Abstract
Interferon induced with helicase C domain 1 (IFIH1) senses and initiates antiviral activity against enteroviruses. Genetic variants of IFIH1, one common and four rare SNPs have been associated with lower risk for type 1 diabetes. Our aim was to test whether these type 1 diabetes-associated IFIH1 polymorphisms are associated with the occurrence of enterovirus infection in the gut of healthy children, or influence the lack of association between gut enterovirus infection and islet autoimmunity.
After testing of 46,939 Norwegian newborns, 421 children carrying the high risk genotype for type 1 diabetes (HLA-DR4-DQ8/DR3-DQ2) as well as 375 children without this genotype were included for monthly fecal collections from 3 to 35 months of age, and genotyped for the IFIH1 polymorphisms. A total of 7,793 fecal samples were tested for presence of enterovirus RNA using real time reverse transcriptase PCR.
We found no association with frequency of enterovirus in the gut for the common IFIH1 polymorphism rs1990760, or either of the rare variants of rs35744605, rs35667974, rs35337543, while the enterovirus prevalence marginally differed in samples from the 8 carriers of a rare allele of rs35732034 (26.1%, 18/69 samples) as compared to wild-type homozygotes (12.4%, 955/7724 samples); odds ratio 2.5, p = 0.06. The association was stronger when infections were restricted to those with high viral loads (odds ratio 3.3, 95% CI 1.3–8.4, p = 0.01). The lack of association between enterovirus frequency and islet autoimmunity reported in our previous study was not materially influenced by the IFIH1 SNPs.
We conclude that the type 1 diabetes-associated IFIH1 polymorphisms have no, or only minor influence on the occurrence, quantity or duration of enterovirus infection in the gut. Its effect on the risk of diabetes is likely to lie elsewhere in the pathogenic process than in the modification of gut infection.
Introduction
Enteroviruses (family Picornaviridae) cause mostly inapparent or subclinical infections; acute disease may range from minor illness to paralytic disease [1]. Enteroviruses are thought to cross the intestinal epithelium via M-cells [2] and to replicate primarily in the lymphoid tissues of the gut [3], [4].
IFIH1 (interferon induced with helicase C domain 1), also known as MDA5 (melanoma differentiation associated gene 5), encodes a cytoplasmic sensor that recognizes certain types of double stranded RNA (dsRNA) molecules, which are commonly produced during the replication of some RNA viruses. Signaling via IFIH1 triggers activation of NF-κB and interferon regulatory pathways, and induces antiviral interferon responses [5]. IFIH1 is triggered by picornaviruses [6], [7], and two recent publications have found in vivo effects of IFIH1 in mda5 knockout mice infected with coxsackievirus [8], [9].
Enteroviruses have been considered as possible environmental triggers or accelerators of islet autoimmunity leading to type 1 diabetes [10]–[12]. The relation seems complex, with partially conflicting results [13]; the research has been going on for more than 30 years now. This hypothesis was greatly sparked when a genome-wide association study of type 1 diabetes identified a significant relation with a common polymorphism in IFIH1 [14]. This association is now convincingly established (odds ratio around 0.85 for the minor allele) [15]–[19].
Recently, deep sequencing of exons and splice sites in the IFIH1 gene revealed that lower risk of type 1 diabetes is also associated with two rare variants with a presumed loss of function (nonsynonymous SNPs rs35744605 (E627X) and rs35667974 (I923V)), and with two noncoding variants affecting conserved splice sites (rs35337543 (1641+1G>C)) and rs35732034 (2807+1G>A)), odds ratios of about 0.5–0.7 [20]. The mechanisms relating IFIH1 polymorphisms to type 1 diabetes, and if enterovirus is involved here, remains unclear and paradoxical. Presumably, variants associated with reduced IFIH1 function would confer the host with a mild antiviral response to enterovirus infection. If so, this would influence the observed association between enterovirus frequency and type 1 diabetes in the population, and failure to account for IFIH1 SNPs in population studies could be hypothesized to “conceal” relations between enterovirus and islet autoimmunity or type 1 diabetes.
The primary aim of the present study was to assess in healthy children whether the IFIH1 polymorphisms associated with type 1 diabetes can predict frequency, viral load, or duration of gut infections with enterovirus. We further aimed to test whether our previously reported lack of association between enterovirus and islet autoimmunity was modified by taking these IFIH1 polymorphisms into account.
