Abstract
Reactive oxygen species (ROS) regulate growth factor receptor signalling at least in part by inhibiting oxidation-sensitive phosphatases. An emerging concept is that ROS act locally to affect signal transduction in different subcellular compartments and that ROS levels are regulated by antioxidant proteins at the same local level. Here, we show that the ER-resident antioxidant peroxiredoxin 4 (Prdx4) interacts with the cytoplasmic domain of the granulocyte colony-stimulating factor receptor (G-CSFR). This interaction occurs when the activated G-CSFR resides in early endosomes. Prdx4 inhibits G-CSF-induced signalling and proliferation in myeloid progenitors, depending on its redox-active cysteine core. Protein tyrosine phosphatase 1b (Ptp1b) appears to be a major downstream effector controlling these responses. Conversely, Ptp1b might keep Prdx4 active by reducing its phosphorylation. These findings unveil a new signal transduction regulatory circuitry involving redox-controlled processes in the ER and activated cytokine receptors in endosomes.
Key words: Granulocyte colony-stimulating factor, Hematopoiesis, Peroxiredoxin, Protein tyrosine phosphatase 1b, Redox signaling
Introduction
Reactive oxygen species (ROS) are generated by nicotinamide adenine dinucleotide phosphate oxidase (Nox) complexes. In phagocytes the Nox system mainly serves to produce high levels of H2O2 for the so-called oxidative burst, a major weapon in host defence against bacteria. In non-phagocytic cells, H2O2 has long been considered as an unwanted by-product of cell metabolism, potentially hazardous because of the damaging effects on proteins, lipids, carbohydrates and nucleic acids (Rhee, 2006; Rhee et al., 2005b). There is increasing evidence that moderate levels of ROS are pivotal for many cellular processes, including the control of cell proliferation, survival and differentiation. For example, H2O2 inactivates enzymes, in particular protein tyrosine and lipid phosphatases, involved in growth factor signalling (Rhee, 2006). As a consequence, the signal magnitude and duration from cell surface receptors, such as platelet-derived growth factor receptor (PDGFR), interleukin 3 receptor (IL-3R), granulocyte-macrophage colony-stimulating factor receptor (GM-CSFR) and G-CSFR is increased (Iiyama et al., 2006; Sundaresan et al., 1995; Zhu et al., 2006). An important new insight that emerges is that ROS are produced in specific subcellular compartments and act locally to modulate signalling responses in different organelles (Rhee, 2006; Terada, 2006).
In view of the local actions of ROS, an attractive hypothesis is that their levels are controlled by nearby antioxidants, but evidence supporting this concept remains limited. Proteins of the peroxiredoxin (Prdx) family are major candidates for such local antioxidant activities (Rhee et al., 2005a). Prdx proteins contain a core of usually two cysteine residues responsible for their redox activity (Rhee et al., 2005a). Prdx1 and Prdx2 were found to associate with PDGFR and to modulate signalling by controlling H2O2 levels at the plasma membrane (Choi et al., 2005; Woo et al., 2010). Importantly, PDGFR-induced tyrosine phosphorylation of Prdx1 temporarily reduced its antioxidant activity, thereby allowing a transient accumulation of H2O2 and inhibition of phosphatase activity in the vicinity of the plasma membrane (Woo et al., 2010). Among the six mammalian Prdx proteins (Rhee et al., 2005a), Prdx1, Prdx 2, and Prdx 6 are cytosolic, Prdx3 mitochondrial and Prdx5 peroxisomal. The exact localization of Prdx4 has been somewhat ambiguous, but recent studies suggest that Prdx4 resides mainly in the endoplasmic reticulum (ER) (Giguere et al., 2007; Sasagawa et al., 2001; Tavender et al., 2008).
Granulocyte colony-stimulating factor (G-CSF) is the major hematopoietic growth factor involved in the production of neutrophils (Demetri and Griffin, 1991; Lieschke et al., 1994; Touw and van de Geijn, 2007). G-CSF induces the proliferation, survival and neutrophilic differentiation of myeloid progenitor cells, which are cellular responses that require a balanced activation and subsequent attenuation of signalling pathways linked to the G-CSFR (Touw and van de Geijn, 2007), a member of the cytokine receptor class I superfamily (Wells and de Vos, 1996). Signal attenuation of the G-CSFR is severely compromised by mutations causing truncations in the cytoplasmic domain of the G-CSFR, as observed in severe congenital neutropenia (SCN) patients showing disease progression to acute myeloid leukaemia (AML) (Dong et al., 1995; Freedman and Alter, 2002; Germeshausen, 2007). A major mechanism implicated in the perturbed signalling functions of these truncated G-CSFR mutants is the loss of appropriate receptor endocytosis and lysosomal routing. These processes are controlled by a dileucine-based internalization motif (Hunter and Avalos, 1999; Ward et al., 1999) and by receptor ubiquitylation involving the suppressor of cytokine signalling protein SOCS3 (Aarts et al., 2004; Hermans et al., 2003; Hortner et al., 2002; Irandoust et al., 2007; Wölfler et al., 2009).
Here, we investigated whether Prdx proteins control signalling from the G-CSFR and in which subcellular compartment this takes place. In contrast to PDGFR, G-CSFR does not bind Prdx1 and Prdx2, but exclusively interacts with Prdx4. This interaction takes place during retrograde routing, when the activated G-CSFR resides in early endosomes. Prdx4 attenuates G-CSF-induced cell proliferation and STAT (signal transducers and activators of transcription) proteins, and this depends on the integrity of its redox-active cysteine core. The tyrosine phosphatase Ptp1b, known to reside in the ER (Eden et al., 2010; Haj et al., 2002), appears the major target for Prdx4-controlled modulation of G-CSF responses. In turn, Ptp1b reduces Prdx4 phosphorylation, which by analogy to Prdx1 might keep Prdx4 in an active state. These findings identify the ER as an important signalling organelle controlling G-CSF responses of myeloid progenitors and provide insight into the complex interplay between redox-controlled processes and receptor signalling at the boundary between the ER and retrograde endocytotic vesicles.
Results
Prdx4 interacts with the G-CSFR
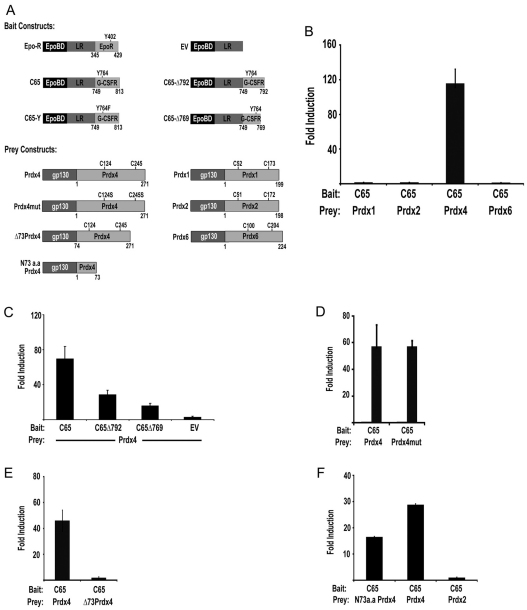
In an earlier yeast two-hybrid screen, we identified peroxiredoxins as putative G-CSFR-interacting proteins. To assess whether these interactions occur in mammalian cells, we used the mammalian protein–protein interaction trap (MAPPIT) assay (Eyckerman et al., 2002). Bait and prey constructs are shown in Fig. 1A. Prdx3 and Prdx5 were excluded from this analysis because of their mitochondrial and peroxisomal localizations, respectively. After a series of standard positive and negative controls to assure specificity of the system (supplementary material Fig. S1), we performed experiments with the Prdx preys and found that Prdx4, but not Prdx1, Prdx2, or Prdx6, interacted with the G-CSFR C-terminus (Fig. 1B). Using additional bait constructs, we identified the distal region spanning amino acids 792–813 as the major PRDX4-binding site (Fig. 1C). This region does not show sequence homology with other cytokine receptors. To assess whether the integrity of the cysteine core of Prdx4 is required for its interaction with G-CSFR, we generated Prdx4 prey constructs in which both cysteines in the active site (Cys124 and Cys245) were changed into serines (Prdx4mut). Prdx4 and Prdx4mut prey constructs showed comparable binding to the G-CSFR baits, implying that the redox status of Prdx4 does not affect this interaction (Fig. 1D). Prdx4 differs from the other family members in that its N-terminus has a 73 amino acid extension, including a predicted signal peptide of 37 amino acids. Deletion of the N-terminal region abolished MAPPIT activity, suggesting that this region is predominantly responsible for Prdx4 binding to the G-CSFR (Fig. 1E). Conversely, a prey construct consisting of only the N-terminal 73 amino acids (N73a.a. Prdx4) interacted with the G-CSFR bait, confirming the importance of the Prdx4 N-terminus for G-CSFR binding (Fig. 1F). In immunoprecipitation experiments, G-CSFR co-precipitated with endogenous Prdx4 from lysates of HEK293T cells ectopically expressing wild-type G-CSFR, confirming the results from the MAPPIT assay. However, under these conditions basal Prdx4 binding to G-CSFR was seen, indicating that the physical interaction between G-CSFR and Prdx4 proteins per se does not depend on receptor activation (supplementary material Fig. S2).
