Abstract
Staphylococcus aureus and Staphylococcus epidermidis ferritin (FtnA and SefA, respectively) homologues are antigenic and highly conserved. A previous study showed that ftnA is a component of the S. aureus PerR regulon with its transcription induced by elevated iron and repressed by PerR, which functions as a manganese-dependent transcriptional repressor. We have further investigated the role of iron and Fur in the regulation of PerR regulon genes ftnA (ferritin), ahpC (alkyl-hydroperoxidase), and mrgA (Dps homologue) and shown that iron has a major role in the regulation of the PerR regulon and hence the oxidative stress response, since in the presence of both iron and manganese, transcription of PerR regulon genes is induced above the repressed levels observed with manganese alone. Furthermore the PerR regulon genes are differentially regulated by metal availability and Fur. First, there is an additional level of PerR-independent regulation of ftnA under low-iron conditions which is not observed with ahpC and mrgA. Second, there is a differential response of these genes to Fur as ftnA expression is constitutive in a fur mutant, while ahpC expression is constitutive under low-Fe/Mn conditions but some repression of ahpC still occurs in the presence of manganese, whereas mrgA expression is still repressed in the fur mutant as in wild-type S. aureus, although there is a decrease in the overall level of mrgA transcription. These studies have also shown that FtnA expression is regulated by growth phase, but maximal transcription of ftnA differs dependent on the growth medium. Moreover, there are significant regulatory differences between the S. aureus and S. epidermidis ferritins, as sefA expression in contrast to that of ftnA is derepressed under low-Fe/Mn ion conditions.
Iron is an essential micronutrient, which is required for many key metabolic processes in both bacterial and mammalian cells. However, since high levels of free intracellular ferric iron are toxic to cells, the amount of free iron in the cytoplasm must be strictly regulated. In bacteria, global regulators such as Fur usually mediate this iron regulation. Three Fur homologues have been identified in Staphylococcus aureus, Fur, PerR, and Zur. Fur is an iron-dependent repressor that tightly regulates the expression of high-affinity iron transporter mechanisms in S. aureus (10, 20), while PerR responds to both metal ion limitation and peroxide stress (10, 11). Analysis of PerR-mediated gene regulation has shown that while there are many similarities in the PerR regulatory pathways in different bacteria, there are also significant differences in the response of PerR to environmental signals. Bacillus subtilis PerR represses the oxidative stress resistance genes coding for alkyl hydroperoxidase (ahpC), the Dps homologue mrgA, and catalase (katA) in response to the presence of manganese and, to a lesser extent, iron (7, 9). In S. aureus PerR similarly mediates the oxidative stress response by repressing katA, ahpC, and mrgA, but unlike B. subtilis, genes within the S. aureus PerR regulon are induced by iron but repressed under iron-deficient and manganese-rich conditions (10, 11). Zur appears to be involved in zinc-mediated gene regulation (14). Staphylococcus epidermidis also appears to have three Fur homologues (8; unpublished observations), but the roles of these proteins in the regulation of iron homeostasis have yet to be determined.
An additional mechanism for overcoming intracellular iron toxicity is to remove free iron from the cytoplasm by compartmentalizing the iron into specialized intracellular iron storage proteins. This results in a reserve of nontoxic iron that can be used as a nutrient source during conditions of iron starvation. Intracellular iron storage has been studied in gram-negative bacteria, but there are still many questions concerning the mechanisms and regulation of the entry and exit of iron into the storage proteins and the contribution of iron storage to virulence.
Ferritins are the major iron storage proteins found in eukaryotes (H chain) and gram-negative prokaryotes (2). They are highly conserved spherical proteins composed of 24 subunits surrounding a central iron storage cavity with the capacity to store up to 4,500 iron atoms. Each subunit is folded into a four-helix bundle typically containing a well-conserved ferroxidase active site. Ferroxidase activity is required for the oxidation of Fe2+ to Fe3+ during the uptake and storage of iron by the ferritin protein. From recent studies it appears that the main functions of ferritins in gram-negative bacteria are iron storage and protection against metal toxicity and oxidative stress, but there is significant variation in the function and regulation of ferritins in different bacterial species. Escherichia coli, Porphyromonas gingivalis, and Campylobacter jejuni ferritins are required for maximal growth under iron-restricted conditions (1, 18, 19), but the C. jejuni ferritin also has an important role in protection against iron-mediated oxidative stress that is not observed in E. coli or P. gingivalis. In contrast, the primary role of the Helicobacter pylori ferritin appears to be in protecting this gastric pathogen against metal toxicity (4). There are also differences in the regulation of ferritin expression between the different organisms. The E. coli ferritin is indirectly induced by Fur under iron-rich conditions, as Fur represses the small RNA RhyB, which directly represses ferritin transcription in the absence of iron (1, 15), while the H. pylori ferritin is repressed by Fur under iron-restricted conditions. Bereswill et al. (4) suggest that in H. pylori Fur has a direct role in the repression of ferritin under low-iron conditions, mediated by an increase in the ratio of iron to other metal ions so that when iron is scarce, other metals become dominant over iron, resulting in Fur-mediated repression. The regulation of the C. jejuni and P. gingivalis ferritins has not yet been determined.
