Abstract
Dental caries is the most common chronic disease in children and a major public health concern due to its increasing incidence, serious health and social co-morbidities, and socio-demographic disparities in disease burden. We performed the first genome-wide association scan for dental caries to identify associated genetic loci and nominate candidate genes affecting tooth decay in 1305 US children ages 3-12 yrs. Affection status was defined as 1 or more primary teeth with evidence of decay based on intra-oral examination. No associations met strict criteria for genome-wide significance (p < 10E-7); however, several loci (ACTN2, MTR, and EDARADD, MPPED2, and LPO) with plausible biological roles in dental caries exhibited suggestive evidence for association. Analyses stratified by home fluoride level yielded additional suggestive loci, including TFIP11 in the low-fluoride group, and EPHA7 and ZMPSTE24 in the sufficient-fluoride group. Suggestive loci were tested but not significantly replicated in an independent sample (N = 1695, ages 2-7 yrs) after adjustment for multiple comparisons. This study reinforces the complexity of dental caries, suggesting that numerous loci, mostly having small effects, are involved in cariogenesis. Verification/replication of suggestive loci may highlight biological mechanisms and/or pathways leading to a fuller understanding of the genetic risks for dental caries.
Keywords: caries, childhood caries, fluoride(s), genetics, genome-wide association study, genomics
Introduction
The importance of genetic factors in dental caries has been recognized for decades (Townsend et al., 1998), with heritability estimated to be from 30% to 60% (Boraas et al., 1988; Wang et al., 2010). However, the specific genes influencing disease are largely unknown. For a disease as widespread and costly as childhood dental caries, surprisingly few studies of specific genetic loci have been reported. Notable associated genes from candidate gene studies include tuftelin (via an environmental interaction) (Slayton et al., 2005), ameloblastin (Deeley et al., 2008) (both involved in tooth enamel composition), and taste genes (Wendell et al., 2010); however, these findings have not been rigorously explored in replication samples. The current lack of understanding regarding the genetic factors leading to dental caries and the paucity of active research in this area showcase the urgent need for more comprehensive investigation of the genetic variants associated with disease.
Toward this end, dental caries was included as one of the outcomes of interest in the GENEVA consortium (Cornelis et al., 2010), a National Institutes of Health initiative aimed at investigating the genes and gene-by-environment interactions that contribute to common complex disease, including tooth decay. Specifically, we performed the first genome-wide association scans (GWAS) for childhood dental caries, including analyses stratified by fluoride exposure, to address the role of genes and gene-by-fluoride interactions in disease.
Materials & Methods
Details regarding sample recruitment, data collection, phenotype assessment, genotyping, and data analysis are available in the online Supplemental Materials. In brief, 1305 white US children aged 3 to 12 yrs were recruited from the Center for Oral Health Research in Appalachia (PITT) (Polk et al., 2008; Wang et al., 2010), the Iowa Fluoride Study (IFS) (Levy et al., 2001, 2003; Marshall et al., 2003; Franzman et al., 2004), and the Iowa Head Start Study (IHS). All participants provided assent with written parental informed consent, and all study procedures were approved by Institutional Review Boards at the pertinent Universities. Primary dentition caries affection status (yes/no for dft score ≥ 1) was assessed by intra-oral examination. DNA was isolated from biological samples (blood, mouthwash, buccal swab, or saliva) and genotyped for 580,000 single-nucleotide polymorphisms (SNPs) on an Illumina platform (Illumina, Inc., San Diego, CA, USA). Additional non-genotyped SNPs were imputed for a total of 1.4 million SNPs. Association between dental caries affection status and each SNP was tested with PLINK (http://pngu.mgh.harvard.edu/~purcell/plink/) (Purcell et al., 2007).
Home water fluoride data were available for a subset of the sample (N = 729). GWAS was performed for stratified low (N = 274; < 0.7 mg/L) and sufficient (N = 455; ≥ 0.7 mg/L) fluoride groups.
We tested 369 SNPs of interest for replication in an independent sample of 1695 white children aged 2 to 7 yrs from the Denmark National Birth Cohort (Olsen et al., 2001). These included the top 54 genotyped SNPs (i.e., 0.01%) from the discovery GWAS, as well as all genotyped SNPs within 50 kb of 8 genes nominated in the US sample based on nearby association signals and plausible biological roles in dental caries. Parental written consent was provided for the Danish sample, and data collection efforts were approved by the Danish Data Protection Agency and Scientific Ethics Committee of the Capital City Region.
