Abstract
The influence of the antibiotic linezolid on the secretion of exotoxins by Staphylococcus aureus was analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis combined with matrix-assisted laser desorption ionization-time of flight mass spectrometry and Western blot analysis. S. aureus suspensions were treated with grading subinhibitory concentrations of linezolid (12.5, 25, 50, and 90% of MIC) at different stages of bacterial growth (i.e., an optical density at 540 nm [OD540] of 0.05 or 0.8). When added to S. aureus cultures at an OD540 of 0.05, linezolid reduced in a dose-dependent manner the secretion of specific virulence factors, including staphylococcal enterotoxin A (SEA) and SEB, bifunctional autolysin, autolysin, protein A, and alpha- and beta-hemolysins. In contrast, other presumably nontoxic exoproteins remained unchanged or even accumulated in supernatants in the presence of linezolid at a 90% MIC. Similarily, when added at OD540 of 0.8, that is, after quorum sensing, linezolid reduced the release of virulence factors, whereas the relative abundance of nontoxic exoproteins such as triacylglycerol lipase, glycerol ester hydrolase, DnaK, or translation elongation factor EF-Tu was found to be increased. Consistently, linezolid reduced in a dose-dependent manner the tumor necrosis factor-inducing activity secreted by S. aureus into the culture supernatants. The results of our study suggest that the expression of virulence factors in S. aureus is especially sensitive to the inhibition of protein synthesis by linezolid, which should be an advantage in the treatment of infections with toxin-producing S. aureus.
Linezolid is a member of the new synthetic class of antibacterial oxazolidinones that inhibit bacterial protein synthesis at the initiation step of protein biosynthesis. Because gram-positive bacteria increasingly develop resistance against currently available antibiotics, e.g., methicillin and vancomycin, linezolid has become a valuable antibiotic for the treatment of nosocomial and community-acquired pneumonia, complicated skin and soft tissue infections caused by methicillin-resistant Staphylococcus aureus, glycopeptide-intermediate S. aureus, methicillin-resistant Staphylococcus epidermidis, vancomycin-resistant enterococci, and penicillin-resistant strains of Streptococcus pneumoniae (11, 27, 33).
The clinical efficacy of antibiotics is not only determined by their respective bactericidal or bacteriostatic activity and pharmacokinetics but also by their action on bacterial virulence factor release, especially at suboptimal concentrations. In principle, antibiotics can both up- and downmodulate the synthesis and release of virulence factors. Protein synthesis-suppressing antibiotics such as clindamycin can induce a general inhibition of exoprotein expression, including virulence factors such as alpha-toxin (19, 24, 35, 40, 44). In contrast, subinhibitory concentrations of the commonly used cell wall affecting β-lactam antibiotics, such as methicillin, lead to an increase of alpha-toxin expression through a stimulatory effect on exoprotein synthesis (16, 17, 30, 39). Although the molecular mode of action of linezolid has been determined, little information was available about the effects of linezolid on bacterial virulence factor production.
The possible up- or downregulation of exoprotein release is especially important for S. aureus infections because S. aureus produces a wide array of toxins that determine, at least in part, the pathogenesis of infection. Thus, antibiotic-induced modulation of virulence factors could lead to either worsening or attenuation of the disease (12, 20, 25).
The production of staphylococcal exoproteins is regulated in a coordinated, growth-phase-dependent manner, occurring preferentially during the postexponential phase of growth (1, 5, 9). When S. aureus organisms reach high cell population densities, they sense a quorum through a cell-cell communication system. Cell-cell communication in bacteria is accomplished through the exchange of signaling molecules called autoinducers in a process referred to as quorum sensing (2, 8, 26, 31). Quorum sensing allows bacterial populations to coordinate gene expression and probably enhance the effectiveness of processes such as virulence factor expression, antibiotic production, and biofilm development (6, 7, 14).
During the postexponential phase of growth, the production of several exoproteins in S. aureus (e.g., alpha-toxin, enterotoxins, toxic shock syndrome toxin 1, and cell wall-associated proteins) is principally regulated by the agr (accessory gene regulator) locus (31, 32). agr acts at the transcriptional level and upregulates alpha-toxin, toxic shock syndrome toxin 1, and other extracellular proteins and downregulates cell wall-associated proteins (31, 32). Besides agr, other pleiotropic regulatory genes such as sarA, sarS, and rot have been identified that transcriptionally control not only virulence factor expression but also cytoplasmic proteins, including catabolic enzymes (6, 8, 10, 18, 42; for a review, see reference 28).
To obtain a comprehensive picture of the antibacterial effects of linezolid, we used the technology of proteomics to analyze the effects of linezolid on virulence factor production by S. aureus. To differentiate the modulation of virulence factor production from effects secondary to quorum-sensing phenomena, we investigated virulence factor secretion by S. aureus exposed to linezolid at distinct growth phases.
MATERIALS AND METHODS
Bacterial growth conditions.
The methicillin-sensitive S. aureus strain ATCC 29213 was obtained from the American Type Culture Collection (ATCC) and used throughout this study. Bacteria were stored as a 20% glycerol stock at −80°C. Bacteria were precultured from the glycerol stock in Luria-Bertani (LB) broth at 37°C with constant shaking to log growth (i.e., an optical density at 540 nm [OD540] of 2). An aliquot of the preculture was inoculated into 200 ml of LB broth to obtain a starting OD540 of 0.05. Bacteria were cultured at 37°C with constant shaking under aerobic conditions. The growth of cells was monitored by reading the OD540 values. The MIC in LB broth was determined by broth microdilution according to the NCCLS standard method M7-A2. The MIC for linezolid was 2.5 mg/liter, and the MIC for erythromycin was 0.15 mg/liter. Solubilized linezolid (2 mg/ml) was commercially obtained from Pharmacia (Pharmacia & Upjohn, Peapark, N.J.), and erythromycin was obtained from Sigma Aldrich (Taufkirchen, Germany).
Exoprotein preparation.
After the indicated growth phases, bacteria cells were pelleted by centrifugation at 8,500 × g for 30 min at 4°C. The culture supernatant was precipitated by adding 100% trichloroacetic acid (Sigma) to a final concentration of 10%. After overnight incubation at 4°C, the precipitate was centrifuged at 8,500 × g for 70 min at 4°C and finally washed three times with ice-cold (−50°C) ethanol. The aggregated proteins were dried by using a Speed-Vac for a few minutes. The protein extracts were dissolved in 0.5 ml of 8 M urea for two-dimensional gel electrophoresis or in 0.5 ml of 0.1 M Tris containing 2 mM phenylmethylsulfonyl fluoride for one-dimensional applications as described previously (3).
The protein concentration was determined with a Bio-Rad (Munich, Germany) protein assay kit according to the instructions of the manufacturer.
SDS-PAGE.