Methods
Subjects and study design
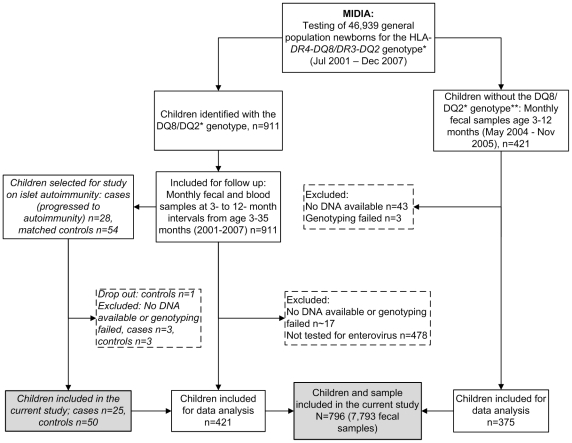
The participants were identified and recruited in the MIDIA-study that during 2001–2007 screened 46,939 newborns from the Norwegian general population for the HLA genotype DRB1*04:01-DQA1*03-DQB1*03:02/DRB1*03-DQA1*05-DQB1*02 (DR4-DQ8/DR3-DQ2) which confers high risk for type 1 diabetes [21]. The present analysis is based on nearly 8,000 fecal samples from 421 children with the HLA DR4-DQ8/DR3-DQ2 genotype and 375 children without this genotype, as described in detail in Figure 1.
Figure 1. Flow chart illustrating the details of inclusion of children and samples in the current analysis.
*HLA-DRB1*04:01-DQA1*03-DQB1*03:02/DRB1*03-DQA1*05-DQB1*02 (DR4-DQ8/DR3-DQ2), conferring high risk for type 1 diabetes. **For each child identified with the high-risk genotype, about 1–4 children without the high risk genotype were enrolled from the same county.
In a separate analysis, we studied whether the IFIH1 polymorphisms modified the risk of type 1 diabetes-related islet autoimmunity among children with the HLA-DR4-DQ8/DR3-DQ2 genotype (Figure 1). Blood samples from these children drawn at ages 3, 6, 9 and 12 months, and annually thereafter were tested for islet autoantibodies (to insulin, glutamic acid decarboxylase, or the protein phosphatase IA2) using radiobinding assays as previously described [21]. The endpoint of islet autoimmunity was defined as repeated positivity for two or three islet autoantibodies. Controls without islet autoimmunity were matched for the HLA DR4-DQ8/DR3-DQ2 genotype, date of birth, length of follow-up and county of residence [13] (Figure 1).
Ethics statement
Written informed consent was provided by participating families. The study was approved by The Regional Committee for Medical Research Ethics (IRB name: “Regional Med Resch Ethics Comm South IRB #2 - South-East A”, IRB00001871) and the Norwegian Data Inspectorate.
Detection of enterovirus RNA in fecal samples
Fecal samples were tested for enterovirus as described earlier [13]. Nucleic acids were co-purified from supernatants of centrifuged feces, and tested for human enterovirus RNA using an internally controlled quantitative reverse transcriptase real-time PCR with primers and probes described in [22]. The median number of fecal samples tested per child was 9 (range 1–36) and the median age at last fecal sample was 12.1 months (range 2.8–37.5).
Genotyping of IFIH1 SNPs
Subjects were genotyped for one common IFIH1 SNP (rs1990760, A946T), and four rare SNPs rs35744605 (E627X), rs35667974 (I923V), rs35732034 (intron 14, 2807+1) and rs35337543 (intron 18, 1641+1). SNP genotyping of rare variant SNPs was performed with the MassARRAY system using iPLEX chemistry (Sequenom, San Diego, CA). The common SNP rs1990760 was genotyped using TaqMan technology with predesigned primer-probe mixes (Applied Biosystems, Foster City, CA, USA). Among children with at least one fecal sample tested for enterovirus (n = 808), genotyping with the MassARRAY system failed in 5–8 DNA samples (<0.1%), and with TaqMan only two samples failed. Duplicate genotyping was performed in 5% randomly chosen samples, with 100% concordance.
Statistical analysis
The final sample for analysis included 796 children and their 7,793 fecal samples (Figure 1). There was no evidence for deviation from Hardy–Weinberg equilibrium (χ2 test) in either of the polymorphisms. Haploview 4.2 was used to assess linkage disequilibria between the alleles of the typed SNPs. Association analyses were performed using logistic regression models with enterovirus infection (yes or no) as the response, individual SNP genotypes as predictors, and a random intercept to account for potential intra-individual correlation (clustering) of infections (xtmelogit in STATA11, StataCorp).
Apart from taking enterovirus positivity as an outcome, in further sensitivity analysis we restricted infections to those with above median quantity of enterovirus RNA among positive samples (defined as high viral load: 10,000 or more virus copies/µl), or to the first enterovirus RNA positive samples among series of two or more consecutively positive samples. Prolonged infection episodes with at least two consecutively positive samples were assessed by excluding other positive samples.