Results
Table 1 summarizes characteristics (age, caries prevalence, and fluoride exposure) for the US discovery sample (including PITT, IFS, and IHS sub-samples) and the Danish replication sample. Differences in sample characteristics between and among sites in this study are expected, due to the differences in recruitment strategies, demography (i.e., population ancestry, socio-economic status), and caries risk factors (i.e., access to oral health care, water source). In general, caries prevalence, defined as 1 or more carious primary tooth surfaces including non-cavitated white spots, cavitated lesions, or fillings, was greater in the US sample (46.9%) compared with the Danish sample (38.1%), and greater in the Appalachia-derived sample (PITT; 57.1%) compared with Iowan samples (IFS, 34.8%; and HIS, 36.4%). Likewise, different proportions of participants were categorized as low-fluoride exposure between PITT and IFS, due to differences in home water sources.
Table 1.
Sample Characteristics
| US |
Denmark |
||||
|---|---|---|---|---|---|
| Study | PITT | IFS | IHS | US total | DNBC |
| Sample size | 724 | 405 | 176 | 1305 | 1695 |
| Mean age (yrs) | 7.3 | 5.1 | 4.1 | 6.2 | 5.3 |
| Age range (yrs) | 3.0 to 12.0 | 3.0 to 8.0 | 3.0 to 8.0 | 3.0 to 12.0 | 2.4 to 7.7 |
| Caries prevalence (%) | 57.1 | 34.8 | 36.4 | 46.9 | 38.1 |
| Low fluoride (%) | 42.8 | 33.0 | NA | 37.6 | NA |
Low fluoride indicates home water source fluoride level < 0.7 mg/L.
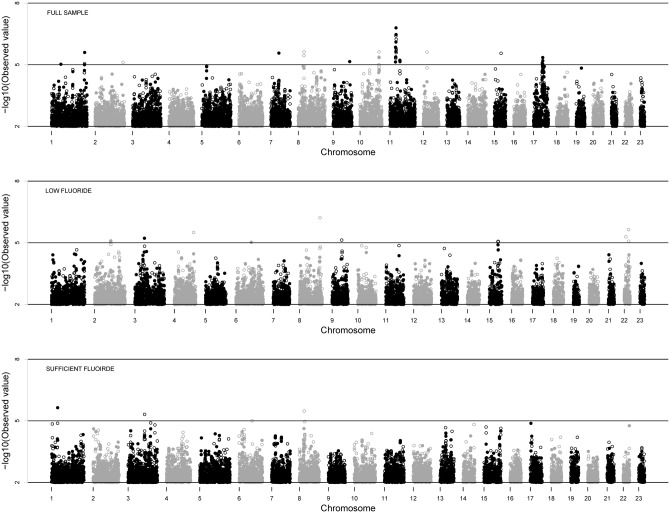
Manhattan plots for GWAS in the US discovery sample and stratified low- and sufficient-fluoride groups are shown in Fig. 1. No associations meeting genome-wide significance (i.e., marginal p-value < 10E-7) were observed. The genomic inflation factor, λ, was 1.065 for GWAS in the full sample, indicating modestly inflated p-values, possibly due in part to the presence of known related individuals in the sample. Adjusted p-values controlling for these family relationships are reported in the text for SNPs of interest. The SNP exhibiting the strongest evidence of association before adjustment was rs11031093 on chromosome 11 (adjusted p-value = 4.9E-6). Permutation methods, which adjust for the multiple tests performed but not for the family structure of the sample, showed that the empirical genome-wide p-value of rs11031093 was 0.071.
Figure 1.
GWAS results in the US discovery sample. From top to bottom, Manhattan plots for the full-sample, low-fluoride, and sufficient-fluoride GWAS are shown, respectively. All p-values are unadjusted and negative log10-transformed. Both genotyped (filled circles) and imputed (open circles) SNPs are shown. Genomic inflation factor was λ = 1.065, SE = 7.85E-6 for the full sample, λ = 0.998, SE = 1.75E-5 for the low-fluoride sample, and λ = 1.014, SE = 1.17E-5 for the sufficient-fluoride sample.
