Abstract
Coccidian parasites are transmitted via a fecal oocyst stage that is exceptionally resistant to environmental stress and harsh chemical treatments, which allows parasites to stably persist outside a host. Because of its oocyst durability Cryptosporidium parvum is a significant water- and food-borne pathogen of humans, as well as animals of agricultural importance. To date, only one apicomplexan oocyst membrane protein has been identified, Cryptosporidium oocyst wall protein 1 (COWP1). COWP1 has a highly cysteine-rich periodicity due to arrays of two apicomplexan-specific motifs, designated the type I and type II domains. In this study, exhaustive BLAST screening of a complete C. parvum genome sequence database resulted in identification of eight additional genes encoding similar arrays of cysteine-rich type I and/or type II domains. Transcript expression analysis revealed that all COWP genes are abundantly expressed at a time when developing oocysts are observed, roughly 48 to 72 h after inoculation of in vitro cultures. A monoclonal antibody recognizing COWP8 specifically localized to the C. parvum oocyst wall, supporting the hypothesis that multiple COWPs play a role in the oocyst wall structure. BLAST screening of the Toxoplasma gondii genome sequence database resulted in identification of a gene encoding at least one COWP homolog (TgOWP1), and this multiexon sequence information was used to isolate a full-length cDNA. Exhaustive screening of Plasmodium sp. genome sequence databases by using COWP genes as BLAST queries failed to detect similar proteins in Plasmodium. We therefore propose that the COWP family of proteins have a structural role in apicomplexan species that produce durable shed cysts capable of surviving environmental stress.
Cryptosporidium parvum is an obligate intracellular parasite that causes severe diarrhea via infection of the lower gastrointestinal tract epithelium of humans and other mammals. Cryptosporidium is one of several genera in the phylum Apicomplexa that have common life cycle and morphological stages, collectively referred to as coccidia (reviewed in reference 5). Coccidian oocyst stages are highly resistant to environmental stress and chemical disinfection (9), and this is attributed to a durable oocyst wall, a complex protective barrier consisting of a double layer of a protein-lipid-carbohydrate matrix (5). Ultrastructural studies have revealed that coccidian oocyst walls appear to form as a result of the interaction of wall-forming bodies and pellicular membranes (7, 14). During oocyst maturation wall-forming bodies degranulate in the cytoplasm, and the vesicular material is delivered into the intermembrane space. Once the construction of the outer layer of the oocyst wall nears completion, a similar process begins for the inner layer. In the case of a related coccidian parasite, Eimeria nieschulzi, seven membranes and two types of wall-forming bodies that appear to be involved in oocyst wall formation have been identified (14).
Despite numerous studies of this unique structure, there has been no detailed description of the C. parvum oocyst wall, particularly at a molecular level. To date, only one apicomplexan oocyst membrane protein has been isolated, Cryptosporidium oocyst wall protein 1 (COWP1) (15). Immunoelectron microscopy demonstrated that COWP1 is present in the inner oocyst wall and inside wall-forming bodies of mature macrogametes. COWP1 has a striking cysteine periodicity, with cysteine residues spaced roughly every 10 to 12 amino acids due to tandem arrays of two cysteine-rich domains, designated type I and type II domains. It has been proposed that an extensive disulfide-bonded globular structure or intermolecular disulfide bonds provide rigidity to the oocyst wall (15).
We were interested in determining if the COWP1 type I and type II domains are present in multiple Cryptosporidium proteins and if these motifs and proteins have a pan-apicomplexan distribution. To address these questions, we performed exhaustive BLAST screening of the recently completed Cryptosporidium genome sequence database and found that type I and type II domains are present as tandem arrays in proteins encoded by eight additional genes. Extensive screening of Plasmodium sp. genome sequence databases failed to detect COWP gene homologs. In contrast, we isolated a complete cDNA for a COWP homolog from Toxoplasma gondii, which is described below and is designated TgOWP1, and therefore we propose that the oocyst wall protein family plays a structural role in apicomplexan genera having externally shed durable oocysts.
MATERIALS AND METHODS
Parasite material.
C. parvum oocysts (Iowa isolate) were purified from calf feces by using a discontinuous sucrose gradient (2). Oocysts were suspended in phosphate-buffered saline (PBS) and were sterilized by incubation in 33% bleach for 7 min on ice. Oocysts were subsequently equilibrated at pH 7.2 by sequential washing and centrifugation at 4°C in Hanks buffered saline solution. Confluent human adenocarcinoma HCT-8 cells were infected with C. parvum oocysts as previously described (1, 18). Total RNA was prepared from mock-infected and C. parvum-infected HCT-8 cultures at 6, 12, 24, 48, and 72 h postinoculation (p.i.) by directly lysing the cells with 4 ml of TRIzol reagent (GIBCO-BRL/Life Technologies, Gaithersburg, Md.). RNA was isolated as described by the manufacturer. Purified RNA was resuspended in RNase-free water, and the integrity of the samples was confirmed by gel electrophoresis (data not shown).
Total protein was prepared from mock-infected and C. parvum-infected HCT-8 cultures by gently scraping cells from a plate, washing them by centrifugation in PBS, and resuspending them in 1× 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS) lysis buffer (10 mM Tris-HCl, 1 mM MgCl2, 1 mM EGTA, 0.1 mM benzamide, 5 mM β-mercaptoethanol, 0.5% CHAPS, 10% [vol/vol] glycerol). Samples were stored at −80°C until they were needed. Following in vitro excystation, C. parvum sporozoites were purified from oocysts walls by DEAE-cellulose chromatography as previously described (12). Purified sporozoites were suspended in 1× CHAPS lysis buffer and stored at −80°C until they were needed. Lysates of C. parvum oocysts were prepared by passing 1 × 109 oocysts (suspended in 2 ml of 1× CHAPS lysis buffer) through a French press at 20,000 lb/in2 and were stored at −80°C until they were needed.
