Abstract
The cytochrome P450 sterol 14α-demethylase enzyme (CYP51) is the target of azole antifungals. Azoles block ergosterol synthesis, and thereby fungal growth, by binding in the active-site cavity of the enzyme and ligating the iron atom of the heme cofactor through a nitrogen atom of the azole. Mutations in and around the CYP51 active site have resulted in azole resistance. In this work, homology models of the CYP51 enzymes from Aspergillus fumigatus and Candida albicans were constructed based on the X-ray crystal structure of CYP51 from Mycobacterium tuberculosis. Using these models, binding modes for voriconazole (VOR), fluconazole (FLZ), itraconazole (ITZ), and posaconazole (POS) were predicted from docking calculations. Previous work had demonstrated that mutations in the vicinity of the heme cofactor had a greater impact on the binding of FLZ and VOR than on the binding of POS and ITZ. Our modeling data suggest that the long side chains of POS and ITZ occupy a specific channel within CYP51 and that this additional interaction, which is not available to VOR and FLZ, serves to stabilize the binding of these azoles to the mutated CYP51 proteins. The model also predicts that mutations that were previously shown to specifically impact POS susceptibility in A. fumigatus and C. albicans act by interfering with the binding of the long side chain.
For over a decade azoles have been a mainstay of the antifungal armamentarium. Azoles inhibit the synthesis of ergosterol, the bulk sterol in fungal membranes, by binding to the heme cofactor located in the active site of the P450-dependent enzyme lanosterol 14α-demethylase (CYP51, also called Erg11p). Ergosterol depletion, coupled with the accumulation of methylated sterol precursors, has been proposed to affect both membrane integrity and the function of some membrane-bound proteins, including chitin synthase (27). The net result is an inhibition of fungal growth.
Resistance to azoles is a concern, particularly during the long-term treatment of AIDS patients with oropharyngeal candidiasis (25). The two most-prevalent causes of resistance are mutations in the target site, resulting in reduced azole binding to CYP51, and decreased intracellular drug accumulation, resulting from increased expression of efflux pump genes (recently reviewed in reference 24). To combat azole resistance, and to extend the spectrum of treatable pathogens, more-potent azoles have been developed. One such agent is posaconazole (POS) (SCH56592), a broad-spectrum triazole in phase III trials. Unlike fluconazole (FLZ) and voriconazole (VOR), POS is not effluxed by the pumps encoded by FLU1 and MDR1. However, POS is a substrate for the ATP-dependent pumps encoded by CDR1 and CDR2. In addition, POS appears to be relatively insensitive to amino acid substitutions in CYP51 (D. Sanglard, F. Ischer, and J. Bille, Abstr. 42nd Intersci. Conf. Antimicrob. Agents Chemother., abstr. M-221, 2002). Finally, POS is active against Aspergillus spp. (17).
The CYP51 proteins belong to the cytochrome P450 superfamily. Unlike the soluble bacterial P450s, all the fungal CYP51 proteins characterized to date are integral membrane proteins, making structural and biophysical characterization more challenging. The publication of the X-ray crystal structure of CYP51 from Mycobacterium tuberculosis (MT-CYP51), which has >25% sequence identity to most fungal CYP51s, provided a new opportunity to study the fungal enzymes (18, 19). In this work, we used the MT-CYP51 structure to construct homology models for the CYP51 proteins from Aspergillus fumigatus (AF-CYP51A) and Candida albicans (CA-CYP51). To gain insight into how the various azoles bind these proteins, we combined docking calculations with information gained from studying substitutions that resulted in azole resistance. The resultant binding models help explain why particular CYP51 mutations have more-profound affects on the binding of some azoles than others. They may also provide insights into the design of azoles with improved affinity for their target site.
MATERIALS AND METHODS
Homology modeling of CYP51s.
The structure of MT-CYP51 complexed with FLZ (PDB code, 1EA1 [18]) was used as the template for the homology models of AF-CYP51A (GenBank accession no. AAK73659) and CA-CYP51 (GenBank accession no. CAA31658). We followed the same naming scheme for secondary structures used by Podust et al. (19). Alignment of the MT-CYP51 sequence with those of AF-CYP51A and CA-CYP51 was performed using ALIGN2D in MODELLER (Accelrys, San Diego, Calif.) (21, 22, 23). From the alignments, spatial restraints (including distance restraints and torsion angle restraints) were derived and used in the three-dimensional model construction with MODELLER. The models were further optimized (including loop refinement) with the internal optimizer of MODELLER and evaluated via MODELLER and Verify3D scores (INSIGHT II; Accelrys).
Docking of ligands.
The torsion angles of several residue side chains around the azole binding cavity in the CYP51 models were manually adjusted and in some cases further minimized using the Tripos force field, Amber charges, and a distance-dependent dielectric model. The three-dimensional structures of all azole ligands were constructed using Sybyl (version 6.9) accessed via CONCORD (Tripos Inc., St. Louis, Mo.). Gasteiger-Huckel charges (Sybyl; Tripos Inc.) were assigned to the azole ligand atoms and heme.
