Abstract
Replacement of the α4 helix of ParC with that of GyrA increased the stability of topoisomerase IV-quinolone-DNA ternary complexes. This mutant topoisomerase IV-mediated cell killing was more efficient than topoisomerase IV-mediated cell killing in Escherichia coli. Thus, the α4 helix plays critical roles in determining the stability and the cytotoxicity of ternary complexes.
DNA gyrase, the first topoisomerase to be identified as a target of quinolone antibacterial drugs (6, 24), is the primary target of quinolone drugs in Escherichia coli (3, 22), whereas topoisomerase IV (Topo IV) is the primary target in some gram-positive bacteria, such as Staphylococcus aureus and Streptococcus pneumoniae (4, 17, 18). These observations have suggested that gyrase and Topo IV are the primary targets in gram-negative and gram-positive bacteria, respectively. However, more recent studies have demonstrated that each quinolone drug has a preferred target and that the target selection can be altered in some bacteria by changes in quinolone structure (5, 19). DNA gyrase remains the primary target of quinolone drugs in E. coli (3, 22). It has been shown that gyrase-mediated cell killing is more efficient than Topo IV-mediated cell killing in E. coli (12). It is not clear what makes ternary complexes formed with E. coli gyrase more cytotoxic than those formed with E. coli Topo IV.
Quinolone resistance-conferring mutations are clustered within a small region (often referred to as the quinolone resistance-determining region) of the gyrA and parC genes (3, 22). Structures of type II topoisomerases have demonstrated that the quinolone resistance-determining region is located in the helix-turn-helix region of the catabolite activator protein-like domain (1, 15). The hot spots for quinolone resistance-conferring mutations, Ser83 and Asp87 of E. coli GyrA, are located in the α4 helix (1, 15), suggesting that the α4 helix directly interacts with the quinolone drug (3, 22). Observations that homologous mutations in the α4 helices of E. coli ParC, the catalytic subunit of Topo IV, and Saccharomyces cerevisiae topoisomerase II alter sensitivity to quinolone drugs support this model (7, 11, 12).
We conducted a domain-swapping experiment to reveal the significance of the different amino acid residues in the α4 helices of E. coli ParC (residues 79 to 90, DSACYEAMVLMA) and E. coli GyrA (residues 82 to 93, DSAVYDTIVRMA). The overlap extension PCR technique (16) was used to replace the entire α4 helix of ParC (5′-gatagcgcctgttatgaagcgatggtcctgatggcg-3′) with that of GyrA (5′-gatagcgccGTttatgaCAcgatCgtccGgatggcg-3′; capital letters represent mutated nucleotides). pET11-parC (14) was used as the template for the PCR, and DNA sequences of the open reading frame were confirmed by dideoxy DNA sequencing. This mutant parC, parC Aα4, was cloned into the pET-11c vector (23). ParC Aα4 was expressed in E. coli HMS174(DE3) (Novagen) and purified according to the method of Hiasa et al. (10). The final preparation of mutant ParC protein was greater than 97% homogeneous for a single band in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (data not shown). This mutant ParC protein was mixed with the wild-type ParE to reconstitute ParC Aα4 Topo IV.