BLAST analysis of apicomplexan genome sequence databases.
The C. parvum genome sequence (type II) is essentially complete (19), and assembly into chromosomal contigs is in the final stages; there are one to three gaps per chromosome (contigs are available at http://www.ncbi.nlm.nih.gov/Microb_blast/unfinishedgenome.htm). The C. parvum genome sequence database was mined for occurrences of COWP type I and type II tandem arrays by using the COWP1 gene (gi:453952) (15) and regions of this gene as BLAST queries. Each additional isolated COWP gene was in turn used to exhaustively screen the C. parvum genome for further occurrences of COWP genes. Gene structure predictions were aided by the fact that Cryptosporidium possesses few introns, and, in fact, introns were observed in only one of the COWP genes (COWP7 gene) isolated in this study. Thus, predictive starts were determined by the presence of methionine residues followed by putative signal peptide sequences. In a similar fashion, exhaustive BLAST screening of the Plasmodium falciparum and Plasmodium yoelii genome sequence databases was conducted by using the PlasmoDB web site (http://plasmodb.org). Screening of the partial T. gondii genome sequence database was performed at the ToxoDB web site (http://www.toxodb.org). Amino acid alignment was performed by using ClustalW (http://www.ebi.ac.uk/clustalw) and the consensus residues calculated by using Consensus (http://www.bork.embl-heidelberg.de/Alignment/consensus.html).
Isolation of TgOWP1 cDNA.
At this point the T. gondii genome sequence database has roughly 2× genome sequence coverage. To determine the complete exon structure of a selected TgOWP gene, designated theTgOWP1 gene, specific primers corresponding to discrete exons were designed and spliced transcripts were isolated by PCR amplification by using a T. gondii sporozoite stage cDNA library (Tsc3ABP; a generous gift from M. White). Sequencing of TgOWP1 was performed directly with the amplified PCR product.
Expression of recombinant COWP8 protein and hybridoma production.
The carboxy-terminal region of the COWP8 coding sequence (nucleotides 631 to 1374) was amplified by using oligonucleotides 102-2F (5′-TCATGGATCCAGCTATGTTGTTCC-3′) and 102-2R (5′-TAAGCTTGATATATCTATCTATATCTG-3′). For directional cloning the amplification primers used included a BamHI recognition sequence (nucleotides 5 to 10) within oligonucleotide 102-2F and a HindIII recognition sequence (nucleotides 2 to 7) within oligonucleotide 102-2R. PCR products were incubated with restriction enzymes BamHI and HindIII and were purified by agarose gel electrophoresis. Cloning, expression, and purification of the recombinant COWP8 protein were carried out by using a QiaExpressionist type IV kit (Qiagen) according to the manufacturer's instructions. Following purification by metal-chelate affinity chromatography with nickel-nitrilotriacetic acid beads (Qiagen), the recombinant protein (containing a six-His tag) was dialyzed against multiple changes of PBS (pH 7.2).
The recombinant protein was emulsified in TiterMax Gold adjuvant (Sigma Chemical Co., St. Louis, Mo.) used according to the manufacturer's directions and was injected interperitoneally into 6-week-old female BALB/c mice. The immunization procedure was repeated at 3- to 4-week intervals, once with and once without the use of an adjuvant. Mice were again boosted with the recombinant protein 4 days prior to harvesting of the spleens. Harvested spleen cells were fused to hypoxanthine-negative, thymidine-negative, and aminopterin-sensitive mouse SP 2/0 myeloma cells (American Type Culture Collection, Rockville, Md.), seeded into 48-well dishes, and cultured for 14 days in HAT media to select for growth of hybridomas. Cultures were switched to HT media and expanded. Supernatants from hybridomas were screened by Western blot analysis (see below). Hybridomas displaying positive results were selected and subcloned by limiting dilution, expanded, and adapted to RPMI 1640 media containing 10 mM HEPES supplemented with 20% fetal calf serum, 2 mM l-glutamine, 1 mM sodium pyruvate, 0.1 mM minimal essential medium (MEM) nonessential amino acids, 1× MEM vitamins and minerals, 50 U of penicillin G per ml, 50 U of streptomycin per ml, and 0.25 μg of amphotericin B (Fungizone; GIBCO-BRL/Life Technologies, Grand Island, N.Y.) per ml (pH 7.2).
Western blot analysis.
For hybridoma screening, lysates containing 5 × 107 oocysts were mixed with an equal amount of 2× reducing sodium dodecyl sulfate (SDS) protein solubilization buffer, resolved on an SDS-12% polyacrylamide gel electrophoresis (PAGE) gel fitted with a single trough preparative comb, and transferred to a nitrocellulose filter. Following blocking of nonspecific binding sites with horse serum for 30 min, the filter was fitted into a 25-lane blotting apparatus, and supernatants from the hybridomas were incubated for 1 h in individual lanes. Serum harvested from immunized mice served as a positive control, while serum harvested from preimmunized mice served as a negative control. Bound antibodies were detected with affinity-purified phosphatase-conjugated goat anti-mouse immunoglobulin G (Jackson ImmunoResearch), and with nitroblue tetrazolium and 5-bromo-4-chloro-3-indolylphosphate (Sigma Chemical Co.).