In the MT-CYP51/FLZ X-ray structure, FLZ binds with its triazole ring perpendicular to the heme porphyrin plane and with N-4 of the triazole coordinated to the heme iron. The other azoles were constrained so that their triazole rings overlapped the FLZ triazole. Since VOR is very similar to FLZ, its initial binding conformation was derived by overlap with FLZ in MT-CYP51/FLZ and then overlap of the AF-CYP51A and CA-CYP51 models with MT-CYP51. The program GOLD (version 2.0; CCDC, Cambridge, United Kingdom), an automated docking program (8), was used to dock POS and ITZ into the binding site in AF-CYP51A and CA-CYP51. The active-site radius was set at 15 Å. For the docking calculations, all residues of the protein and heme were fixed. while the ligands were flexible. A covalent-bond distance of 2 Å from N-4 of the triazole ring to the heme iron was set as a constraint. Twenty-one genetic algorithm runs were performed.
RESULTS
Homology model of AF-CYP51.
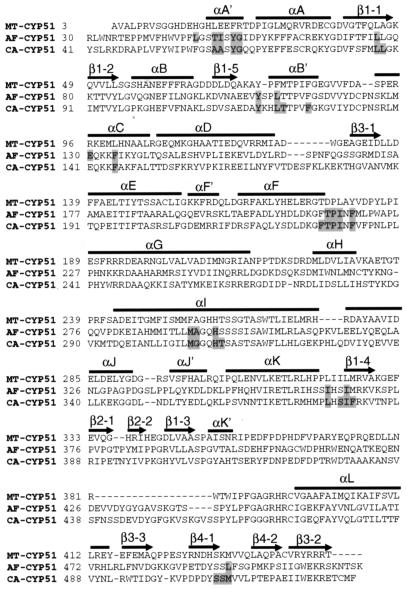
A. fumigatus has two distinct cyp51 genes, cyp51A and cyp51B (14). We focused on cyp51A, which encodes AF-CYP51A, since resistance to VOR, ITZ, and POS has been associated with point mutations in this gene (3, 13, 15; E. K. Manavathu, I. Baskaran, G. J. Alangaden, and P. H. Chandrasekar, Abstr. 41st Intersci. Conf. Antimicrob. Agents Chemother., abstr. J-817, 2001). The sequence identity between AF-CYP51A and MT-CYP51 is 28%. An alignment of the two sequences revealed that there were no insertions or deletions located in any of the secondary-structure regions (Fig. 1). However, MT-CYP51 lacks the N-terminal transmembrane domain found in fungal CYP51 proteins. Therefore, the first 29 residues at the N terminus of AF-CYP51A were deleted since they are predicted to form a transmembrane helix and to fold independently from the catalytic domain. The best model generated with MODELLER exhibited the canonical P450 fold (5, 16, 20) (Fig. 2). The Cα root mean square deviation between the template and the model was 0.5 Å over 442 pairs. The largest insertion, a 15-residue loop (residues 427 to 441), was located far from the active site and was not modeled in detail. Instead, an approximate loop model from MODELLER was used.
FIG. 1.
Alignment of the amino acid sequences of MT-CYP51 with CA-CYP51 and AF-CYP51A. The N-terminal membrane-spanning domains of CA-CYP51 (residues 1 to 40) and AF-CYP51A (residues 1 to 29) were deleted prior to alignment. The secondary structure elements are labeled, and residues involved in binding to POS are highlighted.
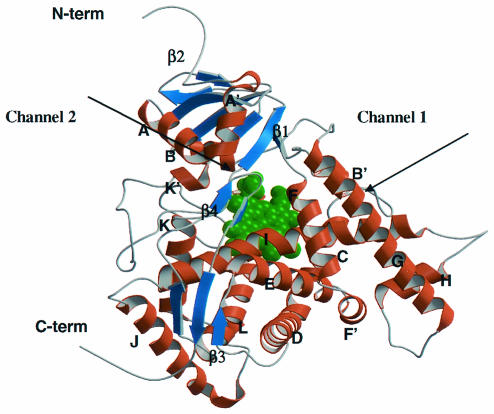
FIG. 2.
Overall structure of the homology model of AF-CYP51A. Helices and strands are shown in brown and blue. The heme cofactor is highlighted in green. Two potential ligand access channels are indicated.
Homology model of CA-CYP51.
As described above, the membrane spanning domain (residues 1 to 40) was deleted from CA-CYP51 prior to modeling. The sequence identity between CA-CYP51 and MT-CYP51 is also 28% (Fig. 1). Similar to AF-CYP51A, the model had a P450 fold (data not shown). The longest loop insertion, 19 residues (residues 439 to 457), was also modeled as a loop and was placed at the same location as in AF-CYP51A.
Identification of potential azole binding sites.
