Abstract
A secreted nuclease, SsnA, was identified in the virulent Streptococcus suis isolate SX332 and subsequently in each of the type strains of capsular serotypes 1 through 9. Screening of 258 porcine clinical isolates from surface (nasal mucosa or palatine tonsil) or internal (joint, brain or other internal organ) locations revealed a significant relationship (P < 0.001) between expression of nuclease and isolation from an internal site. A 3,126-bp gene, ssnA, was identified from a phenotypically nuclease-negative pGh9:ISS1 insertion mutant, and analysis of the predicted SsnA sequence revealed a 35-amino-acid (aa) secretion signal sequence, a 22-aa DNA-binding domain, and a typical gram-positive cell wall sorting motif. A requirement of Ca2+ and Mg2+ for SsnA activity was determined, and the substrate specificity was found to be for single- and double-stranded linear DNA. Reverse transcription-PCR experiments revealed that ssnA is expressed throughout all stages of S. suis growth, and Western blots with porcine anti-S. suis immune sera against a recombinant, truncated SsnA derivative (rSsnAΔ) confirmed that SsnA is expressed in vivo. Furthermore, anti-rSsnAΔ antibodies were sufficient to neutralize SsnA activity. Analyses of subcellular fractions of SX332 and derived mutants, on DNA-containing polyacrylamide gels and by Western blotting, suggest that SsnA is cell wall located.
Streptococcus suis causes several inflammatory porcine diseases, including pneumonia, polyserositis, endocarditis, meningitis, and arthritis (31, 32). Of the 35 capsular serotypes currently described, serotype 2 S. suis strains are most frequently associated with swine disease (21). In addition, humans handling infected pigs or derived products are at risk of infection (39), resulting in potentially fatal diseases such as septicemia, endocarditis, and meningitis (9), as well as postinfection sequalae, including deafness and arthritis (44).
Relatively little is known about the pathogenesis of S. suis disease. Two putative virulence determinants, which have been the focus of several conflicting studies, are the muramidase-released protein (MRP) (35, 41) and extracellular factor (EF) (41). There is a body of work which associates these proteins with the most virulent serotype 2 S. suis strains (41-43, 48). However, other studies have shown that expression of either of these proteins does not appear to affect strain virulence (5, 20, 25). Furthermore, a virulence study in which a well-defined mutant strain lacking both MRP and EF was constructed found the mutant to be equally as virulent as the wild-type parent strain (36). Therefore, the actual importance of these proteins in virulence remains to be elucidated. In MRP+ EF+ serotype 2 strains, the presence of suilysin (SLY), a member of the gram-positive, thiol-activated family of cytolytic toxins (for a review, see reference 2), has been reported to be associated with high virulence (38). However, while a SLY-deficient mutant was found to be completely avirulent in a mouse model of infection, it was only slightly attenuated compared to its isogenic parent in a porcine systemic infection model (1), and no difference in virulence was detected by using an aerosol of S. suis to transmit infection to pigs (24). S. suis also produces a polysaccharide capsule, encoded by the cps genes (34). Acapsular mutants derived from insertional inactivation of the cps locus were found to be highly sensitive to phagocytosis by alveolar lung macrophages (34). Furthermore, these mutants were found to be completely avirulent in germfree pigs following intranasal administration. These results are consistent with another study in which acapsular Tn916-generated mutants were found to be avirulent in both murine and pig infection models (8). Recently, the gene encoding a S. suis serotype 2 fibronectin-binding protein, FBPS, was identified (11). A mutant strain lacking expression of this protein was found to be attenuated in a young piglet infection model with respect to its ability to colonize specific niches, including joints and the central nervous system (11).
Extracellular nucleases are common among prokaryotes and in some instances have been implicated as virulence factors (4, 10, 14). We have determined the presence, in culture supernatants of a pathogenic serotype 2 S. suis strain, of a nuclease which is also produced by all of the pathogenic type strains of serotypes 1 to 9. Clinical field isolates were screened, and the correlation of nuclease production with virulence was examined. Subsequently, a phenotypically nuclease-deficient mutant was isolated following random insertion mutagenesis with pGh9:ISS1 (26), and a gene encoding an exonuclease with a cell wall-anchoring motif was identified.
MATERIALS AND METHODS
Strains, media, and culture conditions.