For examination of COWP8 protein expression, equal amounts of extracts from whole oocysts, purified sporozoites, HCT-8 cells infected for 72 h, and mock-infected HCT-8 cells (representing 1/15 of a 15-cm culture plate) were separated under reducing conditions on an SDS-12% PAGE gel and transferred to nitrocellulose as described above. The filter was washed in 5% acetic acid for 5 min, and this was followed by staining with Ponceau S (0.2% Ponceau S in 2.5% trichloroacetic acid-2.5% acetic acid) for 1 to 3 min. The borders of the resolved proteins were marked, and the filter was destained until it was colorless by washing it in numerous changes of TBS-T (150 mM NaCl, 0.05% Tween 20, 10 mM Tris-HCl; pH 8.0). Following blocking by incubation in horse serum for 30 min, the filter was fitted into a 25-lane blotting apparatus. Bound antibodies were detected with affinity-purified phosphatase-conjugated goat anti-mouse immunoglobulin G (Jackson ImmunoResearch) and with nitroblue tetrazolium and 5-bromo-4-chloro-3-indolylphosphate (Sigma Chemical Co.).
Indirect immunofluorescence assay.
Approximately 2 × 108 unbleached C. parvum oocysts were washed repeatedly in PBS, and one-half of the oocyst sample was ruptured with a French press. Whole-oocyst and ruptured-oocyst samples were washed in PBS, and each sample was divided and placed into eight microcentrifuge tubes. Samples were left untreated or were treated with 1% NP-40, 0.1% SDS, or 0.5% sodium deoxycholate, with a combination of two of the detergents, or with all three detergents for 1 h on ice. Three volumes of PBS were added, and 20 μl of each sample was placed in each well of eight-well Teflon-coated slides. Samples were air dried and fixed in acetone for 5 min. Nonspecific binding sites were blocked for 20 min by using 2.5% goat serum. Slides were incubated for 1 h with either a primary antibody against C. parvum sporozoites, the cognate recombinant protein, or a secondary antibody control. After the slides were washed, they were incubated for 30 min with Cy3-conjugated goat anti-mouse secondary antibody (Jackson ImmunoResearch). The slides were washed and mounted by using Vectorshield (Vector Labs).
Semiquantitative RT-PCR.
To investigate COWP gene expression during C. parvum in vitro development, COWP gene-specific primers (Table 1) were designed, and semiquantitative reverse transcription (RT)-PCR analysis was carried out as previously described (1, 4). Briefly, cDNA was prepared from 2 μg of total RNA by using random hexamer priming and SuperScript II reverse transcriptase (Invitrogen) in a 20-μl reaction mixture. A 2-μl portion of the first-strand cDNA reaction mixture was used in a 20-μl reaction mixture that contained 1× PCR buffer with 1.5 mM MgCl2 (Perkin-Elmer Inc.), each deoxynucleoside triphosphate at a concentration of 200 μM, the forward and reverse primers corresponding to each COWP gene at a concentration of 1 μM, 1.25 U of AmpliTaq DNA polymerase (Perkin-Elmer), and 0.4 μCi of [32P]dCTP (Amersham Pharmacia Biotech Inc., Piscataway, N.J.). After an initial denaturation at 94°C for 2 min, the reaction mixture was subjected to multiple cycles of denaturation at 94°C for 30 s, annealing at 57°C for 30 s, and extension at 72°C for 1 min. The number of cycles was preoptimized so that final accumulation of the PCR product occurred in the exponential phase (27 cycles) (data not shown). After an additional cycle of extension at 72°C for 5 min, the reaction mixtures were kept at 4°C.
TABLE 1.
COWP gene-specific and control primers used in semiquantitative RT-PCR assays
| Gene | Forward primer | Reverse primer |
|---|---|---|
| COWP1 | 5′ -ATATGCAGATAAAGTCTGTCC-3′ | 5′ -AATCTTGCGCATCTATCACCC-3′ |
| COWP2 | 5′ -TTTTGTATCCCCAAGAAGAA-3′ | 5′ -ACTGAGCAATAACCTCCAGA-3′ |
| COWP3 | 5′ -GCCATAATGGTTCACAGTCT-3′ | 5′ -TGGGATACATTTTGGAAGTC-3′ |
| COWP4 | 5′ -CGATTTGGTTGATGAGAGTT-3′ | 5′ -CTCTCGGTTATTTCTGTTGG-3′ |
| COWP5 | 5′ -CAGTCCTGAGCCACGATGTAGA-3′ | 5′ -GGGCACTCCTTCAATGGTTTTT-3′ |
| COWP6 | 5′ -TGAAAAGGGTTTCCAACTTA-3′ | 5′ -ATTGGTCTCTCATCCATACG-3′ |
| COWP7 | 5′ -GAAAGCATGTGATGAAGGTT-3′ | 5′ -TTGTGATATTGGAACTTCGAC-3′ |
| COWP8 | 5′ -ATATGATTGCACTCCAGCTGC-3′ | 5′ -TGGACATCCCAATTCTGCTGG-3′ |
| COWP9 | 5′ -CTGGATTTCCAGGATGTGCT-3′ | 5′ -TGCTTTGTATTCCCGATTCTG-3′ |
| 18s rRNA | 5′ -TAGAGATTGGAGGTTGTTCCT-3′ | 5′ -CTCCACCAACTAAGAACGGCC-3′ |
As the number of developing C. parvum life stages within infected cells changed over time, primers specific for C. parvum 18S rRNA (Table 1) (13) were used to normalize the amounts of cDNA products of the COWP genes to the amount of C. parvum rRNA in the same sample. Since rRNA is much more abundant than any specific mRNA, 2-μl portions of 1:25 dilutions of the cDNA samples were used, and the reactions included only 23 cycles of amplification (1). To monitor PCR efficiency and possible cross-contamination between reaction mixtures, a negative control consisting of 2 μl of cDNA from mock-infected HCT-8 cells and a positive control consisting of 2 ng of C. parvum genomic DNA were included in each batch of PCRs.