Furthermore, cases of islet autoimmunity and matched controls from our previous publication [13] were compared with respect to enterovirus frequency, and the influence of the IFIH1 SNPs evaluated. This was modeled similarly as for the IFIH1 SNP-enterovirus relation, but with autoimmunity status as a covariate and an additional random intercept to specify matched set (three level random intercept model [23]). These analyses were adjusted for, and stratified by the common variants of IFIH1 SNP rs1990760. The impact of the four rare variant IFIH1 SNPs on the relation between enterovirus infections and islet autoimmunity could only be evaluated by exclusion of subjects with rare variant of either SNP. Matched case-control data at the individual level were done using conditional logistic regression, with additional adjustment for cumulative number of enterovirus infections before seroconversion for islet autoantibodies for cases, and the corresponding time for matched controls.
In our pre-study power considerations, we used parameters from our previously published pilot studies [22], [24] and the literature [14], [20], and estimated that we could detect odds ratios of at least 1.3 of the common IFIH1 A946T variant, and odds ratios at about 2–4 for the various rare variants with approximately 80% power.
Results
Cohort study of IFIH1 SNPs and enterovirus frequency
The frequencies of enterovirus in fecal samples by genotypes of the five SNPs are shown in Table 1. While neither the common polymorphism rs1990760, nor the rare rs35744605, rs35337543, rs35667974 of IFIH1 showed any association with presence, quantity or duration of enterovirus RNA in the gut (Table 1 and Table S1), we observed a statistically non-significant tendency towards association for rs35732034 (Table 1). Here the enterovirus prevalence was 26.1% (18/69 samples) among the 8 children carrying the GA genotype, and 12.4% (955/7724 samples) among children with the wild type variant (OR = 2.5, P = 0.06).
Table 1. Prevalence (%) of enterovirus RNA in fecal samples and frequency of positive samples with a high viral load according to IFIH1 genotypes.
| Enterovirus prevalence | Samples of high viral load* | ||||||
| N = 7,793 (796 children) | N = 7,309 (793 children) | ||||||
| IFIH1 SNPs† ‡ | Genotypes | Frequency % | OR | Frequency % | OR | ||
| (n children) | (n samples/n total) | (95% CI) | P-value | (n samples/n total) | (95% CI) | P-value | |
| rs35337543 | GG (788) | 12.5 (963/7,677) | 1.0 (ref) | 6.3 (483/7,676) | 1.0 (ref) | ||
| CG (8) | 8.6 (10/116) | 0.78 (0.30–2.04) | 0.61 | 5.2 (6/116) | 0.89 (0.31–2.51) | 0.82 | |
| rs35744605 | GG (786) | 12.5 (964/7,700) | 1.0 (ref) | 6.3 (483/7,699) | 1.0 (ref) | ||
| GT (10) | 9.7 (9/93) | 0.90 (0.35–2.37) | 0.84 | 6.5 (6/93) | 1.19 (0.43–3.30) | 0.74 | |
| rs35667974 | AA (752) | 12.6 (926/7,376) | 1.0 (ref) | 6.4 (468/7,375) | 1.0 (ref) | ||
| AG (44) | 11.3 (47/417) | 0.86 (0.55–1.36) | 0.53 | 5.0 (21/417) | 0.78 (0.46–1.33) | 0.36 | |
| rs35732034 | GG (788) | 12.4 (955/7,724) | 1.0 (ref) | 6.2 (477/7,723) | 1.0 (ref) | ||
| GA (8) | 26.1 (18/69) | 2.47 (0.97–6.30) | 0.06 | 17.4 (12/69) | 3.35 (1.34–8.38) | 0.01 | |
| rs1990760 | CC (135) | 10.6 (130/1,229) | 1.0 (ref) | 0.72§ | 5.5 (67/1,128) | 1.0 (ref) | 0.91§ |
| CT (386) | 13.1 (493/3,756) | 1.22 (0.91–1.63) | 6.8 (255/3,756) | 1.25 (0.89–1.74) | |||
| TT (275) | 12.5 (350/2,808) | 1.12 (0.82–1.52) | 6.0 (167/2,808) | 1.10 (0.77–1.56) | |||
SNP, single nucleotide polymorphism; OR, odds ratio; CI, confidence interval.
*Excluding enterovirus positive samples with a low-intermediate virus quantity (below 10,000 virus copies/µl enterovirus RNA).
Location of IFIH1 SNPs: rs35337543 (intron 18, 1641+1, G>C), rs35744605 (exon 10, E627X, G>T), rs35667974 (exon 14, I923V, A>G), rs35732034 (intron 14, 2807+1, G>A), rs1990760 (exon 14, A946T, T>C).