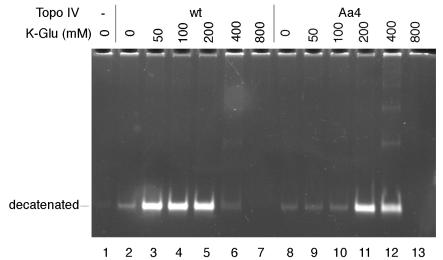
During the initial characterization, the specific activity of ParC Aα4 Topo IV in a kinetoplast DNA (kDNA) decatenation assay appeared to be less than 1% of that of the wild-type Topo IV (data not shown). However, DNA-binding and cleavage assays showed that ParC Aα4 Topo IV could bind and cleave DNA to a similar extent as Topo IV (data not shown; see the following section). The reaction mixtures for the kDNA decatenation assay contained 100 mM potassium glutamate (K-Glu) (7, 14). It was possible that optimal salt concentrations for the catalytic activities of Topo IV and ParC Aα4 Topo IV might be different. To directly examine this possibility, the effects of K-Glu on the decatenation activities of Topo IV and ParC Aα4 Topo IV were assessed (Fig. 1). Topo IV-catalyzed decatenation of kDNA was most efficient in the presence of 50 to 200 mM K-Glu. In contrast, the maximum ParC Aα4 Topo IV-catalyzed decatenation activity was observed when the K-Glu concentration was 200 to 400 mM (Fig. 1). The specific activity of ParC Aα4 Topo IV measured at 300 mM K-Glu was 20 to 30% of that of Topo IV measured at 100 mM K-Glu (data not shown). The inhibitory effects of norfloxacin on the decatenation activities of Topo IV and ParC Aα4 Topo IV were also assessed under these conditions. We found that the quinolone sensitivity of ParC Aα4 Topo IV was similar to that of Topo IV (50% inhibitory concentration, 10 to 15 μM) when the same amounts of enzymes were used in the kDNA decatenation assay. These results suggested that the replacement of the α4 helix of ParC with that of GyrA affected the Topo IV-DNA interaction and altered the catalytic activity, but not the quinolone sensitivity, of Topo IV. The replacement of the α4 helix of ParC with that of GyrA introduced three nonconserved and two conserved changes into the α4 helix of ParC. One of these changes may be responsible for the altered catalytic activity and salt sensitivity of ParC Aα4 Topo IV. Alternatively, these changes may have additive effects on Topo IV. We will conduct a systematic mutational analysis of ParC to identify the amino acid(s) responsible for the altered activity of ParC Aα4 Topo IV.
FIG. 1.
Effects of K-Glu on ParC Aα4 Topo IV-catalyzed decatenation activity. The kDNA decatenation assay was performed in the presence of either 10 fmol (as tetramer) of the wild-type Topo IV (lanes 2 to 7) or 40 fmol (as tetramer) of ParC Aα4 Topo IV (lanes 8 to 13), 0.3 μg of kDNA, and the indicated concentrations of K-Glu. Relative amounts (100% = 0.3 μg) of decatenated kDNA were as follows: lane 1, 0%; lane 2, 12%; lane 3, 64%; lane 4, 58%; lane 5, 58%; lane 6, 5%; lane 7, 0%; lane 8, 3%; lane 9, 4%; lane 10, 6%; lane 11, 57%; lane 12, 49%; and lane 13, 0%. wt, wild-type Topo IV; Aa4, ParC Aα4 Topo IV.
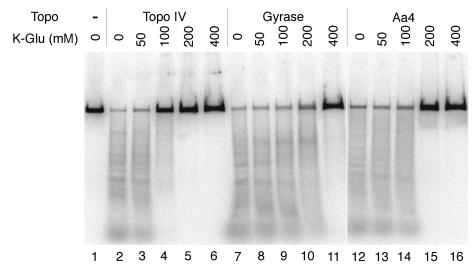
Next, a DNA cleavage assay was used to determine the stability of ternary complexes formed with ParC Aα4 Topo IV, the wild-type Topo IV, or gyrase. The DNA cleavage assay was performed as described previously (7, 9) by using 3′-end-labeled linear pBR322 DNA as a substrate (Fig. 2). In the absence of K-Glu, the extent of norfloxacin-stimulated, ParC Aα4 Topo IV-catalyzed cleavage was similar to that of norfloxacin-stimulated, Topo IV- and gyrase-catalyzed cleavages. These results also showed that the replacement of the α4 helix of ParC with that of GyrA did not affect the quinolone sensitivity of Topo IV. Topo IV-norfloxacin-DNA ternary complexes were more sensitive to salt than gyrase-norfloxacin-DNA ternary complexes, indicating that ternary complexes formed with gyrase were more stable than those formed with Topo IV. Interestingly, ternary complexes formed with ParC Aα4 Topo IV were more stable than those formed with Topo IV (Fig. 2). Thus, the replacement of the α4 helix of ParC with that of GyrA increased the stability of ternary complexes. These results suggested that the amino acid residues in the α4 helix of the catalytic domain of the bacterial type II topoisomerases could modulate the stability of topoisomerase-quinolone-DNA ternary complexes.
FIG. 2.
Ternary complexes formed with ParC Aα4 Topo IV are more stable than those formed with the wild-type Topo IV. The DNA cleavage was performed in the presence of 20 fmol (as molecule) of 32P-labeled linear pBR322 DNA; 50 μM norfloxacin; 100 fmol (as tetramer) of Topo IV (lanes 2 to 6), gyrase (lanes 7 to 11), or ParC Aα4 Topo IV (lanes 12 to 16); and the indicated concentrations of K-Glu. Aa4, ParC Aα4 Topo IV.