PCR products were separated on a 4% nondenaturing polyacrylamide gel, and signals from specific products were captured and quantified by using a phosphorimaging system (Molecular Dynamics Inc., Sunnyvale, Calif.). The level of expression of each COWP gene at each time point was calculated by determining the ratio of its RT-PCR product signal to the signal of the C. parvum 18S rRNA. Three independent time course experiments were used in the analysis. Each of the RNA samples from C. parvum-infected HCT-8 cells was demonstrated to be free of contaminating C. parvum genomic DNA by the lack of amplification product from a reverse transcriptase reaction sham control (data not shown). The mean level of expression of each of the COWP genes was calculated, and the standard error was determined.
Sequence accession numbers.
The complete cDNA and peptide sequences for COWP2 through COWP9 and TgOWP1 have been deposited in the GenBank database under accession numbers AY465051 to AY465058 for COWP2 to COWP9, respectively, and AY465428 for TgOWP1.
RESULTS
Multigene COWP family.
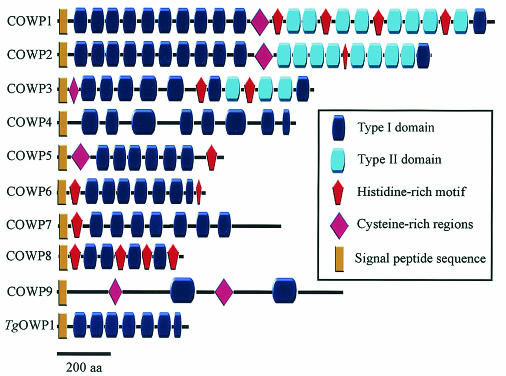
The domain architecture of COWP1 was previously described in detail (15); this protein is composed of tandem arrays of two apicomplexan-specific cysteine-rich motifs, designated the type I and type II domains. The 1,622-amino-acid COWP1 protein is composed of arrays of type I and degenerate type I repeats in the amino-terminal region of the protein, followed by arrays of type II repeats in the carboxy-terminal region (Fig. 1). Because of the recent availability of whole-genome sequence information, we were interested in asking (i) whether type I or type II repeats are present in additional C. parvum genes and (ii) whether COWP-like genes are conserved in other apicomplexan species. The Cryptosporidium genome sequence project is in the final stage; there are one to three gaps per chromosome, and thus by using BLAST screening the presence of genes encoding type I and type II domain-containing proteins in the genome can be readily ascertained. The Cryptosporidium genome database was exhaustively screened for additional genes by using the COWP1 gene sequence and sequences encoding regions in this protein as BLAST queries. Each newly identified gene was subsequently used as a BLAST query to ensure that there was exhaustive mining of the genome sequence. Weighting due to the dramatic cysteine periodicity (a cysteine roughly every 10 to 12 amino acids) lent itself well to identification of additional COWP genes. This analysis revealed eight additional Cryptosporidium genes encoding tandem arrays of type I and/or type II domains. The architectural similarity of these genes to the COWP1 gene indicates that the oocyst wall protein gene belongs to a multigene family (Fig. 1).
FIG. 1.
Schematic diagram showing the domain structure of the COWP family. aa, amino acids.
Analysis of the essentially complete chromosomal contig data indicated that the COWP genes are not clustered in the genome, nor are they localized to telomeres; instead, they are scattered on multiple chromosomes (data not shown). All COWP genes encode short hydrophobic regions at the amino termini that likely represent signal peptide sequences, although it remains to be determined if trafficking of COWPs is via an endoplasmic reticulum pathway. All COWP genes lack apparent transmembrane coding regions and thus are unlikely to encode integral membrane proteins. The striking cysteine periodicity is encoded by all COWP genes and in all instances is due to sequences encoding extensive tandem arrays of the cysteine-rich type I and/or type II domains. Two genes, the COWP2 and COWP3 genes, encode domain architectures similar to that of COWP1, and the proteins have extensive tandem arrays of type I domains in the amino-terminal region, followed by a carboxy-terminal tandem array of type II domains (Fig. 1). This architectural conservation supports the hypothesis that COWP1 through COWP3 have a paralogous relationship due to gene duplication. The remaining genes encode only type I domains and notably do not encode type II domains. A type I domain architecture structure is also present in a Toxoplasma homolog, TgOWP1, as described below. Several of the COWP genes encode histidine-rich motifs previously described in COWP1 (15), and thus the prevalence of this motif is extended to include multiple proteins. The function of histidine repeats is currently unknown, although it has been proposed that they might participate in intermolecular cross-linking that confers stability to the oocyst wall. The COWP7 gene was unique among the COWP genes and rare among Cryptosporidium genes in general in that it contained introns. A comparison of cDNA and genomic sequences resulted in identification of eight introns within the COWP7 gene (data not shown), and therefore this gene is the most highly disrupted gene thus far identified in the Cryptosporidium genome. The COWP9 gene is tenuously related to the COWP gene family, and its divergence is due to its extensive low complexity and to the fact that it encodes less striking cysteine-rich repeats (Fig. 1). Nonetheless, it was included in this analysis based upon (i) its location in the genome sequence 600 bp upstream of and in the same orientation as the COWP8 gene; (ii) the presence of two blocks of cysteine-rich periodicity in COWP9, possibly due to divergent type I domains; (iii) a putative signal peptide sequence in COWP9, suggesting secretory trafficking; and (iv) transcript expression indicative of oocyst-specific expression (see Fig. 3).