Reports of functional effects associated with IFIH1 SNPs: rs35337543 and rs35732034 influences on a putative splice site, rs35744605 is associated with loss of function (ATPase activity, dsRNA binding, truncation of protein), rs35667974 is associated with loss of function (ATPase activity, dsRNA binding), and rs1990760 is not associated with loss of function (reviewed in [29]).
Test for trend (1 d.f.).
This suggestive association was strengthened when the analysis was restricted to infections with high viral loads (OR = 3.4, P = 0.01, Table 1). The association with high viral load remained significant after adjustment for age, calendar year and season of sample collection (p-values 0.01 to 0.025 and ORs 3.35 to 2.97, data not shown). The association with this rare variant of rs35732034 was also consistent (but not statistically significant) when restricting the analysis to infectious episodes (OR = 1.9, P = 0.06) or to prolonged infections (OR = 2.5, P = 0.09) (Table S1). As there were no correlations between the tested SNPs (all r2<0.04), the above association is unlikely to be explained by linkage disequilibrium to other known diabetes-associated variants. The association of each IFIH1 SNP with enterovirus was similar among children with and without the type 1 diabetes-associated HLA DR4-DQ8/DR3-DQ2 high-risk genotype, respectively (data not shown).
Nested case control study of islet autoimmunity
We next assessed whether taking the IFIH1 polymorphisms into account would modify our previously published observation of no association between enterovirus frequency and islet autoimmunity [13]. Adjusting for rs1990760 and excluding the 1 case and 6 controls with a rare variant of rs35744605, rs35667974, rs35732034, or rs35337543 did not influence the results materially (Table 2). There was no significant heterogeneity in the association between enterovirus frequency and islet autoimmunity that would depend on the common IFIH1 rs1990760 (Table 2). Finally, rs1990760 was not significantly associated with persistent autoimmunity before or after adjusting for the cumulative number of enterovirus infections within a child prior to the development of persistent autoimmunity (Table 2).
Table 2. Frequency of enterovirus RNA in faecal samples prior to islet autoimmunity and matched controls and influence of IFIH1 polymorphisms.
| Cases* | Controls* | OR (95% CI)† | OR (95% CI)† | |
| (n = 25 subjects) | (n = 50 subjects) | Unadjusted | Adjusted | |
| EV infection and later development of islet autoimmunity: | ||||
| EV− | 282 | 555 | 1.00 (reference) | 1.00 (reference) |
| EV+ | 42 (13.0%) | 93 (14.4%) | 0.95 (0.58–1.55), P = 0.83 | 0.99 (0.62–1.60)‡ |
| The association of IFIH1 rs1990760 polymorphisms with islet autoimmunity: | ||||
| rs1990760 Ala− | 9 (36.0%) | 20 (40.0%) | 1.00 (reference) | 1.00 (reference) |
| rs1990760 Ala+ | 16 (64.0%) | 30 (60.0%) | 1.18 (0.44–3.14); P = 0.74 | OR = 1.2 (0.44–3.26)§ |
| EV infection and later development of islet autoimmunity, stratified by IFIH1 rs1990760: | ||||
| EV−, rs199076, Ala− | 181 (86.2%) | 300 (86.0%) | 1.00 (reference) | |
| EV+, rs199076, Ala− | 29 (13.8%) | 49 (14.0%) | 0.77 (0.34–1.77) | |
| EV−, rs199076, Ala+ | 101 (88.6%) | 255 (85.3%) | 1.00 (reference) | |
| EV+, rs199076, Ala+ | 13 (11.4%) | 44 (14.7%) | 1.04 (0.50–2.15) | P(interaction) = 0.74∥ |
EV, enterovirus; OR, odds ratio; CI, confidence interval.
*Cases defined as repeated positivity in consecutive blood samples for two or three islet autoantibodies (anti-insulin, anti-GAD, anti-IA2). Controls matched by high risk HLA genotype, date of birth, time of follow-up and county of residence. See reference [13] for more details.
Estimated from a three level random intercept logistic regression model with enterovirus positivity as dependent variable and case/control status and IFIH1 SNPs as independent variables. Nested random effects were specified for individuals (samples within individuals) and for matched set (individuals within matched sets of a case and 1–2 controls).
Enterovirus adjusted for IFIH1 common variant rs1990760 and vice versa. Rare variants could not be adjusted for because of zero observations in one of the comparing groups. Instead subjects/samples with rare variants of these SNPs were excluded from the analysis (1 case, 6 controls; in total 184 samples).
Adjusted for the cumulative number of enterovirus infections within a child until persistent autoimmunity develops.
Testing whether the model stratified for Thr/Thr is significantly different from the model stratified for carriers of at least one Ala allele.