The heme cofactor was extracted from the MT-CYP51 structure and merged into the AF-CYP51A and CA-CYP51 models. To accommodate the heme, Phe134 in AF-CYP51A and Phe145 in CA-CYP51 were both changed to Ala so that only the Cβ side chain atom was retained. Both residues are located in the middle of helix C with their side chains pointing directly at the heme. For the corresponding residue in the MT-CYP51 structure (Leu100) only the Cβ atom coordinates of the side chain were reported, as all the other side chain atoms were disordered. The heme structure and surrounding side chain conformations were energy minimized to avoid steric conflicts. Two CA-CYP51 models, based on MT-CYP51, have been recently published (4, 11). In both models, a two-residue insertion that we placed before helix C (Fig. 1) was placed in the middle of the helix. Helix C is in close proximity to the heme and the ligand binding site. In the MT-CYP51/FLZ X-ray structure, helix C exhibited high levels of B-factors along its entire length, indicating thermal motion or multiple conformations. In addition, some side chain atoms of helix C were disordered. Experimental uncertainties in this region resulted in imprecise models; nevertheless, the helix C conformation of MT-CYP51 was retained in our models.
Through an analysis of all known bacterial and mammalian P450 X-ray structures (11, 18, 28, 29), two putative ligand entry channels were identified that could accommodate the long side chains of POS and ITZ (Fig. 2 and 3). Channel 1 is parallel to the plane of the heme and is formed by the bent helix I, the open BC loop, and helices B′ and G. This channel is apparent in both MT-CYP51 and rabbit P450 2C5 X-ray structures (18, 28). Channel 2, based on bacterial P450BM3 structures (10), is perpendicular to the plane of the heme and is located between the β-sheet and helical domains. The surface entrance of channel 2 in our model is an area surrounded by the FG loop, helix A′, and the β turn connecting strands β4-1 and β4-2. We explored both channels in AF-CYP51A and CA-CYP51 for binding of the long POS side chain. Only the docking results which placed POS in channel 2 are consistent with the reported resistance mutations (see below). Although channel 2 is apparent in the MT-CYP51 X-ray structure, it is not open to the surface. In the model of AF-CYP51A, the conformations of the side chains of residues Tyr107, Phe119, and Leu494 were modified to open an entrance to the surface. In addition, the side chain of Pro216, located in the FG loop, was deleted (specifically, side chain atoms Cβ, Cγ, and Cδ were deleted) to leave enough space for the distal portion of the long side chain of POS. This modification served as an approximation for a possible backbone rearrangement of the FG loop. The latter is plausible since the FG loop has large B factors in the X-ray structure of MT-CYP51 indicating thermal motion or multiple conformations (18). Similar changes for the corresponding side chains in the CA-CYP51 model, including Pro230, were made.
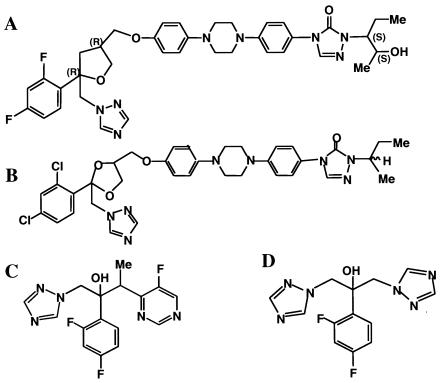
FIG. 3.
Structures of POS (A), ITZ (B), VOR (C), and FLZ (D).
Docking of azole ligands.
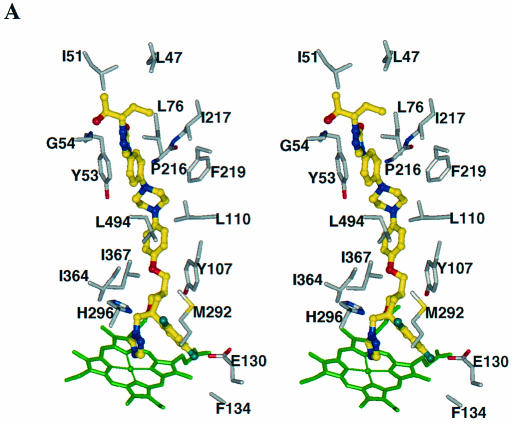
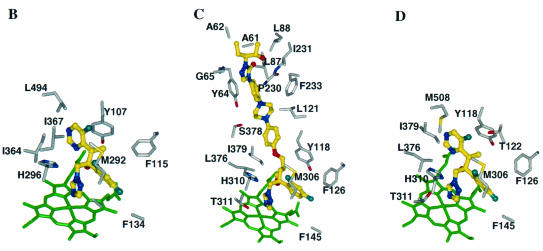
Although POS has four chiral centers, the pharmaceutical product is the enantiomerically pure RRSS isomer. This enantiomer was docked into AF-CYP51A (Fig. 4A). The triazole and the difluoro phenyl rings overlap those of FLZ, and the tetrahydrofuran oxygen of POS superimposes on the hydroxyl oxygen of FLZ. The long side chain of POS is perpendicular to helix I and is surrounded by helix A′, the β turn connecting strands β4-1 to β4-2, the FG loop, helix B′, and β-strand β1-4. The residues 5 Å around POS are predominantly hydrophobic (Leu47, Ile51, Leu76, Tyr107, Leu110, Ile217, Phe219, Met292, Ala293, Ile364, Ile367, Met368, and Leu494). In our model the distal phenyl ring in the POS side chain has aromatic contacts with Tyr53, and there is a hydrogen bond between the urea carbonyl oxygen and Thr50 hydroxyl. Other residues having side chain atoms in contact with POS are His296, Glu130, and Phe134, which are close to the heme, as well as Thr215 and Pro216, which are on the FG loop. The backbone atoms of several residues, including Gly54, also interact with POS.