Khodursky et al. have demonstrated that gyrase-mediated cell killing is more efficient than Topo IV-mediated cell killing in E. coli (12). We found that gyrase-norfloxacin-DNA ternary complexes were more stable than those formed with Topo IV (Fig. 2) and hypothesized that the stability of ternary complexes may correlate with the cytotoxicity of ternary complexes. To examine this possibility, the cytotoxicity of ternary complexes formed with gyrase, Topo IV, or ParC Aα4 Topo IV was assessed in E. coli. We developed an assay system to measure the cytotoxicity of topoisomerase-quinolone-DNA ternary complexes by using E. coli 1596 (C600 with gyrA and parC genes carrying resistance mutations) as a host strain. This strain carries quinolone resistance-conferring mutations corresponding to the conserved Ser in both gyrA and parC genes on the chromosome and exhibits a high level of resistance to quinolone drugs (12). Thus, when strain 1596 is transformed with a plasmid carrying either the gyrA or parC gene, the quinolone sensitivity of the strain depends on the quinolone sensitivity of the topoisomerase gene on the plasmid.
The pET system is not ideal to control the expression level of a cloned protein and perform genetic experiments. However, it has been shown that a pET vector carrying the parC gene, even if its expression is not induced, can complement the temperature-sensitive phenotype of C600parC1215 (14). It is likely that a low level of background expression, in the absence of T7 RNA polymerase, is sufficient to produce enough ParC to substitute for the inactive mutant protein in E. coli. The background expression of the parC gene is very low, which is important because the overproduction of ParC alone is known to be toxic (20). Thus, a pET vector seems to be a sufficient system to measure the cytotoxicity of ternary complexes formed with gyrase, Topo IV, or ParC Aα4 Topo IV.
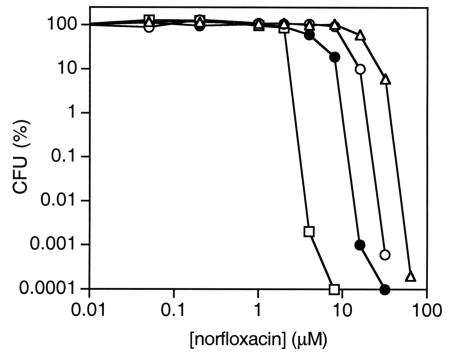
A plating assay (12) was performed to measure the inhibitory effect of norfloxacin on the bacterial growth and determine the effectiveness of gyrase-, Topo IV-, or ParC Aα4 Topo IV-mediated cell killing (Fig. 3). Strain 1596 was transformed with plasmid DNAs, pET-11c, pET11-gyrA, pET11-parC, and pET11-parC Aα4. Transformed cells were grown to mid-logarithmic phase (optical density at 600 nm, 0.5) in LB and plated onto LB agar plates containing various concentrations of norfloxacin. The quinolone sensitivity was determined by comparing the numbers of colonies formed on plates in the presence of various concentrations of norfloxacin to those formed on plates in the absence of norfloxacin (12). As shown in Fig. 3, gyrase-mediated cell killing (99.99% inhibitory concentration [IC99.99] = 4 μM) was more efficient than Topo IV-mediated cell killing (IC99.99 = 24 μM), which coincided well with previous observations (12). Thus, the plating assay using E. coli 1596 transformed with a plasmid carrying a topoisomerase gene could be used to assess the cytotoxicity of ternary complexes formed with the topoisomerase. ParC Aα4 Topo IV-mediated cell killing (IC99.99 = 12 μM) was more efficient than Topo IV-mediated cell killing (Fig. 3). ParC Aα4 Topo IV-norfloxacin-DNA ternary complexes were more cytotoxic than Topo IV-norfloxacin-DNA ternary complexes in E. coli. These results suggested that the stability of a ternary complex may correlate with increased quinolone susceptibility.
FIG. 3.
ParC Aα4 Topo IV-norfloxacin-DNA ternary complexes are more cytotoxic than Topo IV-norfloxacin-DNA ternary complexes. Topo IV-, gyrase-, and ParC Aα4 Topo IV-mediated cell killing was measured in E. coli. The plating assay was performed using strain 1596 carrying pET-11c (open triangles), pET11-parC (open circles), pET11-gyrA (open squares), and pET11-parC Aα4 (closed circles).