The virulent S. suis isolate SX332 was obtained from F. Blecha (College of Veterinary Medicine, Kansas State University), the type strains for serotypes 1 through 9 (19, 27) were obtained from A. Crichlow (Western College of Veterinary Medicine, University of Saskatchewan), and 258 field isolates were collected from swine in North America (primarily western Canada). All strains were stored frozen at −70°C in 15% glycerol. Field strains were classified by recovery from a surface (nasal mucosa or palatine tonsil of clinically normal swine) or internal (joint, brain, or other internal organ of diseased swine) location. Escherichia coli strains were routinely cultured in Luria-Bertani medium (Difco, Detroit, Mich.) supplemented with the appropriate antibiotics at 37°C. S. suis strains were cultured statically in a 5% CO2 atmosphere at 30 or 37°C either in Todd-Hewitt broth (Difco) supplemented with 0.5% (wt/vol) yeast extract (THYB), on Mueller-Hinton agar (MHA) plates (Difco), or on tryptic soy agar plates containing 5% (vol/vol) sheep blood (PML Microbiologicals, Mississauga, Ontario, Canada). Media were supplemented with erythromycin for plasmid-containing S. suis and E. coli strains, to 2 and 350 μg/ml, respectively. For detection of nuclease activity of bacterial colonies on plates, toluidine blue agar (TBA) (28) was used; TBA contains a DNA source that when degraded by nuclease results in a change in pH and a concomitant change in color of TBA from blue to pink.
Molecular biological techniques.
Unless otherwise specified, restriction endonuclease digestions, DNA ligations, agarose and sodium dodecyl sulfate-polyacrylamide (SDS-PA) gel electrophoreses, and Southern and Western blotting were performed according to standard protocols (30). Restriction endonucleases and modifying enzymes were purchased from Amersham Pharmacia Biosciences, Ltd. (Baie d'Urfé, Québec, Canada). The broad-host-range, temperature-sensitive, gram-positive shuttle plasmid pG+host 9 (26) and its derivative pGh9:ISS1 (26) were donated by E. Maguin (Institut National de la Recherche Agronomique, Paris, France). Manipulation of pG+host 9-based plasmids was carried out in E. coli TG1-dev, a recA-deficient derivative of TG1 containing a transposon-borne, chromosomally located copy of the wild-type repA gene (E. Maguin, personal communication). Electrocompetent E. coli was prepared and transformed as described previously (18), and electrocompetent S. suis was prepared and transformed by using a method previously described for Streptococcus agalactiae (17), with the omission of glycine. Genomic DNA was extracted and purified from streptococci by using the NucleoSpin tissue kit (Clontech, Palo Alto, Calif.), and plasmid DNA for miniprep analysis was extracted from E. coli by using the QIAquick spin miniprep kit (Qiagen Inc., Mississauga, Ontario, Canada). Plasmid DNA for sequencing was purified by using the Qiagen plasmid minikit. PCR and sequencing primers are described in Table 1. Automated DNA sequencing was carried out at the National Research Council of Canada sequencing facility (Plant Biotechnology Institute, University of Saskatchewan, Saskatchewan, Canada). For isolation of RNA, S. suis cultures from different time points, corresponding to early exponential, mid-exponential, late exponential, and stationary growth phases (optical densities at 600 nm of ca. 0.2, 0.5, 0.9, and 1.2, respectively), were treated with RNAprotect bacterial reagent (Qiagen), and then total RNA was extracted by using the RNeasy minikit (Qiagen) according to the manufacturer's instructions, following a prelysis incubation of cells in 100 μl of Tris-EDTA (pH 8.0) buffer containing 10 mg of lysozyme per ml and 250 U of mutanolysin at 37°C for 10 min. DNA contamination was removed from RNA during purification by treatment with RNase-free DNase (Qiagen) and following purification by digestion with amplification-grade DNase I (Invitrogen Canada Inc., Burlington, Ontario, Canada). Synthesis of cDNA was performed with SuperScript II RNase H− reverse transcriptase (Invitrogen) according to the manufacturer's supplied protocol, using a random nanomer primer. Reverse transcription-PCR experiments were performed with the ssnA-specific PCR primers XII(HindIII) 01 and XII(HindIII) 02; control PCRs with non-reverse-transcribed RNA samples failed to generate any products, confirming the absence of DNA contamination.
TABLE 1.