FIG. 4.
Azole binding sites in the CYP51 proteins. POS (in stereoview) (A) and VOR (B) binding sites in AF-CYP51A; POS (C) and VOR (D) binding sites in CA-CYP51. For clarity, only selected binding site residues are shown. The heme cofactor is highlighted in green. The azoles are shown as ball-and-stick models with their carbon atoms in yellow. Similarly, for the binding site residues the carbon, oxygen, nitrogen, and fluorine atoms are shown in grey, red, blue, and light blue, respectively.
ITZ bound AF-CYP51A in a similar fashion to POS (data not shown). However, the two chlorine atoms on the phenyl ring of ITZ may lead to steric crowding in the region around Met292 and Ala293.
VOR was docked into AF-CYP51A by overlapping with the X-ray structure of FLZ bound to MT-CYP51. The minimized binding conformation, highlighting active-site residues within 5 Å of VOR, is shown in Fig. 4B. Since VOR is a relatively compact molecule, binding does not involve residues from helix A′ or the FG loop. Instead, the primary binding determinants are hydrophobic interactions between VOR and residues Phe115, Met292, Ala293, Ile364, Ile367, and Leu494. In addition, the Tyr107 side chain displays an aromatic interaction with the pyrimidine ring of VOR. It is noteworthy that VOR and POS appear to have similar interactions with the protein near the heme.
Similar docking strategies were applied to perform docking calculations on POS, ITZ, and VOR in the CA-CYP51 binding site (Fig. 4C and D). Residues that interact with POS are Ala61, Ala62, Tyr64, Gly65, Leu87, Leu88, Tyr118, Leu121, Thr122, Phe126, Phe145, Phe228, Thr229, Pro230, Ile231, Phe233, Met306, Gly307, His310, Thr311, Leu376, Ser378, Ile379, Phe380, Ser506, Ser507, and Met508. Residues that interact with VOR are Tyr118, Thr122, Phe126, Phe145, Phe228, Met306, Gly307, His310, Thr311, Leu376, Ile379, and Met508.
Locations of substitutions in CYP51 resulting in azole resistance. (i) A. fumigatus.
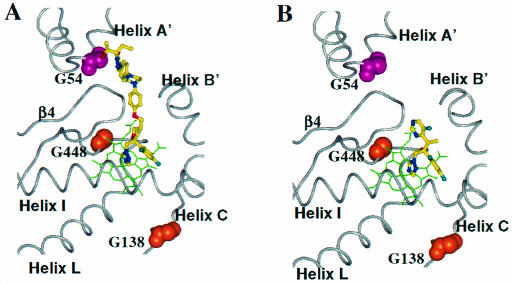
Resistance to POS and ITZ in A. fumigatus results from substitutions at Gly54 in AF-CYP51A (3, 13, 15). Gly54 is located on helix A′ next to the triazole-3-one five-membered ring in the long side chain of POS (Fig. 5). This part of the POS molecule fits snugly inside the pocket formed by helix A′, the FG loop, and the β turn between β-strands β4-1 and β4-2. Replacing Gly54 with amino acids that have side chains would create van der Waals (VDW) conflicts between the side chains and POS. The spectrum of substitutions at Gly54 and the resultant changes in MIC appear to support this prediction (Table 1). Replacing Gly54 with either Glu or Arg resulted in a 30-fold increase in the POS MIC. Introduction of Trp, with its large rigid side chain, resulted in a >250-fold MIC increase (13). ITZ occupies the same binding site as POS; it was therefore not surprising that the substitutions at Gly54 also resulted in large increases in the ITZ MIC.
FIG. 5.
Mapping of substitutions in AF-CYP51A that cause azole resistance. The heme cofactor is highlighted in green. POS (A) and VOR (B) are shown as ball-and-stick models with their carbon atoms in yellow. The atoms of the residues at the sites of substitutions are depicted using space filling models.
TABLE 1.
