Abstract
Escherichia coli O157:H7 carries a chromosomal msbB1 and a plasmid-encoded msbB2 gene. We characterized msbB2 function as a homologue of msbB1 by examination of wild-type organisms and mutant strains that lacked functional msbB1, msbB2, and both msbB1 and msbB2. The msbB double-mutant strain generated pentaacyl lipid A, while the single-mutant strains synthesized hexaacyl lipid A. Complementation with overexpressed msbB2 converted pentaacyl into hexaacyl lipid A in the double-mutant strain. The transcription of both msbB genes occurred simultaneously. Lack of MsbB2 activity slightly increased the microheterogeneity of the lipid A species. These results suggest that the msbB2 gene plays a role not only in the routine generation of fully hexaacylated lipid A but also in suppressing the microheterogeneity of lipid A species, the endotoxic determinant of the organism.
Typical enterohemorrhagic Escherichia coli (EHEC) has the ability to produce Shiga toxin (Stx) and to induce attaching and effacing lesions in the intestine. Among the EHEC strains, serotype O157:H7 is the most frequently encountered in disease outbreaks and in severe sequelae worldwide (3, 23, 31). All O157:H7 strains possess an approximately 92-kb plasmid called pO157, which encodes several virulence-associated genes including the following: ehxA, which encodes an RTX type of hemolysin; katP, which encodes a catalase-peroxidase; espP, which encodes a serine protease; and toxB, whose product confers adhesiveness to cultured host cells (20, 31). The pO157 plasmid also carries a homologue (msbB2) of the chromosomal msbB (multicopy suppressor of htrB) gene, herein designated msbB1. This fact raises questions about the role of msbB2 in lipid A biosynthesis, cell wall composition, and intestinal colonization by EHEC O157:H7. A recent report showed that chromosomal and plasmid-encoded msbB genes are also present in Shigella flexneri and are required for maximal acylation of lipid A and invasion of the intestinal epithelium (10). The msbB2 gene is part of a locus comprising the consecutive open reading frames (ORFs) shf-rfbU-virK-msbB in the virulence plasmid pWR100 of S. flexneri. This locus is also conserved in pO157 (5, 21, 33) and is hereby designated as the shf locus.
The occurrence of the msbB2 gene may imply a role in virulence by modulation of lipid A species that affect the host response to lipopolysaccharide (LPS), but this has not been investigated in E. coli O157:H7. The chromosomal msbB1 gene is 99% identical in nucleotide sequence to the E. coli K-12 msbB gene. The deduced amino acid sequence of the MsbB2 protein is 67% identical to the counterpart MsbB1 protein. The MsbB2 proteins of plasmids pO157 and pWR100 are 69% identical at the amino acid level.
The K-12 msbB gene was originally identified as a multicopy suppressor of the null mutant phenotype of the htrB (high-temperature requirement) gene, which encodes a protein that supports growth of the mutant strain in rich media at temperatures above 33°C (17, 18). Later, the enzymatic functions of the products of the htrB and msbB genes were identified as acyltransferases involved in the addition of secondary acyl chains to the distal glucosamine sugar of lipid A (7). Both HtrB and MsbB depend on the prior addition of 3-deoxy-d-manno-2-octulosonic acid (Kdo) to their precursor substrate and utilize acyl carrier protein as an acyl donor. However, HtrB acts first, adding a 12-carbon laurate chain to the N-linked R-3-hydroxymyristate chain, and is followed by MsbB, which adds a 14-carbon myristate chain to the O-linked R-3-hydroxymyristate chain (7, 26). The htrB and msbB genes are also called lpxL or waaM and lpxM or waaN, respectively, according to their functional relatedness to genes required for LPS biosynthesis (13, 26). In previous studies done with E. coli K-12 msbB mutant strains, lack of MsbB activity caused a dramatic reduction in the bioactivity of the mainly pentaacylated LPS, without affecting viability at any growth temperature (16, 29, 30).
In order to test the hypothesis that the msbB2 gene of plasmid pO157 encodes a protein whose function is similar to that of msbB1, we created mutations in msbB1, msbB2, and both msbB1 and msbB2 in an EHEC O157:H7 strain and determined the effects of these mutations. We showed that the ORF of msbB2 encodes a functional acyltransferase that compensated for the loss of function of msbB1 as judged by lipid A acylation patterns in the mutant strains examined by thin-layer chromatography (TLC). We also demonstrated that the msbB2 gene, cloned in a pBlueScript plasmid, could partially restore the attenuated virulence of an msbB mutant of a septicemic E. coli strain in a mouse lethality model. The TLC assays of lipid A species isolated from msbB mutants of an EHEC O157:H7 strain suggest that MsbB2 acts as a late-functioning acyltransferase involved in lipid A biosynthesis and may suppress minor modifications of lipid A species that occur in the absence of MsbB2.
Creation of msbB mutants.