Discussion
We found a potential association between a presumed loss of function variant in the IFIH1 gene, the rare SNP rs35732034, and a higher frequency of high quantity fecal shedding of enterovirus in infants. Our most important finding however, was the lack or minor influence of the IFIH1 gene variants on the occurrence, quantity or duration of enterovirus gut infections of healthy infants. Furthermore, taking all these five IFIH1 polymorphisms into account did not influence our previously reported lack of association between enterovirus frequency and risk of islet autoimmunity in children with the type 1 diabetes susceptibility HLA-DR4-DQ8/DR3-DQ2 genotype.
Strengths and limitations
To our knowledge, this is the first study investigating the potential impact of diabetes-associated IFIH1 polymorphisms on the frequency and viral load of enterovirus in the gut of healthy infants. Strengths of the present study are the very large number of samples analysed, the population based, longitudinal design, and the rather frequent interval of fecal sample collection. As enteroviruses are thought to replicate primarily in the intestine, and the virus is shed for weeks after an infection, our study is likely to cover the majority of enterovirus infections. We did not study systemic spread of enterovirus, as blood samples would have to be extremely frequent in order to systematically cover most infections: enterovirus is usually present in the blood for a much shorter period than in the gut [1].
We are aware of only two previous studies relating IFIH1 variants to enterovirus RNA in humans, one that investigated the common rs1990760 but which was severely underpowered for genetic association analysis [25] and one relatively large study of young adult patients with established type 1 diabetes (and non-diabetic controls) that investigated two rare type 1 diabetes IFIH1 variants (rs35744605 and rs35667974) [26]. Both of these studies assayed samples from a single time point from each subject, and may consequently have missed important information on the temporal dimension of the infection. Nevertheless, the rare variant of rs35667974 was slightly more frequent among patients positive for enterovirus RNA in blood compared to enterovirus negative patients [26]. These studies [25], [26], like ours, lack information on enterovirus serotype (genotype). The high number of specific types of enterovirus would require a prohibitively large number of samples for reasonable statistical power for individual serotypes. While we had good power to detect potential associations of small to moderate magnitude (OR<2.0) of the common IFIH1 SNP our data suggest that the per allele odds ratio is highly unlikely to be greater than about 1.2 (upper 95% CI for per allele trend OR). We cannot exclude weak to moderate effects of the rare variants because of limited power, but the possible existence of such weak effects of rare IFIH1 variants on enterovirus infections would have limited population impact on the epidemiology of enterovirus infections, if any at all.
Potential explanations of our main findings
If the suggestive association between the rare A variant of rs35732034 and frequency of high quantity enteroviral infections is true, this indicates a dominant or dose-dependent effect of the predicted interference with splicing of the intron [20]. We can only speculate on the potential mechanisms explaining this phenomenon, while the other established type 1 diabetes associated polymorphisms did not predict enterovirus infections. Predictions based on sequence analysis of the rare variants [20], supported by some experimental functional studies [26]–[28], suggest that the rare variants associated with lower risk of type 1 diabetes are loss of function variants (functional studies are reviewed in [29]). Most such functional in vitro studies are limited to one or a few aspects of function in one or a few cell types taken out of their in vivo context, and perhaps with limited or uncertain sensitivity. We must keep in mind that the endpoint observed in the current study, fecal shedding of enteroviral RNA, is the likely the result of a very complex interaction between the human host and the viral agent and probably activation of a number of signaling pathways in multiple cell types. While there is no obvious reason to suspect that the functional consequences of the rare variant of rs35732034 should be more severe than that of the other three rare variant SNPs studied, our results indicate that one normal allele is sufficient for eliciting a physiological normal antiviral host immune control of enterovirus infections in the gut of otherwise healthy infants, at least for the SNPs not associated with enterovirus frequency. One reason for lack of association could be the involvement of complementary innate antiviral pathways, such as those involving the Toll-like receptors (reviewed in [30]), explaining why IFIH1 was not essential for IFN-production after an enterovirus infection in mda5 knockout mice [8]. Another potential explanation could be one of the pathogen survival strategies of host immune evasion involving degradation of host IFIH1 signaling molecules [31]. The two groups who have experimentally infected mda5 knockout mice with coxsackievirus have produced partially conflicting results, but one group showed significantly decreased production of type I IFN, but this was not correlated with increased virus titers [9]. The other group showed that type I IFN levels were not affected, but viruses titers were markedly increased transiently [8].
Enterovirus infections and type 1 diabetes related islet autoimmunity
The evidence for the involvement of the enterovirus genus in type 1 diabetes pathogenesis seem likely, yet the limited numbers of available longitudinal studies of islet autoimmunity where enteroviral RNA have been tested as a risk factor have been inconsistent between populations and sources of the sample [13], [32]–[38]. The evidence seems to be limited to detection in blood or its fractions and predominantly studies done in Finland where serum enterovirus RNA [38] or combined with increases in enterovirus antibodies have been used as indicators of infection [34]–[36]. This is in contrast to studies from other countries (USA and Germany) showing no association [32], [33]. Neither, studies assessing enterovirus RNA in fecal specimens show any association with islet autoimmunity [13], [32], [37].