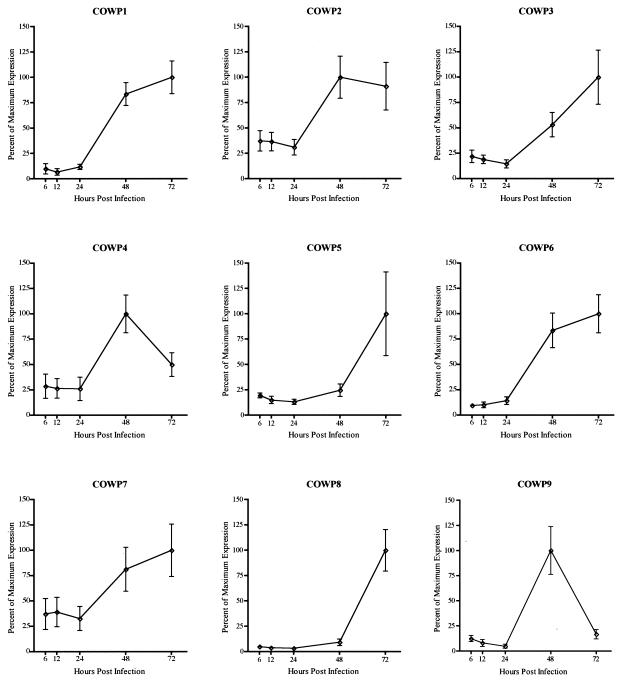
FIG. 3.
Expression profiles of COWP genes during in vitro development. The amount of signal was determined from each of the COWP genes and rRNA by using a phosphorimaging system and ImageQuaNT software. The data are expressed as the ratio of each gene signal to the rRNA signal present in each of the infected samples from three independent time course experiments. The error bars indicate the standard deviations of the means.
Expression analysis of COWP genes.
Transcript expression during in vitro development was determined for the COWP1 gene and compared with that of the newly identified COWP genes (an example of assay results for COWP8 are shown in Fig. 2, and gene expression data are shown in Fig. 3). Due to the semiquantitative nature of the RT-PCR assay, it was not possible to compare the magnitudes of transcript expression (expressed in arbitrary units in Fig. 3) for different genes at a given time, but was is possible to compare developmental profiles (that is, the corresponding stages of expression for a given gene). The results demonstrate that the entire gene family is up-regulated similarly during the later stages of C. parvum in vitro development. As shown in Fig. 3, expression of each of the COWP genes was low to undetectable at 6, 12, and 24 h p.i., but there was a dramatic increase in signal intensity at roughly 48 h p.i. This coincides with the time frame known for parasite sexual reproduction, during which oocyst formation and maturation take place (6, 8). The onset and maximal duration of expression of each COWP gene family member differ somewhat at 48 and 72 h p.i., suggesting that protein expression and perhaps function are manifold in character. Nevertheless, the expression profile for each COWP gene is consistent with a putative role of the protein family in C. parvum oocyst wall structure.
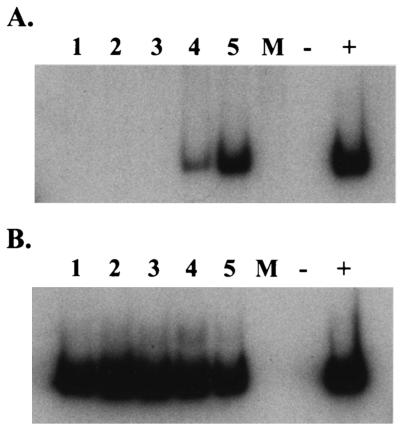
FIG. 2.
COWP mRNA expression during in vitro development as determined by semiquantitative RT-PCR. Portions (2 μg) of total RNA isolated from mock-infected samples (lane M) and C. parvum-infected HCT-8 samples harvested at 6, 12, 24, 48, and 72 h p.i. (lanes 1 to 5, respectively) were reverse transcribed by using random primers in 20-μl (total volume) mixtures. Two microliters of each of the cDNA reaction mixtures was subjected to 27 cycles of amplification in the presence of [32P]dCTP by using primers specific for each gene. Lane −, reverse transcriptase sham control (48- or 72-h-p.i. C. parvum-infected RNA not subjected to RT treatment); lane +, positive control (C. parvum genomic DNA). (A) Representative reaction for assay of COWP8 gene transcript expression. Expression was observed at 48 and 72 h p.i. (lanes 4 and 5). (B) Two microliters of a 1:25 dilution of each of the cDNA reaction mixtures was similarly amplified by using primers specific for C. parvum 18S rRNA.
Expression and immunolocalization of the COWP8 protein.
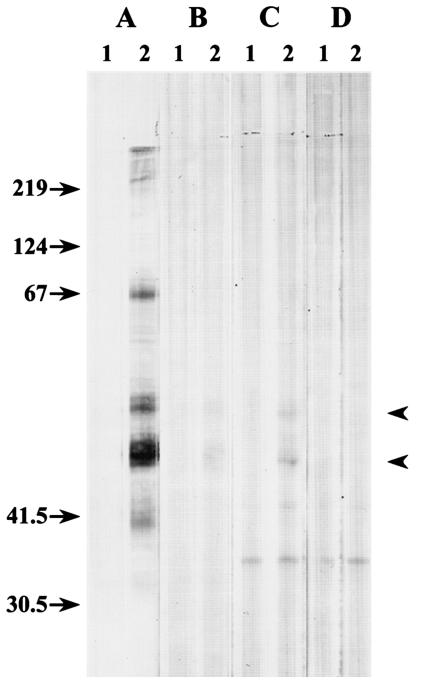
To extend our understanding of COWP expression and localization, we produced a recombinant protein corresponding to COWP8 and generated specific antibody reagents for use in Western blot and immunolocalization studies. Our efforts were focused on COWP8, which initiated this investigation following its annotation from a gene sequence tag analysis before extensive genome sequence data were available (10). Recombinant protein corresponding to the carboxy-terminal region of the protein was expressed in Escherichia coli and was purified as described in Materials and Methods. Following immunization of mice, a monoclonal antibody (Cp8-102) that recognized the recombinant COWP8 protein fragment in Western blots (data not shown) was produced. Cp8-102 did not react to other recombinant poly(His)-tagged proteins, demonstrating the specificity for COWP8. In addition, commercially available antibodies against poly(His)-tagged proteins did not react with the C. parvum lysate, indicating that this motif is not present in C. parvum oocysts (data not shown). Western blot analysis results for oocysts, sporozoites, and C. parvum-infected HCT-8 cells are shown in Fig. 4. A broad set of bands ranging in size from 40 to >200 kDa were recognized by Cp8-102 in the whole-oocyst extract containing oocyst walls and sporozoites (Fig. 4A, lane 2). These bands were absent in extracts prepared from an equivalent