Single msbB1 and msbB2 mutations were achieved by using a modified inverse PCR method (36) and a mutagenic megaprimer-based two-step PCR method (8), respectively, to create in-frame deletion mutants. General gene manipulation methods (28) were exploited for the construction of recombinant vectors, pRE2B1 and pRE7B2 (Table 1), that are sacB-containing allelic exchange vectors carrying the mutated msbB1 and the mutated msbB2 genes, respectively. The construct was moved into streptomycin (Str)-resistant wild-type E. coli O157:H7 strain 4304 (PT14) by mating in order to obtain the desired allelic exchange. The exconjugants were purified on selective media containing appropriate antibiotics and spread on Luria-Bertani (LB) agar containing 10% sucrose for counterselection of double-crossover derivatives. Each potential mutant was isolated, and the mutation was verified by PCR (data not shown). The identified msbB1 mutant was designated strain 4304-M1, and the msbB2 mutant was designated strain 4304-M2 (Table 1). Strain 4304-M2 was then used as recipient for the exchange of the ΔmsbB1 allele carried in plasmid pRE2B1 in order to create a double msbB mutant. The double-crossover colonies were purified, and the mutation was confirmed by PCR (data not shown). The resulting double msbB mutant was designated strain 4304-DM (Table 1).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotype description | Reference or source |
|---|---|---|
| Strains | ||
| E. coli O157:H7 | ||
| 4304 | wild type (phage-type 14), spontaneous Strr strain | This study |
| 4304-M1 | a single msbB1 mutant of wild-type strain (Strr) | This study |
| 4304-M2 | a single msbB2 mutant of wild-type strain (Strr) | This study |
| 4304-DM | a double msbB1/msbB2 mutant strain (Strr) | This study |
| DM-pM2 | 4304-DM transformed with pMsbB2 | This study |
| DM-pB2 | 4304-DM transformed with pBAD-B2 | This study |
| DM-pB2 | 4304-M2 transformed with pBAD-B2 | This study |
| E. coli O18:K1:H7 | ||
| H16 | wild type, a septicemic E. coli isolate | 30 |
| M600 | a msbB mutant of H16 (msbB::Strr) | 30 |
| M600-pBS | M600 carrying intact pBlueScriptII KS+ | This study |
| M600-pM2 | M600 carrying pMsbB2 plasmid | This study |
| M600-pB2 | M600 carrying pBAD-B2 plasmid | This study |
| E. coli JM83 | F−ara D (lac-proAB) rpsL thi [φ80dD(lacZ)M15] | 29 |
| BMS67C12 | a msbB mutant of JM83 (msbB::Tn5), Kmr | 29 |
| DH5αλpir | F−hsdR17 thi-1 gyrA D(lacZYA-argF) supE44 recA1 [φ80dD(lacZ)M15] relA λpir | 11 |
| SM10 (λpir) | thi thr leu tonA lacY supE recA::RP4-2-Tc::Mu λpir Kmr | 11 |
| Plasmids | ||
| pBlueScriptII KS+ | Apr, general cloning, multicopy complementation | Stratagene |
| pUC18 | Apr, general cloning vector | 28 |
| pBAD24 | Apr, arabinose-inducible expression vector | 15 |
| pRE107 | mobilizable suicide vector, Apr, sacB | 11 |
| pRE112 | mobilizable suicide vector, Cmr, sacB | 11 |
| pMsbB2 | recombinant plasmid carrying msbB2 in pBlueScriptII KS+ | This study |
| pRE2B1 | recombinant plasmid carrying mutated msbB1 in pRE112 | This study |
| pRE7B2 | recombinant plasmid carrying mutated msbB2 in pRE107 | This study |
| pBAD-B2 | recombinant plasmid carrying intact msbB2 in pBAD24 | This study |
Complementation of the mutant phenotypes.
The intact msbB2 gene, amplified by PCR using a high-fidelity DNA polymerase (Advantage-HF2 PCR kit; BD Biosciences Clontech, Palo Alto, Calif.), was cloned into an arabinose-inducible expression vector pBAD24 (15). The resulting pBAD-B2 plasmid was introduced into strains 4304-M2, 4304-DM, and M600, and the resulting transformants were named M2-pB2, DM-pB2, and M600-pB2, respectively (Table 1). Septicemic E. coli strain H16 (O18:K1:H7) and its msbB mutant M600 strain were kindly provided by Richard Darveau (University of Washington, Seattle). For high-copy-number vector-based complementation, we cloned the intact msbB2 gene containing its own predicted promoter region into a pBlueScript vector and named the construct pMsbB2. The wild-type H16, M600-pBS, and M600-pM2 strains (Table 1) were tested for growth on MacConkey agar at 37°C, since we found that the double msbB mutant (strain 4304-DM) grown on MacConkey agar showed impairment that was marked at 30°C and moderate at 37°C.
The growth impairment on MacConkey agar was overcome by complementation with plasmid pMsbB2 (data not shown). The MacConkey agar growth impairment phenotype of strain 4304-DM is newly identified in this study, as previous msbB knockout derivatives of E. coli K-12 strains, such as MLK1067 (18), KL423 (22), and BMS67C12 (30), grew normally on MacConkey agar. Interestingly, M600 also showed severe growth impairment on MacConkey agar at 37°C (identified in this study). However, unlike M600 cells that are elongated and/or filamentous (30), no marked alterations in the cell morphology of strain 4304-DM were observed by light microscopy of Gram-stained cells grown at 37°C in LB broth (data not shown). These results suggest that phenotypes of msbB mutants derived from pathogenic E. coli are different from those of msbB knockouts of nonpathogenic K-12 strains, and the growth impairment on MacConkey agar may imply that the function of msbB genes in the pathogenic E. coli might be implicated in overall outer membrane (OM) integrity, which is required for growth under stress conditions and expression of virulence.