PCR and sequencing primers
| Primer | Sequencea | Description |
|---|---|---|
| pGh9 01 | 5′-CCAGTGAGCGCGCGTAATACG-3′ | Anneals to bp 921-901 (− strand) of pGh9:ISS1 |
| pGh9 02 | 5′-GGTATACTACTGACAGCTTCC-3′ | Anneals to bp 4471-4491 (+ strand) of pGh9:ISS1 |
| 5′ISS1(rev) | 5′-CGTAGATAATAACCAACAGCG-3′ | Anneals to bp 151-131 (− strand) of pGh9:ISS1 |
| 3′ISS1(fwd) | 5′-AAGAAATGGAACGCTCTTCGG-3′ | Anneals to bp 695-715 (+ strand) of pGh9:ISS1 |
| XII(EcoRI) 01 | 5′-TGCTTACTTCAAAGGTTCGTGG-3′ | Anneals to bp 584-685 (+ strand) of ssnA contig |
| XII(EcoRI) 02 | 5′-AAGAGTCCGGCTGTTACCGC-3′ | Anneals to bp 1508-1489 (− strand) of ssnA contig |
| XII(HindIII) 01 | 5′-CAAATGTCAAGGGTAGGAGATC-3′ | Anneals to bp 2732-2753 (+ strand) of ssnA contig |
| XII(HindIII) 02 | 5′-GATAGTTTGTGCTAGTATGTGGC-3′ | Anneals to bp 3933-3911 (− strand) of ssnA contig |
| XII(HindIII) 03 | 5′-AGCTGTAAATGCTGATGACCG-3′ | Anneals to bp 4386-4406 (+ strand) of ssnA contig |
| XII(HindIII) 04 | 5′-AGCACAAGTGGAGGTGGAGC-3′ | Anneals to bp 4636-4617 (− strand) of ssnA contig |
| MF1013i 01 | 5′-CCTTACAACTCCATGACACC-3′ | Anneals to bp 5024-5043 (+ strand) of ssnA contig |
| pETssnA 01 | 5′-CATATGACTGAAGTGGCGATAGAGAATTATCC-3′ | Anneals to bp 1549-1574 (+ strand) of ssnA contig; NdeI tag |
| pETssnA 02 | 5′-GGATCCATCTGGTGTGACATCTTGC-3′ | Anneals to bp 2682-2661 (− strand) of ssnA contig; BamH1 tag |
Restriction endonuclease recognition sites are underlined.
Detection of S. suis nuclease activity. (i) Assay of whole cells.
Nuclease activity in whole cells was detected by using a slightly modified version of a previously described method (28). Initially, SX332 was screened for nuclease production; bacteria were streaked on MHA plates and incubated to allow colony formation. The plates were overlaid with 3.5 ml of TBA supplemented with 1 mM (each) CaCl2 and MgCl2. Following incubation at 37°C for no longer than 2 h, nuclease activity was determined as the presence of pink halos surrounding colonies. Subsequently, the nuclease phenotypes of the type strains for S. suis serotypes 1 to 9 and of 258 clinical isolates were evaluated by placing 2 μl of culture of each strain on the surfaces of MHA plates, using a grid template with 50 designated locations per 100-mm-diameter circle. The plates were incubated to allow bacterial growth before the nuclease phenotype was determined as described above.
(ii) Assay of cell fractions.
The nuclease activity of proteins electrophoresed in PA gels containing sonicated, single-stranded salmon sperm DNA (5s DNA) was determined by using a modified version of a previously described method (29). Briefly, denaturing or native PA gels were prepared, with the resolving gel portions supplemented with 5 μg of 5s DNA per ml. Spent media from exponentially growing cultures (optical density at 600 nm of ca. 0.4) were sterilized through 0.2-μm-pore-size syringe filters (Gelman Sciences, Ann Arbor, Mich.) and supplemented with complete EDTA-free protease inhibitor cocktail (Roche Diagnostics, Laval, Québec, Canada) to a 1× final concentration. Supernatants were concentrated ca. 50-fold in Ultrafree-15 BIOMAX-30 K filters (Millipore, Bedford, Mass.) by centrifugation at 2,000 × g at 4°C and buffer exchanged in 40 mM Tris-HCl (pH 7.5). Cell wall proteins were extracted from S. suis as described previously (22) and buffer exchanged as described above. Following electrophoresis of samples, gels were rinsed briefly in water and immersed in 20 mM Tris-HCl (pH 7.5) with or without 1 mM (each) CaCl2 and MgCl2. The gels were then incubated at 37°C for up to 8 h, with occasional replacement of buffer, before addition of ethidium bromide to 1 μg/ml. After continued incubation at 37°C, gels were analyzed with UV transillumination. Proteins with nuclease activity (which degraded surrounding DNA, preventing ethidium bromide staining) appeared as dark bands against the fluorescing background.
Construction of S. suis plasmid insertion mutants and determination of insertion loci.