Substitutions in AF-CYP51A resulting in azole resistance in A. fumigatus isolates
| Isolate | MIC (μg/ml)
|
Substitution in AF-CYP51A | ||
|---|---|---|---|---|
| POS | ITZ | VOR | ||
| ND158a | 0.03 | 0.12 | 0.25 | None (parental strain of MS6, R4-1, and R7-1) |
| MS6a | 0.5 | >16 | 0.12 | Gly54Arg |
| R4-1a | 1 | >16 | 0.25 | Gly54Glu |
| R7-1a | 4 | >16 | 0.25 | Gly54Trp |
| F55064b | 0.25 | 0.25 | 0.25 | None (parental strain of F10 and F33) |
| F10b | 0.25 | 2 | 16 | Gly138Arg |
| F33b | 0.25 | 0.5 | 8 | Gly448Ser |
MICs and data on mutations from Mann et al. (13).
MICs from a personal communication from E. Manavathu; data on mutations as published (Manavathu et. al., 41st ICAAC).
VOR lacks a long side chain that would span channel 2, and therefore substitutions at Gly54 would be predicted to have no impact on VOR binding. This was the case; A. fumigatus isolates with substitutions at Gly54 remained susceptible to VOR (Table 1) (12, 13, 15). In contrast, isolates with amino acid substitutions in AF-CYP51A at either Gly138 or Gly448 were resistant to VOR yet remained susceptible to POS (12; E. K. Manavathu et al., 41st ICAAC). These two residues are located near the heme cofactor (Fig. 5). Replacing Gly138 with Arg and Gly448 with Ser would be predicted to disturb the heme environment. Specifically, the large charged side chain of Arg at position 138 would clash with one of the heme's vinyl side chains and with the side chains of neighboring residues. Similarly, substituting serine at position 448 would result in VDW conflicts between the Ser side chain and the adjacent heme pyrole. Interestingly, neither substitution affected the MIC of POS. From our model we predict that the additional contacts between the long side chain of POS and AF-CYP51A protein compensate for the perturbation in the vicinity of the heme.
(ii) C. albicans.
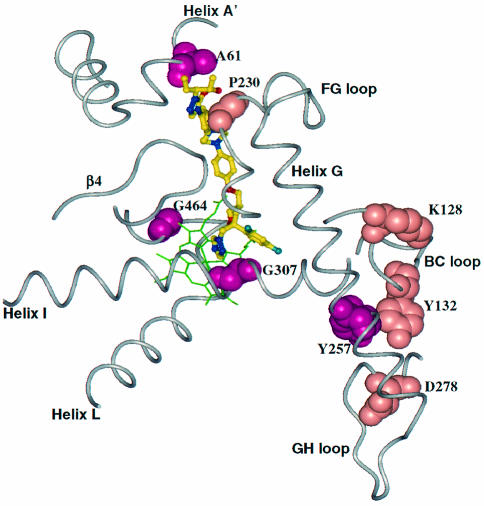
An analysis of two series of C. albicans isolates, from two different patients, revealed that stepwise reductions in azole susceptibility were accompanied by the appearance of specific substitutions in CYP51. The first isolate from patient I had two substitutions in CA-CYP51, Tyr257His and Gly464Ser (Fig. 6). Tyr257 is far from the azole binding site and its effect on azole binding is not immediately obvious (Table 2). Gly464 corresponds to Gly448 in AF-CYP51A and is therefore predicted to disturb the heme environment. As seen for the analogous substitution in A. fumigatus, Gly464Ser resulted in a larger decrease in susceptibility to VOR and FLZ than to POS. The second isolate from patient I acquired a third substitution, Gly307Ser. Since Gly307 is located on helix I and contacts both the triazole and phenyl rings, the substitution would be predicted to negatively impact binding of all azoles through steric conflicts between the polar Ser side chain with the triazole and phenyl rings. In practice, the substitution impacted binding of VOR more than POS. The final isolate from patient I acquired a fourth substitution, Ala61Val. Like Gly54 in AF-CYP51A, Ala61 is located on helix A′. Consequently, the additional increase in the MIC of POS most likely resulted from VDW conflicts between the large Val side chain and the end of the POS side chain.
FIG. 6.
Mapping of substitutions in CA-CYP51 that cause azole resistance. The heme cofactor is highlighted in green. The azoles are shown as ball-and-stick models with their carbon atoms in yellow. The atoms of the residues at the sites of substitutions are depicted using space filling models.
TABLE 2.
Substitutions in CA-CYP51 resulting in azole resistance in clinical C. albicans isolates
| Patient and isolate | MIC (μg/ml)
|
Substitutions in CA-CYP51 | |||
|---|---|---|---|---|---|
| POS | FLZ | ITZ | VOR | ||
| C43c | 0.03 | 0.125 | 0.06 | 0.03 | None (azole-sensitive reference strain) |
| Ia | |||||
| C438 | 0.5 | 128 | 0.5 | 2 | Tyr257His, Gly464Ser |
| C439 | 1 | >256 | 4 | 16 | Tyr257His, Gly464Ser, Gly307Ser |
| C440 | 4 | >256 | 8 | >16.0 | Tyr257His, Gly464Ser, Gly307Ser, Ala61Val |
| IIb | |||||
| C369 | 0.25 | 32 | 1 | 4 | Lys128Thr, Tyr132His, Asp278Asn, Gly464Ser |
| C375 | 2 | 48 | 16 | 4 | Lys128Thr, Tyr132His, Asp278Asn, Gly464Ser, Pro230Leu |
MICs and data on mutations from A. Chau et al., unpublished data.