Transcriptional analysis of the two msbB genes.
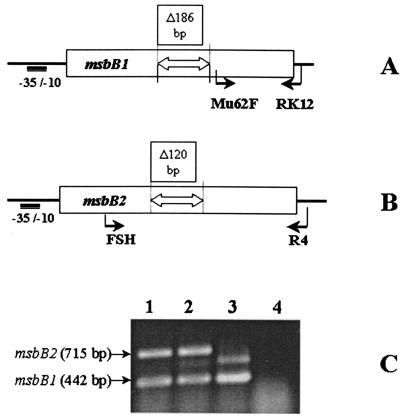
Since the EHEC O157 strain has two msbB genes, it was interesting to see if transcription of both msbB genes occurred constitutively in the wild type and single mutants of the O157 strain. Total RNA was extracted from the mid-log phase bacterial cultures and reverse transcriptase PCR (RT PCR) was conducted by using the SuperScript one-step RT PCR kit (Invitrogen Canada Inc., Burlington, Ontario, Canada) as described in the manual, with two gene-specific primer sets: Mu62F (TTCATCCAGTCGGTACGTCA) and RK12 (AACTTGAAGCTTATCATCAGGCGAAG) for msbB1 and FSH (GAGAATATCGGTACCGCCATGTTTGC) and R4 (TATTGCTGGGTGAGCTCATTATCCTG) for msbB2. The RT PCR mixture was prepared, containing a 15 pM concentration of each primer and 1 μg of total RNA extracted from the wild-type 4304, 4304-M1, and 4304-M2 strains, respectively. The priming sites of Mu62F and RK12 are located outside the region of the 186-bp internal deletion in the msbB1 mutant (Fig. 1A), and the size of cDNA amplicons (Mu62F-RK12) for msbB1 alleles in the wild-type 4304, 4304-M1, and 4304-M2 strains was 442 bp, as shown by RT PCR (Fig. 1C). The RT PCR products for msbB2 (FSH-R4; 715 bp) amplified in the wild-type 4304, 4304-M1, and 4304-M2 strains were exactly matched to the expected product sizes of the msbB2 alleles. Because the priming sites of the FSH and R4 primers encompass the deleted region for the msbB2 mutation (Fig. 1B), the msbB2 amplicon of strain 4304-M2 (lane 3) was smaller, due to the internal 120-bp deletion, than the amplicons of the intact msbB2 gene of the wild-type 4304 (lane 1) and 4304-M1 (lane 2) strains. Since we used the same amounts of total RNA extract and gene-specific primers in the one-tube RT PCRs, either the msbB1 or msbB2 amplicons served as an internal standard for the semiquantitative comparison of the other's mRNA transcription in the strains grown in LB at 37°C. The RT PCR results (Fig. 1C) showed a marked increase in intact msbB1 transcription in strain 4304-M2 (lane 3, lower band) and a decrease in the transcription of the nonfunctional msbB2 (lane 3, upper band). However, there were no distinct changes in the relative amounts of msbB transcription between the wild-type and 4304-M1 strains. It appears that the transcription of both msbB genes in the wild-type and 4304-M1 strains takes place simultaneously, regardless of the nonfunctional ΔmsbB1 (lane 2, lower band) in strain 4304-M1, grown at 37°C in LB broth.
FIG. 1.
RT PCR detection of mRNA transcripts for msbB genes in E. coli O157:H7 strain 4304 and its msbB mutants. Primer pairs Mu62F and RK12 (A) and FSH and R4 (B) were used for RT PCR detection of mRNA transcripts of the msbB1 and msbB2 genes, respectively. The proposed promoter regions and internal deletions in the msbB1 and msbB2 genes are shown. RT PCR results (C) show that the FSH-R4 amplicon in strain 4304-M2 (lane 3) is consistent with the internal 120-bp deletion of the msbB2 allele, compared with amplicons (715 bp) of the wild-type (lane 1) and 4304-M1 (lane 2) strains. A Taq-amplified (no-RT) control was loaded in lane 4.
Analysis of the lipid A profiles.
For analysis of the fatty acid acylation patterns of lipid A species generated by the msbB mutants and the wild-type strains, a mild-acid hydrolysis method was used to extract lipid A species as described elsewhere (2, 37). The mild-acid hydrolysis procedure allows cleavage of the ketosidic linkage between Kdo and the distal glucosamine sugar of the lipid A molecule without disturbing the acyl chains of lipid A. However, this protocol generates a minor fraction of lipid A species dephosphorylated at position 1 (37). TLC was first performed with the lipid A species isolated from the two single msbB mutant strains (4304-M1 and 4304-M2), in order to compare acylation patterns of the single mutants with those of the wild-type 4304 and the M600 strains (Fig. 2A).