number of purified sporozoites (Fig. 4B, lane 2), indicating that the Cp8-102 antigens are specific for the oocyst wall. The size of the lower bands (47 to 50 kDa) is similar to the predicted molecular mass of COWP8 (49.9 kDa). Analysis of extracts from HCT-8 cells infected for 72 h with C. parvum resulted in identification of three bands, and the sizes of the two largest bands were similar to the sizes of major bands detected in whole-oocyst extracts (Fig. 4C, lane 2). These bands were absent in HCT-8 cells that were mock infected for 72 h (Fig. 4D, lane 2) or HCT-8 cells infected with C. parvum for less than 72 h (data not shown); thus, COWP8 expression correlated with stages during which mRNA is abundantly expressed (Fig. 2A and 3). A band at 67 kDa was also observed in the whole-oocyst extract sample, and this band may have resulted from dimerization of the lower-molecular-weight proteins or from posttranslational modification. It is interesting that this band was not detected with the sample of cells infected for 72 h with C. parvum, which may support the idea that there is posttranslational modification during or after wall structure assembly. A third smaller band in the C. parvum- and mock-infected HCT-8 cells was recognized by the secondary control antibody (Fig. 4C and D, lane 1), indicating that this band was not specifically recognized by Cp8-102. Although Cp8-102 recognized abundant antigens present in whole-oocyst extracts, repeated attempts with this reagent failed to immunolocalize COWP8 in intact, unbleached oocysts when an indirect fluorescence assay was used. To determine if the location of the Cp8-102 epitope was a location other than the outside surface of the oocyst wall, unbleached C. parvum oocysts were either left intact or ruptured by passing them through a French press. Aliquots of both samples were treated with one detergent or a combination of detergents to determine if the epitope was concealed within the intact oocyst wall structure or was exposed following solubilization. Indirect immunofluorescence assay analysis of C. parvum oocysts processed with the French press and treated with each detergent (1% NP-40, 0.1% SDS, or 0.5% sodium deoxycholate) revealed staining of the entire oocyst wall (data not shown). Treating the oocysts with a combination of two detergents increased the staining (data not shown), and staining was maximized by the use of all three detergents (Fig. 5A). Preparations of intact oocysts with or without detergent treatment did not react with Cp8-102 (data not shown). As shown in Fig. 5, oocysts that remained intact did not react with Cp8-102, while ruptured oocysts displayed various degrees of staining. The variability of staining may have been related to the difference efficiencies of detergent extraction of individual oocysts and to the thickness of the oocysts relative to the focal plane of the objective. These analyses indicate that the COWP8 epitope is restricted to the inside of the C. parvum oocyst wall and requires disruption for exposure. Consistent with the Western blot analysis (Fig. 4B lane 2), Cp8-102 did not stain any of the free sporozoites or sporozoites within ruptured oocysts (data not shown).
FIG. 4.
Expression of COWP8 as determined by immunoblotting. Protein lysates from C. parvum oocysts (A), from purified sporozoites (B), from HCT-8 cells infected for 72 h with C. parvum (C), and from HCT-8 cells mock infected for 72 h (D) were resolved on an SDS-12% PAGE gel under reducing conditions, transferred to nitrocellulose, and probed with either an isotype-matched secondary control monoclonal antibody (lanes 1) or monoclonal antibody Cp8-102 (lanes 2). The arrowheads indicate the presence of oocyst wall protein in the sample obtained 72 h p.i..
FIG. 5.
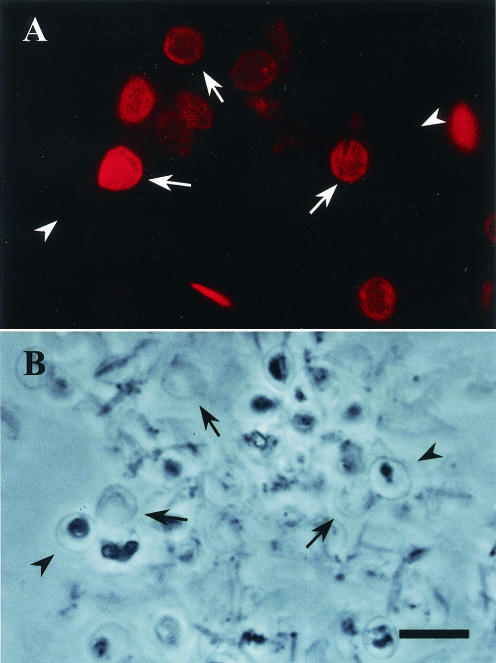
Immunofluorescence staining of C. parvum oocysts with monoclonal antibody Cp8-102. C. parvum oocysts were mechanically lysed with a French press and treated with NP-40, SDS, and sodium deoxycholate. Oocysts were dried onto a glass slide, fixed with acetone, and probed with monoclonal antibody Cp8-102 and a Cy3-conjugated secondary antibody. (A) Oocysts viewed by fluorescence microscopy; (B) same image viewed by phase-contrast microscopy. The arrows indicate ruptured oocysts, and the arrowheads indicate intact oocysts. Bar = 10 μm.
Oocyst wall protein homolog in T. gondii.