Fig. 1.
MAPPIT assay of interactions between Prdx and G-CSFR. (A) Bait and prey MAPPIT constructs. Epo-R, erythropoietin receptor; LR, leptin receptor. (B) MAPPIT with G-CSFR-C65 (amino acids 749–813) bait and Prdx1, Prdx2, Prdx4 and Prdx6 prey constructs showing specific interaction with Prdx4. (C) Mapping of domains of the G-CSFR involved in Prdx4 binding. Mutants C65Δ792 and C65Δ769 represent C-terminal deletions of 21 and 44 amino acids of C-CSFR-C65 bait, respectively; EV, empty bait vector control. (D) MAPPIT showing that cysteines in the active sites of Prdx4 (Cys124 and Cys245) are not involved in interaction of Prdx4 with G-CSFR-C65. (E) Loss of Prdx4 binding to G-CSFR upon deletion of the specific extended N-terminus of Prdx4 (Δ73Prdx4). (F) Binding of the N-terminal region of Prdx4 (N73a.a Prdx4) to C65-G-CSFR. Full-length Prdx4- and Prdx2-containing preys were included as positive and negative controls, respectively. MAPPIT experiments were performed in HEK293T cells as detailed in Materials and Methods.
Colocalization of endocytosed G-CSFR with Prdx4 residing in the ER/ERGIC
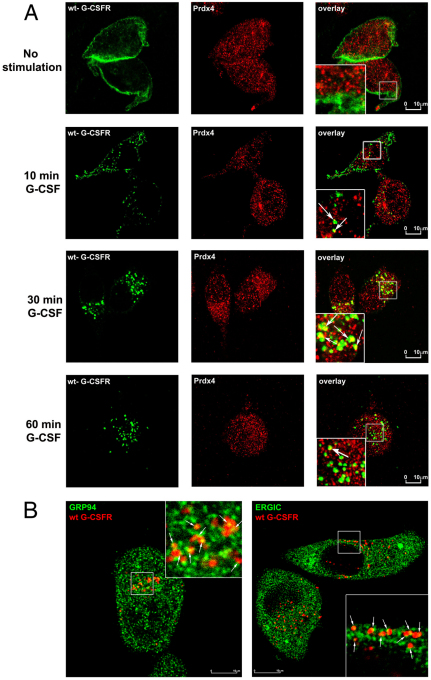
We subsequently used confocal microscopy to study where Prdx4 and G-CSFR interact in intact cells. Prdx4 did not colocalize with G-CSFR at the cell surface. Thirty minutes after G-CSF treatment, when G-CSFR resided in early endosome antigen 1 (EEA1)-positive early endosomes, colocalization with Prdx4 was maximal and declined after 1 hour, when the G-CSFR was present in late endosomes and lysosomes (Fig. 2A and supplementary material Fig. S3). By contrast, colocalization of the lysosomal routing defective G-CSFR mutant K5R persisted at 60 minutes after G-CSF stimulation, confirming that Prdx4 interacts with G-CSFR localized in early endosomes (Irandoust et al., 2007; Wölfler et al., 2009) (supplementary material Fig. S4a). Quantification of these data is shown in supplementary material Fig. S4b. As expected based on the MAPPIT experiments, no significant colocalization of Prdx2 and G-CSFR was detectable, neither at the plasma membrane nor in endocytotic vesicles (supplementary material Fig. S5). Because Prdx4 was detected mainly in the ER and ER–Golgi intermediate compartment (ERGIC), with only a minor fraction in the Golgi (supplementary material Fig. S6), these findings imply that the interaction between G-CSFR and Prdx4 takes place when the endocytosed G-CSFR complexes are in proximity of the ER or ERGIC. Supporting this, G-CSFR also co-stained with the ER marker Grp94 and with the ERGIC marker ERGIC-53 after ligand-induced internalization (Fig. 2B). In situ proximity ligation analysis (in situ-PLA) is a recently developed tool to demonstrate the close proximity of proteins in situ. Using this technique, we could confirm the results obtained by confocal laser scanning microscopy (CLSM) showing colocalization of G-CSFR, Ptp1b and Prdx4 (supplementary material Figs S7 and S8).
Fig. 2.
G-CSFR and Prdx4 interaction and colocalization. (A) HeLa cells ectopically expressing wild-type G-CSFR were deprived of growth factor for 4 hours. Surface membrane G-CSFRs were labelled with anti-G-CSFR antibody prior to stimulation with G-CSF for different times. Cells were permeabilized, fixed and stained with anti-Prdx4 antibody, followed by secondary anti-rabbit Cy3 and anti-mouse Cy5 antibodies, and then analysed by CLSM. (B) Colocalization of endocytosed G-CSFR (arrows) with Grp94-stained ER (left) and with ERGIC-53-stained ERGIC (right) 30 minutes after ligand stimulation. Experimental conditions were the same as for A.