A random pGh9:ISS1 insertion library of S. suis SX332 was constructed as described previously for other gram-positive species (26, 37), and targeted chromosomal integration of plasmids (other than pGh9:ISS1) by homologous recombination was also performed as described previously (7). Colonies resistant to erythromycin at 37°C (the nonpermissive temperature for plasmid replication) were considered likely to have undergone plasmid integration into the chromosome. Sequences flanking the sites of chromosomally integrated plasmids were obtained by using the sequence rescue (SR) technique, essentially as described previously (26). Briefly, genomic DNAs from integrant strains were digested with restriction endonucleases for which a single recognition site was present within the inserted vector (Fig. 1). Following ligation, circularized genomic fragments flanked by vector sequences (containing the erythromycin resistance gene) were recovered after transformation of E. coli TG1-dev and selection in the presence of erythromycin.
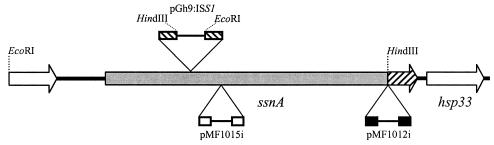
FIG. 1.
Schematic representation of the ssnA locus. The ca. 4.5-kb region between the EcoRI and HindIII recognition sites was obtained by SR from integrated pGh9:ISS1. The ca. 1.3-kb region between the HindIII site and the 3′ end, including the hatched portion of ssnA, was obtained by SR from integrated pMF1012i. Nuclease-deficient mutants were constructed by disruption of ssnA by random insertion of pGh9:ISS1 (SX435), and targeted insertion of pMF1012i (SX436) and pMF1015i (SX437).
SR was performed from the pGh9:ISS1 integrant mutant, SX435, using EcoRI for upstream and HindIII for downstream sequence retrieval. Subsequently, ca. 1.6 kb of the upstream flanking region was recovered in a ca. 6.2-kb plasmid, designated pXII(EcoRI), and ca. 2.9 kb of the downstream flanking region was recovered in a ca. 7.5-kb plasmid, designated pXII(HindIII). The junctions between pGh9:ISS1 and cloned inserts were initially determined with the primers 5′ISS1(rev) plus pGh9 02 for pXII(EcoRI) and 3′ISS1(fwd) plus pGh9 01 for pXII(HindIII). The pXII(EcoRI) sequence was completed with primers XII(EcoRI) 01 and XII(EcoRI) 02, and the pXII(HindIII) sequence was completedwith the primers XII(HindIII) 01 and XII(HindIII) 02. Sequence data determined the presence of an 8-bp duplicated sequence (ATATCTAC) on either side of the pGh9:ISS1 insertion site, allowing the upstream and downstream sequences to be joined in frame, resulting in a contiguous sequence (contig) of 4,499 bp.
To determine chromosomal sequences downstream of the region obtained by SR from pXII(HindIII), an internal fragment of ssnA was cloned into pG+host 9. PCR with the primers XII(HindIII) 01 plus pGh9 01 amplified a DNA product from pXII(HindIII) which was digested with PvuI and HindIII to liberate a 414-bp fragment corresponding to the central portion of the ssnA gene. The ssnA fragment was blunt ended by using the PCR polishing kit (Stratagene, La Jolla, Calif.) and cloned into the SmaI site of pG+host 9. Subsequently, a clone containing the ssnA fragment in the opposite orientation to the pG+host 9 repA(Ts) gene was identified and designated pMF1012i. A mutant, designated SX436, was identified following homologous recombination of pMF1012i into the SX332 chromosome (Fig. 1). Plasmid integration at the desired locus was confirmed by PCR with the primers XII(HindIII) 01 plus XII(HindIII) 02 and XII(HindIII) 01 plus pGh9 01 (data not shown); as expected, the former control PCR amplified a fragment of ca. 1.2 kb from the ssnA gene of SX436 and wild-type SX332 DNA (the amplified region lay upstream of the pMF1012i insertion site), while the latter PCR amplified a product of ca. 1.85 kb from SX436 but failed to amplify any product from SX332, due to the pGh9 01 primer's specificity for the integrated plasmid. SR was performed from SX436 with EcoRI, and a clone containing a plasmid of ca. 5.6 kb was identified and designated pMF1013i. pMF1013i was sequenced with XII(HindIII) 03 and MF1013i 01; when combined with the above sequence, a 5,821-bp contig was obtained.