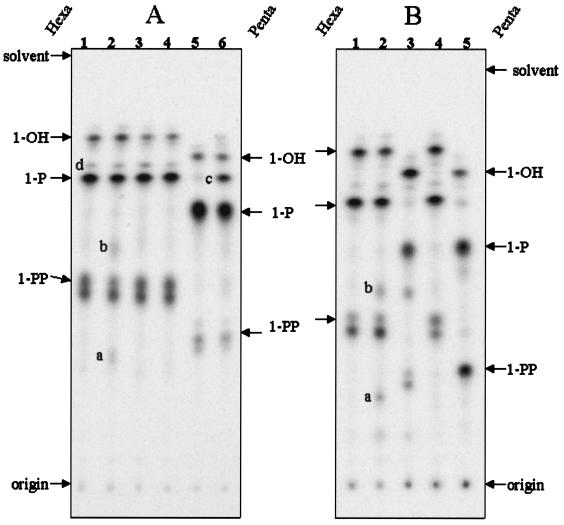
FIG. 2.
Thin-layer chromatograms of 32P-labeled lipid A species generated in various msbB mutant strains. Hexaacylated lipid A species are indicated to the left of each panel (Hexa), while pentaacylated species (Penta) are indicated on the right. (A) The lipid A species of E. coli O157:H7 strains 4304 (lane 1), 4304-M2 (lane 2), 4304-M1 (lane 3), and M2-pB2 (lane 4) occurred as hexaacyl forms, with small amounts of heptaacyl forms (spot d). Two unknown small spots (a and b) were present in the lipid A species of strain 4304-M2. The lipid A species of M600 (lane 5) were mainly pentaacylated, and the uninduced complemented mutant (lane 6) partially restored hexaacyl lipid A species (spot c). (B) Lipid A species were isolated from wild-type strain 4304 (lane 1), 4304-M2 (lane 2), 4304-DM (lane 3), DM-pB2 (lane 4), and BMS67C12 (lane 5). The lipid A species in lane 5 were used as a marker for pentaacyl lipid A species. The unknown spots (lane 2, a and b) occurred consistently in strain 4304-M2. The pentaacyl lipid A species of strain 4304-DM were converted completely into hexaacyl lipid A species in DM-pB2 grown in the presence of 0.2% arabinose. The three major hexaacylated and pentaacylated lipid A species that differ in their phosphorylation state at the anomeric carbon are designated as 1-hydroxyl (1-OH), 1-monophosphoryl (1-P), and 1-PP.
The lipid A species of the wild-type strain 4304 (Fig. 2, lane 1) contained mainly hexaacyl residues with a small amount of heptaacyl forms (Fig. 2, spot d), which are presumably due to modification by the PagP enzyme after the newly synthesized hexaacyl lipid A species are translocated into the OM (1). The lipid A species extracted from strain 4304-M2 showed an acylation pattern which was very similar to that of the wild type, except that two additional small spots (Fig. 2, spots a and b) were evident, which may indicate a modification of lipid A species produced from this mutant strain possibly by substitution of one of the phosphate groups in lipid A with an unknown hydrophilic molecule. These spots were absent in the lipid A species of strain 4304-M2 complemented with plasmid pBAD-B2 (lane 4). These results may indicate that inactivation of the msbB2 gene causes minor modifications in the lipid A species of the EHEC O157:H7 strain, and the modifications are suppressed by introduction of a functional MsbB2. Interestingly, the TLC pattern with the lipid A species of strain 4304-M1 (lane 3) was identical to that of the wild type, indicating that the intact msbB2 carried in plasmid pO157 may fully complement the loss of function of MsbB1 (a known myristoyl-transferase). As expected, the lipid A species of M600 (lane 5) were mainly pentaacylated, and M600-pB2, the complemented strain of M600 carrying pBAD-B2, showed a partial restoration of hexaacyl lipid A species (lane 6, spot c) when grown under uninduced culture conditions.
Because we were unable to find distinct msbB knockout phenotypes (pentaacyl lipid A species) in TLC assays for the single mutants (Fig. 2A), we created a double msbB mutant strain (4304-DM) for the assessment of the lipid A phenotypes by TLC (Fig. 2B). Strain 4304-DM (Fig. 2B, lane 3) showed pentaacyl lipid A species, compared with the wild-type 4304 (lane 1), 4304-M2 (lane 2), and DM-pB2 (lane 4) strains. The lipid A species (lane 5) extracted from BMS67C12 (msbB::Tn5) derived from a K-12 strain JM 83 (29) was used as a marker for pentaacyl lipid A species in the TLC assay (Fig. 2B). Interestingly, strain DM-pB2 showed an identical lipid A pattern to that of the wild-type 4304 strain when the culture was induced with 0.2% arabinose. The two unknown spots (a and b) occurred reproducibly in strain 4304-M2 but not in strain DM-pB2. These results reconfirmed that inactivation of the msbB2 gene causes minor modifications in lipid A species of EHEC O157:H7 strain. Consistently, the pentaacyl lipid A species of strain 4304-DM were converted completely into the hexaacyl forms in DM-pB2 grown in the presence of arabinose, implying that the overexpressed MsbB2 alone could generate a lipid A pattern identical to that of the wild-type strain 4304. Two unknown spots (a and b) occurred consistently in strain 4304-M2, implying that the intact msbB1 gene alone in the ΔmsbB2 mutant may cause the unknown modifications in the lipid A species. Determination of the structure of the unknown lipid A spots is required to address whether these subtle changes in the lipid A species of strains 4304-M2 and 4304-DM are due to the reduced gene dosage effect or to the absence of the MsbB2 activity. The 4304 strains showed an additional spot below the 1-diphosphoryl (1-PP) lipid A spot. The lower spot of the 1-PP doublet was purified, analyzed by mass spectrometry, and determined to be lipid A substituted with pyrophosphoryl-ethanolamine (data not shown). Collectively, the TLC assays suggest that MsbB2 acts as a late-functioning acyltransferase involved in lipid A biosynthesis, in concert with MsbB1, and may suppress minor modifications of lipid A species that occur in the absence of MsbB2.