Prdx4 inhibition of G-CSF-induced proliferation of myeloid progenitor cells requires the integrity of its cysteine core
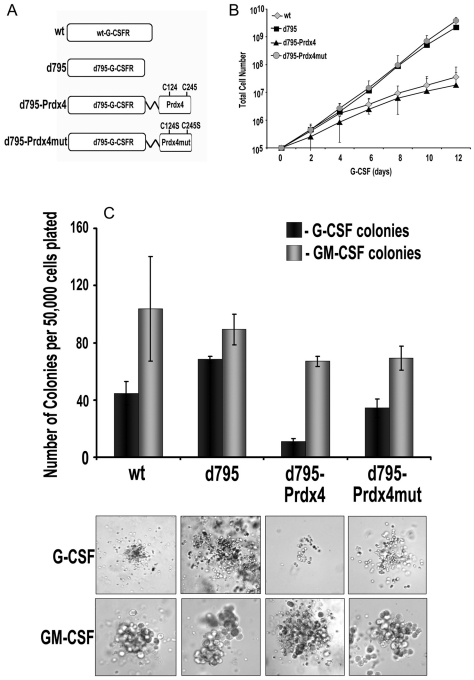
To directly study the functional implications of Prdx4 binding on G-CSFR signalling and to determine the role of the redox-active cysteines, we fused the catalytic domain of Prdx4 and the mutant domain missing the crucial cysteine residues directly to G-CSFR-Δ795 (to give G-CSFR-Δ795-Prdx4 and G-CSFR-Δ795-Prdx4mut, respectively). G-CSFR-Δ795 was chosen because it lacks a major C-terminal domain that is responsible for endogenous Prdx4 binding, as identified by MAPPIT (Fig. 3A). Although a drawback of this approach is that the interaction with Prdx4 is constitutive, as opposed to temporal and depending on endocytotic routing, the routing of the G-CSFR towards the ER and ERGIC was not affected by the Prdx4 fusion (supplementary material Fig. S9A). Hence, Prdx4 is still able to function in this setting at the ER–early endosome interface. We first transduced 32D cells with these constructs and selected clones with comparable expression of wild-type G-CSFR (32D/wt), G-CSFR-Δ795 (32D/Δ795), G-CSFR-Δ795-Prdx4 (32D/Δ795-Prdx4) and G-CSFR-Δ795-Prdx4mut (32D/Δ795-Prdx4mut) at the plasma membrane (supplementary material Fig. S9B), allowing comparisons of their signalling abilities at equal receptor densities. Relative to 32D/wt cells, the 32D/Δ795 clones showed increased proliferation in response to G-CSF, whereas the proliferation rate of 32D/Δ795-Prdx4 clones was similar to that of 32D/wt clones (Fig. 3B). By contrast, 32D/Δ795-Prdx4mut clones showed the same elevated proliferation rate as and 32D/Δ795 clones, indicating that the redox-active thiol group of Prdx4 confers growth inhibition (Fig. 3B). No differences in proliferation rates of these clones were seen in IL-3-containing cultures (not shown). To extend these observations to primary myeloid progenitors, we transduced bone marrow cells from Csf3r−/− mice with the above-mentioned G-CSFR constructs using the BABE retroviral vector (conferring puromycin resistance), and cultured these cells in colony cultures supplemented with G-CSF or GM-CSF and puromycin. GM-CSF-induced colony formation did not differ significantly in the presence of puromycin (Fig. 3C), which is indicative of the comparable transduction efficiencies of the G-CSFR–puromycin expression constructs. In line with the findings in 32D cells, G-CSFR-Δ795-expressing bone marrow cells produced higher numbers and greater-sized colonies, whereas G-CSFR-Δ795-Prdx4-expressing bone marrow cells generated fewer and smaller colonies in response to G-CSF compared to wild-type G-CSFR-expressing bone marrow cells (Fig. 3C). As in 32D cells, this effect of Prdx4 depended on the integrity of the redox active cysteines.
Fig. 3.
Prdx4 attenuates G-CSF-induced proliferation in myeloid progenitors depending on its active cysteine core. (A) G-CSFR–Prdx4 fusion constructs. For details see Materials and Methods. (B) Proliferation of 32D cells expressing constructs shown in A. Cells were cultured in 10 ng/ml G-CSF. Data represent the means ± s.d. of five independent clones for each transfectant. (C) Colony assay of csf3r−/− bone marrow cells transduced with the constructs shown in A. Transduced bone marrow cells (50,000) were plated in triplicate in medium containing either G-CSF or GM-CSF and puromycin as a selection marker. Colonies were counted on day 7 of culture (upper panel). Data are expressed as the mean of triplicate cultures ± s.d. Lower panel: representative photomicrographs showing differences in colony size.
Prdx4 attenuates G-CSF-induced STAT3 activation
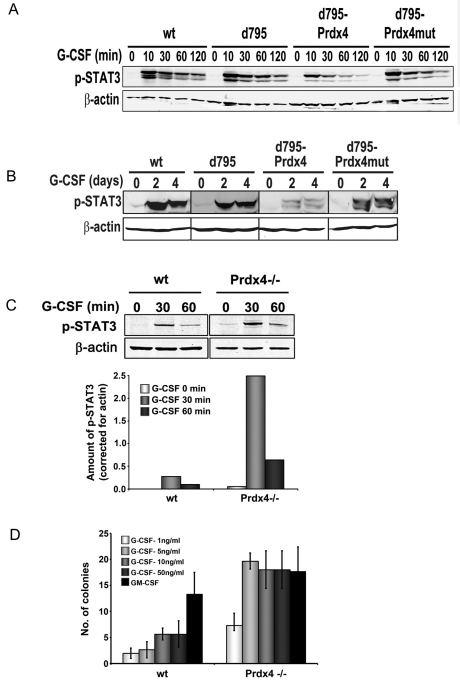
To identify the signalling pathways that are modulated by Prdx4, lysates of 32D clones expressing the different G-CSFR forms were subjected to western blotting with phospho-specific antibodies to detect activation of STAT3, STAT5 and Akt. In experiments in which cells were first deprived of growth factor and serum and then stimulated with G-CSF for up to 120 minutes, fusion of active Prdx4, but not the inactive mutant, inhibited STAT3 phosphorylation (Fig. 4A). In long-term cultures, in which cells were switched from IL-3- to G-CSF-containing culture medium and lysates made every 2 days, phospho-STAT3 levels were again markedly reduced in 32D/Δ795-Prdx4 clones compared to 32D/Δ795-Prdx4mut clones (Fig. 4B). By contrast, no significant differences were seen in STAT5 and Akt phosphorylation (not shown). A possible explanation for this difference is that G-CSF-induced STAT3 activation requires recruitment of STAT3 to phosphotyrosines in the G-CSFR membrane distal cytoplasmic region, whereas STAT5 and Akt activation occurs through the membrane proximal G-CSFR region without the involvement of receptor tyrosines (de Koning et al., 1996; Dong and Larner, 2000; Dong et al., 1998). Moreover, Akt phosphorylation is already downregulated early in G-CSFR endocytosis, i.e. before endosome–ER contacts take place and Prdx4 modulates G-CSFR signalling (Irandoust et al., 2007).
Fig. 4.
Prdx4 and Ptp1b attenuate G-CSFR signalling. (A) 32D cells expressing G-CSFR-Prdx4 fusion and control constructs (see Fig. 3A) were deprived of growth factor for 4 hours and then stimulated with 100 ng/ml G-CSF for the indicated times. β-actin was used as loading control. (B) Western blot analysis of phospho-STAT3 levels in the same 32D cell transfectants at 0, 2 and 4 days of culture with 100 ng/ml G-CSF. (C) Western blot analysis of phospho-STAT3 in wild-type and Prdx4−/− MEFs stably expressing G-CSFR after serum deprivation followed by G-CSF stimulation for indicated times. Histograms show quantification of bands using Odyssey application software 3.0 (Li-Cor). (D) Colony assay of Prdx4−/− and wild-type littermate control bone marrow cells. Cells (50,000) were plated in triplicate in Methocult containing either G-CSF or GM-CSF and puromycin as a selection marker. Colonies were counted on day 7 of culture. Data are expressed as the mean of triplicate cultures ± s.d.
Increased G-CSF-induced STAT3 activation and myeloid colony formation in Prdx4−/− cells
Having shown that enforced interaction of Prdx4 with G-CSFR attenuates G-CSF signalling in myeloid progenitors, we investigated how depletion of Prdx4 affects G-CSF signalling. To this end, we transduced Prdx4-deficient and Prdx4-proficient mouse embryonic fibroblasts (MEFs) with the G-CSFR and assessed phosphorylation of STAT3, STAT5 and Akt. As illustrated in supplementary material Fig. S10, Prdx4−/− cells showed a significantly increased accumulation of ROS relative to MEFs expressing Prdx4, supporting the major role of ER-resident Prdx4 in controlling intracellular ROS levels. Phospho-STAT3 levels were significantly increased in Prdx4−/− cells relative to wild-type controls (Fig. 4C). Again, phospho-Akt levels did not significantly differ between Prdx4-deficient and Prdx4-proficient cells, whereas phospho-STAT5 was below detection level in these cells (data not shown). To assess whether Prdx4 deficiency affects G-CSF signalling in primary hematopoietic cells, we performed colony cultures with increasing concentrations of G-CSF. Prdx4−/− bone marrow cells yielded significantly higher numbers of colonies relative to wild-type littermate control cells at different concentrations of G-CSF (Fig. 4D). By contrast, GM-CSF-induced colony growth did not differ between Prdx4-deficient and wild-type bone marrow cells, showing that G-CSF, but not GM-CSF, signalling is controlled by Prdx4.