If the COWPs function in the formation of a rigid oocyst wall and if the genes have an ancient origin prior to the divergence of the extant Apicomplexa, then it can be proposed that coccidian parasites, such as Toxoplasma (16), should also possess oocyst wall protein genes. In contrast, hemospororidians, such as Plasmodium and Babesia, might be expected to lack members of this gene family since these organisms do not shed oocysts having a rigid cyst wall. Indeed, after exhaustive screening of the Plasmodium sp. genome sequence databases, including the complete P. falciparum genome sequence database, we failed to identify COWP gene homologs or genes encoding recognizable tandem repeats of the cysteine-rich type I or type II domains. In contrast, BLAST screening of the Toxoplasma genome sequence database by using regions of COWP genes as queries revealed numerous open reading frames on distinct assembly contigs encoding type I domain-like cysteine periodicity (data not shown). All open reading frames were fragmented, presumably due to the presence of multiple introns and, in part, due to the current incompleteness of the genome assembly. To confirm the structure of a selected Toxoplasma gene, designated the TgOWP1 gene, we designed exon-specific PCR primers and isolated a putative full-length TgOWP1 gene by using a Toxoplasma cDNA library as an amplification template. Comparison of this spliced transcript with genomic sequence data confirmed that the TgOWP1 gene possesses three introns (lengths, 206, 85, and 393 nucleotides), including an amino-terminal intron that delineates a putative signal peptide sequence present on a short exon (Fig. 6).
FIG. 6.
Amino acid alignment of TgOWP1 and COWP6. Cysteine residues are shaded. The bottom line indicates the consensus residues, abbreviated as follows: h, hydrophobic residues (L, I, Y, F, M, W, A, C, V); l, aliphatic residues (L, I, A, V); c, charged residues (K, E, R, D, H); p, polar residues (S, T, E, D, R, K, H, N, Q); s, small residues (S, A, G, D, N, P, V, T). Conserved residues in the consensus sequence are indicated by uppercase letters. The arrowheads indicate the locations of splice sites in the corresponding TgOWP1 transcript, delineating the four-exon gene structure.
The 499-amino-acid TgOWP1 possesses a putative signal peptide sequence, followed by six type I (six-cysteine) domains and then by a single four-cysteine type I domain at the carboxy terminus (Fig. 1). Note that an alternative delineation is to split the six-cysteine type I domains into three tandem repeats consisting of two-cysteine motifs (15), in which case TgOWP1 contains a tandem array of 20 type I domains. The absence of type II domains in TgOWP1 suggests that its structure is similar to the structure of COWP4 through COWP9, which also lack type II domains, and TgOWP1 exhibits the greatest sequence similarity to COWP6 (Fig. 6). Isolation of additional TgOWP genes should aid in confirming possible orthologous relationships in the oocyst wall protein gene family. Conservation of orthologs across the apicomplexan clade would support the hypothesis that each oocyst wall protein has a discrete, evolutionarily conserved function. TgOWP1 does not possess a histidine-rich motif, which is found in the majority of the COWPs, including COWP6. In order to determine the possible presence of histidine-rich motifs and type II domains, additional putative members of the Toxoplasma oocyst wall protein family must be isolated.
DISCUSSION
Externally shed durable cysts are parasite stages that are present in a subset of the Apicomplexa, including the coccidians Toxoplasma, Sarcocystis, Eimeria, Cyclospora, and Cryptosporidium. Other apicomplexan species, such as Plasmodium, Babesia, and Theileria species, either do not have a cyst stage or do not shed oocysts into the external environment. Due to its highly effective protective function, the oocyst wall is perhaps the single most important structure ensuring survival and propagation of coccidian cysts under environmental stress conditions. As such, it represents a great obstacle to safe disinfection of drinking water and surfaces contaminated with coccidian parasites. The protein complement of the multiple outer membranes of Cryptosporidium and Toxoplasma oocysts likely contributes to oocyst durability. Cryptosporidium oocysts are known to exist in two forms, thin walled and thick walled. The role of these alternative structures remains unclear; however, it is believed that thin-walled oocysts reinitiate the parasite developmental cycle within the same host, while thick-walled oocysts are the major form shed into the environment. It has not been determined if the two oocyst walls have different compositions. The presence of an extended family of COWP genes indicates that multiple proteins are probably used to construct the C. parvum oocyst shell, and it is possible that the type and therefore the fate of developing oocysts are determined by differential usage of multiple COWPs during oocyst formation.
The uniqueness of the coccidian oocyst wall is shown by the fact that BLAST screening of the GenBank nonredundant (nr) database by using COWP amino acid sequences as queries did not identify any obvious nonapicomplexan homologs of the Cryptosporidium COWP family. The best hits obtained are instead proteins that have a marked cysteine periodicity, and the high e values associated with these hits are due to weighting of the cysteine identities. Notably, several of the hits are expressed sequence tags from Anopheles gambiae (e value, 2 × 10−26) and reveal a family of mosquito genes (e.g., gi:21295519 and gi:21298058) that encode a cysteine periodicity strikingly similar to that of COWPs that is possibly due to convergence of protein structure. It is possible that the proteins have similar roles, and it is therefore of interest to study the localization and function of the mosquito proteins. A similar convergence of structure and possible function might also underlie the cysteine periodicity in cell wall protein QID74 of filamentous fungi (gi:3250920) (11), the BR3 salivary protein of the dipteran insect Chironomus (gi:7058) (11), and the multigene family of surface antigenic proteins in Paramecium (gi:3250920) (17). Many proteins have a cysteine periodicity deriving from domain repetition, such as the extensive tandem arrays of the six-cysteine epidermal growth factor-like domain found in proteins with widespread phylogenetic distribution, including numerous proteins encoded by the Caenorhabditis elegans genome and several proteins encoded by the Plasmodium genome. In this regard the cysteine periodicity in the COWPs is simply repetition of a cysteine-rich module, and it is therefore reasonable to assume that repetition of an unrelated cysteine-rich module in a different organism might suggest convergence rather than ancestral homology.