Until recently, no iron storage proteins had been identified in staphylococci. However, during their analysis of the Per regulon in S. aureus, Horsburgh et al. (11) identified a ferritin homologue by N-terminal amino acid sequencing of a protein which was upregulated in a PerR mutant. This study showed that transcription of ftnA and other PerR-regulated genes was induced by iron and repressed by the manganese-dependent PerR transcriptional repressor as elevated levels of manganese repressed transcription of the PerR regulon genes in wild-type S. aureus 8325-4 but not in the isogenic perR mutant (11). However, in this report we show that PerR regulation is far more complex than previously reported; iron plays a key role in PerR-mediated regulation, and the PerR regulon genes are differentially regulated by metal ion availability and by Fur. Moreover, we describe the identification of a ferritin homologue in S. epidermidis and show that there are significant regulatory differences between the S. aureus and S. epidermidis ferritins in response to metal ions.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
S. aureus strains RN6390B and BB and S. epidermidis strains 901 and 570 were from our laboratory culture collections. S. epidermidis strains 100721 and 81/1157 were fresh clinical isolates obtained from Public Health Laboratory Service, London, United Kingdom. S. epidermidis Tu3298 was a kind gift from F. Gotz. S. aureus wild-type strain 8325-4 and isogenic mutant strains MJH001 (8325-4 per::kan) and MJH010 (8325-4 fur::tet) were kind gifts from M. Horsburgh and S. Foster, Sheffield, United Kingdom (10, 11). All staphylococcal strains were cultured aerobically at 37°C in Luria-Bertani (LB) broth or agar, with tetracycline (5 μg/ml) or kanamycin (50 μg/ml) added when required. Staphylococci were routinely cultured under iron-restricted conditions by growing statically for 18 h at 37°C in RPMI 1640 tissue culture medium, containing 2 mg of NaHCO3 per ml, which had been depleted of iron by batch incubation with 6% (wt/vol) Chelex 100 (Sigma Ltd.) and then supplemented with 10% RPMI 1640 to provide trace elements required for growth (CRPMI) (16). CRPMI cultures were incubated in 5% CO2 in air (16). The concentrations of iron and manganese in CRPMI are <1 μM (unpublished observations). Where indicated the medium was supplemented with 10 μM ferric citrate and/or 10 μM MnCl2.
In some experiments S. aureus was grown in a two-stage protocol to obtain logarithmically growing cells. Initially 10-ml volumes of bacterial culture were grown for 18 h as described above in RPMI 1640 tissue culture medium before pelleting of bacteria at 3,500 × g for 5 min and resuspension in 1 ml of CRPMI. One-hundred-microliter volumes of the bacterial suspension were then used to inoculate fresh 10-ml volumes of CRPMI, and cultures were incubated at 37°C in 5% CO2 in air for 4 h. Ferric citrate and/or MnCl2 (10 μM each) was then added to the appropriate cultures, and incubation continued for a further 3 h before bacteria were harvested for analysis. Alternatively, to assess ferritin expression at different times during growth, bacteria were grown overnight in RPMI and resuspended in CRPMI as described above before inoculation into CRPMI-10 μM ferric citrate. Cultures were incubated at 37°C in 5% CO2 in air and harvested at 4 h (log phase), 8 h (late log phase), or 24 h (stationary phase) post inoculation.
E. coli XL1-Blue or BL21 DE3 (Novagen) was cultured at 37°C in LB broth or agar. Following transformation with recombinant plasmids, kanamycin (50 μg/ml) was added to the medium for growth of these strains.
Long-term stocks of bacterial strains were stored at −80°C in 10% (vol/vol) glycerol in LB broth.
DNA preparation and manipulation.
Genomic and plasmid DNA was prepared and manipulated as described by Morrissey et al. (16).
Northern blot analysis.
Staphylococcal RNA was extracted using a Qiagen RNeasy total RNA kit, but with lysostaphin (100 μg/ml) added to the initial cell lysis step. Sample loadings were adjusted and equally loaded based on optical density measurements of the bacterial cultures. Equally loaded RNA samples and RNA markers (Promega) were electrophoresed on 1.5% agarose-formaldehyde gels and then transferred to Hybond N+ membrane as described in Promega's protocols and applications guide (3rd ed.). The Northern blots were incubated overnight at 50οC with digoxigenin-labeled DNA probes (Boehringer Mannheim) obtained by random priming of PCR products from S. aureus RN6390-B and S. epidermidis Tu3298 genomic DNA. Primers used for generation of DNA probes were ftnA (F2pF 5′-CACCTGCAGGAGGTGTATCAAAATGTTAAG-3′ and FerSR 5′-TAACCCGGGCTTCGTCGAATGTACGAG-3′), ahpC (ahpCf 5′-AGAAGGATCCGTTGAGAATACAAATCTTC-3′ and ahpCR 5′-AGCAGAATTCCTTCTTCCCATTTAGCTG-3′), mrgA (mrgAf 5′-GGAGTGTATTAAATATGAG-3′ and mrgAR 5′-CTACTGATGTTTGCATAC-3′), and sefA (sefAf 5′-CACCTGCAGCGGGGTGATTGAAGATG-3′ and sefAR 5′-TAACCCGGGGAACGCGCTGCTAATTC). The hybridized filter was washed sequentially in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.1% (wt/vol) sodium dodecyl sulfate (SDS) at room temperature for 15 min and in 0.5× SSC-0.1% (wt/vol) SDS at 68°C for 15 min. The bound probe was visualized using CDP-star (Boehringer Mannheim) according to manufacturer's protocol. Each experiment was repeated at least three times with equivalent RNA loadings of gels. Transcript profiles were similar in each case, and this was supported by densitometry of autoradiographs. Densitometric analysis of autoradiographs was performed using Bio-Rad Quantity One software.
Expression of the S. aureus ferritin homologue in E. coli.