To allow targeted disruption of ssnA, a 1,143-bp PCR fragment corresponding to the region directly downstream of the predicted secretion signal peptide-encoding sequence of ssnA was amplified from SX332 genomic DNA with the primers pETssnA 01 and pETssnA 02. The fragment was ligated into pPCR-SCRIPT by using the PCR-SCRIPT cloning kit (Stratagene), and a recombinant plasmid containing the ssnA fragment in the opposite orientation to the pPCR-SCRIPT lacZ gene was identified and designated pMF1005i. Following NotI and XhoI digestion of pMF1005i, a fragment containing bp 106 to 1236 of ssnA and adjacent vector sequences was cloned into pG+host 9 to create plasmid pMF1015i. Following chromosomal integration of pMF1015i (Fig. 1), confirmation of integration at the desired locus was obtained by PCR with XII(HindIII) 01 plus pGh9 01 and pGh9 02 plus XII(HindIII) 04. Both pG+host 9 primers are specific for the pG+host 9 backbone of pGh9:ISS1, and as expected, the former primer pair amplified a ca. 2.2-kb fragment (corresponding to the region directly upstream of the inserted plasmid) from SX437, whereas no product was obtained from wild-type SX332 DNA (data not shown). The second primer pair amplified an expected product of ca. 3.2 kb (corresponding to the region directly downstream of the inserted plasmid) from SX437, while the control SX332 PCR was negative (data not shown). SX437 was screened with TBA and found to be nuclease deficient.
Expression and purification of a truncated SsnA derivative.
A DNA fragment, corresponding to the sequence encoding amino acid residues Thr36 to Asp412 of SsnA (377 amino acids [aa] immediately following the predicted signal peptide cleavage site), was liberated from pMF1005i with NdeI and BamHI and cloned into the pET-15b expression plasmid (Novagen, Madison, Wis.). The resulting construct, designated pMF1025, was used to transform E. coli BL21(DE3). Expression of the N-terminally six-histidine-residue-tagged, truncated SsnA protein (rSsnAΔ) was induced by addition of IPTG (isopropyl-β-d-thiogalactopyranoside) to exponentially growing cultures to a concentration of 1 mM. Following induction, cultures were incubated at 30°C for 1 h, and then rifampin was added to 400 μg/ml. Incubation was continued for a further 3 h, prior to protein purification by immobilized metal affinity chromatography with Ni-nitrilotriacetic acid agarose (Qiagen) according to the manufacturer's instructions.
Generation of immune sera.
Anti-SsnA polyclonal antibodies were raised in New Zealand White rabbits. An initial subcutaneous immunization (day 0) was performed with 1 ml of PBS containing 25 μg of rSsnAΔ and 30% oil-in-water emulsion (VSA3; MVP Laboratories, Ralston, Nebr.). Booster vaccinations were performed, as described above, at days 20 and 27, and rabbits were bled out to obtain immune serum on day 33. Porcine anti-S. suis SX323 immune sera were obtained from pigs challenged intrapharyngeally with live SX332 (performed as part of an unrelated study [24]). Nonimmune sera were also obtained from unchallenged control animals from the same experiment.
Computational analyses.
Sequence manipulations were performed with the Clone Manager Suite software (Scientific & Educational Software, Durham, N.C.). Translated sequences were identified by comparison with protein sequences in the National Center for Biotechnology Information database, using the Clone Manager Blastx interface. Further sequence analyses were performed with the Prosite database of families and domains (http://www.expasy.ch/prosite) and applications of the European Molecular Biology Open Software Suite package (http://www.hgmp.mrc.ac.uk/Software/EMBOSS/) via the Canadian Bioinformatics Resource web interface (http://bioinfo.pbi.nrc.ca/). Analysis of the statistical significance of nuclease phenotypes in clinical field isolates was performed by using cross-tabulation in the SYSTAT 10 software package (SPSS Science, Chicago, Ill.).
Nucleotide sequence accession number.
The 5,821-bp contig containing the S. suis SX332 ssnA gene was submitted to GenBank under accession number AY254322.
RESULTS
Screening of S. suis isolates for nuclease activity.
Pink halos were observed surrounding growth of the virulent S. suis isolate SX332 and each of the type strains of serotypes 1 through 9, indicating expression of nuclease (data not shown). As shown in Table 2, most of the field strains isolated from internal sites (87.5%) expressed a nuclease phenotype, whereas fewer than half (40.2%) of the surface isolates displayed an equivalent phenotype. This difference in proportion was found to be highly statistically significant (P < 0.001), with an odds ratio of 10.4 (95% confidence interval, 4.7 to 23.0).