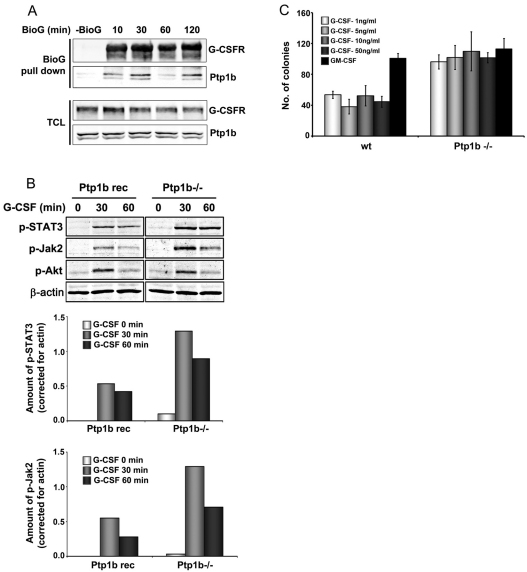
Ptp1b inhibits G-CSF-induced signalling and proliferation of myeloid progenitors
Ptp1b resides at the ER (Frangioni et al., 1992; Haj et al., 2002), is highly sensitive to oxidation (Groen et al., 2005) and is therefore a likely downstream effector of Prdx4-modulated signalling. We confirmed the colocalization of Ptp1b and Prdx4 in the ER and ERGIC compartments in primary bone marrow cells (supplementary material Fig. S11). Biotinylated G-CSF pull-down showed that Ptp1b interacts with G-CSFR (Fig. 5A). Western blot analysis in Ptp1b−/− and Ptp1b reconstituted MEFs transduced with G-CSFR showed significantly elevated G-CSF-induced Jak2 and STAT3 phosphorylation in the Ptp1b−/−cells (Fig. 5B). Similar to Prdx4-deficient cells, phospho-Akt levels were unaffected by Ptp1b depletion (Fig. 5B). Colony cultures with Ptp1b−/− bone marrow cells also yielded results comparable to those obtained with Prdx4-deficient cells, showing elevated numbers and greater-sized colonies in response to G-CSF, with GM-CSF-induced colony formation unchanged relative to wild-type Ptp1b controls (Fig. 5C).
Fig. 5.
Ptp1b interacts with G-CSFR and attenuates signalling. (A) Ptp1b immunoprecipitation from HEK293T cells transfected with lysine-less pBABE-K5R-G-CSFR and pJ3H-Ptp1b-HA constructs. Cells were deprived of growth factor for 4 hours, followed by stimulation with biotinylated G-CSF (BioG) for the indicated times. Precipitates were collected on streptavidin-coated beads. Blots were stained for G-CSFR and Ptp1b; TCL total cell lysate. (B) Western blot analysis of phospho-STAT3, phospho-Jak2 and phospho-Akt in Ptp1b−/− and reconstituted MEFs stably expressing G-CSFR (Ptp1b rec); stimulation conditions as for Fig. 4C. β-actin served as loading control in all experiments. Ptp1b−/− and reconstituted MEFs expressed comparable G-CSFR expression levels, as determined by flow cytometry. Histograms show quantifications of phospho-STAT3 and phospho-JAK2 (Odyssey 3.0). (C) Colony assays of Ptp1b−/− and wild-type littermate control bone marrow cells. Culture conditions were similar to those described for A.
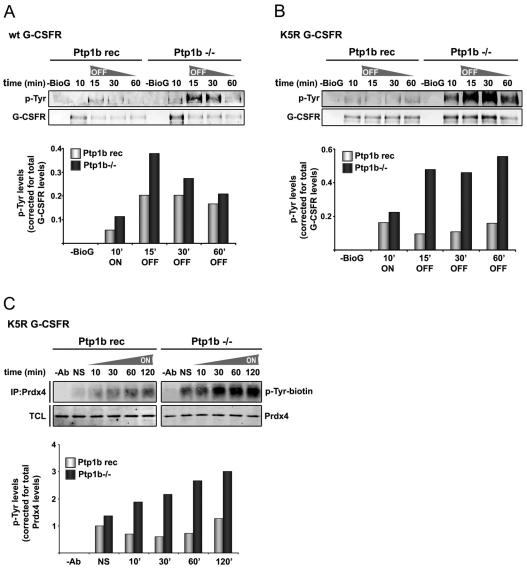
Ptp1b reduces phosphorylation of both G-CSFR and Prdx4
We then asked whether Ptp1b controls ligand-induced G-CSFR phosphorylation. For this, we performed off-rate experiments in which cells were stimulated for 10 minutes with G-CSF, then washed and further cultured without growth factor. Tyrosine phosphorylation of G-CSFR was elevated in Ptp1b−/− MEFs compared to the reconstituted control cells (Fig. 6A). Supporting the notion that Ptp1b reduces G-CSFR phosphorylation in early endosomes, this difference was more pronounced in cells expressing the G-CSFR mutant K5R (Fig. 6B). Notably, because the K5R mutant is hampered in its routing to late endosomes and lysosomes, increased phosphorylation levels of G-CSFR persisted at 30 minutes and 60 minutes after G-CSF washout. Finally, because phosphorylation of Prdx1 has been reported to decrease its activity (Woo et al., 2010), we investigated whether Ptp1b controls the phosphorylation status of Prdx4. Prdx4 was clearly hyperphosphorylated in Ptp1b-deficient MEFs compared to reconstituted cells (Fig. 6C). This was also evident in the absence of G-CSF stimulation (Fig. 6C), indicating that receptor signalling is not required for Ptp1b-controlled phosphorylation of Prdx4.
Fig. 6.
Increased tyrosine phosphorylation of G-CSFR and Prdx4 in Ptp1b-deficient cells. (A) Phosphotyrosine analysis of wild-type G-CSFR after biotinylated G-CSF (BioG) pull-down in Ptp1b−/− or Ptp1b reconstituted MEFs expressing wild-type G-CSFR (Ptp1b rec). Ten minutes after addition of biotinylated G-CSF, cells were washed and further cultured without growth factor. Histograms show quantifications of Tyr-P levels relative to total G-CSFR protein in biotinylated G-CSF pull-downs at the indicated times. Data shown are representative of three independent experiments. (B) Similar to A, with lysine-less G-CSFR mutant K5R that accumulates in early endosomes. (C) Phosphotyrosine analysis of Prdx4 immunoprecipitates in Ptp1b−/− or reconstituted control MEFs at various times after stimulation with G-CSF. Histograms show quantification of Tyr-P (Odyssey) levels relative to immunoprecipitated Prdx4. Data are representative of two independent experiments. TCL total cell lysates, NS not stimulated.