In vivo assessment of msbB2 function in a mouse lethality model.
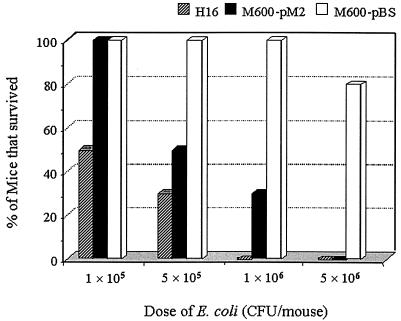
A mouse lethality test (30) was conducted to assess whether the msbB2 gene could compensate for the loss of virulence of a msbB mutant (M600) of a septicemic E. coli strain (H16). The wild-type H16, M600-pBS, and M600-pM2 strains (Table 1) were prepared as four dosages (1 × 105, 5 × 105, 1 × 106, and 5 × 106 CFU/mouse) and were administered in a 0.2-ml inoculum by the intraperitoneal route to 8- to 10-week-old BALB/c mice. Each dose was administered to a group of five mice, and the experiment was duplicated. Mortality was monitored for 5 days, and the numbers of mice that survived were recorded, although the majority of the deaths occurred within 48 h postinoculation. Data obtained from two independent experiments were combined for the assessment of msbB2-mediated virulence. At the lowest challenge dose (105 CFU) 50% (5 of 10) of the mice survived after receiving the wild-type septicemic E. coli strain (H16) (Fig. 3), whereas 100% of the mice survived after receiving the msbB mutant (M600-pBS) or the high-copy-number msbB2-complemented mutant (M600-pM2) strain. As the dosage of the E. coli strains increased to 106 CFU, the percentages of surviving mice challenged with the wild type and the M600-pM2 declined steadily (Fig. 3). The percentage of surviving mice challenged with M600-pBS did not change from 100% until the challenge dose was 5 ×106 CFU. The data from each of the four dose groups were compared for statistical significance by two-tailed t tests. The wild-type strain (H16) was significantly more virulent than the mutant organism (M600-pBS) at all doses (P < 0.05). The virulence of the complemented mutant strain (M600-pM2) was significantly greater than that of the mutant strain (P < 0.05) and was similar to that of the wild-type strain (P > 0.05) at the 1 × 106 and 5 × 106 CFU/mouse doses.
FIG. 3.
Survival rate (%) of mice challenged with E. coli H16 (O18:K1:H7) and its msbB mutants carrying either the pBlueScript vector control (M600-pBS) or pMsbB2 (M600-pM2). A mouse lethality test (30) was used in groups of five 8- to 10-week old female BALB/c mice. Each mouse was inoculated intraperitoneally with a 0.2-ml inoculum. The data on survival of the mice were combined from two independent experiments.
The results of the mouse studies showed that the pO157-derived msbB2 clone (pMsbB2) partially restored the attenuated virulence of an msbB mutant strain (M600) of a K1-encapsulated septicemic E. coli strain in a murine model. The partial restoration of mouse lethality was consistent with the lipid A acylation pattern of the complemented mutant strain revealed by TLC analysis. The lipid A species isolated from M600-pM2 showed the presence of a hexaacylated lipid A spot as well as a pentaacylated spot (data not shown). Because the mutant strain complemented with pBAD-B2 (Table 1) showed fully hexaacylated lipid A species in the presence of 0.2% arabinose (data not shown), it appears that expression of the msbB2 gene (pMsbB2) in the complemented mutant strain was poorly induced in vivo. These results suggest that MsbB2 of pO157 has the ability to restore the compromised OM integrity that occurs in M600, derived from a different pathotype of E. coli (O18:K1:H7). A previous report (30) noted that M600 showed pleiotropic phenotypes, including loss of K1 capsulation, which might contribute to loss of virulence in mice. The partial restoration of mouse lethality by M600-pM2 may indicate that the complemented strain might be encapsulated by the correction of the compromised OM functions caused by the endogeneous msbB gene mutation. A recent study (24) conducted with Neisseria meningitidis (serogroup B) showed that surface expression of lipooligosaccharide and porins (PorA and PorB) in the msbB mutant of the encapsulated Neisseria meningitidis (serogroup B) strain was markedly reduced, presumably due to the modification of lipid A structure. These results strongly support the hypothesis that the lipid A structure of gram-negative bacteria plays a significant role in determining whether some structures associated with the OM are attached to the bacteria.
The shf locus.