One-dimensional denaturating sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed with 10% polyacrylamide gels according to the method of Schägger and Jagow (34) in a Bio-Rad Protean-II electrophoresis system. The gels were stained overnight in 0.2% Coomassie brilliant blue R250 with 45% ethanol and 15% acetic acid. Alternatively, silver staining was performed according to the method described by Shevchenko et al. (38).
Two-dimensional gel electrophoresis was performed according to the method of O'Farrell (29) by using the Multiphor II (Pharmacia-FRG) system according to the instructions of the manufacturer. Protein samples were separated by using immobilized pH gradient (IPG) strips in a nonlinear pH range of 3 to 10. Isoelectric focusing was performed as described by Görg et al. (15) with 8 M urea, 2 M thiourea, 2% CHAPS {3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate}, and 0.05% bromophenol blue. Isoelectric focusing was carried out with the same buffer with increasing voltage levels (i.e., 1 h at 100 V, 1 h at 200 V, 1 h at 500 V, 1 h at 1,000 V, 1 h at 2,000 V, and 14 h at 3,500 V). Rod gels were soaked for 15 min at an ambient temperature in equilibration buffer (50 mM Tris-HCl [pH 8.8], 8 M urea, 2 M thiourea, 30% glycerol, 2% SDS, 0.05% bromophenol blue, 10 mg of dithiothreitol/ml). A second equilibration was performed for a further 15 min in equilibration buffer containing 25 mg of iodoacetamide/ml instead of dithiothreitol and applied to a second dimension by using a 12.5% Tris-glycine SDS gel (25 cm by 20 cm by 1.0 mm) with the Ettan Dalt II system (Pharmacia).
In-gel preparation of tryptic peptides.
In-gel digestion with trypsin was performed according to standard protocols (21, 38) with minor modifications. Coomassie blue-stained protein bands were excised from the gel, washed three times for 10 min with water (high-pressure liquid chromatography grade; Merck, Darmstadt, Germany), equilibrated with 100 μl of 50 mM NH4HCO3 (pH 7.8), shrunk with acetonitrile, rehydrated with 100 μl of 50 mM NH4HCO3 (pH 7.8), and finally shrunk again with acetonitrile. The gel pieces were reswollen in a digestion buffer containing 50 mM NH4HCO3 and treated with 0.2 μg of trypsin (Promega) at 37°C for 16 h. Peptides were extracted as described previously (3). The pellet was dissolved in 10 μl of 0.1% trifluoroacetic acid (TFA).
MALDI-TOF/MS.
Aliquots of 0.5 μl of the combined extract were used for matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF/MS) to obtain MS fingerprints. Mass spectra were obtained with the Bruker REFLEX IV mass spectrometer (Bruker-Daltonik, Leipzig, Germany). The validation of all data obtained, including averaging of the TOF data, recalibration on trypsin signals, and all further data processing, was carried out by using XMASS 5.1.1 postanalysis software.
LC-MS.
Liquid chromatography (LC)-MS data were aquired on a Q-Tof II quadrupole-TOF mass spectrometer (Micromass, Manchester, United Kingdom) equipped with a Z spray source. Samples were introduced by using the Ultimate Nano-LC system (LC Packings, Amsterdam, The Netherlands) equipped with the Famos autosampler and the Switchos column switching module. The column setup comprises a 0.3-mm-by-1-mm trap column and a 0.075-by-150-mm analytical column, both packed with 3 μm PepMap C18 (LC Packings, Amsterdam, The Netherlands). Samples were diluted 1:10 in 0.1% TFA. A total of 10 μl was injected onto the trap column and desalted for 3 min with 0.1% TFA and a flow rate of 30 μl/min. The 10 port valve switched the trap column into the analytical flowpath, and the peptides were eluted onto the analytical column by using a gradient of 5% acetonitrile (ACN) in 0.1% TFA to 40% ACN in 0.1% TFA over 20 min and a column flow rate of ca. 200 nl/min, resulting from a 1:1,000 split of the 200 μl/min flow delivered by the pump. The electrospray ionization (ESI) interface comprised a metal-coated PicoTip spray emitter (New Objective, Woburn, Mass.) mounted onto the PicoTip holder assembly (New Objective). Stable nanospray was established by the application of 2.5 to 3.0 kV to the distal end of the PicoTip and a nitrogen counter flow rate of ∼40 liter/min. The data-dependent acquisition of MS and tandem MS (MS/MS) spectra was controlled by the Masslynx software. Survey scans of 1 s covered the range from m/z 400 to 1,200. Doubly and triply charged ions rising above a given threshold were selected for MS/MS experiments. In MS/MS mode the mass range from m/z 40 to 1,400 was scanned in 1 s, and 10 scans were added up for each experiment. Micromass-formated peaklists were generated from the raw data by using the Proteinlynx software module.
Database searching.
Proteins were identified from MALDI fingerprint data by using MASCOT for websearch (http://www.matrixscience.com) against a public database (National Center for Biotechnology Information) or a locally installed protein prospector algorithm (http://prospector.ucsf.edu) with sequence data obtained from The Institute for Genomic Research (http://www.tigr.org), Oklahoma University, or the N315 database (23). To identify proteins with the LC-MS peaklists, we performed MS/MS ion searches by using a local installation of Mascot 7.0 and the sequence databases mentioned above.
Western blot.
Western blot analysis was performed under the conditions described by Towbin et al. (41). Antibodies to S. aureus enterotoxin A (SEA), SEB, and protein A were purchased from Sigma Aldrich.
Tumor necrosis factor (TNF) release assay. (i) Preparation of bacterial exotoxins.
An overnight culture of ATCC 29213 in Dulbecco modified Eagle medium (DMEM; Biochrom AG, Berlin, Germany) was diluted 30-fold in 1,000 ml of prewarmed DMEM, incubated for 30 min at 37°C with constant shaking, and divided into aliquots of 200 ml. Graded concentrations of linezolid (12.5, 25, 50 and 90% MIC) were added to the diluted bacterial suspensions before incubation for further 4 h. S. aureus supernatants without antibiotic treatment served as controls. Proteins secreted into the supernatants were filtered through a 0.2-μm-pore-size filter (Braun Melsungen AG, Melsungen, Germany) and immediately analyzed as described below.
(ii) Preparation of spleen cells and macrophages.
C57BL/6 mice were obtained from Charles River Wiga, Sulzfeld, Germany. Mice were kept under barrier conditions and used when 6 to 10 weeks old. Mice were euthanized by cervical dislocation. Resident peritoneal macrophages were harvested by rinsing the peritoneal cavity with chilled 0.9% NaCl. Single spleen cell suspensions were prepared by passing the spleen through cell strainer meshes of 100-μm pore size. Cells were seeded at a density of 106/ml in DMEM-5% fetal calf serum without antibiotics into 96-well flat tissue culture plates (Nunc, Kamstrup, Denmark) and incubated in 5% CO2 at 37°C for 1 h to allow adherence. Nonadherent cells were removed by aspiration before bacterial filtrates were added to the adherent cells. After incubation for 16 h the supernatants were collected, centrifuged (1,000 × g for 5 min), and stored at −70°C until TNF was measured by enzyme-linked immunosorbent assay (ELISA).