TABLE 2.
Nuclease phenotypes of S. suis field isolates
| Phenotype | No. of S. suis isolates from:
|
|
|---|---|---|
| Surface location | Internal organ | |
| Nuclease positive | 78 | 56 |
| Nuclease negative | 116 | 8 |
| Total | 194 | 64 |
Generation and screening of a nuclease-deficient mutant.
A pGh9:ISS1 random insertion library of SX332 was constructed, and comparison of total CFU per milliliter with erythromycin-resistant CFU per milliliter at 37°C revealed a pGh9:ISS1 chromosomal integration frequency of 5 × 10−2. Approximately 10,000 individual colonies were screened for nuclease activity by TBA overlay. A single colony which was not surrounded by a pink halo was identified, indicating the absence of nuclease activity. Subsequently, this clone, designated SX435, was restreaked on selective agar plates, tested for nuclease activity, and confirmed to be nuclease deficient. Southern blot analysis of SX435 with a probe corresponding to the ermAM gene of pGh9:ISS1 determined the presence of a single band (data not shown), confirming that a single integration of pGh9:ISS1 had occurred.
Analysis of the region containing the nuclease-encoding gene.
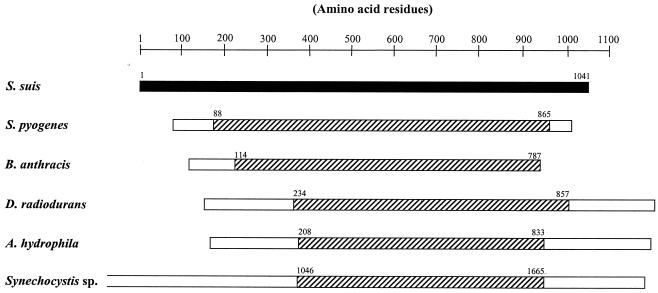
The locus of pGh9:ISS1 insertion in SX435 was determined following SR (Fig. 1). The presence of coding regions within the 4,499-bp contig was determined, and translated open reading frames (ORFs) were compared with the GenBank sequence database. Homologues of genes described previously for other species were annotated accordingly. At the extreme 5′ end of the chromosomal fragment, a 558-bp partial reading frame was found, encoded by the positive strand. The translated 186-aa product showed 73% identity and 82% similarity to aa 566 to 750 of a 754-aa putative glutamate-cysteine ligase enzyme of Streptococcus mutans UA159 (accession no. AAN58035). Approximately 0.8 kb further downstream lay a second ORF encoded by the positive DNA strand, starting at bp 1384 and extending throughout the remainder of the sequence. The translated 1,057-aa product showed the most extensive homology, at 59% identity and 73% similarity, to a 910-aa conserved hypothetical protein of Streptococcus pyogenes (group A Streptococcus [GAS]) M1 (accession no. AAK33693) and MGAS8232 (accession no. AAL97472). The protein also shared 38% identity and 52% similarity over aa 114 to 787 of a putative member of the Bacillus anthracis AP endonuclease 1 family (accession no. NP_654986). Furthermore, regions of the predicted ORFs in the S. suis sequence shared homology with regions of extracellular nuclease proteins of several bacteria (Fig. 2), including those of a Synechosystis sp. (accession no. BAA16955), Aeromonas hydrophila (accession no. AAB39273), Vibrio cholerae (accession no. AAF95762), and Deinococcus radiodurans (accession no. AAF12592). This gene, apparently encoding an S. suis nuclease, was designated ssnA (for S. suis secreted nuclease A).
FIG. 2.
Schematic representation of homology between S. suis SsnA and nucleases from other species. The numbers shown with each protein denote the amino acid residues at which regions of homology (hatched boxes) begin and end.
To determine the remaining ssnA sequence, SR was performed from chromosomally integrated pMF1012i. Analysis of the resulting sequence revealed that the ssnA ORF extended a further 67 bp downstream of the previously determined sequence (Fig. 1). In addition, a second, 867-bp ORF, encoded by the positive DNA strand, was identified 80 bp downstream of ssnA, the translated 288-aa product of which shared 79% identity and 90% similarity over its entire length to the 290-aa, 33-kDa heat shock protein (Hsp33) of Streptococcus pneumoniae (accession no. AAK76239). The S. suis gene has been designated hsp33.