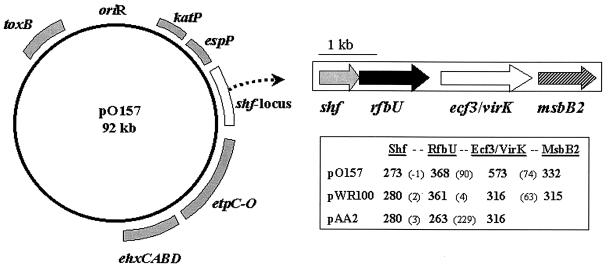
In both S. flexneri and O157 EHEC strains, the plasmid-encoded msbB2 gene is part of a conserved shf locus, illustrated in Fig. 4. The loci consist of four ORFs (shf-rfbU-virK/ecf3-msbB2) in the virulence-associated plasmids (4, 5, 21, 33). There is heterogeneity of individual ORFs clustered in this locus in terms of distribution and proposed protein sizes (Fig. 4). The 100-kb plasmid pAA2 of an enteroaggregative and diffusely adherent E. coli strain possesses a homologue of the shf locus, without the msbB2 gene (9). The occurrence of the shf loci in the virulence plasmids suggests that msbB2 may have been transferred along with the upstream genes during the evolution of these plasmids and that there may be functional associations among the proteins encoded in this locus. Intriguingly, homologues of segments (shf or shf-rfbU) of the locus are found in chromosomes of various bacteria, including S. flexneri 2a (21, 27), Salmonella enterica serovar Typhimurium (21, 27), and Staphylococcus epidermidis (14, 27).
FIG. 4.
Genetic organization of msbB2-bearing shf locus in plasmid pO157 and other virulence plasmids. The msbB2-bearing shf loci consist of ORFs shf-rfbU-ecf3/virK-msbB2 in virulence plasmids pWR100 and pO157 (4, 5, 33). The pAA2 of an enteroaggregative and diffusely adherent E. coli strain possesses the shf locus, without the msbB2 gene (9). In S. flexneri, the third gene in the locus encodes functional VirK (316 aa), which is different from Ecf3 of pO157. Ecf3 is 80% identical toYijP (f577; GenBank accession no. U00006), which has been associated with virulence in E. coli K1 meningitis (34). Several putative virulence-associated genes are marked as gray boxes around the map of the pO157 plasmid (5). The numbers in bold within the box located below the illustrated ORFs are the amino acid numbers comprising each ORF in the shf locus. The numbers in parentheses are nucleotide base pairs in the gaps between the ORFs.
A BLAST program (National Center for Biotechnology Information-GenBank) homology search revealed that the proposed Shf protein has 25% identity with a region of IcaB of S. epidermidis (56 of 216 amino acids [aa]; GenBank accession no. U43366). The icaB gene is in an operon involved in production of polysaccharide intercellular adhesion (14). The second ORF (rfbU) of the shf locus encodes a glycosyltransferase showing a motif shared with genes for α-glycosyltransferase involved in LPS biosynthesis. The corresponding ORF (capU) in pAA2 showed 52% identity (134 of 253 aa) with rfbU of plasmid pO157 (9, 21, 33). The third ORF (Ecf3) of the pO157 shf-locus (3) encodes a hypothetical protein consisting of 573 aa and belongs to a multigene membrane protein (YijP/YhbX/YhjW/YjdB) family encoded in the E. coli chromosome (S.-H. Kim and C. Gyles, unpublished data). In S. flexneri, however, the third gene in the locus is functional VirK (316 aa), which appears to be different from Ecf3 of pO157. The Ecf3 homologues (grouped in the YijP/YhbX/YhjW/YjdB family) are scattered in the chromosomes of E. coli K-12 and O157:H7 EHEC. Among them, E. coli K-12 YijP encoded in f577 (GenBank accession no. U00006) and its counterpart in the chromosome of O157:H7 EHEC (ORF577; accession no. AE005627) are 80% identical to Ecf3 and have been associated with virulence in E. coli K1 meningitis (34). The YijP protein family, whose functions are at present unknown, has similar properties in that they are proposed as integral membrane proteins consisting of 500 to 600 aa and containing 5 to 6 potential transmembrane domains in the N-terminal portion (data not shown).
The significance of the redundancy and association of genes comprising the shf locus in EHEC strain O157:H7 remains unknown. A DNA hybridization study (3) showed that the shf locus was restricted to virulence plasmids of eae-positive Shiga-toxigenic E. coli strains and was not found in the large plasmids from eae-negative Shiga-toxigenic E. coli strains. Thus, it would be interesting to assess whether there are functional associations between the proteins of the locus for enterocyte effacement and the shf locus. We found that there is no apparent transcription termination signal (ρ independent) between the genes of the pO157-encoded shf locus, implying that the entire locus is organized as one transcriptional unit. However, the regulation of individual genes and their functional associations remain elusive.
Conclusions.
The msbB2 gene of pO157 is able to function as a lipid A acyltransferase and may play a role in suppressing minor modification of lipid A species. Suppression of such microheterogeniety of lipid A species may be required for expression of some structures associated with the OM, which may be of benefit to EHEC O157:H7 under certain circumstances. Further studies on the possible roles of the msbB2-bearing shf locus may enhance our understanding of msbB2 functions in cell wall integrity and its possible contribution to the overall function of msbB2-bearing plasmids in pathogenesis. Previous reports have shown that the virulence of some pathogenic gram-negative bacteria was attenuated when the msbB gene was inactivated (10, 19, 25, 29, 30, 32). Because msbB gene inactivation in E. coli K-12 leads to generation of non- and/or less-endotoxic pentaacyl lipid A molecules instead of the fully endotoxic hexaacyl lipid A, we speculate that the plasmid-encoded msbB2 gene in plasmid pO157 may play a role in the modulation of lipid A species that affects the host response to LPS, possibly by direct interactions of the host innate immune system and the modified LPS (6, 12, 29, 32). Furthermore, msbB mutant strains of clinical isolates showed pleiotropic phenotypes, such as loss of K1 encapsulation, growth arrest due to septation defects during cell division, reduction of secretion function of a type III secretion system, and even decrease in expression of lipooligosaccharide and porins in the OM (22, 24, 30, 35). Thus, the carriage of msbB2 in plasmid pO157 may provide the OM functions required for secretion and/or attachment of OM-associated virulence factors.