(iii) ELISA.
TNF in the supernatants was determined by using the Mouse TNF-α DuoSet ELISA (R&D Systems, Inc., Minneapolis, Minn.). The mininum detection level of the test was 31.25 pg/ml.
RESULTS
Effects of linezolid on S. aureus growth.
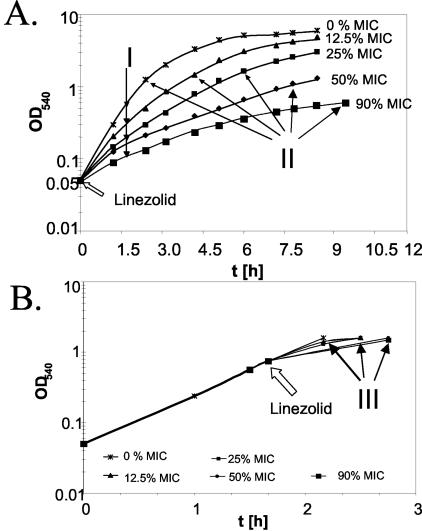
To examine the influence of linezolid on the bacterial production of virulence factors prior to or after quorum sensing, S. aureus strain ATCC 29213 was treated with linezolid at different stages of growth. Treatment of S. aureus from the start of growth with subinhibitory concentrations of linezolid (e.g., 12.5, 25, 50, and 90% MIC) attenuated the growth rate in a dose-dependent manner (Fig. 1).
FIG. 1.
Effect of linezolid on growth of S. aureus. S. aureus strain ATCC 29213 was grown at 37°C. At indicated time points, the OD540 was measured. (A) Bacteria were grown in the presence of grading concentrations of linezolid from the start of growth (OD540 ∼0.05) and harvested after 2 h (I) or after they reached an OD540 of ∼1.0 (II). (B) Bacteria were grown first to an OD540 of 0.8; thereafter, growth was continued in the presence of linezolid with increasing concentrations and harvested at an OD540 of ∼1.0 (III). Arrows indicate the time point of harvest.
For the analysis of protein profiles, supernatants were harvested after 2 h (Fig. 1A, I), which reveals protein secretion within a constant period of time and yet at various densities. Alternatively, supernatants were harvested at similar OD values (Fig. 1A, II), which allows protein secretion at similar bacterial densities over different periods of time. When S. aureus was first cultured to an OD540 of 0.8 and then exposed to linezolid, only slightly retarded growth rates were observed (Fig. 1). Supernatants were harvested at an OD540 of ∼1.3 (Fig. 1B, III), which was achieved within a narrow period of time. As depicted in Table 1, the overall secretion of exoproteins remained largely constant. As expected, a slight decrease of protein secretion was observed in the presence of linezolid at a concentration of 90% MIC.
TABLE 1.
Protein contents in supernatants of S. aureus
| Antibiotica | Protein content (mg/ml) at antibiotic concn (% MIC) of:
|
||||
|---|---|---|---|---|---|
| 0 | 12.5 | 25 | 50 | 90 | |
| Linezolid I | 4.89 | 4.23 | 4.13 | 3.71 | 2.77 |
| Linezolid II | 4.33 | 3.92 | 3.57 | 3.14 | 2.71 |
| Linezolid III | 4.31 | 5.06 | 4.06 | 4.27 | 2.92 |
| Erythromycin II | 4.48 | 4.49 | 4.16 | 3.80 | 2.48 |
I to III refer to the culture conditions described in the legend to Fig. 1.
Effects of linezolid on protein secretion by S. aureus.
To monitor the changes in the pattern of secreted proteins, constant amounts of proteins within culture supernatants of S. aureus treated with subinhibitory concentrations of linezolid were analyzed by SDS-PAGE.
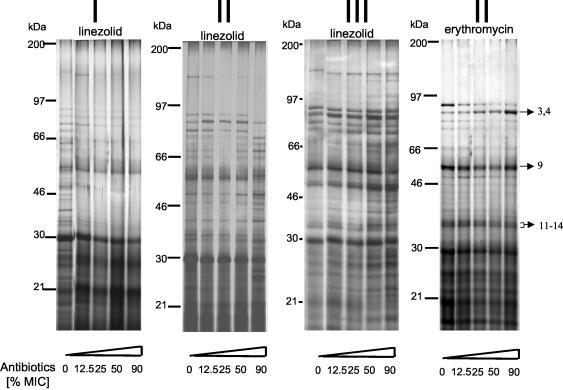
When S. aureus was exposed to linezolid for 2 h, linezolid reduced the secretion of higher-molecular-weight proteins into the supernatants in a dose-dependent manner (Fig. 2I, linezolid). This result is not necessarily secondary to decreased bacterial cell densities because equal amounts of proteins were loaded onto the gels. When linezolid-treated S. aureus cultures were harvested at similar cell densities (OD540 of 0.8 to 1.0), significant changes of the exoprotein patterns were observed, especially when linezolid was added at a 90% MIC (Fig. 2II, linezolid).
FIG. 2.
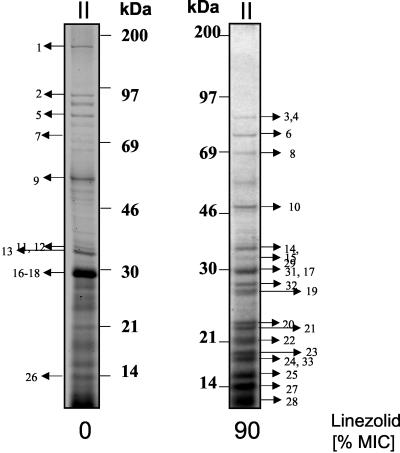
SDS-PAGE of exotoxin production by S. aureus in the presence of antibiotics. Culture supernatants of S. aureus treated with different concentrations of linezolid or erythromycin were analyzed by SDS-PAGE as described in Materials and Methods. Protein bands were visualized by silver staining (sample of 10 μg of protein). Panels I to III correspond to the conditions described in the legend to Fig. 1. The arrows indicate the protein bands subjected to MALDI-TOF/MS and/or LC-MS analysis.
The effects of linezolid on protein secretion by S. aureus were compared to the effects of erythromycin, a macrolide with protein inhibitory activity. S. aureus was grown from the beginning of the culture in the presence of grading subinhibitory concentrations of erythromycin and harvested at a cell OD540 of 1.3. Compared to linezolid, erythromycin induced less-pronounced changes in protein patterns. Notably lipase and glycerol ester hydrolase (Fig. 2, proteins “3,4”) appeared slightly enhanced.
When linezolid was added to S. aureus at a density of OD540 of 0.8 and supernatants were harvested at an OD540 of ∼1.3, a significant increase of select exoproteins was discovered (Fig. 2III, linezolid).
Because of the limitation in the resolution of the one-dimensional gel electrophoresis, secreted proteins were also analyzed by two-dimensional SDS-PAGE analysis.