MICs and data on mutations from reference 10a.
Reference strain (not a patient isolate).
The first isolate from patient II had four substitutions in CA-CYP51 and exhibited a larger decrease in susceptibility to VOR and FLZ than for POS and ITZ (Table 2). The MIC changes can most likely be attributed to the Gly464Ser substitution, as the other three substitutions (Lys128Thr, Asp278Asn, and Tyr132His) are relatively far from the drug binding site (Fig. 6). The next isolate from patient II acquired a fifth substitution, Pro230Leu, which primarily impacted the MICs of POS and ITZ. Pro230 is on the FG loop and is predicted to be in close contact with the POS side chain. The substitution of Leu for Pro at this position would likely affect the binding of POS and ITZ, but not VOR and FLZ, through VDW conflicts between the Leu side chain and POS and ITZ long side chains.
DISCUSSION
The discovery and refinement of azole drugs has relied entirely on whole-cell testing (2). Structure based approaches to designing more efficacious azoles have been stymied by the fact that the drug target site, CYP51, is an integral membrane protein and has proven difficult to purify and crystallize. Biostructural assessments of the drug-binding determinants have been limited to what could be inferred from more-distant relatives in the P450 superfamily. However, the recent elucidation of the crystal structure of the P450 14α-sterol demethylase from M. tuberculosis, coupled with the identification of substitutions in fungal CYP51 enzymes that confer resistance to specific structural classes of azoles, has provided an opportunity to construct homology models that are potentially superior to those based on other bacterial P450 structures (1, 6, 7, 9, 18, 26).
This study provides a model for the binding of azoles with extended side chains such as POZ and ITZ. The key finding from our work is that the long side chains of POS and ITZ appear to be contained within channel 2 and consequently make extensive hydrophobic contacts along their entire lengths. This channel is evident in the X-ray structure of MT-CYP51 as a cavity of 2,600 Å3 (18). The long-chain azoles span channel 2, extending from the heme to the surface of the protein, ending in a region surrounded by helix A′, the β-turn connecting β4-1 and β4-2, and the FG loop (compare Fig. 2 and 4). The model elegantly explains how a substitution, at Gly54 in the A′ helix of AF-CYP51A, confers resistance to POS and ITZ by perturbing the binding of the long side chain in channel 2. Similarly, substitutions in either the A′ helix (at residue Ala61) or the FG loop (at residue Pro230) of CA-CYP51, in combination with substitutions near the heme site, specifically impact resistance to POS by the same mechanism. However, unlike Gly54 in AF-CYP51A, substitutions in CA-CYP51 at Ala61 or Pro230 alone did not confer reduced susceptibility to POS when the corresponding CYP51 allele was expressed in Saccharomyces cerevisiae (10a; A. Chau et al., unpublished data).
From the model, it seems probable that the added interactions furnished by the long side chain of POS and ITZ would result in inherently tighter binding affinities compared with azoles lacking such substituents. Although precise biophysical measurements are lacking to prove this hypothesis, the resistance data appear to support this contention. Specifically, mutations near the heme site, which result in significant levels of resistance to FLZ and VOR in C. albicans and A. fumigatus, have far less impact on the susceptibility of the organisms to POS and ITZ (this study; Sanglard et al., 42nd ICAAC; Manavathu et al, 41st ICAAC). We propose that the lack of impact of these mutations on POS and ITZ results from inherently tighter affinity and/or reflects compensatory adjustments for binding made along channel 2 by these drugs. This hypothesis is in direct contrast to the work of Ji et al. (7). These workers constructed a CA-CYP51 model based on the bacterial P450BM3 X-ray structure and evaluated the binding of ITZ. Although they also placed the long side chain of ITZ in a channel, the side chain reached the surface of the protein at a different location (surrounded by helix A, β1-1, β1-2, and the FG loop); their alignment for the A′ helix also differed from the one described here. These are both features that in the present model help explain how particular mutations specifically impact the binding of POS and ITZ. Based on their model, Ji et al. (7) asserted that “the side chains of the inhibitors were not the determinants of activity” but merely adjusted physicochemical or pharmacokinetic attributes. Our model for POS binding, along with the supporting resistance data, argues for a much more significant role of the side chains in determining activity. Furthermore, our model may explain differential effects of substituents on the phenyl side chain close to the heme. The MIC data suggested that the Glu and Arg substitutions of A. fumigatus residue Gly54 impact the binding of ITZ more than POS. This may be because the larger substituents on the phenyl ring of ITZ, in a sterically restricted pocket, exacerbate the effects of the substitutions at Gly54.