The IFIH1 alleles that had been previously associated with reduced type 1 diabetes risk were predicted to be loss of function variants, and may thus confer increased risk for enterovirus infection. We therefore reasoned that our previously published lack of association between enterovirus RNA and islet autoimmunity [13] could be influenced by adjustment for the IFIH1 SNPs. However, adjusting for IFIH1 (rs1990760) and the exclusion of children with rare IFIH1 alleles did not influence the lack of association. A prospective cohort study recently reported lack of association with islet autoantibody development for genotypes of the IFIH1 SNP rs2111485, a SNP in close proximity and in strong linkage disequilibrium with rs1990760. While this may partially be explained by limited statistical power, their study showed an association with faster progression from autoimmunity to type 1 diabetes [39]. This may hint towards a potential role in a different immunological context, but which would be very demanding to study in humans.
In conclusion, despite the predictions that rare IFIH1 variants previously associated with type 1 diabetes are loss of function variants, and experimental data showing an important role of IFIH1 in antiviral immunity to enteroviruses, out data suggests that these variants have no or just minor impact on the frequency and duration of gut infection with enterovirus in healthy infants. Its effect on the risk of diabetes is likely to lie elsewhere in the pathogenic process than in the modification of gut infection frequency.
Supporting Information
The effect of IFIH1 genotypes on different measures of enterovirus infection in longitudinal fecal samples.
(DOC)
Acknowledgments
We thank the public health care nurses, in particular Turid Wetlesen and Lene Gustavsen, for their effort in the recruitment to the MIDIA study and follow-up of participating families. We also thank Trond Rasmussen for excellent data base management, and Caroline Anna Brorsson, Regine Bergholdt, Bodil Bosmann Jørgensen, Rikke Bonne and others at Hagedorn Research Institute, Novo Nordisk, Denmark, for help with planning the genotyping experiments and for valuable inputs. Peter A. Torjussen was responsible for islet autoantibody testing. The staff at the Biobank, Norwegian Institute of Public Health, helped with sample registration and initial handling. We express our gratitude to all the parents and children for participating in the study.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This study and the MIDIA project were funded by the Norwegian Organization for Health and Rehabilitation (2008/0182), the Ministry of Education of the Czech Republic (grant MSM0021620814) and the Ministry of Health of the Czech Republic (IGA MZ 11465-5), the Research Council of Norway (grants 135893/330, 155300/320, 156477/730, and 166515/V50), the Norwegian Diabetes Association and Sigurd K. Thoresen's Legacy. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Pallansch MA, Roos RP. Enteroviruses: polioviruses, coxsackieviruses, echoviruses, and newer enteroviruses. In: Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, et al., editors. Fields Virology. Philadelphia: Lippincott Williams & Wilkins; 2007. pp. 839–894. [Google Scholar]
- 2.Ouzilou L, Caliot E, Pelletier I, Prevost MC, Pringault E, et al. Poliovirus transcytosis through M-like cells. J Gen Virol. 2002;83:2177–2182. doi: 10.1099/0022-1317-83-9-2177. [DOI] [PubMed] [Google Scholar]
- 3.Minor PD. Poliovirus. In: Nathanson N, editor. Viral Pathogenesis. Philadelphia: Lippincott-Raven; 1997. pp. 555–574. [Google Scholar]
- 4.Bodian D. Emerging concept of poliomyelitis infection. Science. 1955;122:105–108. doi: 10.1126/science.122.3159.105. [DOI] [PubMed] [Google Scholar]