Discussion
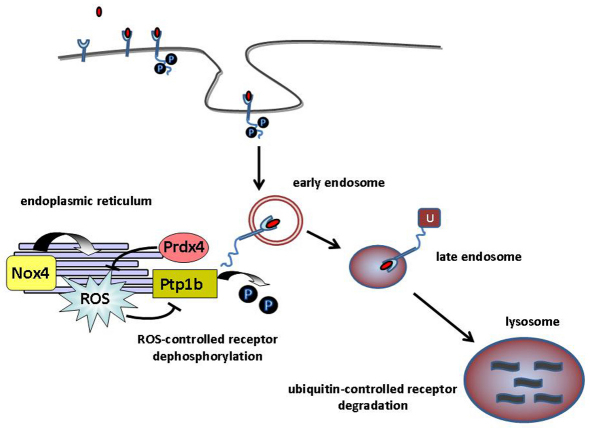
The key finding reported here is that the antioxidant protein Prdx4, localized in the ER, attenuates G-CSFR signalling from early endosomes. In addition, we provide evidence to suggest that this is achieved by preventing the loss of activity of the ER-resident tyrosine phosphatase Ptp1b. Recent studies have shown that different Nox complexes allocate to specific subcellular compartments (Hilenski et al., 2004; Lambeth, 2004; Ushio-Fukai, 2006; Van Buul et al., 2005; Wu et al.) and act in spatially restricted microdomains, which is thought to be essential for specificity of ROS-mediated signalling (Terada, 2006). For instance, the local action of Nox2 in a signalling module was demonstrated for interleukin-1 (IL-1) signalling: after activation of the IL-1 receptor, Nox2 controlled the binding of TRAF6 to the IL-1R–MyD88 complex in early endosomes (Li et al., 2006). Nox4, on the other hand has been shown to drive ROS signalling from the ER (Chen et al., 2008; Wu et al., 2010). A recent study on the interleukin-4 (IL-4) receptor showed that ROS promotes IL-4 signalling by inhibition of Ptp1b (Sharma et al., 2008). Furthermore, Nox4 was shown to be responsible for ROS production in the ER and to be crucial for the regulation of Ptp1b, further illustrating that the specificity of intracellular ROS-controlled signalling depends on the localization of Nox isoforms within particular subcellular compartments (Chen et al., 2008). Based on our findings and these recent reports, we propose a model in which Prdx4 negatively controls G-CSF signalling by neutralizing ROS produced by Nox4 in the ER, thereby keeping Ptp1b in an active state (Fig. 7). Conversely, Ptp1b inhibits tyrosine phosphorylation of Prdx4, which by analogy to Prdx1, might increase its activity (Woo et al., 2010).
Fig. 7.
Model of Prdx4-controlled G-CSF signalling. Activation of G-CSFR leads to internalization of G-CSF (red ovals) and entry into early endosomes. G-CSFR dephosphorylation is then mediated by Ptp1b, which requires endosomal trafficking of the G-CSFR towards the ER, where Ptp1b resides. Ptp1b activity is inhibited by ROS, which is locally produced in the ER by Nox4 (Chen et al., 2008). ER-resident Prdx4 reduces ROS, thereby elevating Ptp1b activity. After or during dephosphorylation in early endosomes, G-CSFR ubiquitylation takes place (Wölfler et al., 2009), which triggers routing to late endosomes and lysosomes, where G-CSFR are degraded. P, phosphate; U, ubiquitin.
Whether G-CSFR and Prdx4 are direct substrates of Ptp1b or whether Ptp1b acts mainly through dephosphorylation and inhibition of JAKs (Fig. 5B) as also shown previously for interferon and leptin signalling (Myers et al., 2001; Zabolotny et al., 2002) is presently unknown. Neither the G-CSFR cytoplasmic domain nor Prdx4 contains the Ptp1b recognition motif [E/D]Y(P)Y(P)[R/K] identified in JAK2 and TYK2 (Myers et al., 2001; Salmeen et al., 2000). However, this does not preclude that tyrosine-based motifs in G-CSFR are a direct substrate for Ptp1b. Another unresolved issue is whether Prdx4, in addition to its antioxidant function, acts as a tethering molecule and thereby stabilizes the interaction between G-CSFR and Ptp1b at the ER–early endosome interface. The finding that Prdx4 is localized at the ER and ERGIC is in agreement with an earlier report (Tavender et al., 2008). Based on structure prediction and resistance to proteinase K digestion, this study also suggests that Prdx4 resides inside the ER lumen (Tavender et al., 2008). Although this would be in apparent conflict with an interaction of Prdx4 with the G-CSFR cytoplasmic domain, proteins residing in the ER might retrotranslocate from the lumen and enter the cytoplasm. For instance, this was demonstrated for the protein chaperone calreticulin (Afshar et al., 2005; Frangioni et al., 1992). A similar process could explain how Prdx4 exits the ER lumen and binds to the cytosolic tail of G-CSFR. Possibly, an interaction with the ER lipid bilayer or with binding proteins located therein would keep Prdx4 in proximity of the ER. Notably, in immunoprecipitation experiments, G-CSFR also co-precipitated with Prdx4 in the absence of growth factor (supplementary material Fig. S2). This interaction might occur either after ligand-independent constitutive retrograde routing of G-CSFR or when G-CSFR passes through the ER in the biosynthetic pathway. Whether either of these would have functional significance is unknown.
In conclusion, our study suggests that the antioxidant protein Prdx4 negatively controls G-CSF signalling in the early endosome compartment by reducing ROS levels in proximity of the ER, thereby keeping Ptp1b active. As a consequence, STAT3 phosphorylation is reduced, either by direct (receptor dephosphorylation) or indirect (reduced JAK activity) mechanisms. Intriguingly, the G-CSFR truncation mutants found in SCN and AML respond differentially to suppressor of cytokine signaling 3 (SOCS3) in terms of STAT3 versus STAT5 inhibition: SOCS3-mediated inhibition of STAT5 activation is abolished as a result of the G-CSFR truncation, whereas inhibition of STAT3 remained largely intact (van de Geijn et al., 2004). This discrepancy relates to the fact that SOCS3-induced STAT5 inhibition entirely depends on SOCS box-mediated ubiquitylation of G-CSFR, whereas SOCS3-induced STAT3 inhibition is less dependent on this process (Irandoust et al., 2007; van de Geijn et al., 2004; Wölfler et al., 2009). Together with these previous results our current data suggest that, whereas attenuation of STAT5 mainly depends on lysosomal degradation of the activated G-CSFR, STAT3 inhibition is mediated mainly by dephosphorylation of STAT3-binding tyrosine motifs controlled by the kinase inhibitory region of SOCS3 and by Ptp1b, when the G-CSFR resides in the early endosome. These findings support the idea that signal diversification from certain growth factor receptors is to a major extent determined by routing dynamics, a concept that becomes increasingly attractive to explain how growth factor receptors exert specific functions while activating largely overlapping signalling pathways. The dynamic interplay between ER-localized enzyme systems and activated receptors in endocytotic vesicles turns out to be a key event in this process.
Deregulation of redox-controlled signalling pathways is increasingly implicated in a variety of diseases, including leukaemia. For instance, the gene encoding Thioredoxin inhibitory protein (Txnip), a common target for retroviral integration in murine leukaemia virus-induced mouse leukaemia, appears to be significantly upregulated in a subgroup of human AML patients (Erkeland et al., 2009). A case of AML with a chromosomal translocation t(X;21)(p22;q22) has been reported in which the PRDX4 gene located on Xp22 was fused to RUNX1 at 21q22, resulting in a RUNX1-PRDX4 fusion transcript (Zhang et al., 2004). We screened 65 myelodysplastic syndrome (MDS) patients and 113 AML patients for possible mutations, but no mutations in the PRDX4 coding region were detected, suggesting that genomic aberrations affecting PRDX4 are rare in MDS and AML (Palande et al., 2011). Intriguingly, this study also showed that PRDX4 expression is significantly downregulated in acute promyelocytic leukaemia (APL) cells, involving H3K27 tri-methylation as a mechanism of histone-mediated gene silencing (Palande et al., 2011). Although the role of PRDX4 downregulation in primary APL stem and progenitor cells remains to be established, predictably the loss of PRDX4 might lead to reduced ER-linked PTP1B activity, providing an explanation for the increased responsiveness of APL clonogenic precursors to G-CSF (Pebusque et al., 1988).
Materials and Methods
PCR primers
Primers used for the preparation of constructs are listed in supplementary material Table S1. All PCR products were checked for correct nucleotide sequences.