The intact ssnA ORF was observed to be 3,186 bp, encoding a predicted protein of 1,061 aa; however, within the first 70 bp of the DNA sequence were two other potential ATG codons, each in frame with the first. Only the last of the three start codons exhibited a potential ribosome-binding site (GGAG), 10 bp immediately upstream. Analysis of the predicted SsnA sequence, translated from the first start codon, by using the SPScan application predicted three different putative signal peptides, with identical cleavage sites but with lengths of 55, 46, and 35 aa, respectively, corresponding to each of the three translational start codons. Typical gram-positive signal peptides are usually approximately 30 aa in length, so it is likely that the last of the three start codons corresponds to the translational start site of SsnA. While a longer signal peptide is possible (for example, the staphylococcal nuclease gene, nuc, encodes a 60-aa signal peptide [33]), the 35-aa sequence is in keeping with the observed ribosome-binding site. Therefore, the 1,041-aa SsnA is encoded by a 3,126-bp ORF, with a predicted 35-aa signal peptide cleavage site between amino acid residues Ala35 and Thr36. Sequence data revealed the pGh9:ISS1 insertion locus to be downstream of bp 634 of ssnA. A putative nucleic acid binding domain (ITETNIAQLATQAQATLVSLKN) was predicted within SsnA between aa 256 and 277 by using the Helixturnhelix application (12). Comparison of SsnA with the Prosite database revealed a typical gram-positive cell wall sorting signal between aa 1011 and 1041. This sequence fulfilled all criteria proposed for gram-positive surface proteins, including a conserved hexapeptide (LPKTG) motif followed by a hydrophobic domain and finally a cluster of basic residues (16). Nonquantitative reverse transcription-PCR analysis consistently generated a DNA fragment of the expected size of ca. 1.2 kb, confirming that the ssnA gene is expressed throughout all growth stages examined (data not shown).
Porcine immune recognition of SsnA.
Western blotting was performed to determine whether SsnA was expressed during in vivo infection with S. suis. Pooled immune sera, obtained from pigs challenged experimentally with S. suis, were used to probe nitrocellulose-bound rSsnAΔ. A strong band of ca. 44 kDa was observed, corresponding to rSsnAΔ (data not shown). Conversely, sera from naive pigs failed to detect rSsnAΔ, confirming the specificity of the immune reaction. These combined data confirmed that SsnA is expressed by S. suis in vivo and that it appears to be a strongly immunogenic protein.
Serum neutralization of SsnA.
The ability of rabbit anti-rSsnAΔ antibodies to neutralize SsnA activity was determined. Single colonies of SX332 on THYB agar plates were overlaid with TBA, and 25-μl drops of anti-rSsnAΔ serum were placed directly on the TBA at different locations on the plates. After incubation at 37°C for up to 18 h, no nuclease activity was observed surrounding colonies in the immediate vicinity of the antiserum drops, whereas pink halos surrounded all outlying colonies (data not shown). Similar assays performed with nonimmune rabbit serum failed to inhibit SsnA activity, confirming that SsnA is specifically neutralized by anti-rSsnAΔ antibodies.
Targeted disruption of ssnA.
To confirm that ssnA was in fact the nuclease-encoding gene, targeted disruption was carried out by homologous recombination of pMF1015i into the chromosome (Fig. 1), and a nuclease-deficient mutant, designated SX437, was identified following screening with TBA. Interestingly, the mutant SX436 (see above) was derived from the insertion of pMF1012i ca. 0.3 kb upstream of the putative cell wall sorting signal; conveniently, this mutant was theoretically deficient in its capacity to be cell wall anchored. Screening with TBA confirmed a nuclease-positive phenotype, and thus the final ca. 100 aa of SsnA is not required for enzymatic activity.
Analysis of nuclease in cell fractions.
