EV, Dunn AK. The heme-uptake gene cluster in Vibrio fischeri is regulated by Fur and contributes to symbiotic colonization. Environ. Microbiol., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hansson M, Gustafsson MC, Kannangara CG, Hederstedt L. 1997. Isolated Bacillus subtilis HemY has coproporphyrinogen III to coproporphyrin III oxidase activity. Biochim. Biophys. Acta 1340:97–104 [DOI] [PubMed] [Google Scholar]
- 20. Massé E, Vanderpool CK, Gottesman S. 2005. Effect of RyhB small RNA on global iron use in Escherichia coli. J. Bacteriol. 187:6962–6971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. McHugh JP, et al. 2003. Global iron-dependent gene regulation in Escherichia coli. A new mechanism for iron homeostasis. J. Biol. Chem. 278:29478–29486 [DOI] [PubMed] [Google Scholar]
- 22. Mey AR, Craig SA, Payne SM. 2005. Characterization of Vibrio cholerae RyhB: the RyhB regulon and role of ryhB in biofilm formation. Infect. Immun. 73:5706–5719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mey AR, Wyckoff EE, Kanukurthy V, Fisher CR, Payne SM. 2005. Iron and fur regulation in Vibrio cholerae and the role of fur in virulence. Infect. Immun. 73:8167–8178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ochsner UA, Wilderman PJ, Vasil AI, Vasil ML. 2002. GeneChip expression analysis of the iron starvation response in Pseudomonas aeruginosa: identification of novel pyoverdine biosynthesis genes. Mol. Microbiol. 45:1277–1287 [DOI] [PubMed] [Google Scholar]
- 25. Frustaci JM, O’Brian MR. 1993. The Escherichia coli visA gene encodes ferrochelatase, the final enzyme of the heme biosynthetic pathway. J. Bacteriol. 175:2154–2156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Matringe M, Camadro JM, Labbe P, Scalla R. 1989. Protoporphyrinogen oxidase as a molecular target for diphenyl ether herbicides. Biochem. J. 260:231–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cox TM. 2003. Protoporphyria, p 121–150 In Kadish KM, Smith KM, Guilard R, The porphyrin handbook, vol 14 Academic Press, New York, NY. [Google Scholar]
- 28. O’Connor AE, Gallagher WM, Byrne AT. 2009. Porphyrin and nonporphyrin photosensitizers in oncology: preclinical and clinical advances in photodynamic therapy. Photochem. Photobiol. 85:1053–1074 [DOI] [PubMed] [Google Scholar]
- 29. Zoładek T, Nguyen BN, Rytka J. 1996. Saccharomyces cerevisiae mutants defective in heme biosynthesis as a tool for studying the mechanism of phototoxicity of porphyrins. Photochem. Photobiol. 64:957–962 [DOI] [PubMed] [Google Scholar]
- 30. Cooperman SS, et al. 2005. Microcytic anemia, erythropoietic protoporphyria, and neurodegeneration in mice with targeted deletion of iron-regulatory protein 2. Blood 106:1084–1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Galy B, et al. 2010. Iron regulatory proteins secure mitochondrial iron sufficiency and function. Cell Metab. 12:194–201 [DOI] [PubMed] [Google Scholar]
- 32. Dunn AK, Martin MO, Stabb EV. 2005. Characterization of pES213, a small mobilizable plasmid from Vibrio fischeri. Plasmid 54:114–134 [DOI] [PubMed] [Google Scholar]
- 33. Zubieta, et al. 2007. Crystal structures of two novel dye-decolorizing peroxidases reveal a beta-barrel fold with a conserved heme-binding motif. Proteins 69:223–233 [DOI] [PubMed] [Google Scholar]
- 34. Kranz RG, Richard-Fogal C, Taylor JS, Frawley ER. 2009. Cytochrome c biogenesis: mechanisms for covalent modifications and trafficking of heme and for heme-iron redox control. Microbiol. Mol. Biol. Rev. 73:510–528 , Table of Contents. doi: 10.1128/MMBR.00001-09. PubMed [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Richard-Fogal CL, et al. 2009. A conserved haem redox and trafficking pathway for cofactor attachment. EMBO J. 28:2349–2359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Rezuchova B, Miticka H, Homerova D, Roberts M, Kormanec J. 2003. New members of the Escherichia coli sigmaE regulon identified by a two-plasmid system. FEMS Microbiol. Lett. 225:1–7 [DOI] [PubMed] [Google Scholar]
- 37. Rhodius VA, Suh WC, Nonaka G, West J, Gross CA. 2006. Conserved and variable functions of the sigmaE stress response in related genomes. PLoS Biol. 4:e2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wyckoff EE, Schmitt M, Wilks A, Payne SM. 2004. HutZ is required for efficient heme utilization in Vibrio cholerae. J. Bacteriol. 186:4142–4151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sellers VM, Wang KF, Johnson MK, Dailey HA. 1998. Evidence that the fourth ligand to the [2Fe-2S] cluster in animal ferrochelatase is a cysteine. Characterization of the enzyme from Drosophila melanogaster. J. Biol. Chem. 273:22311–22316 [DOI] [PubMed] [Google Scholar]
- 40. Bose JL, Rosenberg CS, Stabb EV. 2008. Effects of luxCDABEG induction in Vibrio fischeri: enhancement of symbiotic colonization and conditional attenuation of growth in culture. Arch. Microbiol. 190:169–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Dunn AK, et al. 2010. The alternative oxidase (AOX) gene in Vibrio fischeri is controlled by NsrR and upregulated in response to nitric oxide. Mol. Microbiol. 77:44–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Dunn AK, Millikan DS, Adin DM, Bose JL, Stabb EV. 2006. New rfp- and pES213-derived tools for analyzing symbiotic Vibrio fischeri reveal patterns of infection and lux expression in situ. Appl. Environ. Microbiol. 72:802–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Brenner DA, Bloomer JR. 1980. A fluorometric assay for measurement of protoporphyrinogen oxidase activity in mammalian tissue. Clin. Chim. Acta 100:259–266 [DOI] [PubMed] [Google Scholar]
- 44. Dailey TA, Meissner P, Dailey HA. 1994. Expression of a cloned protoporphyrinogen oxidase. J. Biol. Chem. 269:813–815 [PubMed] [Google Scholar]
- 45. Franklin MR, Phillips JD, Kushner JP. 1997. Cytochrome P450 induction, uroporphyrinogen decarboxylase depression, porphyrin accumulation and excretion, and gender influence in a 3-week rat model of porphyria cutanea tarda. Toxicol. Appl. Pharmacol. 147:289–299 [DOI] [PubMed] [Google Scholar]
- 46. Stabb EV, Reich KA, Ruby EG. 2001. Vibrio fischeri genes hvnA and hvnB encode secreted NAD(+)-glycohydrolases. J. Bacteriol. 183:309–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Overbeek R, et al. 2005. The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res. 33:5691–5702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bateman A, et al. 2004. The Pfam protein families database. Nucleic Acids Res. 32:D138–D141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Saitou N, Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406–425 [DOI] [PubMed] [Google Scholar]