Editor: V. J. DiRita
REFERENCES
- 1.Bishop, R. E., H. Gibbons, T. Guina, M. S. Trent, S. I. Miller, and C. R. H. Raetz. 2000. Transfer of palmitate from phospholipids to lipid A in outer membranes of gram-negative bacteria. EMBO J. 19:5071-5080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bligh, E. G., and W. J. Dyer. 1959. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37:911-917. [DOI] [PubMed] [Google Scholar]
- 3.Boerlin, P., S. Chen, J. K. Colbourne, R. Johnson, S. de Grandis, and C. Gyles. 1998. Evolution of enterohemorrhagic Escherichia coli hemolysin plasmids and the locus for enterocyte effacement in Shiga toxin-producing E. coli. Infect. Immun. 66:2553-2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buchrieser, C., P. Glaser, C. Rusniok, H. Nedjari, H. d'Hauteville, F. Kunst, P. Sansonetti, and C. Parsot. 2000. The virulence plasmid pWR100 and the repertoire of proteins secreted by the type III secretion apparatus of Shigella flexneri. Mol. Microbiol. 38:760-771. [DOI] [PubMed] [Google Scholar]
- 5.Burland, V., Y. Shao, N. T. Perna, G. Plunkett, H. J. Sofia, and F. R. Blattner. 1998. The complete DNA sequence and analysis of the large virulence plasmid of Escherichia coli O157:H7. Nucleic Acids Res. 26:4196-4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cario, E., D. Brown, M. McKee, K. Lynch-Devaney, G. Gerken, and D. K. Podolsky. 2002. Commensal-associated molecular patterns induce selective Toll-like receptor-trafficking from apical membrane to cytoplasmic compartments in polarized intestinal epithelium. Am. J. Pathol. 160:165-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clementz, T., Z. Zhou, and C. R. H. Raetz. 1997. Function of the Escherichia coli msbB gene, a multicopy suppressor of htrB knockouts, in the acylation of lipid A. J. Biol. Chem. 272:10353-10360. [DOI] [PubMed] [Google Scholar]
- 8.Colosimo, A., Z. Xu, G. Novelli, B. Dallapiccola, and D. C. Gruenert. 1999. Simple version of “megaprimer” PCR for site-directed mutagenesis. BioTechniques 26:870-873. [DOI] [PubMed] [Google Scholar]
- 9.Czeczulin, J. R., T. S. Whittam, I. R. Henderson, F. Navarro-Garcia, and J. P. Nataro. 1999. Phylogenetic analysis of enteroaggregative and diffusely adherent Escherichia coli. Infect. Immun. 67:2692-2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.d'Hauteville, H., S. Khan, D. J. Maskell, A. Kussak, A. Weintraub, J. Mathison, R. J. Ulevitch, N. Wuscher, C. Parsot, and P. J. Sansonetti. 2002. Two msbB genes encoding maximal acylation of lipid A are required for invasive Shigella flexneri to mediate inflammatory rupture and destruction of the intestinal epithelium. J. Immunol. 168:5240-5251. [DOI] [PubMed] [Google Scholar]
- 11.Edwards, R. A., L. H. Keller, and D. M. Schifferli. 1998. Improved allelic exchange vectors and their use to analyze 987P fimbria gene expression. Gene 207:149-157. [DOI] [PubMed] [Google Scholar]
- 12.Ernst, R. K., T. Guina, and S. I. Miller. 2001. Salmonella typhimurium outer membrane remodeling: role in resistance to host innate immunity. Microbes Infect. 3:1327-1334. [DOI] [PubMed] [Google Scholar]
- 13.Erridge, C., E. Bennett-Guerrero, and I. R. Poxton. 2002. Structure and function of lipopolysaccharides. Microbes Infect. 4:837-851. [DOI] [PubMed] [Google Scholar]
- 14.Gerke, C., A. Kraft, R. Süβmuth, O. Schweitzer, and F. Götz. 1998. Characterization of the N-acetylglucosaminyltransferase activity involved in the biosynthesis of the Staphylococcus epidermidis polysaccharide intercellular adhesion. J. Biol. Chem. 273:18586-18593. [DOI] [PubMed] [Google Scholar]
- 15.Guzman, L.-M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hedlund, M., C. Wachtler, E. Johansson, L. Hang, J. E. Somerville, R. P. Darveau, and C. Svanborg. 1999. P fimbriae-dependent, lipopolysaccharide-independent activation of epithelial cytokine responses. Mol. Microbiol. 33:693-703. [DOI] [PubMed] [Google Scholar]
- 17.Karow, M., O. Fayet, A. Cegielska, T. Ziegelhoffer, and C. Georgopoulos. 1991. Isolation and characterization of the Escherichia coli htrB gene, whose product is essential for bacterial viability above 33°C in rich media. J. Bacteriol. 173:741-750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karow, M., and C. Georgopoulos. 1992. Isolation and characterization of the Escherichia coli msbB gene, a multicopy suppressor of null mutations in the high-temperature requirement gene htrB. J. Bacteriol. 174:702-710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Khan, S. A., P. Everest, S. Servos, N. Foxwell, U. Zähringer, H. Brade, E. Th. Rietschel, G. Dougan, I. G. Charles, and D. J. Maskell. 1998. A lethal role for lipid A in Salmonella infections. Mol. Microbiol. 29:571-579. [DOI] [PubMed] [Google Scholar]