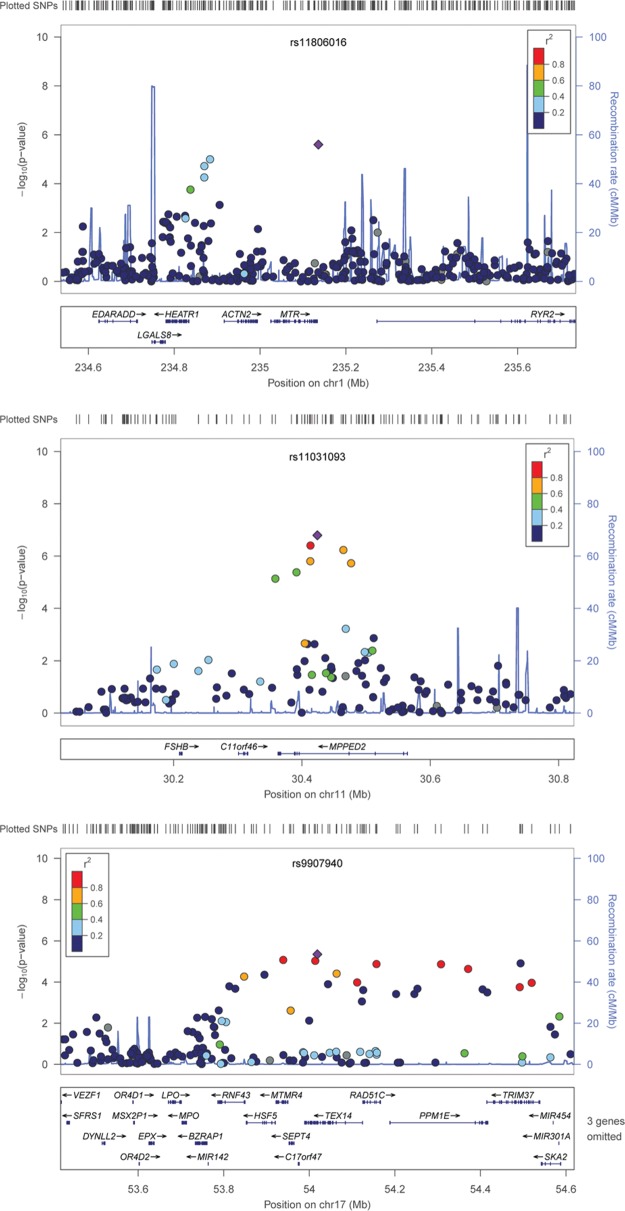
Although no SNPs met conservative criteria for genome-wide significance, multiple suggestive loci, represented by one or more associated SNPs with p-values of between 10E-5 and 10E-7, were observed in the discovery sample. Association results for noteworthy suggestive signals on chromosomes 1, 11, and 17 are detailed in Fig. 2. We assessed the plausible roles of “nearby” genes based on linkage disequilibrium (LD, measured as r2) and recombination rate as defined by the samples of European ancestry in the HapMap project (The International HapMap Project, 2003), as well as physical proximity.
Figure 2.
Nominated loci of interest. Association results from the GWAS for 3 suggestive loci. Top panel: suggestive locus near ACTN2, MTR, and EDARADD genes on chromosome 1. Middle panel: suggestive locus near MPPED2 on chromosome 11. Bottom panel: suggestive locus near LPO on chromosome 17. Negative log10-transformed p-values and physical positions for SNPs in the region are shown. Colors indicate linkage disequilibrium between the top SNP (diamond) and other SNPs based on HapMap CEU data. The rug plot indicates regional SNP density. The recombination rate overlay is based on HapMap CEU data. Gene positions and directions of transcription are annotated.
SNPs in an LD block on chromosome 1 that includes the ACTN2 and MTR genes, and is < 100 kb from EDARADD, showed modest association to dental caries (rs11806016, adjusted p-value = 1.4E-5). ACTN2 (actinin alpha 2) is found in the cytoskeletal microfilament bundles of ameloblasts, with enamel-depositing cells present only during tooth development. A study of gene expression and enamel formation in mice showed that ACTN2 (along with other actin- and cytoskeletal-related genes) may be involved in organizing ameloblasts during tooth enamel formation (Sehic et al., 2010). Genes affecting tooth enamel, such as ACTN2, may play a role in caries susceptibility. MTR (methionine synthase), the gene responsible for methionine and homocysteine production, has not been previously implicated in dental caries, although maternal MTR may be associated with non-syndromic cleft lip and palate (Mostowska et al., 2006), which in turn is associated with dental caries (Al-Dajani, 2009; Parapanisiou et al., 2009; Britton and Welbury, 2010). Genes involved in the structure and development of orofacial features may provide the basis for the link between clefting and dental caries. Mutations in EDARADD (ectodysplasin-A receptor-associated adapter protein), positioned about 100-500 kb from associated SNPs on chromosome 1, cause hypohidrotic ectodermal dysplasia, a Mendelian syndrome that results in abnormal development of teeth, skin, hair, nails, and sweat glands. It is unknown if EDARADD is also involved in non-syndromic variation in dental phenotypes.