The S. aureus ferritin homologue, ftnA was amplified by PCR using primers (FtnAXf 5′-CAGCATATCAAGAGGTGTATCAAAATGTTA-3′ and FtnAXr 5′-CACAAGCTTCCCAAATGCCTATCATGT-3′) and RN6390B genomic DNA as template. The resulting PCR fragment was digested with EcoRV and HindIII, purified with a PCR purification kit (Qiagen), and ligated into similarly digested and purified pET30a (Novagen). The resulting recombinant plasmid, pET30a/FtnA, was transformed firstly into E. coli XL1-Blue and subsequently into E. coli BL21 DE3. Protein expression was induced by addition of IPTG (isopropyl-d-thiogalactopyranoside) as previously described (16), and the recombinant protein was purified by electroelution from SDS-polyacrylamide gels for 18 h at 10 mA in 50 mM Tris-50 mM glycine-0.1% (wt/vol) SDS buffer (pH 8.8) using a Bio-Rad electroelutor.
Polyclonal antibody production.
Antibody to electroeluted FtnA was produced in male Wistar rats. Electroeluted antigen was resuspended in sterile phosphate-buffered saline (PBS) and administered subcutaneously initially in Freund's complete adjuvant and 2 and 4 weeks later in Freund's incomplete adjuvant. Serum was collected 2 weeks after the third immunization.
SDS-polyacrylamide gel electrophoresis and immunoblotting.
All samples were solubilized by boiling in Laemmli sample buffer (12) for 5 min. Whole-cell lysates of staphylococci were prepared by lysostaphin digestion as previously described (6), and quantities of bacteria for digestion were standardized on the basis of measurement of optical density at 600 nm. Polypeptides were separated by SDS-polyacrylamide gel electrophoresis using a 4% (wt/vol) acrylamide stacking gel and 10% (wt/vol) resolving gel in a Bio-Rad Mini Protean II gel apparatus as previously described (3). For immunoblotting, polypeptides were transferred to BioTrace NT membrane (Gelman) followed by blocking in 3% bovine serum albumin as previously described (3). Membranes were incubated for 1 h in preimmune or immune rat serum diluted 1/1,000 (vol/vol) in PBS-0.1% (wt/vol) bovine serum albumin and 0.1% (vol/vol) Tween 20. After thorough washing in PBS, blots were incubated for 1 h with anti-rat horseradish peroxidase-labeled conjugate (Dako) (1/5,000 dilution in the above-described buffer). After washing in PBS, bound conjugate was detected using an ECL system (Amersham Pharmacia Biotech) according to the manufacturer's instructions. All experiments were repeated at least three times, and the protein profiles were identical in each case.
RESULTS
Identification of ferritin homologues in S. aureus and S. epidermidis.
Using a TBLASTN search and the C. jejuni ferritin polypeptide sequence (accession number Q46106) we have identified a single open reading frame encoding an S. aureus ferritin homologue in the S. aureus 8325 genome database (http://www.ncbi.nlm.nih.gov/Microb_blast/unfinishedgenome.html). The single open reading frame identified in the S. aureus genome database has a predicted 48% identity and 67% amino acid similarity to the Bacillus halodurans ferritin protein and 33% identity and 54% amino acid similarity to the C. jejuni ferritin protein and was designated ftnA. The S. aureus ftnA sequence was then used to identify a single ferritin gene (sefA, for S. epidermidis ferritin) in the S. epidermidis unpublished genome database.
The staphylococcal ftnA and sefA open reading frames encode putative polypeptides of 19.5 kDa, which are highly conserved between the two bacteria (Fig. 1). The staphylococcal ferritin homologues retain many of the conserved structural features of previously identified ferritins, including those amino acids identified in the eukaryotic ferritin H as participating in iron chelation and thus forming the ferroxidase center (Glu-17, Tyr-24, Glu-49, Glu-50, His-53, Glu-94, and Gln 127) (2), suggesting that the staphylococcal ftnA and sefA genes encode functional ferritin proteins (Fig. 1). Our sequence analysis has shown that the ftnA and sefA genes appear to be monocistronic and are preceded by significantly different upstream sequences. These contain two putative Per boxes that are 104 bp apart in S. aureus, with nucleotide identity (14 and 16 of 17 nucleotides, respectively) to the S. aureus Per box consensus sequence, and two sequences that are 45 bp apart in S. epidermidis, with nucleotide identity (14 and 12 of 17 nucleotides, respectively) to the Per box consensus sequence (Fig. 2). Most significantly, however, there is also a sequence with nucleotide identity (13 of 19 nucleotides) to the S. aureus Fur box consensus sequence in the S. aureus ferritin promoter that is not found in any of the other S. aureus and putative S. epidermidis PerR-regulated promoter sequences, including katA, a gene positively regulated by Fur (10). These observations suggest that ferritin regulation is likely to be more complex than previously reported (11) with possibly both Fur and PerR being involved in the regulation of S. aureus but only PerR regulating S. epidermidis ferritin expression.
FIG. 1.
Alignment of the amino acid sequences of the ferritin proteins of S. aureus (S. aur), S. epidermidis (S. epi), and C. jejuni (C. jeu). Boxed amino acids indicate residues conserved for iron chelation and ferroxidase activity (Glu-17, Tyr-24, Glu-49, Glu-50, His-53, Glu-94, and Gln 127) (2).
FIG. 2.
Alignment of the S. aureus (ftnA) and S. epidermidis (sefA) 5′ promoter sequences. Boxed nucleotides indicate putative PerR boxes, and the dotted box indicates a putative Fur box (5′ AAAAATGATATTTATTCTC 3′) in the ftnA sequence. The translational start point is indicated in boldface type, while the putative ribosomal binding site is underlined.
Iron has an important role in the regulation of S. aureus ferritin expression.