Ternary complexes formed with ParC Aα4 Topo IV were more stable than those formed with Topo IV but less stable than those formed with gyrase (Fig. 2). Similarly, ParC Aα4 Topo IV-mediated cell killing was more efficient than Topo IV-mediated cell killing but less efficient than gyrase-mediated cell killing (Fig. 3). These results indicate that, although the α4 helix modulates both the stability and the cytotoxicity of topoisomerase-quinolone-DNA ternary complexes, there must be other factors that also influence the stability and the cytotoxicity of ternary complexes. It is not clear what these factors may be. One model, proposed based on the action of quinolone drugs against E. coli (10, 12), is that the locations of gyrase and Topo IV, relative to advancing replication forks, may affect the effectiveness of cell killing. In E. coli, gyrase is thought to act in front of replication forks, whereas Topo IV acts behind forks (8, 21). Thus, ternary complexes formed with gyrase more frequently collide with replication forks and trigger the cytotoxic events than those formed with Topo IV. Another possibility is that subtle differences in amino acid residues around the catalytic site Tyr, including those in the α4 helix, between GyrA and ParC may have additive effects on the interactions among the topoisomerase, the quinolone drug, and the DNA in a ternary complex and affect the stability and the cytotoxicity of the ternary complex (7). Structural studies of type II topoisomerases also support the model that the α4 helix may directly interact with the DNA (1, 15). It seems reasonable to assume that the protein level of topoisomerases in vivo is a critical factor (13) which may not only determine the number of ternary complexes formed on the chromosome but also influence the superhelicity of the chromosome. Further studies are necessary to examine these possibilities.
DNA gyrase and Topo IV are highly homologous with each other (2, 25). However, results described here suggest that gyrase and Topo IV may interact, at their catalytic domains, with the DNA and/or the quinolone drug in distinct manners. Amino acid residues near the active site, especially those in the α4 helix, are likely to be responsible for the drug sensitivity of a topoisomerase and the cytotoxicity of a topoisomerase-quinolone-DNA ternary complex. Thus, subtle differences in amino acid residues between GyrA and ParC may influence the selection of the primary target by a quinolone drug in vivo.
Acknowledgments
We thank Nicholas Cozzarelli for providing E. coli strains and Arkady Khodursky for comments on these studies.
This work was supported by grants from the National Institutes of Health (GM59465), the University of Minnesota Graduate School Grant-in-Aid Program, and the Minnesota Medical Foundation.
REFERENCES
- 1.Berger, J. M., S. J. Gamblin, S. C. Harrison, and J. C. Wang. 1996. Structure and mechanism of DNA topoisomerase II. Nature 379:225-232. [DOI] [PubMed] [Google Scholar]
- 2.Champoux, J. J. 2001. DNA topoisomerases: structure, function, and mechanism. Annu. Rev. Biochem. 70:369-413. [DOI] [PubMed] [Google Scholar]
- 3.Drlica, K., and M. Malik. 2003. Fluoroquinolones: action and resistance. Curr. Top. Med. Chem. 3:249-282. [DOI] [PubMed] [Google Scholar]
- 4.Ferrero, L., B. Cameron, B. Manse, D. Lagneaux, J. Crouzet, A. Famechon, and F. Blanche. 1994. Cloning and primary structure of Staphylococcus aureus DNA topoisomerase IV: a primary target of fluoroquinolones. Mol. Microbiol. 13:641-653. [DOI] [PubMed] [Google Scholar]
- 5.Fournier, B., X. Zhao, T. Lu, K. Drlica, and D. C. Hooper. 2000. Selective targeting of topoisomerase IV and gyrase in Staphylococcus aureus: different patterns of quinolone-induced inhibition of DNA synthesis. Antimicrob. Agents Chemother. 44:2160-2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gellert, M., K. Mizuuchi, M. H. O'Dea, T. Itoh, and J. Tomizawa. 1977. Nalidixic acid resistance: a second genetic character involved in DNA gyrase activity. Proc. Natl. Acad. Sci. USA 74:4772-4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hiasa, H. 2002. The Glu-84 of the ParC subunit plays critical roles in both topoisomerase IV-quinolone and topoisomerase IV-DNA interactions. Biochemistry 41:11779-11785. [DOI] [PubMed] [Google Scholar]