In addition to chromosome 1, other association signals were observed on chromosomes 11 and 17. One of the most significant SNPs associated with dental caries in this study was rs11031093 on chromosome 11 (adjusted p-value = 4.9E-6) in the MPPED2 gene (metallophosphoesterase domain containing 2). This gene plays no known role in dental caries; however, one study showed that expression of MPPED2 was decreased by a factor of 5 in oral epithelial cell lines exposed to bacterial pathogens (Milward et al., 2007), indicating a possible role in response to oral bacterial colonization. Likewise, many SNPs within a broad genomic region (about 800 kb) on chromosome 17 also demonstrated modest association with dental caries. At the boundary of the associated region is LPO (lactoperoxidase; although no associated SNPs were actually within the gene), which codes a salivary enzyme playing a key role in oral bacterial metabolism, and inhibits plaque formation and gingivitis (Tenovuo and Kurkijarvi, 1981; Tenovuo et al., 1981).
Our GWAS of stratified US samples yielded notable differences in suggestive loci between low- and sufficient-fluoride groups (as well as GWAS in the full sample), though no SNPs met criteria for genome-wide significance. However, in the low- fluoride group, a suggestive association was observed on chromosome 22 near tuftelin-interacting protein 11, TFIP11 (rs2071862, p-value = 5.1E-6). TFIP11 interacts with tuftelin, an enamel gene, and mouse experiments have shown that TFIP11 is expressed in secretory ameloblasts and odontoblasts, co-localizing with tuftelin in the extracellular enamel matrix (Paine et al., 2000). Although a previous candidate gene study did not observe main effects of SNPs in either TFIP11 or TUFT1 (tuftelin) with dental caries (Slayton et al., 2005), the known biological role of TFIP11 suggests that it may contribute to susceptibility to tooth decay, possibly through an interaction with fluoride. In contrast, this locus did not yield evidence for association in the sufficient-fluoride group. However, 2 other suggestive signals near plausible candidate genes were observed in the sufficient-fluoride group, including EPHA7 (ephrin type-A receptor 7), the nearest gene (approximately 100 kb away) to a suggestive signal on chromosome 6 (rs12191601, p-value = 9.9E-6). EPHA7, along with other members of the ephrin-Eph signaling system, is involved in the development of tooth and supporting tissues in the mouse (Luukko et al., 2005). Another suggestive signal (rs6691598, p-value = 2.3E-6) in the ZMPSTE24 gene (a zinc metallopeptidase) on chromosome 1 was observed in the sufficient-fluoride group. Mutations in ZMPSTE24 cause mandibuloacral dysplasia, a Mendelian disorder characterized by craniofacial and mandible anomalies (including tooth abnormalities), as well as other skeletal and skin abnormalities (Agarwal et al., 2003). In contrast, in the low-fluoride group, these 2 genes did not show association with dental caries. Though neither EPHA7 nor ZMPSTE24 has been previously implicated in dental caries, their known involvement in tooth development and mandibular anomalies, respectively, suggests that they may also be involved in cariogenesis in persons with sufficient fluoride intake.
Two (partially overlapping) sets of suggestive SNPs identified in the US sample were tested for replication in the Danish sample. The first set tested for SNP-wise replication included the top 54 (i.e., 0.01%) genotyped SNPs from the GWAS. Several of these 54 SNPs fell into groups of complete or very high LD, and therefore altogether represent 31 roughly independent association signals. Association results in the replication sample for these 54 SNPs are shown in Appendix Table 1. None of the top 54 SNPs yielded significant evidence for replication after Bonferroni correction for the 31 association signals (all p-values > 0.0016). However, at the suggestive signal on chromosome 17, 8 SNPs (of 9) in a high-LD block showed nominal replication (p-values = 0.011 to 0.037). Similarly, a SNP on chromosome 10 (rs7095537) showed nominal significance (p-value = 0.008) in the replication set, although the effect was in the opposite direction.