To generate an anti-FtnA antiserum for investigation of staphylococcal ferritin expression, the ftnA open reading frame was cloned into pET30a and expressed by induction with IPTG. A major polypeptide of 21 kDa was induced following addition of IPTG. Cell fractionation studies (data not shown) indicated that the polypeptide pelleted with the E. coli membrane fraction, suggesting membrane association or protein insolubility. The 21-kDa polypeptide was purified from SDS-polyacrylamide gels by electroelution and used to generate polyclonal antibody in rats.
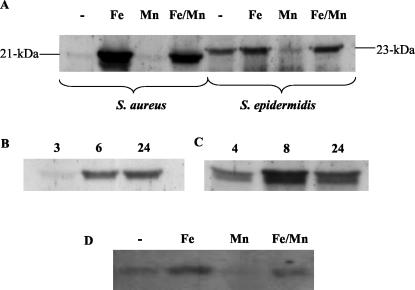
Immunoblot analysis of staphylococcal cells grown for 18 h in CRPMI with anti-FtnA rat serum identified an antigenic polypeptide of approximately 21 kDa in S. aureus 8325-4 (Fig. 3A) and the S. aureus strains BB and RN6390B (data not shown), which was upregulated in response to the addition of ferric citrate to the growth medium (Fig. 3A). Ferritin expression was observed to be dependent on the growth phase of the cells, but interestingly, the maximal ferritin polypeptide levels detected changed depending on the growth medium used. In the relatively rich growth medium LB, maximum ferritin polypeptide levels were observed in 24-h stationary-phase cells (Fig. 3B). However, in the severely iron-restricted CRPMI medium supplemented with 10 μM ferric citrate, maximum ferritin levels were detected in cells after 8 h rather than 24 h of incubation.
FIG. 3.
Immunoblots with anti-FtnA serum of soluble extracts from S. aureus 8325-4 and S. epidermidis 901 grown in CRPMI (-) and CRPMI with the addition of iron (Fe) and/or manganese (Fe/Mn; Mn), demonstrating the variation in ferritin expression between S. aureus and S. epidermidis (A); S. aureus 8325-4 grown in LB for 3 h (log phase), 6 h (late log phase), or 24 h (stationary phase) (B); S. aureus 8325-4 grown in CRPMI-10 μM ferric citrate for 4 h (log phase), 8 h (late log phase), or 24 h (stationary phase) (C); and Northern blot analysis of S. epidermidis Tu3298 ferritin transcripts showing that the variation in ferritin expression seen between S. aureus and S. epidermidis occurs at the transcriptional level (D). Total RNA was prepared from logarithmic S. epidermidis cells grown in CRPMI (-) and CRPMI with the addition of iron (Fe) and/or manganese (Fe/Mn; Mn), the Northern blot was hybridized with digoxigenin-labeled sefA PCR product. The blots shown are examples of replicate experiments all showing similar transcript profiles.
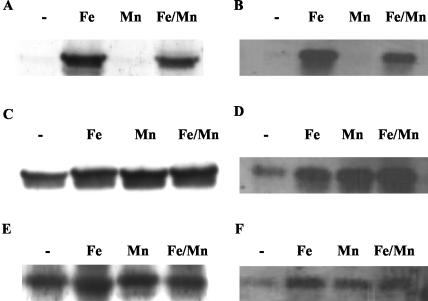
It has previously been reported that ftnA is regulated by the manganese-dependent PerR repressor (11). Since our previous studies have shown that expression of some S. aureus genes can vary significantly between strains grown in different media (17), as part of our present investigation into further defining the role of iron and Fur in S. aureus ferritin regulation we have repeated the investigation of the effect of iron and manganese on ftnA expression using our culture condition; moreover, we have extended the previous investigation by determining the effect of the simultaneous addition of both iron and manganese on FtnA expression. Furthermore, we have used immunoblot and Northern blot analysis as our methods of assaying gene expression in S. aureus, as these complementary methods of assaying RNA transcript levels and translated proteins allow us to demonstrate if there is any posttranscriptional or posttranslational regulation of the ferritin polypeptide and show transcript levels directly instead of being reliant on the expression of a heterologous gene. Figures 3A and 4A and B show that ftnA expression is repressed in metal-deficient CRPMI growth medium and CRPMI with 10 μM manganese added, as previously reported with CLR medium (11). However, our studies demonstrate that the addition of both iron and manganese together resulted in induced levels of ferritin (Fig. 3A), although iron alone results in a higher level of induction. This suggests either that iron relieves the PerR-manganese repression of FtnA or that manganese represses the activation effect of iron when iron and manganese are present together. Northern blot analysis of S. aureus total RNA has demonstrated that this effect of iron on the regulation of ferritin expression is occurring at the transcriptional level, as a 480-bp ftnA transcript was present in cells grown in the presence of iron or iron and manganese together, but not in cells grown under iron-restricted conditions or with manganese alone (Fig. 4B), suggesting that in S. aureus, iron has a major role in the regulation of ferritin expression. Furthermore, we show that there is no posttranscriptional or posttranslational regulation of the ferritin polypeptide since there is no apparent difference in the regulation of ferritin at the transcript or translational level (Fig. 4).
FIG. 4.
Immunoblots with anti-Ftn serum of whole-cell extracts (A, C, and E) and Northern blot analysis (B, D, and F) of total RNA prepared from S. aureus 8325-4 (A and B), perR mutant MJH001 (C and D) and fur mutant MJH010 (E and F) grown in CRPMI (-) or CRPMI with iron (Fe) and/or manganese (Fe/Mn; Mn) showing partial derepression of ferritin in MJH001 and MJH010 under iron limitation (-) and enhanced expression in the presence of added metal ions (Fe, Mn, Fe/Mn). Northern blots were hybridized with digoxigenin-labeled ftnA PCR product. The blots shown are examples of replicate experiments all showing similar transcript profiles.