- 8.Hiasa, H., and K. J. Marians. 1996. Two distinct modes of strand unlinking during θ-type DNA replication. J. Biol. Chem. 271:21529-21535. [DOI] [PubMed] [Google Scholar]
- 9.Hiasa, H., and M. E. Shea. 2000. DNA gyrase-mediated wrapping of the DNA strand is required for the replication fork arrest by the DNA gyrase-quinolone-DNA ternary complex. J. Biol. Chem. 275:34780-34786. [DOI] [PubMed] [Google Scholar]
- 10.Hiasa, H., D. O. Yousef, and K. J. Marians. 1996. DNA strand cleavage is required for replication fork arrest by a frozen topoisomerase-quinolone-DNA ternary complex. J. Biol. Chem. 271:26424-26429. [DOI] [PubMed] [Google Scholar]
- 11.Hsiung, Y., S. H. Elsea, N. Osheroff, and J. L. Nitiss. 1995. A mutation in yeast TOP2 homologous to a quinolone-resistant mutation in bacteria: mutation of the amino acid homologous to Ser83 of Escherichia coli gyrA alters sensitivity to eukaryotic topoisomerase inhibitors. J. Biol. Chem. 270:20359-20364. [DOI] [PubMed] [Google Scholar]
- 12.Khodursky, A. B., E. L. Zechiedrich, and N. R. Cozzarelli. 1995. Topoisomerase IV is a target of quinolones in Esherichia coli. Proc. Natl. Acad. Sci. USA 92:11801-11805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kreuzer, K. N., and N. R. Cozzarelli. 1979. Escherichia coli mutants thermosensitive for deoxyribonucleic acid gyrase subunit A: effects on deoxyribonucleic acid replication, transcription, and bacteriophage growth. J. Bacteriol. 140:424-435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lavasani, L. S., and H. Hiasa. 2001. A ParE-ParC fusion protein is a functional topoisomerase. Biochemistry 40:8438-8443. [DOI] [PubMed] [Google Scholar]
- 15.Morais Cabral, J. H., A. P. Jackson, C. V. Smith, N. Shikotra, A. Maxwell, and R. C. Liddington. 1997. Crystal structure of the breakage-reunion domain of DNA gyrase. Nature 388:903-906. [DOI] [PubMed] [Google Scholar]
- 16.Morton, R. M., H. D. Hunt, S. N. Ho, J. K. Pullen, and L. R. Pease. 1993. Gene splicing by overlap extension. Methods Enzymol. 217:270-279. [DOI] [PubMed] [Google Scholar]
- 17.Pan, X.-S., J. Ambler, S. Mehtar, and L. M. Fisher. 1996. Involvement of topoisomerase IV and DNA gyrase as ciprofloxacin target in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 40:2321-2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pan, X.-S., and L. M. Fisher. 1996. Cloning and characterization of the parC and parE genes of Streptococcus pneumoniae encoding DNA topoisomerase IV: role in fluoroquinolone resistance. J. Bacteriol. 178:4060-4069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pan, X.-S., and L. M. Fisher. 1997. Targeting of DNA gyrase in Streptococcus pneumoniae by sparfloxacin: selective targeting of gyrase or topoisomerase IV by quinolones. Antimicrob. Agents Chemother. 41:471-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peng, H., and K. J. Marians. 1993. Escherichia coli topoisomerase IV: purification, characterization, subunit structure, and subunit interactions. J. Biol. Chem. 268:24481-24490. [PubMed] [Google Scholar]
- 21.Peter, B. J., C. Ullsperger, H. Hiasa, K. J. Marians, and N. R. Cozzarelli. 1998. The structure of supercoiled intermediates in DNA replication. Cell 94:819-827. [DOI] [PubMed] [Google Scholar]
- 22.Reece, R. J., and A. Maxwell. 1991. DNA gyrase: structure and function. Crit. Rev. Biochem. Mol. Biol. 26:335-375. [DOI] [PubMed] [Google Scholar]
- 23.Studier, W. F., A. H. Rosenberg, and J. J. Dunn. 1990. Use of T7 RNA polymerase to direct the expression of cloned genes. Methods Enzymol. 185:60-89. [DOI] [PubMed] [Google Scholar]
- 24.Sugino, A., C. L. Peebles, K. N. Kreuzer, and N. R. Cozzarelli. 1977. Mechanism of action of nalidixic acid: purification of Escherichia coli nalA gene product and its relationship to DNA gyrase and a novel nicking-closing enzyme. Proc. Natl. Acad. Sci. USA 74:4767-4771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang, J. C. 1996. DNA topoisomerases. Annu. Rev. Biochem. 65:635-692. [DOI] [PubMed] [Google Scholar]