The second set tested represented gene-wise replication, i.e., 369 SNPs within 50 kb of the 8 above-mentioned genes nominated by the US GWAS: ACTN2, MTR, EDARADD, MPPED2, LPO, TFIP11, EPHA7, and ZMPSTE24. Note that these genes were chosen based on proximity to suggestive associations and plausible cariogenic roles; however, the suggestive SNPs from the GWAS were not in all cases < 50 kb from the gene. The number of SNPs in these regions ranged in size from 15 SNPs (124 kb) to 67 SNPs (256 kb). Due to gradients of LD within each gene, the number of independent tests performed is unclear, and therefore the level of significance required for successful replication is difficult to gauge. Nevertheless, none of the SNPs in these 8 gene regions yielded p-values < 0.001 in the replication sample (Table 2), and therefore did not meet the significance level required for successful replication after any reasonable adjustment for multiple testing. All genes showed at least one SNP at significance approximately equivalent to p-value = 0.05, which was expected by chance alone. However, TFIP11 showed nominal significance (p-value < 0.05) for 9 SNPs (of 57) spread across the 118-kb region, many of which were in very low LD, thus representing roughly independent tests of association. While no single SNP in TFIP11 was statistically significant, given the total number of SNPs tested for replication, we conclude that the occurrence of several independent, nominally significant SNPs in this gene, in conjunction with the known role of this gene in enamel formation, lends additional support for its role in susceptibility to caries.
Table 2.
Tests for Replication in Nominated Genes of Interest
| Gene | chr. | Position from (BP) | to (BP) | Size of Region (kb) |
No. SNPs | No. SNPs p < 0.05 |
min p | SNP |
|---|---|---|---|---|---|---|---|---|
| ACTN2 | 1 | 234867370 | 235030633 | 163.2 | 55 | 1 | 0.04718 | rs7556238 |
| MTR | 1 | 234977486 | 235180911 | 203.4 | 44 | 1 | 0.04718 | rs7556238 |
| EDARADD | 1 | 234575167 | 234752676 | 177.5 | 65 | 1 | 0.04923 | rs10489788 |
| MPPED2 | 11 | 30353586 | 30604421 | 250.9 | 58 | 0 | 0.05473 | rs538811 |
| LPO | 17 | 53621805 | 53750756 | 128.9 | 26 | 1 | 0.00875 | rs3744103 |
| TFIP11 | 22 | 25170477 | 25288331 | 117.8 | 57 | 9 | 0.00914 | rs3752523 |
| EPHA7 | 6 | 93963094 | 94218919 | 255.9 | 67 | 3 | 0.01523 | rs1983722 |
| ZMPSTE24 | 1 | 40452296 | 40576442 | 124.1 | 15 | 1 | 0.04805 | rs10489431 |
no. SNPs = number of SNPs in the genomic region of interest (i.e., within 50 kb of gene).
no. SNPs p < 0.05 = number of SNPs in the genomic region yielding p-values < 0.05.
min p = most significant p-value for any SNP in the genomic region of interest.
SNP = SNP showing strongest association in the genomic region of interest.
Loci implicated in published candidate gene studies, such as enamel genes AMBN (Deeley et al., 2008) and TUFT1 (Slayton et al., 2005), and taste response genes TAS1R2, TAS2R38, and GNAT3 (Wendell et al., 2010) did not meet genome-wide significance in this study. However, nominally significant genotyped SNPs in or around these genes were observed for AMBN (rs4694075, p-value = 0.01), TAS1R2 (rs4912075, p-value = 0.04), and GNAT3 (rs704871, p-value = 0.004) in the US sample. See Appendix for details.
Discussion
We performed the first GWAS for childhood dental caries. While no single SNP exhibited association at genome-wide significance, several genomic regions showed suggestive evidence for association. In particular, associations on chromosomes 1, 11, and 17 nominated several novel candidate genes with plausible roles on dental caries, including ACTN2, MTR, EDARADD, MPPED2, and LPO. Moreover, 3 of 5 previously studied candidate genes, including enamel- and taste-related genes, showed nominal p-values. Indicative of possible gene-environment interactions, findings from fluoride-stratified GWAS identified putative caries gene TFIP11 and also nominated novel gene candidates, EPHA7 and ZMPSTE24.