G-CSFR expression constructs
The G-CSFR wild-type and K5R expression constructs have been described previously (Irandoust et al., 2007). To create G-CSFR-Prdx4 fusions, the G-CSFR part was amplified using primers Fw7 G-CSFR and Δ73Prdx4-Δ795GR Rv (supplementary material Table S1). A glycine–glycine–serine (GGS) flexible linker was introduced between G-CSFR and Prdx4. For amplification of Δ73 Prdx4, the primers Δ73Prdx4-Δ795GR Fw and Rv Prdx4 EcoRV (supplementary material Table S1) were used. These fragments were used as a template for the fusion PCR, performed with primers Fw7 G-CSFR and Rv Prdx4 EcoRV. The fusion product was cloned as an EcoRV-HpaI fragment into the pBABE/G-CSFR (wild-type) vector. The multisite-directed mutagenesis kit from Stratagene (Huissen, The Netherlands) was used to mutate both cysteines in the active site of Prdx4, using Δ795-G-CSFR-Δ73Prdx4 as a template.
Cells, retroviral transduction and transfection
Mouse bone marrow progenitor cells were isolated as described (Erkeland et al., 2007) and transduced with pBABE virus generated in Phoenix E cells expressing the different G-CSFR constructs. Cells were pre-cultured for 2 days in CellGro medium supplemented with IL-3 (10 ng/ml), Flt3-ligand (50 ng/ml), stem cell factor (10 ng/ml) and thrombopoietin (10 ng/ml) (Hermans et al., 2003). 32D cells were transduced with the same retroviral constructs, as described (Erkeland et al., 2007). For each construct, multiple independent clones were expanded for further analysis. Mouse embryonic fibroblasts were transduced with virus generated by transfection of Phoenix E cells with pBABE/G-CSFR (wild-type). Cells expressing wild-type GCSFR were selected using puromycin (1.5 μg/ml) selection. HEK293T cells were transiently transfected using calcium phosphate precipitation, and HeLa cells were transfected using lipofectamine (Invitrogen, Breda, The Netherlands).
Mammalian protein–protein interaction trap assay
Bait constructs
G-CSFR fragments were cloned in-frame with the MAPPIT bait receptor, consisting of the extracellular domain of the erythropoietin receptor and the cytoplasmic domain of leptin receptor lacking STAT3-binding sites, as described (Erkeland et al., 2007).
Prey constructs
Prdx1, Prdx 2, Prdx 4 and Prdx 6 sequences were amplified from HL60 cells using forward primers with a 5′ EcoRI site, followed by the respective Prdx sequence. The reverse primers were designed with an XhoI restriction site 3′ of the STOP codon. The Prdx fragments were cloned into the pMG2 prey vector (Eyckerman et al., 2002), thus generating the FLAG-tagged Prdx–gp130 fusion constructs. Prey–bait interactions were quantified in STAT3 luciferase assays as described (Erkeland et al., 2007; Eyckerman et al., 2002). In brief, HEK293T cells (2×105) were transfected with bait and prey constructs along with a luciferase reporter (pXP2d2-rPAP-Luci). At 48 hours after transfection, the chimeric bait receptors were activated with erythropoietin (0.5 U/ml) for 24 hours or left unstimulated. Luciferase activity from STAT3 luciferase reporter was determined using the Steady Glo luciferase assay system (Promega, Leiden, The Netherlands).
Antibodies and fluorescent reagents
Prdx4 rabbit polyclonal (Ab15574), Prdx4 mouse monoclonal (Ab16943) and GRP94 rabbit polyclonal (Ab3674) antibodies were purchased from Abcam (Cambridge, UK). Goat polyclonal antibodies against EEA1 (sc6414) and β-actin (sc1616) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Mouse anti-human G-CSFR (CD114) was from Becton Dickinson (Franklin Lakes, NJ); phospho-STAT3 (Tyr705 and Ser727), Akt and phospho-JAK2 (Y1007/1008) antibodies were from Cell Signaling (Danvers, MA), anti phospho-STAT5 (STAT5A-pY694, STAT5B-pY699); and anti-phosphotyrosine 4G10 and 4G10-biotin antibodies were from Millipore (Billerica, MA). The latter were visualized on western blots using Streptavidin-IRDye800CW (LI-COR, Lincoln, NE). Secondary donkey anti-rabbit Cy3 and donkey anti-mouse Cy5 antibodies used for confocal imaging were from Jackson ImmunoResearch. ERGIC-53 rabbit polyclonal antibody (E1031) was purchased from Sigma-Aldrich.
Immunoprecipitations, Bio-G-CSF pull-downs and western blotting
HEK293T cells transfected with pBABE/G-CSFR were placed for 4 hours in DMEM without foetal calf serum and then stimulated with G-CSF (100 ng/ml). After stimulation, cells were lysed at 4°C in lysis buffer (50 mM Tris-HCl pH 7.4, 50 mM NaCl, 10% glycerol, 1% NP40, 0.5% sodium deoxycholate, 20 mM NaF) containing a cocktail of protease inhibitors. Lysates were incubated overnight at 4°C with pre-washed Protein G Dynabeads (Invitrogen/DYNAL) coated with Prdx4 mouse monoclonal antibody purchased from Abcam. Immune complexes were visualized by western blotting. To study tyrosine phosphorylation of the G-CSFR, pull-down of G-CSFR using biotinylated G-CSF and streptavidin-coated beads was performed as described (Wölfler et al., 2009).
Confocal laser scanning microscopy
HeLa cells transiently transfected with pLNCX2/G-CSFR (wild-type) were used for CLSM. At 48 hours after transfection, HeLa cells were deprived of growth factor and serum by incubation for 4 hours in DMEM. G-CSFR antibody (2.5 μg/ml) was added to the medium and incubated at room temperature for 20 minutes to allow binding of the antibody to the extracellular domain of the G-CSFR. Excess antibody was washed off and cells stimulated with G-CSF for different time periods. Immunostaining was performed as described previously (Irandoust et al., 2007). Cells were imaged using the multitrack detection mode on a Zeiss LSM 519 confocal microscope equipped with Argon and HeNe lasers using a 63× Planochromat oil immersion objective.
Colony assays
Retrovirally transduced Csf3r−/− bone marrow cells
Cells were harvested 48 hours after retroviral infection and placed in Methocult (M3231; Stem Cell Technologies, Vancouver, Canada) containing puromycin (1.5 μg/ml) and either human recombinant G-CSF (100 ng/ml, Amgen, Breda, The Netherlands) or mouse GM-CSF (20 U/ml, Peprotech, Rocky Hill). All cultures were done in triplicate and colonies were counted on day 7 of culture.
Prdx4−/− and Ptp1b−/− bone marrow cells
Femurs, tibiae and sterna from Prdx4−/− (Iuchi et al., 2009), Ptp1b−/− and age- and sex-matched control mice (Klaman LD, 2000) were shipped on ice from the laboratories of JF and BGN. Bone marrow mononuclear cells were obtained as described (Erkeland et al., 2003) and cultured in colony assays with different concentrations of G-CSF or with one standard concentration of GM-CSF (10 ng/ml). All animal experiments were performed according to the relevant regulatory standards.
Proximity ligation assay and ROS measurements
In situ-PLA was performed using the Duolink II PLA kit (Olink Bioscience, Uppsala, Sweden; http://www.olink.com/products-services/duolink/situ-pla-technology). ROS levels were detected using CM-H2DCFDA (Invitrogen, Carlsbad, CA). Details on the antibodies and incubation conditions used for these experiments are given in supplementary material Figs S7 and S8.
Supplementary Material
Acknowledgements
We thank Marieke von Lindern for many fruitful discussions and comments on the manuscript.
Footnotes
Funding
This work was supported by the Dutch Cancer Society ‘KWFkankerbestrijding’; and the National Institutes of Health [grant number R37 CA49132 to B.G.N.]. Deposited in PMC for release after 12 months.