Cryptosporidium has recently been determined to be more closely related to gregarine parasites and thus diverged prior to the common ancestors of both coccidians and the Haemospororidae (3, 20). Identification of a putative oocyst wall protein in T. gondii, TgOWP1, indicates that the type I domain is relatively ancient in the apicomplexan lineage and was lost in parasites lacking an external shed oocyst stage. With the imminent availability of additional alveolate protozoan genome sequences, such as the sequence of the ciliate Tetrahymena, the origin of the oocyst wall protein as a structural unit in rigid cyst walls can be investigated.
Acknowledgments
We thank Michael W. White for the generous gift of a T. gondii sporozoite cDNA library (Tsc3ABP).
Editor: S. H. E. Kaufmann
REFERENCES
- 1.Abrahamsen, M. S., and A. A. Schroeder. 1999. Characterization of intracellular Cryptosporidium parvum gene expression. Mol. Biochem. Parasitol. 104:141-146. [DOI] [PubMed] [Google Scholar]
- 2.Arrowood, M. J., and C. K. Sterling. 1987. Isolation of Cryptosporidium oocysts and parasites using discontinuous sucrose and isopycnic Percoll gradients. J. Parasitol. 73:314-319. [PubMed] [Google Scholar]
- 3.Carreno, R. A., D. S. Martin, and J. R. Barta. 1999. Cryptosporidium is more closely related to the gregarines than to coccidian as shown by phylogenetic analysis of apicomplexan parasites inferred using small-subunit ribosomal RNA gene sequences. Parasitol. Res. 85:899-904. [DOI] [PubMed] [Google Scholar]
- 4.Deng, M., T. J. Templeton, N. R. London, C. Bauer, A. A. Schroeder, and M. S. Abrahamsen. 2002. Cryptosporidium parvum genes containing thrombospondin type 1 domains. Infect. Immun. 70:6987-6995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fayer, R. 1997. Cryptosporidium and cryptosporidiosis. CRC Press, Inc., Boca Raton, Fla.
- 6.Gut, J., C. Petersen, R. Nelson, and J. Leech. 1991. Cryptosporidium parvum: in vitro cultivation in Madin-Darby canine kidney cells. J. Protozool. 38:72S-73S. [PubMed] [Google Scholar]
- 7.Harris, J. R., and F. Petry. 1999. Cryptosporidium parvum: structural components of the oocyst wall. J. Parasitol. 85:839-849. [PubMed] [Google Scholar]
- 8.Hijjawi, N. S., B. P. Meloni, U. M. Morgan, and R. C. A. Thompson. 2001. Complete development and long-term maintenance of Cryptosporidium parvum human and cattle genotypes in cell culture. Int. J. Parasitol. 31:1048-1055. [DOI] [PubMed] [Google Scholar]
- 9.Korich, D. G., J. R. Mead, M. S. Madore, N. A. Sinclair, and C. R. Sterling. 1990. Effects of ozone, chlorine dioxide, chlorine, and monochloramine on Cryptosporidium parvum oocyst viability. App. Environ. Microbiol. 56:1423-1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu, C., V. Vigdorovich, V. Kapur, and M. S. Abrahamsen. 1999. A random survey of the Cryptospordium parvum genome. Infect. Immun. 67:3960-3969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rey, M., S. Ohno, J. A. Pintor-Toro, A. Llobell, and T. Benitez. 1998. Unexpected homology between inducible cell wall protein QID74 of filamentous fungi and BR3 salivary protein of the insect Chironomus. Proc. Natl. Acad. Sci. 95:6212-6216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Riggs, M. W., and L. E. Perryman. 1987. Infectivity and neutralization of Cryptosporidium parvum sporozoites. Infect. Immun. 55:2081-2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rochelle, P. A., R. DeLeon, M. H. Stewart, and R. L. Wolfe. 1997. Comparison of primers and optimization of PCR conditions for detection of Cryptosporidium parvum and Giardia lamblia in water. Appl. Environ. Microbiol. 63:106-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sibert, G. J., and C. A. Speer. 1980. Fine structure of zygotes and oocysts of Eimeria nieschulzi. J. Protozool. 27:374-379. [DOI] [PubMed] [Google Scholar]
- 15.Spano, R., C. Puri, L. Ranucci, L. Putignani, and A. Crisanti. 1997. Cloning of the entire COWP gene of Cryptosporidium parvum and ultrastructural localization of the protein during sexual stage development. Parasitology 114:427-437. [DOI] [PubMed] [Google Scholar]
- 16.Speer, C. A., S. Clark, and J. P. Dubey. 1998. Ultrastructure of the oocysts, sporocysts, and sporozoites of Toxoplasma gondii. J. Parasitol. 84:505-512. [PubMed] [Google Scholar]
- 17.Thai, K. Y., and J. D. Forney. 2000. Analysis of the conserved cysteine periodicity of Paramecium variable surface antigens. J. Eukaryot. Microbiol. 47:242-248. [DOI] [PubMed] [Google Scholar]
- 18.Upton, S. J., M. Tilley, and D. B. Brillhart. 1995. Effects of select medium supplements on in vitro development of Cryptosporidium parvum in HCT-8 cells. J. Clin. Microbiol. 33:371-375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Widmer, G., L. Lin, V. Kapur, and M. S. Abrahamsen. 2002. Genomics and genetics of Cryptosporidium parvum: the key to understanding cryptosporidiosis. Microbes Infect. 4:1081-1090. [DOI] [PubMed] [Google Scholar]
- 20.Zhu, G., J. S. Keithly, and H. Philippe. 2000. What is the phylogenetic position of Cryptosporidium? Int. J. Syst. Evol. Microbiol. 50:1673-1681. [DOI] [PubMed] [Google Scholar]