Results from the US GWAS did not replicate in the Danish sample after adjustment for multiple comparisons. While true positive variants may not have reached sufficiently low p-values in one or both samples due to the limited sample sizes, especially for stratified cohorts, differences in demography, socio-economic status, health care structure, environmental exposures, and behaviors may also partly explain the incongruent results. The Danish sample exhibits lower prevalence of dental caries (38.1%) than the US sample (46.9%). Perhaps the effects of genetic risk factors in the Danish sample are counterbalanced by protective non-genetic factors to a greater extent than in the US sample. In other words, caries liability due to risk genes may be environment-specific. The notion that gene-environment interactions are important in caries pathogenesis is especially appealing, given the complex interplay of bacterial, dietary, salivary, morphological, hygienic, and exposure-related factors leading to disease.
This study benefits from several strengths, including genome-wide SNP data on both independent samples, and rigorous and thorough assessment of dental phenotypes. Genotyping and quality control/quality assurance, conducted by CIDR and the GENEVA coordinating center, yielded data of exceptional quality. Moreover, as the first GWAS for dental caries reported to date, this study accomplished the principal goal (of the non-hypothesis-based GWAS study design) of generating interest in genes and genomic regions previously unstudied in the context of oral health. Moreover, this study highlights the challenges of identifying genes involved in common complex disease, namely, that numerous genes, mostly of small effect sizes, are likely to contribute to cariogenesis, and that discovery of individual variants may be exceedingly difficult. While research into the genetics of dental caries lags behind many other prominent common complex diseases, this study provides a launching pad for future candidate gene and functional studies of tooth decay and insight into possible genetic interactions with fluoride exposure. The public availability of these data via dbGaP will facilitate the utility of this study in designing future efforts and cross-study collaborations to understand the genetics of dental caries.
Footnotes
A supplemental appendix to this article is published electronically only at http://jdr.sagepub.com/supplemental.
This study is supported by the following NIH grants: U01- DE018903, U01-HG004438, U01-HG004423, U01-HG004446, R01-DE014899, R01-DE0 9551, R01-DE12101, R03-DE021425, and P60-DE-13076, and by NIH contract HHSN268200782-096C. Other support was provided by the Danish NRF, Danish Pharmacists' Fund, Egmont Foundation, March of Dimes, Augustinus Foundation, and Health Fund of the Danish Health Insurance Societies.
The authors declare no potential conflicts of interest with respect to the authorship and/or publication of this article.
References
- Agarwal AK, Fryns JP, Auchus RJ, Garg A. (2003). Zinc metalloproteinase, ZMPSTE24, is mutated in mandibuloacral dysplasia. Hum Mol Genet 12:1995-2001 [DOI] [PubMed] [Google Scholar]
- Al-Dajani M. (2009). Comparison of dental caries prevalence in patients with cleft lip and/or palate and their sibling controls. Cleft Palate Craniofac J 46:529-531 [DOI] [PubMed] [Google Scholar]
- Boraas JC, Messer LB, Till MJ. (1988). A genetic contribution to dental caries, occlusion, and morphology as demonstrated by twins reared apart. J Dent Res 67:1150-1155 [DOI] [PubMed] [Google Scholar]
- Britton KF, Welbury RR. (2010). Dental caries prevalence in children with cleft lip/palate aged between 6 months and 6 years in the West of Scotland. Eur Arch Paediatr Dent 11:236-241 [DOI] [PubMed] [Google Scholar]