Supplementary material available online at http://jcs.biologists.org/lookup/suppl/doi:10.1242/jcs.089656/-/DC1
References
- Aarts L. H., Roovers O., Ward A. C., Touw I. P. (2004). Receptor activation and 2 distinct COOH-terminal motifs control G-CSF receptor distribution and internalization kinetics. Blood 103, 571-579 [DOI] [PubMed] [Google Scholar]
- Afshar N., Black B. E., Paschal B. M. (2005). Retrotranslocation of the chaperone calreticulin from the endoplasmic reticulum lumen to the cytosol. Mol. Cell. Biol. 25, 8844-8853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K., Kirber M. T., Xiao H., Yang Y., Keaney J. F. J. (2008). Regulation of ROS signal transduction by NADPH oxidase 4 localization. J. Cell Biol. 181, 1129-1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi M. H., Lee I. K., Kim G. W., Kim B. U., Han Y. H., Yu D. Y., Park H. S., Kim K. Y., Lee J. S., Choi C., et al. (2005). Regulation of PDGF signalling and vascular remodelling by peroxiredoxin II. Nature 435, 347-353 [DOI] [PubMed] [Google Scholar]
- de Koning J. P., Dong F., Smith L., Schelen A. M., Barge R. M., van der Plas D. C., Hoefsloot L. H., Lowenberg B., Touw I. P. (1996). The membrane-distal cytoplasmic region of human granulocyte colony-stimulating factor receptor is required for STAT3 but not STAT1 homodimer formation. Blood 87, 1335-1342 [PubMed] [Google Scholar]
- Demetri G. D., Griffin J. D. (1991). Granulocyte colony-stimulating factor and its receptor. Blood 78, 2791-2808 [PubMed] [Google Scholar]
- Dong F., Larner A. C. (2000). Activation of Akt kinase by granulocyte colony-stimulating factor (G-CSF): evidence for the role of a tyrosine kinase activity distinct from the Janus kinases. Blood 95, 1656-1662 [PubMed] [Google Scholar]
- Dong F., Brynes R. K., Tidow N., Welte K., Löwenberg B., Touw I. P. (1995). Mutations in the gene for the granulocyte colony-stimulating-factor receptor in patients with acute myeloid leukemia preceded by severe congenital neutropenia. N. Engl. J. Med. 333, 487-493 [DOI] [PubMed] [Google Scholar]
- Dong F., Liu X., de Koning J. P., Touw I. P., Hennighausen L., Larner A., Grimley P. M. (1998). Stimulation of Stat5 by granulocyte colony-stimulating factor (G-CSF) is modulated by two distinct cytoplasmic regions of the G-CSF receptor. J. Immunol. 161, 6503-6509 [PubMed] [Google Scholar]
- Eden E. R., White I. J., Tsapara A., Futter C. E. (2010). Membrane contacts between endosomes and ER provide sites for PTP1B-epidermal growth factor receptor interaction. Nat. Cell Biol. 12, 267-272 [DOI] [PubMed] [Google Scholar]
- Erkeland S. J., Valkhof M., Heijmans-Antonissen C., Delwel R., Valk P. J., Hermans M. H., Touw I. P. (2003). The gene encoding the transcriptional regulator Yin Yang 1 (YY1) is a myeloid transforming gene interfering with neutrophilic differentiation. Blood 101, 1111-1117 [DOI] [PubMed] [Google Scholar]
- Erkeland S. J., Aarts L. H., Irandoust M., Roovers O., Klomp A., Valkhof M., Gits J., Eyckerman S., Tavernier J., Touw I. P. (2007). Novel role of WD40 and SOCS box protein-2 in steady-state distribution of granulocyte colony-stimulating factor receptor and G-CSF-controlled proliferation and differentiation signaling. Oncogene 26, 1985-1994 [DOI] [PubMed] [Google Scholar]
- Erkeland S. J., Palande K. K., Valkhof M., Gits J., Danen-van Oorschot A., Touw I. P. (2009). The gene encoding thioredoxin-interacting protein (TXNIP) is a frequent virus integration site in virus-induced mouse leukemia and is overexpressed in a subset of AML patients. Leuk. Res. 33, 1367-1371 [DOI] [PubMed] [Google Scholar]
- Eyckerman S., Lemmens I., Lievens S., Van der Heyden J., Verhee A., Vandekerckhove J., Tavernier J. (2002). Design and use of a mammalian protein-protein interaction trap (MAPPIT). Sci. STKE 2002, PL18 [DOI] [PubMed] [Google Scholar]
- Frangioni J. V., Beahm P. H., Shifrin V., Jost C. A., Neel B. G. (1992). The nontransmembrane tyrosine phosphatase PTP-1B localizes to the endoplasmic reticulum via its 35 amino acid C-terminal sequence. Cell 68, 545-560 [DOI] [PubMed] [Google Scholar]
- Freedman M. H., Alter B. P. (2002). Risk of myelodysplastic syndrome and acute myeloid leukemia in congenital neutropenias. Semin. Hematol. 39, 128-133 [DOI] [PubMed] [Google Scholar]
- Germeshausen M., Ballmaier M., Welte K. (2007). Incidence of CSF3R mutations in severe congenital neutropenia and relevance for leukemogenesis: Results of a long-term survey. Blood 109, 93-99 [DOI] [PubMed] [Google Scholar]
- Giguere P., Turcotte M. E., Hamelin E., Parent A., Brisson J., Laroche G., Labrecque P., Dupuis G., Parent J. L. (2007). Peroxiredoxin-4 interacts with and regulates the thromboxane A(2) receptor. FEBS Lett. 581, 3863-3868 [DOI] [PubMed] [Google Scholar]
- Groen A., Lemeer S., van der Wijk T., Overvoorde J., Heck A. J. R., Ostman A., Barford D., Slijper M., den Hertog J. (2005). Differential oxidation of protein-tyrosine phosphatases. J. Biol. Chem. 280, 10298-10304 [DOI] [PubMed] [Google Scholar]
- Haj F. G., Verveer P. J., Squire A., Neel B. G., Bastiaens P. (2002). Imaging sites of receptor dephosphorylation by PTP1B on the surface of the endoplasmic reticulum. Science 295, 1708-1711 [DOI] [PubMed] [Google Scholar]
- Hermans M. H., van de Geijn G. J., Antonissen C., Gits J., van Leeuwen D., Ward A. C., Touw I. P. (2003). Signaling mechanisms coupled to tyrosines in the granulocyte colony-stimulating factor receptor orchestrate G-CSF-induced expansion of myeloid progenitor cells. Blood 101, 2584-2590 [DOI] [PubMed] [Google Scholar]
- Hilenski L. L., Clempus R. E., Quinn M. T., Lambeth J. D., Griendling K. K. (2004). Distinct subcellular localizations of Nox1 and Nox4 in vascular smooth muscle cells. Arterioscler. Thromb. Vasc. Biol. 24, 677-683 [DOI] [PubMed] [Google Scholar]
- Hortner M., Nielsch U., Mayr L. M., Johnston J. A., Heinrich P. C., Haan S. (2002). Suppressor of cytokine signaling-3 is recruited to the activated granulocyte-colony stimulating factor receptor and modulates its signal transduction. J. Immunol. 169, 1219-1227 [DOI] [PubMed] [Google Scholar]
- Hunter M. G., Avalos B. R. (1999). Deletion of a critical internalization domain in the G-CSFR in acute myelogenous leukemia preceded by severe congenital neutropenia. Blood 93, 440-446 [PubMed] [Google Scholar]
- Iiyama M., Kakihana K., Kurosu T., Miura O. (2006). Reactive oxygen species generated by hematopoietic cytokines play roles in activation of receptor-mediated signaling and in cell cycle progression. Cell. Signal. 18, 174-182 [DOI] [PubMed] [Google Scholar]
- Irandoust M. I., Aarts L. H., Roovers O., Gits J., Erkeland S. J., Touw I. P. (2007). Suppressor of cytokine signaling 3 controls lysosomal routing of G-CSF receptor. EMBO J. 26, 1782-1793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iuchi Y., Okada F., Tsunoda S., Kibe N., Shirasawa N., Ikawa M., Okabe M., Ikeda Y., Fujii J. (2009). Peroxiredoxin 4 knockout results in elevated spermatogenic cell death via oxidative stress. Biochem. J. 419, 149-158 [DOI] [PubMed] [Google Scholar]