The regulation of S. epidermidis ferritin expression in response to metals is significantly different from S. aureus ferritin regulation.
The S. aureus anti-FtnA serum also detected a 23-kDa antigenic ferritin homologue in S. epidermidis 901 (Fig. 3A) and a variety of other S. epidermidis strains (data not shown). Interestingly immunoblot analysis indicated that unlike S. aureus, S. epidermidis ferritin expression is not fully repressed under iron-restricted conditions, although its repression in response to manganese appears to be similar to that of S. aureus. In addition, although the S. epidermidis ferritin is induced in response to iron, this is to a much lower extent than that observed for S. aureus. All five laboratory and clinical isolates of S. epidermidis analyzed showed identical patterns of ferritin regulation to S. epidermidis 901 in the presence of iron and/or manganese (data not shown). Northern blot analysis of logarithmic S. epidermidis Tu3298 cells exposed to iron and/or manganese for 3 h before analysis shows that the variation in ferritin regulation and the lower level of sefA expression compared to ftnA is occurring at the transcriptional level (Fig. 3D) and is not due to the S. aureus FtnA antisera reacting less well with the SefA polypeptide.
Repression of S. aureus ferritin appears to require both PerR and Fur.
Our analysis of the ftnA promoter sequences (Fig. 2) identified putative consensus PerR- and Fur-binding sites, suggesting that both regulators may be involved in regulation of ftnA. To determine whether both PerR and Fur are involved in the iron- and manganese-mediated regulation of ferritin, we compared ferritin expression in an S. aureus wild-type strain (8325-4), an S. aureus 8325-4 perR mutant (MJH001), and an S. aureus 8325-4 fur mutant (MJH010). The wild-type strain 8325-4 and the isogenic mutants were grown in the presence and absence of iron, manganese, or both ions. Densitometric analysis of the autoradiographs of Northern blots of total RNA obtained from logarithmically growing cells shows that in comparison with wild-type S. aureus results there is a sixfold increase in ftnA transcription in a perR mutant in the presence of manganese (Fig. 4C and D) and a twofold increase under metal-depleted (unsupplemented CRPMI) growth conditions. Moreover, ftnA expression in a perR mutant in the presence of both iron and manganese is comparable with that seen with iron alone, whereas in wild-type S. aureus a lower level of ftnA expression is observed under iron- and manganese-rich conditions compared to that seen with to growth in the presence of iron alone. Thus, these results suggest that PerR regulates ftnA under these growth conditions.
Interestingly, there appears to be additional PerR-independent regulation of ftnA under metal-depleted conditions since in a perR mutant, addition of iron and/or manganese resulted in a fourfold-higher level of ftnA transcription in comparison to the results seen with the unsupplemented medium (Fig. 4C and D, compare lane 1 with lanes 2 to 4). Thus, these results suggest that ferritin regulation is far more complex than previously reported (11) and factors other than PerR and manganese have a role in the transcriptional regulation of ferritin in S. aureus.
Furthermore, these studies show (Fig. 4E and F) that ftnA expression is also constitutive in an S. aureus fur mutant (MJH010) although there is still a fourfold-lower level of ftnA transcription under metal-deficient growth conditions compared to that seen when iron and/or manganese is present. Immunoblot analysis with anti-FtnA serum (Fig. 4A, C, and E) reflected the overall pattern of ftnA expression detected by Northern blotting. Therefore, we have shown that both PerR and Fur play a role in the transcriptional regulation of ftnA in S. aureus and that there is additional regulation of ftnA under metal-depleted conditions.
Members of the S. aureus PerR regulon are differentially regulated in response to iron and manganese availability.
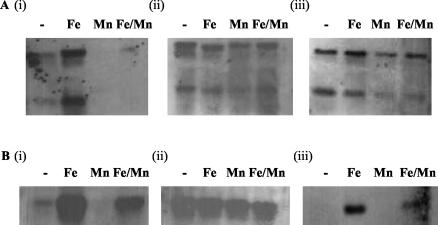
It has previously been demonstrated that there are PerR consensus binding sequences in the promoters of ftnA, ahpC, and mrgA (11). However, our sequence analysis of the promoter sequences of ftnA, ahpC, and mrgA has shown that there are significant sequence differences in these promoter regions. Most importantly, ftnA, as well as having two PerR consensus binding sequences, also appears to have a putative Fur binding consensus sequence, in contrast to the other PerR regulon components which only have PerR boxes. Moreover, there are differences in the number of putative PerR boxes, as ahpC only has a single PerR box whereas mrgA has two PerR boxes. These differences in the promoter sequences of the genes may indicate differential regulation of these genes by PerR and Fur. To investigate the role of Fur in controlling mrgA and ahpC expression and to determine whether other members of the PerR regulon are regulated differently from ftnA, Northern blot analysis was used. Figure 5 (panels i) shows that in S. aureus 8325-4 and in common with ftnA, the transcription of both ahpC and mrgA is repressed under manganese-rich growth conditions and induced in the presence of iron. However, there are differences between the transcription results seen with the ftnA, ahpC, and mrgA genes under other growth conditions. All genes are repressed under metal-depleted growth conditions, but there is a twofold-higher level of ahpC and mrgA transcripts than ftnA transcripts, indicating continued repression of ftnA under metal-depleted conditions that is not observed with ahpC and mrgA. In addition, although the presence of both iron and manganese results in induced levels of transcription of all three genes, there are significant differences in the level of induction observed between the different genes. There is a fourfold increase in mrgA and ftnA transcription in the presence of manganese and iron compared with that observed under metal-depleted conditions; however, there is only a twofold increase in transcription of ahpC in the presence of iron and manganese (Fig. 4B; Fig. 5, panels i).