- Cornelis MC, Agrawal A, Cole JW, Hansel NN, Barnes KC, Beaty TH, et al. (2010). The Gene, Environment Association Studies consortium (GENEVA): maximizing the knowledge obtained from GWAS by collaboration across studies of multiple conditions. Genet Epidemiol 34:364-372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deeley K, Letra A, Rose EK, Brandon CA, Resick JM, Marazita ML, et al. (2008). Possible association of amelogenin to high caries experience in a Guatemalan-Mayan population. Caries Res 42:8-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzman MR, Levy SM, Warren JJ, Broffitt B. (2004). Tooth-brushing and dentifrice use among children ages 6 to 60 months. Pediatr Dent 26:87-92 [PubMed] [Google Scholar]
- International HapMap Consortium (2003). The International HapMap Project. Nature 426:789-796 [DOI] [PubMed] [Google Scholar]
- Levy SM, Warren JJ, Davis CS, Kirchner HL, Kanellis MJ, Wefel JS. (2001). Patterns of fluoride intake from birth to 36 months. J Public Health Dent 61:70-77 [DOI] [PubMed] [Google Scholar]
- Levy SM, Warren JJ, Broffitt B. (2003). Patterns of fluoride intake from 36 to 72 months of age. J Public Health Dent 63:211-220 [DOI] [PubMed] [Google Scholar]
- Luukko K, Loes S, Kvinnsland IH, Kettunen P. (2005). Expression of ephrin-A ligands and EphA receptors in the developing mouse tooth and its supporting tissues. Cell Tissue Res 319:143-152 [DOI] [PubMed] [Google Scholar]
- Marshall TA, Levy SM, Broffitt B, Warren JJ, Eichenberger-Gilmore JM, Burns TL, et al. (2003). Dental caries and beverage consumption in young children. Pediatrics 112(3 Pt 1):e184-e191 [DOI] [PubMed] [Google Scholar]
- Milward MR, Chapple IL, Wright HJ, Millard JL, Matthews JB, Cooper PR. (2007). Differential activation of NF-kappaB and gene expression in oral epithelial cells by periodontal pathogens. Clin Exp Immunol 148:307-324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostowska A, Hozyasz KK, Jagodzinski PP. (2006). Maternal MTR genotype contributes to the risk of non-syndromic cleft lip and palate in the Polish population. Clin Genet 69:512-517 [DOI] [PubMed] [Google Scholar]
- Olsen J, Melbye M, Olsen SF, Sorensen TI, Aaby P, Andersen AM, et al. (2001). The Danish National Birth Cohort—its background, structure and aim. Scand J Public Health 29:300-307 [DOI] [PubMed] [Google Scholar]
- Paine CT, Paine ML, Luo W, Okamoto CT, Lyngstadaas SP, Snead ML. (2000). A tuftelin-interacting protein (TIP39) localizes to the apical secretory pole of mouse ameloblasts. J Biol Chem 275:22284-22292 [DOI] [PubMed] [Google Scholar]
- Parapanisiou V, Gizani S, Makou M, Papagiannoulis L. (2009). Oral health status and behaviour of Greek patients with cleft lip and palate. Eur Arch Paediatr Dent 10:85-89 [DOI] [PubMed] [Google Scholar]
- Polk DE, Weyant RJ, Crout RJ, McNeil DW, Tarter RE, Thomas JG, et al. (2008). Study protocol of the Center for Oral Health Research in Appalachia (COHRA) etiology study. BMC Oral Health 8:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. (2007). PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81:559-575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sehic A, Risnes S, Khan QE, Khuu C, Osmundsen H. (2010). Gene expression and dental enamel structure in developing mouse incisor. Eur J Oral Sci 118:118-130 [DOI] [PubMed] [Google Scholar]
- Slayton RL, Cooper ME, Marazita ML. (2005). Tuftelin, mutans streptococci, and dental caries susceptibility. J Dent Res 84:711-714 [DOI] [PubMed] [Google Scholar]
- Tenovuo J, Kurkijarvi K. (1981). Immobilized lactoperoxidase as a biologically active and stable form of an antimicrobial enzyme. Arch Oral Biol 26:309-314 [DOI] [PubMed] [Google Scholar]
- Tenovuo J, Mansson-Rahemtulla B, Pruitt KM, Arnold R. (1981). Inhibition of dental plaque acid production by the salivary lactoperoxidase antimicrobial system. Infect Immun 34:208-214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townsend GC, Aldred MJ, Bartold PM. (1998). Genetic aspects of dental disorders. Aust Dent J 43:269-286 [DOI] [PubMed] [Google Scholar]
- Wang X, Shaffer JR, Weyant RJ, Cuenco KT, Desensi RS, Crout R, et al. (2010). Genes and their effects on dental caries may differ between primary and permanent dentitions. Caries Res 44:277-284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendell S, Wang X, Brown M, Cooper ME, DeSensi RS, Weyant RJ, et al. (2010). Taste genes associated with dental caries. J Dent Res 89: 1198-1202 [DOI] [PMC free article] [PubMed] [Google Scholar]