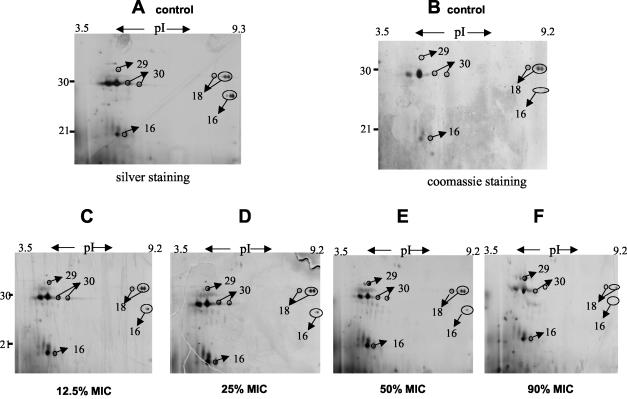
A typical set of experiments is depicted in Fig. 3 (upper panels, A to F). S. aureus cultures were grown in the presence of grading concentrations of linezolid. At a cell OD540 of 1.0, supernatants were harvested and subjected to two-dimensional gel electrophoresis. As shown in Fig. 3, discrete changes in protein profiles can be detected, a finding consistent with the results obtained by one-dimensional SDS-PAGE (Fig. 2II, linezolid).
FIG. 3.
Two-dimensional patterns of exoproteins in the presence of graded concentrations of linezolid. (Upper panels [A to F]) Partial view (pI 3 to 10 and 10 to 40 kDa); (lower panels [A to D]) partial view (pI 3.2 to 6.9 and 46 to 66 kDa). S. aureus ATCC 29213 was grown at 37°C with increasing concentrations of linezolid from the start of growth (OD540 ∼0.05) and harvested at an OD540 of ∼1.0. Supernatants of S. aureus cultures were subjected to two-dimensional gel electrophoresis. Lettered panels: A and B, untreated; C to F treated with linezolid at 12.5, 25, 50, and 90% MICs, respectively. Protein spots were visualized by silver staining (A and C to F [sample of 100 μg of protein]) or Coomassie blue stain (B [sample of 500 μg of protein]). The arrows indicate the protein bands subjected to MALDI-TOF/MS and/or LC-MS analysis.
Identification of linezolid-sensitive proteins.
To identify individual linezolid-sensitive proteins, both one- and two-dimensional gels were visualized by Coomassie blue staining, and stained bands were subjected to MALDI-TOF/MS and/or LC-MS/MS analysis as a complementary technique (Fig. 3 and 4). Protein profiles were subjected to the genome sequence databases as described in Materials and Methods. The peptide matches and the sequence coverage allowed the identification of 33 proteins. The changes in the secreted protein patterns of S. aureus treated with linezolid are illustrated in Tables 2 and 3.
FIG. 4.
Identification of exoproteins of S. aureus treated with linezolid. Supernatants of S. aureus cultures left untreated (left) or treated with linezolid at 90% MIC from the start of growth (OD540 ∼0.05) (right) were harvested at an OD540 of ∼0.8. Protein bands were visualized by coomassie staining. The arrows indicate the protein bands subjected to MALDI-TOF/MS or LC-MS/MS analysis.
TABLE 2.
S. aureus exoproteins identified by MALDI-MS fingerprints
| Spot no. | GenBank accession no. | Z scorea | Sequence coverageb (%) | Molecular mass (Da) measured | Molecular mass (Da)/pI (theoretical) | Linezolid changec | Putative identification |
|---|---|---|---|---|---|---|---|
| 1 | 1703465 | 1.77e+007A | 20 | 145,000 | 137,385/9.60 | ↓ | Bifunctional autolysin |
| 2 | 15924043 | 171B | 56 | 97,000 | 102,656/9.65 | ↓↓ | Autolysin |
| 3 | 13702629 | 1.98e+023A | 77 | 90,000 | 76,662/6.58 | ↑↑ | Triacylglycerol lipase |
| 4 | 13700235 | 7.16e+013A | 34 | 90,000 | 76,543/8.99 | ↑↑ | Glycerol ester hydrolase |
| 5 | 15925634 | 101B | 21 | 80,000 | 69,186/5.96 | ↓↓ | Hypothetical protein ORFID:SA2437 (similar to autolysin) |
| 6 | 15924570 | 122B | 30 | 75,000 | 66,321/4.65 | ↑↑ | DnaK protein |
| 8 | 15926694 | 1.59e+003A | 19 | 72,000 | 69,196/4.9 | ↑ | GTP-binding elongation factor homolog ORFID:SA0959 |
| 9 | 225821 | 101B | 49 | 60,000 | 57,939/5.48 | ↓↓ | Protein A |
| 10 | 15923538 | 175B | 65 | 48,000 | 43,134/4.74 | ↑ | Translational elongation factor TU |
| 11 | 21283669 | 74B | 41 | 36,000 | 31,350/7.68 | ↓↓ | Truncated beta-hemolysin |
| 12 | 15926142 | 72B | 47 | 36,000 | 35,871/9.67 | ↓↓ | Hypothetical protein ORFID:SA0423 (similar to autolysin) |
| 13 | 15924153 | 6e+012A | 34 | 33,000 | 35,953/8.70 | ↓↓ | Alpha-hemolysin |
| 14 | 15923949 | 1.31e+012A | 50 | 33,000 | 35,289/8.67 | ↑↑ | Glycerophosphoryl diester phosphodiesterase |
| 16 | 119131 | 116B | 45 | 28,000 | 27,663/9.15 | ↓↓ | Epidermal cell differentiation inhibitor precursor (EDIN) |
| 18 | 15925289 | 64B | 51 | 31,000 | 29,366/8.96 | ↓↓ | Secretory antigen precursor SsaA homolog ORFID:SA2093 |
| 19 | 15923528 | 122B | 58 | 26,000 | 24,693/9.00 | ↑↑ | 50S ribosomal protein L1 |
| 20 | 15925225 | 243B | 74 | 24,000 | 19,774/9.54 | ↑↑ | 50S ribosomal protein L6 |
| 21 | 15927829 | 112B | 56 | 23,000 | 22,451/9.90 | ↑↑ | 50S ribosomal protein L4 |
| 22 | 15925208 | 180B | 72 | 21,000 | 16,323/9.3 | ↑↑ | 50S ribosomal protein L13 |
| 23 | 2500269 | 106B | 57 | 20,000 | 15,587/10.28 | ↑↑ | 50S ribosomal protein L15 |
| 24 | 15927797 | 80B | 43 | 16,000 | 14,607/10.56 | ↑↑ | 30S ribosomal protein S9 |
| 25 | 15925235 | 65B | 40 | 14,000 | 12,827/9.92 | ↑↑ | 50S ribosomal protein L22 |
| 26 | 15926570 | 3.56e+007A | 84 | 12,000 | 15,898/9.28 | +/− | Hypothetical protein (similar to cell surface protein Map-w) |
| 27 | 15927227 | 74B | 66 | 13,000 | 11,326/9.84 | ↑ | 50S ribosomal protein L21 |
| 28 | 14029570 | 54B | 32 | 9,000 | 7,990/10.16 | ↑ | 50S ribosomal protein L27 |
| 29 | 15925085 | 101B | 36 | 32,000 | 24,063/5.53 | ↑↑ | Hypothetical protein (similar to SceD precursor ORFID:SA1898) |
| 30 | 15923655 | 3.45e+004A | 40 | 29,000 | 28,169/6.12 | ↓ | Secretory antigen precursor SsaA homolog ORFID:SA0620 |
Superscripts: A, Z-score value from Protein Prospector; B, Z-score value from Mascot.