- 5.Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140:805–820. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- 6.Gitlin L, Barchet W, Gilfillan S, Cella M, Beutler B, et al. Essential role of mda-5 in type I IFN responses to polyriboinosinic:polyribocytidylic acid and encephalomyocarditis picornavirus. Proc Natl Acad Sci U S A. 2006;103:8459–8464. doi: 10.1073/pnas.0603082103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kato H, Takeuchi O, Sato S, Yoneyama M, Yamamoto M, et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441:101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- 8.Huhn MH, McCartney SA, Lind K, Svedin E, Colonna M, et al. Melanoma differentiation-associated protein-5 (MDA-5) limits early viral replication but is not essential for the induction of type 1 interferons after Coxsackievirus infection. Virology. 2010;401:42–48. doi: 10.1016/j.virol.2010.02.010. [DOI] [PubMed] [Google Scholar]
- 9.Wang JP, Cerny A, Asher DR, Kurt-Jones EA, Bronson RT, et al. MDA5 and MAVS mediate type I interferon responses to coxsackie B virus. J Virol. 2010;84:254–260. doi: 10.1128/JVI.00631-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hyöty H, Taylor KW. The role of viruses in human diabetes. Diabetologia. 2002;45:1353–1361. doi: 10.1007/s00125-002-0852-3. [DOI] [PubMed] [Google Scholar]
- 11.Richardson SJ, Willcox A, Bone AJ, Foulis AK, Morgan NG. The prevalence of enteroviral capsid protein vp1 immunostaining in pancreatic islets in human type 1 diabetes. Diabetologia. 2009;52:1143–1151. doi: 10.1007/s00125-009-1276-0. [DOI] [PubMed] [Google Scholar]
- 12.Roivainen M. Enteroviruses: new findings on the role of enteroviruses in type 1 diabetes. Int J Biochem Cell Biol. 2006;38:721–725. doi: 10.1016/j.biocel.2005.08.019. [DOI] [PubMed] [Google Scholar]
- 13.Tapia G, Cinek O, Rasmussen T, Witsø E, Grinde B, et al. Human enterovirus RNA in monthly fecal samples and islet autoimmunity in Norwegian children with high genetic risk for type 1 diabetes: the MIDIA study. Diabetes Care. 2011;34:151–155. doi: 10.2337/dc10-1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smyth DJ, Cooper JD, Bailey R, Field S, Burren O, et al. A genome-wide association study of nonsynonymous SNPs identifies a type 1 diabetes locus in the interferon-induced helicase (IFIH1) region. Nat Genet. 2006;38:617–619. doi: 10.1038/ng1800. [DOI] [PubMed] [Google Scholar]
- 15.Barrett JC, Clayton DG, Concannon P, Akolkar B, Cooper JD, et al. Genome-wide association study and meta-analysis find that over 40 loci affect risk of type 1 diabetes. Nat Genet. 2009 doi: 10.1038/ng.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Concannon P, Onengut-Gumuscu S, Todd JA, Smyth DJ, Pociot F, et al. A human type 1 diabetes susceptibility locus maps to chromosome 21q22.3. Diabetes. 2008;57:2858–2861. doi: 10.2337/db08-0753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klinker MW, Schiller JJ, Magnuson VL, Wang T, Basken J, et al. Single-nucleotide polymorphisms in the IL2RA gene are associated with age at diagnosis in late-onset Finnish type 1 diabetes subjects. Immunogenetics. 2010;62:101–107. doi: 10.1007/s00251-009-0417-4. [DOI] [PubMed] [Google Scholar]
- 18.Qu HQ, Marchand L, Grabs R, Polychronakos C. The association between the IFIH1 locus and type 1 diabetes. Diabetologia. 2008;51:473–475. doi: 10.1007/s00125-007-0895-6. [DOI] [PubMed] [Google Scholar]
- 19.Todd JA, Walker NM, Cooper JD, Smyth DJ, Downes K, et al. Robust associations of four new chromosome regions from genome-wide analyses of type 1 diabetes. Nat Genet. 2007;39:857–864. doi: 10.1038/ng2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nejentsev S, Walker N, Riches D, Egholm M, Todd JA. Rare variants of IFIH1, a gene implicated in antiviral responses, protect against type 1 diabetes. Science. 2009;324:387–389. doi: 10.1126/science.1167728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stene LC, Witsø E, Torjesen PA, Rasmussen T, Magnus P, et al. Islet autoantibody development during follow-up of high-risk children from the general Norwegian population from three months of age: design and early results from the MIDIA study. J Autoimmun. 2007;29:44–51. doi: 10.1016/j.jaut.2007.04.003. [DOI] [PubMed] [Google Scholar]
- 22.Cinek O, Witsø E, Jeansson S, Rasmussen T, Drevinek P, et al. Longitudinal observation of enterovirus and adenovirus in stool samples from Norwegian infants with the highest genetic risk of type 1 diabetes. J Clin Virol. 2006;35:33–40. doi: 10.1016/j.jcv.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 23.Rabe-Hesketh S, Skrondal A. Multilevel and longitudinal modelling using Stata. 2008. Stata Press, College station, Texas, USA.