FIG. 5.
Northern blot analysis of total RNA prepared from S. aureus 8325-4 (i), perR mutant MJH001 (ii), and fur mutant MJH010 (iii) grown in CRPMI (-) or CRPMI with iron (Fe) and/or manganese (Fe/Mn; Mn), showing derepression of ahpC (A) and mrgA (B) in MJH001 under conditions of iron limitation and in the presence of manganese and differential regulation of ahpC and mrgA in MJH010. Northern blots were hybridized with digoxigenin-labeled ahpC (A) and mrgA (B) PCR product. Two transcripts of approximately 2.1 and 0.6 kb were detected with the ahpC DNA probe; the 2.1-kb transcript is likely to correspond to a full-length operon transcript containing the ahpC and ahpF genes, with the 0.6-kb transcript encoding ahpC, the first gene of the operon. The blots shown are examples of replicate experiments, all showing similar transcript profiles.
Moreover, there is a differential response of these genes to both PerR and Fur. As described above, ftnA transcription is derepressed under both low-ion (CRPMI) and manganese-rich growth conditions in both perR and fur mutant strains, although there is still some repression of ftnA under low-ion conditions (Fig. 4D and F). In comparison, transcription of mrgA and ahpC is constitutive in a perR mutant strain under all growth conditions (Fig. 5, panels ii). However, the most-significant difference is in the response of the genes to Fur. Transcription of ahpC is fully derepressed in a fur mutant under all growth conditions, although there is a twofold reduction in transcription when manganese is present (Fig. 5A, panel iii). In contrast, mrgA transcription is not constitutive in the fur mutant, since mrgA is still repressed in a fur mutant after growth in either CRPMI (no iron or manganese) or CRPMI supplemented with 10 μM manganese (Fig. 5B, panel iii). However, there is a twofold decrease in the level of mrgA transcripts in the fur mutant compared with that seen with the wild type. Consequently, our data suggest that PerR regulon genes are differentially regulated by metal availability and that Fur is involved in the regulation of ftnA, ahpC and possibly mrgA. These results indicate that PerR regulation is far more complex than the perceived consensus (11), as iron and Fur significantly influence PerR repression.
DISCUSSION
Iron storage systems have been well studied in eukaryotes and some gram-negative bacteria, but much less is known about the iron storage mechanisms of gram-positive bacteria. Ferritin-like proteins have been described for Listeria innocua (5), but these proteins share homology with the mammalian L chain ferritins and the DPS family of stress-induced DNA binding proteins. In contrast to other members of the DPS family, the L. innocua ferritin-like proteins are able to store iron but are unable to bind DNA. This is consistent with a role for these proteins in iron storage rather than in protecting DNA against oxidative stress. S. aureus has an open reading frame with significant homology to the L. innocua Dps-like ferritin, MrgA, but comparison of the S. aureus amino acid sequence with both DPS ferritin-like and DNA-binding proteins suggests that in S. aureus MrgA is likely to function as a DNA-binding protein with an important role in oxidative stress resistance rather than a ferritin-like iron storage protein. The S. aureus and S. epidermidis ferritin proteins are highly conserved and have all the structural features of the eukaryotic heavy chain ferritins and the gram-negative ferritin molecules, suggesting that they function as staphylococcal iron storage proteins.
A previous report (11) has shown that in S. aureus ftnA, like other components of the PerR regulon, is induced by the presence of iron and is repressed under both metal-ion-deficient and manganese-rich growth conditions. The authors concluded that PerR was acting as a manganese-dependent transcriptional repressor of the oxidative stress response. However, our data show that PerR regulation is far more complex than described previously (11), with our results showing that iron has a significant role in PerR regulation of ftnA, ahpC, and mrgA. The importance of iron in the regulation of ftnA transcription is indicated by the fact that when S. aureus cells are grown in the presence of both iron and manganese, ftnA expression is actually induced, although not to the same level as that observed when S. aureus is grown with iron alone. This suggests that there is still some PerR-dependent repression of ftnA when both iron and manganese are present or that manganese is repressing iron activation. However, Northern blot analysis of the perR mutant MJH001 demonstrates that transcription of ftnA is completely derepressed in iron- and manganese-grown cells compared with that seen with the wild type, suggesting that it is in fact continued manganese-PerR-mediated repression occurring under iron and manganese growth conditions. This influence of iron is also observed with other members of the PerR regulon, since both mrgA and ahpC are transcribed in the presence of iron and manganese although ahpC is not induced to the same degree as mrgA and ftnA, suggesting stronger manganese-mediated repression of ahpC than mrgA or ftnA or increased induction of mrgA and ftnA by iron. This influence of iron in the regulation of ftnA, mrgA, and ahpC suggests that there may be competition between iron and manganese for binding sites on PerR, such that when iron displaces or prevents manganese from binding to PerR, it prevents PerR from binding DNA. Thus, it is possible that the PerR-dependent repression of ftnA is dependent on the balance of intracellular iron and manganese concentrations, with increasing iron and decreasing manganese levels leading to increased expression of ftnA. At present, however, it is not known which metal forms of PerB bind DNA in S. aureus. It is also possible that changes in manganese and iron content may affect intracellular peroxide levels which in turn may influence S. aureus PerB binding of promoter DNA since the PerR regulon is also known to be induced by oxidative stress (11).