Sequence coverage of total amino acid numbers.
↑↑, Great accumulation; ↑, weak accumulation; ↓, weak inhibition; ↓↓, complete inhibition; +/−, no effect.
TABLE 3.
S. aureus exoproteins identified by LC-MS/MS analysis
| Spot no. | GenBank accession no. | Z scorea | Molecular mass (Da) measured | Molecular mass (Da)/ pI (theoretical) | Linezolid changeb | Putative identification |
|---|---|---|---|---|---|---|
| 1 | 1703465 | 995 | 145,000 | 137,385/9.59 | ↓↓ | Bifunctional autolysin precursor |
| 2 | 15924043 | 1260 | 97,000 | 102,593/9.65 | ↓↓ | Autolysin |
| 3 | 15925661 | 1690 | 90,000 | 76,616/6.58 | ↑↑ | Triacylglycerol lipase |
| 4 | 15923310 | 1520 | 90,000 | 76,496/8.99 | ↑↑ | Glycerol ester hydrolase |
| 5 | 15925634 | 1150 | 80,000 | 69,186/5.96 | ↓↓ | Hypothetical protein ORFID:SA2437 (similar to autolysin) |
| 7 | 15923709 | 942 | 70,000 | 74,353/9.04 | ↓ | Hypothetical protein ORFID:SA0674 |
| 9 | 225821 | 1130 | 60,000 | 57,939/5.48 | ↓↓ | Protein A |
| 13 | 15924153 | 1010 | 33,000 | 35,953/8.7 | ↓↓ | Alpha-hemolysin precursor |
| 14 | 15923949 | 759 | 33,000 | 35,289/8.67 | ↑↑ | Glycerophosphoryl diester phosphodiesterase |
| 15 | 15923085 | 387 | 32,000 | 37,064/7.71 | ND | 1-Phosphatidylinositol phosphodiesterase precursor |
| 16 | 119131 | 550 | 32,000 | 27,663/9.15 | ↓↓ | Epidermal cell differentiation inhibitor precursor (EDIN) |
| 17 | 21284219 | 283 | 30,000 | 24,188/6.11 | ↓↓ | Immunodominant antigen A |
| 18 | 15925289 | 551 | 30,000 | 29,366/8.96 | ↓↓ | Secretory antigen precursor SsaA homolog ORFID:SA2093 |
| 20 | 15925225 | 380 | 24,000 | 19,774/9.54 | ↑↑ | 50S ribosomal protein L6 |
| 25 | 15925235 | 206 | 14,000 | 12,827/9.92 | ↑↑ | 50S ribosomal protein L22 |
| 26 | 15926570 | 137 | 12,000 | 15,898/9.28 | +/− | Hypothetical protein (similar to cell surface protein Map-w) |
| 28 | 14029570 | 131 | 9,000 | 7,990/10.16 | ↑ | 50S ribosomal protein L27 |
| 29 | 15925085 | 192 | 32,000 | 24,063/5.53 | ND | Hypothetical protein (similar to SceD precursor ORFID:SA1898) |
| 31 | 15924246 | 424 | 29,000 | 29,133/5.44 | ↑↑ | 30S ribosomal protein S2 |
| 32 | 15927830 | 232 | 28,000 | 23,703/9.8 | ↑↑ | 50S ribosomal protein L3 |
| 33 | 3024540 | 199 | 16,000 | 14,922/9.04 | ↑↑ | 50S ribosomal protein L11 |
Z-score value from Mascot.
↑↑, Great accumulation; ↑, weak accumulation; ↓, weak inhibition; ↓↓, complete inhibition; ND, not determined; +/−, no effect.
At 90% MIC, linezolid enhanced the relative expression of lipases; these included triacylglycerol lipase and glycerol ester hydrolase (Fig. 4, proteins 3 and 4, and Table 3), glycerophosphoryl diester phosphodiesterase (Fig. 4, protein 14, and Table 3), and stress-induced proteins such as the DnaK (Fig. 4, protein 6, and Table 2); a hypothetical protein similar to SceD (Fig. 4, protein 29 and Table 2); and proteins involved in the protein synthesis, such as the ribosomal proteins (Fig. 4, proteins 19 to 25 and 27 to 28, and Table 2) and the translational-elongation factor EF-TU (Fig. 4, protein 10, and Table 2).
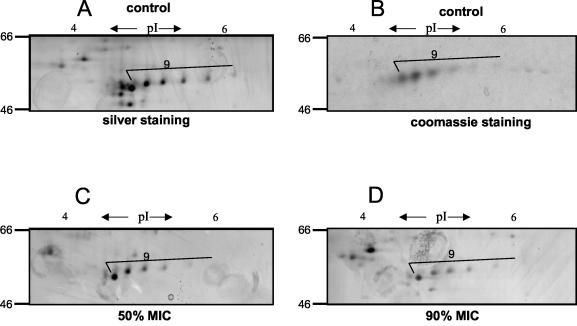
In contrast, linezolid reduced the expression of virulence factors or proteins such as alpha- and beta-hemolysin (Fig. 4, proteins 11 and 13, and Table 2), members of the autolysin family (e.g., autolysin, bifunctional autolysin, and the hypothetical protein similar to autolysin) (Fig. 4, proteins 1, 2, 5, and 12, and Table 2), EDIN (Fig. 4, protein 16, and Table 3), and secretory antigen precursor SsaA homolog (Fig. 4, proteins 18 and 30, and Table 2). A decrease of protein A was also detected (Fig. 4, protein 9; Table 1; and Fig. 3, lower panels [A to D]). Although protein A appeared as a single band in one-dimensional gels, multiple protein A species were resolved by two-dimensional gel analysis (Fig. 3, lower panels [A to D]). Notably, the abundance of some proteins remained unchanged (e.g., the hypothetical protein, similar to cell surface protein Map-w [Fig. 4, protein 26, and Table 2]).