- 24.Witsø E, Palacios G, Cinek O, Stene LC, Grinde B, et al. High prevalence of human enterovirus a infections in natural circulation of human enteroviruses. J Clin Microbiol. 2006;44:4095–4100. doi: 10.1128/JCM.00653-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schulte BM, Bakkers J, Lanke KH, Melchers WJ, Westerlaken C, et al. Detection of enterovirus RNA in peripheral blood mononuclear cells of type 1 diabetic patients beyond the stage of acute infection. Viral Immunol. 2010;23:99–104. doi: 10.1089/vim.2009.0072. [DOI] [PubMed] [Google Scholar]
- 26.Chistiakov DA, Voronova NV, Savost'Anov KV, Turakulov RI. Loss-of-function mutations E6 27X and I923V of IFIH1 are associated with lower poly(I:C)-induced interferon-beta production in peripheral blood mononuclear cells of type 1 diabetes patients. Hum Immunol. 2010;71:1128–1134. doi: 10.1016/j.humimm.2010.08.005. [DOI] [PubMed] [Google Scholar]
- 27.Downes K, Pekalski M, Angus KL, Hardy M, Nutland S, et al. Reduced expression of IFIH1 is protective for type 1 diabetes. PLoS ONE. 2010;5 doi: 10.1371/journal.pone.0012646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shigemoto T, Kageyama M, Hirai R, Zheng J, Yoneyama M, et al. Identification of loss of function mutations in human genes encoding RIG-I and MDA5: implications for resistance to type I diabetes. J Biol Chem. 2009;284:13348–13354. doi: 10.1074/jbc.M809449200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chistiakov DA. Interferon induced with helicase C domain 1 (IFIH1) and virus-induced autoimmunity: a review. Viral Immunol. 2010;23:3–15. doi: 10.1089/vim.2009.0071. [DOI] [PubMed] [Google Scholar]
- 30.Kemball CC, Alirezaei M, Whitton JL. Type B coxsackieviruses and their interactions with the innate and adaptive immune systems. Future Microbiol. 2010;5:1329–1347. doi: 10.2217/fmb.10.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mukherjee A, Morosky SA, Delorme-Axford E, Dybdahl-Sissoko N, Oberste MS, et al. The coxsackievirus B 3C protease cleaves MAVS and TRIF to attenuate host type I interferon and apoptotic signaling. PLoS Pathog. 2011;7:e1001311. doi: 10.1371/journal.ppat.1001311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Graves PM, Rotbart HA, Nix WA, Pallansch MA, Erlich HA, et al. Prospective study of enteroviral infections and development of beta-cell autoimmunity. Diabetes autoimmunity study in the young (DAISY). Diabetes Res Clin Pract. 2003;59:51–61. doi: 10.1016/s0168-8227(02)00198-5. [DOI] [PubMed] [Google Scholar]
- 33.Fuchtenbusch M, Irnstetter A, Jager G, Ziegler AG. No evidence for an association of coxsackie virus infections during pregnancy and early childhood with development of islet autoantibodies in offspring of mothers or fathers with type 1 diabetes. J Autoimmun. 2001;17:333–340. doi: 10.1006/jaut.2001.0550. [DOI] [PubMed] [Google Scholar]
- 34.Lönnrot M, Salminen K, Knip M, Savola K, Kulmala P, et al. Enterovirus RNA in serum is a risk factor for beta-cell autoimmunity and clinical type 1 diabetes: a prospective study. Childhood Diabetes in Finland (DiMe) Study Group. J Med Virol. 2000;61:214–220. [PubMed] [Google Scholar]
- 35.Sadeharju K, Hamalainen AM, Knip M, Lönnrot M, Koskela P, et al. Enterovirus infections as a risk factor for type I diabetes: virus analyses in a dietary intervention trial. Clin Exp Immunol. 2003;132:271–277. doi: 10.1046/j.1365-2249.2003.02147.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Salminen K, Sadeharju K, Lönnrot M, Vahasalo P, Kupila A, et al. Enterovirus infections are associated with the induction of beta-cell autoimmunity in a prospective birth cohort study. J Med Virol. 2003;69:91–98. doi: 10.1002/jmv.10260. [DOI] [PubMed] [Google Scholar]
- 37.Salminen KK, Vuorinen T, Oikarinen S, Helminen M, Simell S, et al. Isolation of enterovirus strains from children with preclinical Type 1 diabetes. Diabet Med. 2004;21:156–164. doi: 10.1111/j.1464-5491.2004.01097.x. [DOI] [PubMed] [Google Scholar]
- 38.Oikarinen S, Martiskainen M, Tauriainen S, Huhtala H, Ilonen J, et al. Enterovirus RNA in blood is linked to the development of type 1 diabetes. Diabetes. 2011;60:276–279. doi: 10.2337/db10-0186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Winkler C, Lauber C, Adler K, Grallert H, Illig T, et al. An Interferon-Induced Helicase (IFIH1) Gene Polymorphism Associates With Different Rates of Progression From Autoimmunity to Type 1 Diabetes. Diabetes. 2011;60:685–690. doi: 10.2337/db10-1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The effect of IFIH1 genotypes on different measures of enterovirus infection in longitudinal fecal samples.
(DOC)