During these investigations we have provided evidence to suggest that there is additional PerR-independent regulation of ftnA, as in the perR and fur mutant strains repression of ftnA still occurs under iron-deficient growth conditions. This additional regulation appears to be ferritin specific, as there is a low level of transcription of mrgA and ahpC under metal-depleted conditions in wild-type S. aureus that is not observed with ftnA and expression of mrgA and ahpC is fully derepressed in a perR mutant strain under all growth conditions. This additional level of regulation of ftnA may have physiological significance in that the presence of ferritin when iron conditions are low may be deleterious to the cell, as the ferritin may sequester iron, thereby preventing it from being freely accessible for important metabolic functions. This is supported by the observation that in a relatively iron-rich medium (LB), maximal FtnA polypeptide levels are observed in stationary-phase cells; however, in a severely iron-restricted medium (CRPMI) with only 10 μM iron citrate there is a decrease in ferritin polypeptide after 24 h, indicating a possible depletion of extracellular iron resulting in a repression of ferritin expression.
Importantly we have shown that Fur is involved in the regulation of ftnA, ahpC, and mrgA expression, but interestingly the three genes appear to be differentially regulated by Fur. ftnA and ahpC expression is constitutive under manganese-rich and iron-restricted growth conditions in a fur mutant even with the presence of functional PerR although there appears to still be some repression of ahpC under manganese-rich conditions, which may correspond to a strong level of PerR regulation as indicated above. Interestingly, although mrgA expression is not constitutive in the fur mutant, there appears to be a decrease in the level of mrgA transcription, suggesting that Fur is involved in the positive regulation of mrgA. Thus, these results indicate that there is differential regulation of the PerR regulon components in response to iron and manganese availability mediated via PerR and Fur.
If both PerR and Fur regulate ftnA and ahpC, it is not yet clear how ferritin expression is constitutive in both perR and fur mutants when there are still functional Fur and PerR proteins present in the respective mutant strains. Moreover, if Fur is involved in the repression of these genes this is occurring under iron-restricted conditions, and this may seem unlikely as Fur is usually considered an iron-dependent repressor, although recently it has become apparent that Fur is responsible for the negative regulation of H. pylori ferritin under iron-restricted conditions (4). It is also possible that Fur is indirectly repressing transcription through the action of a small RNA such as RyhB in E. coli (15).
Differential regulation of genes in the B. subtilis PerR regulon has also been reported recently (7). Most components of the B. subtilis PerR regulon are repressed in response to the presence of manganese and iron, while some are manganese specific (7, 9). As with S. aureus it is not clear which factors are important for the differences in regulation between B. subtilis PerR components, but it has been suggested that there are distinct metal forms of PerR (Fe-PerR and Mn-PerR) that may differ in DNA target selectivity. Our sequence analysis has shown that there are significant sequence differences in the promoter regions of ftnA, ahpC, and mrgA. Most importantly ftnA appears to have a putative Fur binding consensus sequence in contrast to the other PerR regulon components. These have only PerR binding consensus sequences; ahpC has only a single PerR consensus binding sequence in both S. aureus and S. epidermidis, and mrgA has only one PerR box in S. epidermidis but two PerR boxes in S. aureus. It could be that Fur directly represses ftnA expression under low-ion conditions but may have an indirect role in the regulation of ahpC. It is also possible that the induction of ftnA and ahpC we noted in the fur mutant is due to increased intracellular iron levels affecting PerR repression and that the differential response of ahpC and mrgA to Fur may be due to the metal dependency of PerR binding and promoter sequences having a major role in defining PerR binding specificity to the different PerR regulon genes. It is clear from our studies that both Fur and PerR regulation is extremely complex and that we need more information to clarify their relative roles in regulating ftnA, ahpC, and mrgA expression. It will be important to establish if Fur is directly involved in ftnA and ahpC regulation, and it will be necessary to determine which features of PerR-regulated promoter sequences are important for differential regulation of these genes.
Our sequence analysis also revealed significant differences between the S. aureus and S. epidermidis ferritin promoters, with only the S. aureus promoter containing a consensus Fur binding sequence. The results presented here show that there are indeed significant differences in the pattern of expression of the S. aureus and S. epidermidis ferritins, as unlike ftnA, sefA is derepressed under iron-restricted conditions. Manganese repression is similar in both bacteria, as is the induction of ferritin under iron-rich conditions, although sefA is expressed at a significantly lower level than ftnA. It is presently unclear why there is this difference in ferritin expression and whether this has any implications for staphylococcal physiology and virulence. In fact S. aureus and S. epidermidis have very different iron requirements, with S. epidermidis requiring significantly higher iron levels to sustain growth in vitro than S. aureus (13; unpublished observations). This may be due in part to the derepressed levels of ferritin present in S. epidermidis under iron-restricted conditions storing free intracellular iron so that it is unable to be utilized for metabolism. If this were the case then it could also explain why there is an additional mechanism of regulation of ftnA under low-ion conditions, and this physiological difference between the two bacteria may be a significant factor contributing to the ability of S. aureus to more readily cause serious systemic infections where iron restriction may be further exacerbated.
The results reported here show that iron-responsive gene regulation in S. aureus and S. epidermidis is very complex and that the roles of Fur and PerR in gene regulation in S. aureus are not as simple as previously reported, and therefore these results have major implications on the currently perceived consensus of PerR and Fur regulation in S. aureus. Most importantly, we have shown that iron plays the key role in PerR regulation of ftnA, ahpC, and mrgA and hence the S. aureus oxidative stress response.