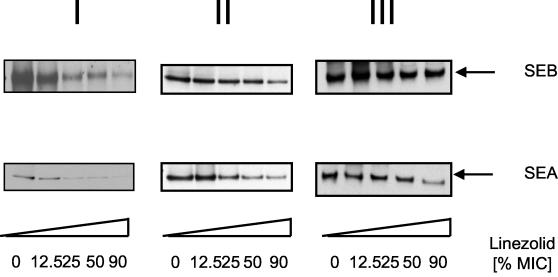
Due to their low concentration in the S. aureus supernatants, SEA and SEB could not be identified by SDS-PAGE and/or MALDI-TOF/MS (3). To visualize the effects of linezolid on SEA and SEB secretion, these virulence factors were analyzed by Western blotting. As shown in Fig. 5, linezolid reduced in a dose-dependent manner the secretion of virulence factors SEA and SEB.
FIG. 5.
Western blot analysis of exotoxins of S. aureus treated with linezolid. S. aureus ATCC 29213 was grown at 37°C and treated with linezolid at various growth phases. Panels I to III correspond to the conditions described in the legend to Fig. 1. Supernatants were subjected to SDS-PAGE. After transfer to nitrocellulose, proteins were stained specifically with the indicated antibodies against SEA and SEB. A horseradish peroxidase-conjugated goat anti-rabbit antibody was used as second reagent and visualized by using an enhanced chemiluminescence detection kit (Amersham-Pharmacia).
Linezolid reduces the proinflammatory activity of S. aureus supernatants.
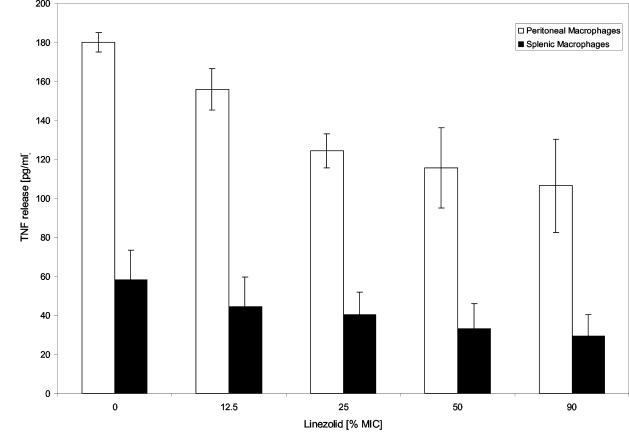
To elucidate the biological relevance of changes in the protein profiles induced by linezolid, supernatants were added to murine splenic macrophages or peritoneal macrophages. Linezolid dissolved in DMEM alone did not induce TNF production by isolated peritoneal macrophage cells (data not shown). The culture supernatants of S. aureus previously treated with grading subinhibitory concentrations of linezolid (OD540 of 0.05) elicited significantly lower amounts of TNF generated by either spleen or peritoneal macrophages (Fig. 6). Apparently, linezolid reduced the TNF-inducing activity in a dose-dependent manner.
FIG. 6.
TNF release by macrophages stimulated with supernatants of S. aureus. S. aureus strains were cultured in DMEM and treated with the indicated concentrations of linezolid as described in the text. Secreted proteins present in the supernatant were collected, followed by incubation for 20 h at 37°C with mouse splenic macrophages or peritoneal macrophages. The TNF levels were measured by ELISA. Linezolid itself (1× MIC and 2× MIC) did not induce TNF (data not shown).
DISCUSSION
The numerous virulence factors produced by S. aureus play an important role for the pathogenesis of infection. Therefore, the clinical performance of antibiotics used for the treatment of S. aureus infections not only depends on the respective bacteriostatic or bactericidal effects but also on the ability to prevent virulence factor release by dying or stressed bacteria. We report here that linezolid at subinhibitory concentrations potently inhibits the secretion of S. aureus virulence factors. Notably, the prevention of virulence production was also observed when other strains of S. aureus were investigated (data not shown). Our observations suggest that adverse effects might not be expected during linezolid treatment at any stage of infection.
The effects of antibiotics on virulence factor synthesis by S. aureus have been intensively studied in the past. As a paradigm of a protein synthesis inhibitor targeting the 50S rRNA, the clindamycin effect on extracellular proteins has been well documented. Clindamycin at concentrations of 12.5% MIC has been shown to decrease the expression of virulence factors, such as alpha- and delta-hemolysin, and coagulase (35). Clindamycin also blocked production of several of the individual exoprotein genes (e.g., spa, hla, and spr), suggesting that the primary effect must be differential inhibition of the synthesis of one or more regulatory proteins (19).
Indeed, many genes encoding virulence factors are coordinately regulated in response to a variety of intra- and extracellular signals. Octapaptide signaling molecules reaching a threshold at increased cell densities mediate a transcriptional switch from genes encoding surface-expressed proteins such as adhesins to genes encoding exoproteins (21). The best-studied regulatory loci are the accessory gene regulator agr and staphylococcal accessory regulator sarA loci (7, 8, 18). It is tempting to speculate that linezolid-induced inhibition of global regulators might result in the decreased virulence factor secretion observed in our study. However, the pattern of individual exoproteins reduced in the presence of linezolid is not consistent with the pattern to be expected after inactivation of a specific global regulator. For example, protein A is a prototypical surface protein anchored into the cell wall via glycine cross-links (36). As shown in that study and recently by Gemmell and Ford (13), linezolid reduces protein A expression. If the primary effect of linezolid was to decrease expression of the octapeptide, then protein A expression was expected to be increased rather than decreased. Similarily, lipase (glycerol ester hydrolase) belongs to the group of agr- and sarA-upregulated genes (10, 45) The finding that lipase was increased in the supernatants of linezolid-treated S. aureus also indicates that linezolid inhibits virulence factor expression in an agr- and sarA-independent manner.
Alternatively, the differential effects of linezolid on virulence factor secretion could be due to different half-lives of the respective protein and/or mRNA. Although 80% of all mRNAs had half-lives of between 3 and 8 min, a wide range of stabilities was reported for individual mRNAs of Escherichia coli (4, 37). It might be predicted that transcripts of housekeeping genes and proteins would have longer half-lives than transcripts and proteins synthesized in response to acute stimuli. A shorter half-life of virulence factors would explain why virulence factors were more sensitive to linezolid than other proteins. However, little is known about possible relationships between gene function and protein or mRNA half-life or abundance, and the apparent sensitivity of virulence factors for the action of linezolid requires further investigations.
Remarkably, using oligonucleotide microarrays for the study of global RNA degradation in wild-type E. coli, Selinger et al. reported that a single operon (tdcABCDEFG) was relatively rifampin insensitive (37). All seven open reading frames of this operon that encode a pathway for the transport and anaerobic degradation of l-threonine were significantly upregulated at 2.5 min after rifampin addition. Although the precise mechanism of rifampin-induced upregulation of the tdcABCDEFG operon needs to be determined, differential sensitivity to rifampin can be brought about by specific structural features of the RNAP holoenzyme (43).