Two other CA-CYP51 models based on MT-CYP51 were recently published (4, 11). Neither model addressed the binding of long-chain azoles. In addition, both publications used sequence alignments that differ from ours, which resulted in differences in both the pockets close to the heme, as well as in the remote binding determinants for POS and ITZ.
The present study provides the first description of a potential binding mode for azoles in CYP51A from A. fumigatus and may explain how specific mutations in AF-CYP51A confer resistance to one structural group of azoles (e.g., the compact azoles FLZ and VOR) but not to another (e.g., extended azoles ITZ and POS) and vice versa. The model also provides a working framework for the rational design of other azoles with extended substituents that could similarly exploit channel 2 for added binding interactions.
Acknowledgments
We thank Zhuyan Guo (SPRI) for many helpful discussions, Eduardo Zaborowski (SPRI) for providing computer application software support, Elias Manavathu (Wayne State University, Detroit, Mich.) for communicating unpublished data, and Todd Black (SPRI) and Michelle Trietel (SPRI) for editing suggestions.
REFERENCES
- 1.Boscott, P. E., and G. H. Grant. 1994. Modeling cytochrome P450 14 alpha demethylase (Candida albicans) from P450cam. J. Mol. Graph. 12:185-195. [DOI] [PubMed] [Google Scholar]
- 2.D'Arcy, P. F., and E. M. Scott. 1978. Antifungal agents. Prog. Drug Res. 22:93-147. [DOI] [PubMed] [Google Scholar]
- 3.Diaz-Guerra, T. M., E. Mellado, M. Cuenca-Estrella, and J. L. Rodriguez-Tudela. 2003. A point mutation in the 14α-sterol demethylase gene cyp51A contributes to itraconazole resistance in Aspergillus fumigatus. Antimicrob. Agents Chemother. 47:1120-1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fukuoka, T., D. A. Johnston, C. A. Winslow, M. J. de Groot, C. A. Hitchcock, and S. G. Filler. 2003. Genetic basis for differential activities of fluconazole and voriconazole against Candida krusei. Antimicrob. Agents Chemother. 47:1213-1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Graham, S. E., and J. A. Peterson. 1999. How similar are P450s and what can their differences teach us? Arch. Biochem. Biophys. 369:24-29. [DOI] [PubMed] [Google Scholar]
- 6.Holtje, H. D., and C. Fattorusso. 1998. Construction of a model of the Candida albicans lanosterol 14-alpha-demethylase active site using the homology modeling technique. Pharm. Acta Helv. 72:271-277. [DOI] [PubMed] [Google Scholar]
- 7.Ji, H., W. Zhang, Y. Zhou, M. Zhang, J. Zhu, Y. Song, J. Lu, and J. Zhu. 2000. A three-dimensional model of lanosterol 14α-demthylase of Candida albicans and its interaction with azole antifungals. J. Med. Chem. 43:2493-2505. [DOI] [PubMed] [Google Scholar]
- 8.Jones, G., P. Willet, R. C. Glen, A. R. Leach, and R. Taylor. 1997. Development and validation of a genetic algorithm for flexible docking. J. Mol. Biol. 267:727-748. [DOI] [PubMed] [Google Scholar]
- 9.Lewis, D. F., A. Wiseman, and M. H. Tarbit. 1999. Molecular modeling of lanosterol 14 alpha-demethylase (CYP51) from Saccharomyces cerevisiae via homology with CYP102, a unique bacterial cytochrome P450 isoform: quantitative structure-activity relationships (QSARs) within two related series of antifungal azole derivatives. J. Enzyme Inhib. 14:175-192. [DOI] [PubMed] [Google Scholar]
- 10.Li, H., and T. L. Poulos. 1997. The structure of the cytochrome p450BM-3 haem domain complexed with the fatty acid substrate, palmitoleic acid. Nat. Struct. Biol. 4:140-146. [DOI] [PubMed] [Google Scholar]
- 10a.Li, X., N. Brown, A. S. Chau, J. L. López-Ribot, M. T. Ruesga, G. Quindos, C. A. Mendrick, R. S. Hare, D. Loebenberg, B. DiDomenico, and P. M. McNicholas. 4December2003. Changes in susceptibility to posaconazole in clinical isolates of Candida albicans. J. Antimicrob. Chemother. 10.1093/jac/dkh027.. [DOI] [PubMed]
- 11.Macchiarulo, A., G. A. Costantino, D. Fringuelli, A. Vecchiarelli, F. Schiaffella, and R. Fringuelli. 2002. 1,4-Benzothiazine and 1,4-benzoxazine imidazole derivatives with antifungal activity: a docking study. Bioorg. Med. Chem. 10:3415-3423. [DOI] [PubMed] [Google Scholar]
- 12.Manavathu, E. K., J. L. Cutright, D. Loebenberg, and P. H. Chandrasekar. 2000. A comparative study of the in vitro susceptibilities of clinical and laboratory-selected resistant isolates of Aspergillus spp. to amphotericin B, itraconazole, voriconazole and posaconazole (SCH 56592). J. Antimicrob. Chemother. 46:229-234. [DOI] [PubMed] [Google Scholar]