Acknowledgments
This work was supported by program grant G9219778 from the Medical Research Council.
We thank Malcolm Horsburgh and Simon Foster for 8325-4 and the fur (MJH010) and perR (MJH001) mutants.
Editor: V. J. DiRita
REFERENCES
- 1.Abdul-Tehrani, H., A. J. Hudson, Y-S Chang, A. R. Timms, C. Hawkins, J. M. Williams, P. M. Harrison, J. R. Guest, and S. C. Andrews. 1999. Ferritin mutants of Escherichia coli are iron deficient and growth impaired and fur mutants are iron deficient. J. Bacteriol. 181:1415-1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andrews, S. C. 1998. Iron storage in bacteria. Adv. Microbiol. Physiol. 40:281-351. [DOI] [PubMed] [Google Scholar]
- 3.Arbuthnott, J. P., E. Arbuthnott, A. D. J. Arbuthnott, W. J. Pike, and A. Cockayne. 1992. Investigation of microbial growth in vivo: evaluation of a novel in vivo chamber implant system. FEMS Microbiol. Lett. 100:75-80. [DOI] [PubMed] [Google Scholar]
- 4.Bereswill, S., S. Greiner, A. H. M. Van Vliet, B. Waidner, F. Fassbinder, E. Schiltz, J. G. Kusters, and M. Kist. 2000. Regulation of ferritin-mediated cytoplasmic iron storage by the ferric uptake regulator homolog (Fur) of Helicobacter pylori. J. Bacteriol. 182:5948-5953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bozzi, M., G. Mignogna, S. Stefanini, D. Barra, C. Longhi, P. Valenti, E. Chiancone. 1997. A novel non-heme iron-binding ferritin related to the DNA-binding proteins of the Dps family in Listeria innocua. J. Biol. Chem. 272:3259-3265. [DOI] [PubMed] [Google Scholar]
- 6.Cockayne, A., P. J. Hill, N. B. L. Powell, K. Bishop, C. M. Sims, and P. Williams. 1998. Molecular cloning of a 32k-Da lipoprotein component of a novel iron-regulated Staphylococcus epidermidis ABC transporter. Infect. Immun. 66:3767-3774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fuangthong, M., A. F. Herbig, N. Bsat, and J. D. Helmann. 2002. Regulation of the Bacillus subtilis fur and perR genes by PerR: not all members of the PerR regulon are peroxide inducible. J. Bacteriol. 184:3276-3286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heidrich, C., K. Hantke, G. Bierbaum, and H.-G. Sahl. 1996. Identification and analysis of a gene encoding a Fur-like protein of Staphylococcus epidermidis. FEMS Microbiol. Lett. 140:253-259. [DOI] [PubMed] [Google Scholar]
- 9.Herbig, A. F., and J. D. Helmann. 2001. Roles of metal ions and hydrogen peroxide in modulating the interaction of the Bacillus subtilis PerR peroxide regulon repressor with operator DNA. Mol. Microbiol. 41:849-859. [DOI] [PubMed] [Google Scholar]
- 10.Horsburgh, M. J., E. Ingham, and S. J. Foster. 2001. In Staphylococcus aureus, Fur is an interactive regulator with PerR, contributes to virulence, and is necessary for oxidative stress resistance through positive regulation of catalase and iron homeostasis. J. Bacteriol. 183:468-475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Horsburgh, M. J., M. O. Clements, H. Crossley, E. Ingham, and S. J. Foster. 2001. PerR controls oxidative stress resistance and iron storage proteins and is required for virulence in Staphylococcus aureus. Infect. Immun. 69:3744-3754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 13.Lindsay, J., and T. V. Riley. 1994. Staphylococcal iron requirements, siderophore production, and iron-regulated protein expression. Infect. Immun. 62:2309-2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lindsay, J. A., and S. J. Foster. 2001. zur: a Zn2+ responsive regulatory element of Staphylococcus aureus. Microbiology 147:1259-1266. [DOI] [PubMed] [Google Scholar]
- 15.Masse, E., and S. Gottesman. 2002. A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. Proc. Natl. Acad. Sci. USA 99:4620-4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morrissey, J. A., A. Cockayne, P. J. Hill, and P. Williams. 2000. Molecular cloning and analysis of a putative siderophore ABC transporter from Staphylococcus aureus. Infect. Immun. 68:6281-6288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morrissey, J. A., A. Cockayne, J. Hammacott, K. Bishop, A. Denman-Johnson, P. J. Hill, and P. Williams. 2002. Conservation, surface exposure and in vivo expression of the Frp family of iron-regulated cell wall proteins in Staphylococcus aureus. Infect. Immun. 70:2399-2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ratnayake, D. B., S. N. Wai, Y. Shi, K. Amako, H. Nakayama, and K. Nakayama. 2000. Ferritin form the obligate anaerobe Porphyromonas gingivalis: purification, gene cloning and mutant studies. Microbiology 146:1119-1127. [DOI] [PubMed] [Google Scholar]
- 19.Wai, S. N., K. Nakayama, K. Umene, T. Moriya, and K. Amako. 1996. Construction of a ferritin-deficient mutant of Campylobacter jejuni: contribution of ferritin to iron storage and protection against oxidative stress. Mol. Microbiol. 20:1127-1134. [DOI] [PubMed] [Google Scholar]
- 20.Xiong, A., V. K. Singh, G. Cabrera, and R. K. Jayaswal. 2000. Molecular characterisation of the ferric regulator, Fur, from Staphylococcus aureus. Microbiology 146:659-668. [DOI] [PubMed] [Google Scholar]