The unexpected detection of the rifampin-insensitive tdc operon underlines the necessity to study the effects of an antibiotic in a comprehensive manner. The high-resolution two-dimensional protein gel electrophoresis is a well-established technique for visualizing a very large set of proteins secreted by a bacterial cell (22, 45). A combination of two-dimensional gelelectrophoresis and MALDI/TOF analysis or N-terminal Edman sequencing analysis had already led to the identification of 18 differentially regulated exoproteins by SarA and σB mutants of S. aureus (45). In addition to two-dimensional analysis, we also used one-dimensional protein gels combined with the identification of protein by MALDI-TOF/MS, which allowed the assignment of 31 exoproteins, including basic proteins that were not resolved by conventional two-dimensional analysis. Using this methodology of proteomics, we detected some specific effects of linezolid that were not observed with other protein synthesis-inhibiting antibiotics. For example, linezolid induced the secretion of ribosomal proteins into bacterial supernatants (Fig. 2 and Table 2). In contrast, both erythromycin (Fig. 2) and tetracycline (data not shown) do not induce the accumulation of ribosomal proteins in S. aureus culture supernatants. Linezolid prevents the formation of the formylmethionyl-tRNA:mRNA:30S subunit ternary complex. Thus, the linezolid-induced failure of ribosomal assembly might promote the leakage of small-sized individual ribosomal proteins.
Taken together, the results of our study provide comprehensive analysis of the effects of linezolid on virulence factor release by S. aureus. The methods presented here establish a framework for further investigation of the mode of action of antibiotics on a proteome-wide basis.
Acknowledgments
These investigations received support from a Maria Pesch grant from the University of Cologne.
REFERENCES
- 1.Arvidson, A., and K. Tegmark. 2001. Regulation of virulence determinants in Staphylococcus aureus. Int. J. Med. Microbiol. 291:159-170. [DOI] [PubMed] [Google Scholar]
- 2.Bassler, B. L. 1999. How bacteria talk to each other: regulation of gene expression by quorum sensing. Curr. Opin. Microbiol. 2:582-587. [DOI] [PubMed] [Google Scholar]
- 3.Bernardo, K., S. Fleer, N. Pakulat, O. Krut, F. Hünger, and M. Krönke. 2002. Identification of Staphylococcus aureus exotoxins by combined SDS gel electrophoresis and MALDI-TOF-MS. Proteomics 2:740-746. [DOI] [PubMed] [Google Scholar]
- 4.Bernstein, J. A., A. B. Khodursky, P. H. Lin, S. Lin-Chao, and S. N. Cohen. 2002. Global analysis of mRNA decay and abundance in Escherichia coli at single-gene resolution using two-color fluorescent DNA microarrays. Proc. Natl. Acad. Sci. USA 99:9697-9702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bjorklind, A., and A. Arvidson. 1980. Mutants of Staphylococcus aureus affected in the regulation of exoprotein synthesis. FEMS Microbiol. Lett. 7:203-206. [Google Scholar]
- 6.Booth, M. C., A. L. Cheung, K. L. Hatter, B. D. Jett, M. C. Callegan, and M. S Gilmore. 1997. Staphylococcal accessory regulator (sar) in conjunction with agr contributes to Staphylococcus aureus virulence in endophthalmitis. Infect. Immun. 65:1550-1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheung, A. L., K. J. Eberhardt, E. Cheung, M. R. Yeaman, P. M. Sullam, M. Ramos, and A. S. Bayer. 1994. Diminished virulence of sar−/agr− mutant of Staphylococcus aureus in the rabbit model of endocarditis. J. Clin. Investig. 94:1815-1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheung, A. L., M. Koomey, C. A. Butler, J. Projan, and V. A. Fischetti. 1992. Regulation of exoprotein expression in Staphylococcus aureus by a locus (sar) distinct from agr. Proc. Natl. Acad. Sci. USA 89:6462-6466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Czop, J. K., and M. S. Bergdoll. 1974. Staphylococcal enterotoxin synthesis during the exponential, transitional, and stationary growth phases. Infect. Immun. 9:229-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dunman, P. M., E. Murphy, S. Haney, D. Palacios, G. Tucker-Kellogg, S. Wu, E. L. Brown, R. J. Zagursky, D. Shlaes, and S. J. Projan. 2001. Transcription profiling-based identification of Staphylococcus aureus genes regulated by the agr and/or sarA loci. J. Bacteriol. 183:7341-7353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ford, C. W., J. C. Hamel, D. M. Wilson, J. K. Moerman, D. Stapert, R. J. Yancey, Jr., D. K. Hutchinson, M. R. Barbachyn, and S. J. Brickner. 1996. In vivo activities of U-100592 and U-100766, novel oxazolidinone antimicrobial agents against experimental bacterial infections. Antimicrob. Agents Chemother. 40:1508-1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gemmell, C. G. 1991. Antibiotics and expression of microbial virulence factors: implications for host defense. J. Chemother. 3(Suppl. 1):105-111. [PubMed] [Google Scholar]
- 13.Gemmell, C. G., and C. W. Ford. 2002. Virulence factor expression by gram-positive cocci exposed to subinhibitory concentrations of linezolid. J. Antimicrob. Chemother. 50:665-672. [DOI] [PubMed] [Google Scholar]
- 14.Gillaspy, A. F., S. G. Hickmon, R. A. Skinner, J. R. Thomas, C. L. Nelson, and M. S. Smeltzer. 1995. Role of the accessory gene regulator (agr) in pathogenesis of staphylococcal osteomyelitis. Infect. Immun. 63:3373-3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Görg, A., W. Postel, and S. Günther. 1988. The current state of two-dimensionalelectrophoresis with immobilized pH gradients. Electrophoresis 9:531-546. [DOI] [PubMed] [Google Scholar]
- 16.Hacker, J., M. Ott, and H. Hof. 1993. Effects of low, subinhibitory concentrations of antibiotics on expression of virulence gene cluster of pathogenic Escherichia coli by using a wild-type gene fusion. Int. J. Antimicrob. Agents 2:263-270. [DOI] [PubMed] [Google Scholar]
- 17.Hallander, H. O., G. Laurell, and G. Lofstrom. 1966. Stimulation of staphylococcal haemolysin production by low concentrations of penicillin. Acta Pathol. Microbiol. Scand. 68:142-148. [DOI] [PubMed] [Google Scholar]