- 20.Law, D. 2000. Virulence factors of Escherichia coli O157 and other Shiga toxin-producing E. coli. J. Appl. Microbiol. 88:729-745. [DOI] [PubMed] [Google Scholar]
- 21.Luck, S. N., S. A. Turner, K. Rajakumar, H. Sakellaris, and B. Adler. 2001. Ferric dicitrate transport system (Fec) of Shigella flexneri 2a YSH6000 is encoded on a novel pathogenicity island carrying multiple antibiotic resistance genes. Infect. Immun. 69:6012-6021.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murray, S. R., D. Bermudes, K. Suwwan DE Felipe, and K. B. Low. 2001. Extragenic suppressors of growth defects in msbB Salmonella. J. Bacteriol. 183:5554-5561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.O'Loughlin, E. V., and R. M. Robins-Browne. 2001. Effect of Shiga toxin and Shiga-like toxins on eukaryotic cells. Microbes Infect. 3:493-507. [DOI] [PubMed] [Google Scholar]
- 24.Post, D. M. B., M. R. Ketterer, N. J. Phillips, B. W. Gibson, and M. A. Apicella. 2003. The msbB mutant of Neisseria meningitidis strain NMB has a defect in lipooligosaccharide assembly and transport to the outer membrane. Infect. Immun. 71:647-655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Post, D. M. B., N. J. Phillips, J. Q. Shao, D. D. Entz, B. W. Gibson, and M. A. Apicella. 2002. Intracellular survival of Neisseria gonorrhoeae in male urethral epithelial cells: importance of a hexaacyl lipid A. Infect. Immun. 70:909-920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Raetz, C. R. H., and C. Whitfield. 2002. Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 71:635-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rajakumar, K., F. Luo, C. Sasakawa, and B. Adler. 1996. Evolutionary perspective on a composite Shigella flexneri 2a virulence plasmid-borne locus comprising three distinct genetic elements. FEMS Microbiol. Lett. 144:13-20. [DOI] [PubMed] [Google Scholar]
- 28.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd edition. Cold Spring Harbor Press, Cold Spring Harbor, N.Y.
- 29.Somerville, J. E., Jr., L. Cassiano, B. Bainbridge, M. D. Cunningham, and R. P. Darveau. 1996. A novel Escherichia coli lipid A mutant that produces an anti-inflammatory lipopolysaccharide. J. Clin. Investig. 97:359-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Somerville, J. E., Jr., L. Cassiano, and R. P. Darveau. 1999. Escherichia coli msbB gene as a virulence factor and a therapeutic target. Infect. Immun. 67:6583-6590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tatsuno, I., M. Horie, H. Abe, T. Miki, K. Makino, H. Shinagawa, H. Taguchi, S. Kamiya, T. Hayashi, and C. Sasakawa. 2001. toxB gene on pO157 of enterohemorrhagic Escherichia coli O157:H7 is required for full epithelial cell adherence phenotype. Infect. Immun. 69:6660-6669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van der Ley, P., L. Steeghs, H. J. Hamstra, J. ten Hove, B. Zomer, and L. van Alphen. 2001. Modification of lipid A biosynthesis in Neisseria meningitidis lpxL mutants: influence on lipopolysaccharide structure, toxicity, and adjuvant activity. Infect. Immun. 69:5981-5990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Venkatesan, M. M., M. B. Goldberg, D. J. Rose, E. J. Grotbeck, V. Burland, and F. R. Blattner. 2001. Complete DNA sequence and analysis of the large virulence plasmid of Shigella flexneri. Infect. Immun. 69:3271-3285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang, Y., S.-H. Huang, C. A. Wass, M. F. Stins, and K. S. Kim. 1999. The gene locus yijP contributes to Escherichia coli K1 invasion of brain microvascular endothelial cells. Infect. Immun. 67:4751-4756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Watson, P. R., A. Benmore, S. A. Khan, P. W. Jones, D. J. Maskell, and T. S. Wallis. 2000. Mutation of waaN reduces Salmonella enterica serovar Typhimurium-induced enteritis and net secretion of type III secretion system 1-dependent proteins. Infect. Immun. 68:3768-3771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu, Y., and Z. Gong. 1999. Adaptation of inverse PCR to generate an internal deletion. BioTechniques 26:639-641. [DOI] [PubMed] [Google Scholar]
- 37.Zhou, Z., S. Lin, R. J. Cotter, and C. R. H. Raetz. 1999. Lipid A modifications characteristic of Salmonella typhimurium are induced by NH4VO3 in Escherichia coli K12. J. Biol. Chem. 274:18503-18514. [DOI] [PubMed] [Google Scholar]