- Klaman L. D., Boss O., Peroni O. D., Kim J. K., Martino J. L., Zabolotny J. M., Moghal N., Lubkin M., Kim Y.-B., Sharpe A. H., et al. (2000). Increased energy expenditure, decreased adiposity, and tissue-specific insulin sensitivity in protein-tyrosine phosphatase 1B-deficient mice. Mol. Cell. Biol. 20, 5479-5489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambeth J. D. (2004). NOX enzymes and the biology of reactive oxygen. Nat. Rev. Immunol. 4, 181-189 [DOI] [PubMed] [Google Scholar]
- Li Q., Harraz M. M., Zhou W., Zhang L. N., Ding W., Zhang Y., Eggleston T., Yeaman C., Banfi B., Engelhardt J. F. (2006). Nox2 and Rac1 regulate H2O2-dependent recruitment of TRAF6 to endosomal interleukin-1 receptor complexes. Mol. Cell. Biol. 26, 140-154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieschke G. J., Grail D., Hodgson G., Metcalf D., Stanley E., Cheers C., Fowler K. J., Basu S., Zhan Y. F., Dunn A. R. (1994). Mice lacking granulocyte colony-stimulating factor have chronic neutropenia, granulocyte and macrophage progenitor cell deficiency, and impaired neutrophil mobilization. Blood 84, 1737-1746 [PubMed] [Google Scholar]
- Myers M. P., Andersen J. N., Cheng A., Tremblay M. L., Horvath C. M., Parisien J. P., Salmeen A., Barford D., Tonks N. K. (2001). TYK2 and JAK2 are substrates of protein-tyrosine phosphatase 1B. J. Biol. Chem. 276, 47771-47774 [DOI] [PubMed] [Google Scholar]
- Palande K. K., Beekman R., van der Meeren L. E., Beverloo H. B., Valk P. J., Touw I. P. (2011). The antioxidant protein peroxiredoxin 4 is epigenetically down regulated in acute promyelocytic leukemia. PLoS One 6, e16340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pebusque M. J., Lafage M., Lopez M., Mannoni P. (1988). Preferential response of acute myeloid leukemias with translocation involving chromosome 17 to human recombinant granulocyte colony-stimulating factor. Blood 72, 257-265 [PubMed] [Google Scholar]
- Rhee S. G. (2006). H2O2, a necessary evil for cell signaling. Science 312, 1882-1883 [DOI] [PubMed] [Google Scholar]
- Rhee S. G., Chae H. Z., Kim K. (2005a). Peroxiredoxins: a historical overview and speculative preview of novel mechanisms and emerging concepts in cell signaling. Free Radic. Biol. Med. 38, 1543-1552 [DOI] [PubMed] [Google Scholar]
- Rhee S. G., Kang S. W., Jeong W., Chang T. S., Yang K. S., Woo H. A. (2005b). Intracellular messenger function of hydrogen peroxide and its regulation by peroxiredoxins. Curr. Opin. Cell Biol. 17, 183-189 [DOI] [PubMed] [Google Scholar]
- Salmeen A., Andersen J. N., Myers M. P., Tonks N. K., Barford D. (2000). Molecular basis for the dephosphorylation of the activation segment of the insulin receptor by protein tyrosine phosphatase 1B. Mol. Cell 6, 1401-1412 [DOI] [PubMed] [Google Scholar]
- Sasagawa I., Matsuki S., Suzuki Y., Iuchi Y., Tohya K., Kimura M., Nakada T., Fujii J. (2001). Possible involvement of the membrane-bound form of peroxiredoxin 4 in acrosome formation during spermiogenesis of rats. Eur. J. Biochem. 268, 3053-3061 [DOI] [PubMed] [Google Scholar]
- Sharma P., Chakraborty R., Wang L., Min B., Tremblay M. L., Kawahara T., Lambeth J. D., Haque S. J. (2008). Redox regulation of interleukin-4 signaling. Immunity 29, 551-564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaresan M., Yu Z. X., Ferrans V. J., Irani K., Finkel T. (1995). Requirement for generation of H2O2 for platelet-derived growth factor signal transduction. Science 270, 296-299 [DOI] [PubMed] [Google Scholar]
- Tavender T. J., Sheppard A. M., Bulleid N. J. (2008). Peroxiredoxin IV is an endoplasmic reticulum-localized enzyme forming oligomeric complexes in human cells. Biochem. J. 411, 191-199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terada L. S. (2006). Specificity in reactive oxidant signaling: think globally, act locally. J. Cell Biol. 174, 615-623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touw I. P., van de Geijn G. J. (2007). Granulocyte colony-stimulating factor and its receptor in normal myeloid cell development, leukemia and related blood cell disorders. Front. Biosci. 12, 800-815 [DOI] [PubMed] [Google Scholar]
- Ushio-Fukai M. (2006). Localizing NADPH oxidase-derived ROS. Sci. STKE 349, re8 [DOI] [PubMed] [Google Scholar]
- Van Buul J. D., Fernandez-Borja M., Anthony E. C., Hordijk P. L. (2005). Expression and localization of NOX2 and NOX4 in primary human endothelial cells. Antioxid. Redox Signal. 7, 308-317 [DOI] [PubMed] [Google Scholar]
- van de Geijn G.-J. M., Gits J., Aarts L. H. J., Heijmans-Antonissen C., Touw I. P. (2004). G-CSF receptor truncations found in SCN/AML relieve SOCS3-controlled inhibition of STAT5 but leave suppression of STAT3 intact. Blood 104, 667-674 [DOI] [PubMed] [Google Scholar]
- Ward A. C., van Aesch Y. M., Schelen A. M., Touw I. P. (1999). Defective internalization and sustained activation of truncated granulocyte colony-stimulating factor receptor found in severe congenital neutropenia/acute myeloid leukemia. Blood 93, 447-458 [PubMed] [Google Scholar]
- Wells J. A., de Vos A. M. (1996). Hematopoietic receptor complexes. Annu. Rev. Biochem. 65, 609-634 [DOI] [PubMed] [Google Scholar]
- Wölfler A., Irandoust M., Meenhuis A., Gits J., Roovers O., Touw I. P. (2009). Site-specific ubiquitination determines lysosomal sorting and signal attenuation of the granulocyte colony-stimulating factor receptor. Traffic 10, 1168-1179 [DOI] [PubMed] [Google Scholar]
- Woo H. A., Yim S. H., Shin D. H., Kang D., Yu D. Y., Rhee S. G. (2010). Inactivation of peroxiredoxin I by phosphorylation allows localized H2O2 accumulation for cell signaling. Cell 140, 517-528 [DOI] [PubMed] [Google Scholar]
- Wu R. F., Ma Z., Liu Z., Terada L. S. (2010). Nox4-derived H2O2 mediates endoplasmic reticulum signaling through local Ras activation. Mol. Cell. Biol. 30, 3553-3568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zabolotny J. M., Bence-Hanulec K. K., Stricker-Krongrad A., Haj F., Wang Y., Minokoshi Y., Kim Y. B., Elmquist J. K., Tartaglia L. A., Kahn B. B., et al. (2002). PTP1B regulates leptin signal transduction in vivo. Dev. Cell 2, 489-495 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Emmanuel N., Kamboj G., Chen J., Shurafa M., Van Dyke D. L., Wiktor A., Rowley J. D. (2004). PRDX4, a member of the peroxiredoxin family, is fused to AML1 (RUNX1) in an acute myeloid leukemia patient with a t(X;21)(p22;q22). Genes Chromosomes Cancer 40, 365-370 [DOI] [PubMed] [Google Scholar]
- Zhu Q. S., Xia L., Mills G. B., Lowell C. A., Touw I. P., Corey S. J. (2006). G-CSF induced reactive oxygen species involves Lyn-PI3-kinase-Akt and contributes to myeloid cell growth. Blood 107, 1847-1856 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.