- 18.Heinrichs J. H., M. G. Bayer, and A. L. Cheung. 1996. Characterization of the sar locus and its interaction with agr in Staphylococcus aureus. J. Bacteriol. 178:418-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Herbert, S., P. Barry, and R. P. Novick. 2001. Subinhibitory clindamycin differentially inhibits transcription of exoprotein genes in Staphylococcus aureus. Infect. Immun. 69:2996-3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jenö, P., T. Mini, S. Moes, E. Hintermann, and M. Horst. 1995. Internal sequences from proteins digested in polyacrylamide gels. Anal. Biochem. 224:75-82. [DOI] [PubMed] [Google Scholar]
- 21.Ji, G., R. C. Beavis, and R. P. Novick. 1995. Cell density control of staphylococcal virulence mediated by an octapeptide pheromone. Proc. Natl. Acad. Sci. USA 92:12055-12059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jungblut, P. R., U. E. Schaible, H.-J. Mollenkopf, U. Zimny-Arndt, B. Raupach, J. Mattow, P. Halada, S. Lamer, K. Hagens, and S. H. E. Kaufmann. 1999. Comparative proteome analysis of Mycobacterium tuberculosis and Mycobacterium bovis BCG strains: toward functional genomics of microbial pathogens. Mol. Microbiol. 33:1103-1117. [DOI] [PubMed] [Google Scholar]
- 23.Kuroda, M., T. Ohta, I. Uchiyama, T. Baba, H. Yuzawa, I. Kobayashi, L. Cui, A. Oguchi, K. Aoki, Y. Nagai, J. Lian, T. Ito, M. Kanamori, H. Matsumaru, A. Maruyama, H. Murakami, Hosoyama, A., Y. Mizutani-Ui, N. K. Takahashi, T. Sawano, R. Inoue, C. Kaito, K. Sekimizu, H. Hirakawa, S. Kuhara, S. Goto, J. Yabuzaki, M. Kanehisa, A. Yamashita, K. Oshima, K. Furuya, C. Yoshino, T. Shiba, M. Hattori, N. Ogasawara, H. Hayashi, and K. Hiramatsu. 2001. Whole genome sequencing of methicillin-resistant Staphylococcus aureus. Lancet 357:1225-1240. [DOI] [PubMed] [Google Scholar]
- 24.Lagrou, K., W. E. Peetermans, M. Jorissen, J. Verhaegen, J. Van Damme, and J. Van Eldere. 2000. Subinhibitory concentrations of erythromycin reduce pneumococcal adherence to respiratory epithelial cells in vitro. J. Antimicrob. Chemother. 46:717-723. [DOI] [PubMed] [Google Scholar]
- 25.Lowy, F. D. 1998. Staphylococcus aureus infections. N. Engl. J. Med. 339:520-532. [DOI] [PubMed] [Google Scholar]
- 26.Miller, M. B., and B. L. Bassler. 2001. Quorum sensing in bacteria. Annu. Rev. Microbiol. 55:165-199. [DOI] [PubMed] [Google Scholar]
- 27.Munckhof, W. J., C. Giles, and J. D. Turnidge. 2001. Post-antibiotic growth suppression of linezolid against gram-positive bacteria. J. Antimicrob. Chemother. 47:879-883. [DOI] [PubMed] [Google Scholar]
- 28.Novick, R. P. 2003. Autoinduction and signal transduction in the regulation of staphylococcal virulence. Mol. Microbiol. 48:1429-1449. [DOI] [PubMed] [Google Scholar]
- 29.O'Farrell, P. H. 1975. High-resolution two-dimensional electrophoresis of proteins. J. Biol. Chem. 250:4007-4021. [PMC free article] [PubMed] [Google Scholar]
- 30.Ohlsen, K., W. Ziebuhr, K. P. Koller, W. Hell, A. Wichelhaus, and J. Hacker. 1998. Effects of subinhibitory concentrations of antibiotics on alpha-toxin (hla) gene expression of methicillin-sensitive and methicillin-resistant Staphylococcus aureus isolates. Antimicrob. Agents Chemother. 42:2817-2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peng, H.-L., R. P. Novick, B. Kreiswirth, J. Kornblum, and P. Schlievert. 1988. Cloning, characterization, and sequencing of an accessory gene regulator (agr) in Staphylococcus aureus. J. Bacteriol. 170:4365-4372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rescei, P., B. Kreiswirth, M. O'Reilly, P. Schlievert, A. Gruss, and R. P. Novick. 1986. Regulation of exoprotein gene expression in Staphylococcus aureus by agr. Mol. Gen. Genet. 202:58-61. [DOI] [PubMed] [Google Scholar]
- 33.Rybak, M. J., D. M. Cappelletty, T. Moldovan, J. R. Aeschlimann, and G. W. Kaatz. 1998. Comparative in vitro activities and post-antibiotic effects of the oxazolidinone compounds eperozolid and linezolid versus vancomycin against Staphylococcus aureus, coagulase-negative staphylococci, Enterococcus faecalis, and E. faecium. Antimicrob. Agents Chemother. 42:721-724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schägger, H., and G. V. Jagow. 1987. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166:368-379. [DOI] [PubMed] [Google Scholar]
- 35.Schlievert, P. M., and J. A. Kelly. 1984. Clindamycin-induced suppression of toxic shock syndrome-associated exotoxin production. J. Infect. Dis. 149:471. [DOI] [PubMed] [Google Scholar]
- 36.Schneewind, O., A. Fowler, and K. F. Faull. 1995. Structure of the cell wall anchor of surface proteins in Staphylococcus aureus. Science 268:103-106. [DOI] [PubMed] [Google Scholar]
- 37.Selinger, D. W., R. M. Saxena, K. J. Cheung, G. M. Church, and C. Rosenow. 2003. Global RNA half-life analysis in Escherichia coli reveals positional patterns of transcript degradation. Genome Res. 13:216-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shevchenko, A., M. Wilm, O. Vorm, and M. Mann. 1996. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 68:850-858. [DOI] [PubMed] [Google Scholar]
- 39.Shibl, A. M. 1983. Effect of antibiotics on production of enzymes and toxins by microorganisms. Rev. Infect. Dis. 5:865-875. [DOI] [PubMed] [Google Scholar]
- 40.Sofer, D., N. Gilboa-Garber, A. Belz, and N. C. Garber. 1999. Subinhibitory erythromycin represses production of Pseudomonas aeruginosa lectins, autoinducer, and virulence factors. Chemotherapy 45:335-341. [DOI] [PubMed] [Google Scholar]
- 41.Towbin, H., T. Staehelin, and J. Gordon. 1979. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. USA 76:4350-4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vandenesch, F., J. Kornblum, and R. P. Novick. 1991. A temporal signal, independent of agr, is required for hla but not spa transcription in Staphylococcus aureus. J. Bacteriol. 173:6313-6320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wegrzyn, A., A. Szalewska-Palasz, A. Blaszczak, K. Liberek, and G. Wegrzyn. 1998. Differential inhibition of transcription from sigma70- and sigma32-dependent promoters by rifampicin. FEBS Lett. 440:172-174. [DOI] [PubMed] [Google Scholar]
- 44.Yoh, M., E. K. Frimpong, S. P. Voravuthikunchai, and T. Honda. 1999. Effect of subinhibitory concentrations of antimicrobial agents (quinolones and macrolide) on the production of verotoxin by enterohemorrhagic Escherichia coli O157:H7. Can. J. Microbiol. 45:732-739. [PubMed] [Google Scholar]
- 45.Ziebandt, A. K., H. Weber, J. Rudolph, R. Schmid, D. Hoper, S. Engelmann, and M. Hecker. 2001. Extracellular proteins of Staphylococcus aureus and the role of SarA and sigma B. Proteomics 1:480-493. [DOI] [PubMed] [Google Scholar]