- 13.Mann, P. A., R. Parmegiani, S.-Q. Wei, C. A. Mendrick, X. Li, D. Loebenberg, R. S. Hare, S. S. Walker, B. J. DiDomenico, and P. M. McNicholas. 2003. Mutations in Aspergillus fumigatus resulting in reduced susceptibility to posaconazole appear to be restricted to a single amino acid in the cytochrome P450 14α-demethylase. Antimicrob. Agents Chemother. 47:577-581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mellado, E., T. M. Diaz-Guerra, M. Cuenca-Estrella, and J. L Rodriguez-Tudela. 2001. Identification of two different 14-alpha sterol demethylase-related genes (cyp51A and cyp51B) in Aspergillus fumigatus and other Aspergillus species. J. Clin. Microbiol. 39:2431-2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nascimento, A. M., G. H. Goldman, S. Park, S. A. Marras, G. Delmas, U. Oza, K. Lolans, M. N. Dudley, P. A. Mann, and D. S. Perlin. 2003. Multiple resistance mechanisms among Aspergillus fumigatus mutants with high-level resistance to itraconazole. Antimicrob. Agents Chemother. 47:1719-1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peterson, J. A., and S. E. Graham. 1998. A close family resemblance: the importance of structure in understanding cytochromes P450. Structure 6:1079-1085. [DOI] [PubMed] [Google Scholar]
- 17.Pfaller, M. A., S. A. Messer, R. J. Hollis, and R. N. Jones. 2001. In vitro activities of posaconazole (SCH 56592) compared with those of itraconazole and fluconazole against 3685 clinical isolates of Candida spp. and Cryptococcus neoformans. Antimicrob. Agents Chemother. 45:2862-2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Podust, L. M., T. L. Poulos, and M. R. Waterman. 2001. Crystal structure of cytochrome P450 14α-sterol demethylase (CYP51) from Mycobacterium tuberculosis in complex with azole inhibitors. Proc. Natl. Acad. Sci. USA 98:3068-3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Podust, L. M., J. Stojan, T. L. Poulos, and M. R. Waterman. 2001. Substrate recognition sites in 14α-sterol demethylase from comparative analysis of amino acid sequences and X-ray structure of Mycobacterium tuberculosis CYP51. J. Inorg. Biochem. 87:227-235. [DOI] [PubMed] [Google Scholar]
- 20.Poulos, T. L. 1995. Cytochrome P450. Curr. Opin. Struct. Biol. 5:767-774. [DOI] [PubMed] [Google Scholar]
- 21.Sali, A., and T. L. Blundell. 1993. Comparative protein modeling by satisfaction of spatial restraints. J. Mol. Biol. 234:779-815. [DOI] [PubMed] [Google Scholar]
- 22.Sanchez, R., and A. Sali. 1997. Advances in comparative protein-structure modeling. Curr. Opin. Struct. Biol. 7:206-214. [DOI] [PubMed] [Google Scholar]
- 23.Sanchez, R., and A. Sali. 1998. Large-scale protein structure modeling of the Saccharomyces cerevisiae genome. Proc. Natl. Acad. Sci. USA 95:13597-13602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sanglard, D., and F. C. Odds. 2002. Resistance of Candida species to antifungal agents: molecular mechanisms and clinical consequences. Lancet ii:73-85. [DOI] [PubMed] [Google Scholar]
- 25.Sheehan, D. J., C. A. Hitchcock, and C. M. Sibley. 1999. Current and emerging azole antifungal agents. Clin. Microbiol. Rev. 12:40-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tsukuda, T., Y. Shiratori, M. Watanabe, H. Ontsuka, K. Hattori, M. Shirai, and N. Shimma. 1998. Modeling, synthesis and biological activity of novel antifungal agents. Bioorg. Med. Chem. Lett. 8:1819-1824. [DOI] [PubMed] [Google Scholar]
- 27.Vanden Bossche, H., G. Willemsens, and P. Marichal. 1987. Anti-Candida drugs-the biochemical basis for their activity. Crit. Rev. Microbiol. 15:57-72. [DOI] [PubMed] [Google Scholar]
- 28.Wester, M. R., E. F. Johnson, C. Marques-Soares, P. M. Dansette, D. Mansuy, and C. D. Stout. 2003. Structure of a substrate complex of mammalian cytochrome P450 2C5 at 2.3 Å resolution: evidence for multiple substrate binding modes. Biochemistry 42:6370-6379. [DOI] [PubMed] [Google Scholar]
- 29.Williams, P., J. Cosme, V. Sridhar, E. F. Johnson, and D. E. McRee. 2000. Mammalian microsomal cytochrome P450 monooxygenase: structural adaptations for membrane binding and functional diversity. Mol. Cell 5:121-131. [DOI] [PubMed] [Google Scholar]