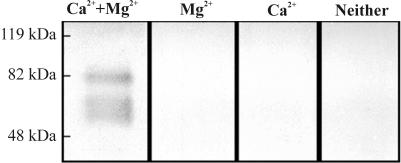
Concentrated culture supernatants of SX332 were prepared in denaturing sample loading buffer (containing SDS and β-mercaptoethanol) or in native loading buffer (without SDS or β-mercaptoethanol). Duplicate samples were prepared in each buffer, and one of each was boiled for 3 min prior to gel loading (the experiment was repeated on several occasions). Following electrophoresis of proteins on 5s DNA-containing SDS-PA gels, nuclease activity was restored by incubation in buffer containing MgCl2 and CaCl2. Ethidium bromide staining at 37°C was performed for a minimum of 1 h; however, it was occasionally necessary to continue incubation for up to 8 h to allow visualization of nuclease activity. Samples loaded in nondenaturing buffer revealed numerous high-molecular-mass dark bands, visible as a smear against the fluorescent background of the stained gel (data not shown); this was attributed to inadequate disruption of protein aggregates in the native buffer. Samples loaded in denaturing sample buffer revealed a major band with an apparent molecular mass of ca. 80 kDa and two lower-molecular-mass bands of ca. 68 and 63 kDa (Fig. 3), indicating that all had nuclease activity; the two lower-molecular-mass bands most likely derived from proteolytic degradation of the higher-molecular-mass moiety (see Discussion). Boiling of SX332 samples failed to reduce the extent of the observed nuclease activity, confirming the heat-stable nature of SsnA (data not shown); however, samples prepared in denaturing buffer revealed less nuclease activity than those prepared in native buffer (data not shown), suggesting incomplete refolding prior to detection of activity. Nuclease activity was detected only in the presence of Mg2+ and Ca2+ together (Fig. 3), suggesting that SsnA has an absolute requirement for both ions for activity. Furthermore, in addition to 5s DNA, SDS-PA gels were also prepared with either 5 μg of linear, double-stranded DNA (unboiled, sonicated salmon sperm DNA) per ml, 20 μg of yeast tRNA per ml, or 10 μg of double-stranded, closed circular plasmid DNA (pPCR-SCRIPT) per ml. Nuclease activity equivalent to that in 5s DNA was observed in gels containing linear, double-stranded DNA fragments; however, with all other substrates no nuclease activity was evident (data not shown), even following extended incubation (>48 h) prior to staining.
FIG. 3.
Nuclease in S. suis supernatant extracts was detected in 5s DNA-containing SDS-PA gels. In the presence of Ca2+ and Mg2+, three protein bands of ca. 80, 68, and 63 kDa were observed to possess nuclease activity.
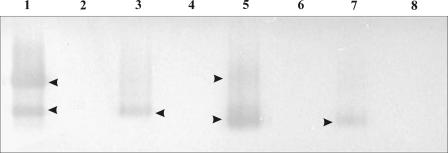
Equivalent quantities of cell wall extracts, prepared from SX332 and derived mutants, were analyzed on native 5s DNA-containing PA gels (Fig. 4). Samples were not boiled prior to electrophoresis, and nondenaturing sample loading buffer was used. Nuclease activity was detected for SX332 in the form of two distinct bands. No activity was observed for SX435 and SX437, while a faint, single band was observed for SX436, corresponding approximately in size to the lower-molecular-mass band of SX332. Western blot analysis of cell wall fractions was also performed (Fig. 5). SX332 samples revealed two dominant bands of ca. 112 kDa and 108 kDa, corresponding to the predicted sizes of SsnA with and without signal peptide. Equivalent bands were completely absent from SX435 and SX437 samples. The ca. 112-kDa band was also absent from SX436 samples; however, there was the suggestion of an extremely faint band, of ca. 100 kDa, in place of the 108-kDa band of SX332, corresponding to the predicted 98-kDa truncated SsnA with an intact signal peptide.
FIG. 4.
Determination of nuclease activity in S. suis subcellular fractions. Proteins were analyzed on 5s DNA-containing native PA gels, and samples correspond to supernatant fractions (lanes 1 to 4) and cell wall fractions (lanes 5 to 8). Samples 1 and 5 derive from SX332, 2 and 6 derive from SX435, 3 and 7 derive from SX436, and 4 and 8 derive from SX437. Arrowheads indicate bands with nuclease activity. No activity was observed for SX435 and SX437, while the upper nuclease band was absent in SX436 samples.
FIG. 5.
Western blot analysis of mutanolysin-extracted S. suis cell wall proteins. Two nuclease bands were detected in SX332 which were absent from SX435 and SX437. A very faint band of approximately 100 kDa was observed for SX436.
DISCUSSION
S. suis causes infections of both porcine and human hosts (9, 31, 32, 44). Despite numerous studies, in which factors reportedly associated with virulence were identified, relatively little is actually known about the pathogenesis of S. suis disease. To a large extent, this is due to conflicting reports of the importance of putative virulence factors during infection. This confusion is most notable with the proteins MRP (35, 41) and EF (41), for which similar numbers of studies have reported that they either contribute (41-43, 48), or do not contribute (5, 20, 25, 36) to strain virulence. The pathogenesis of S. suis disease would appear to involve an interplay between multiple virulence factors, including MRP, EF, suilysin, capsule, host protein ligands, and other bacterial attributes, all of which interact with host resistance.
In a search for novel factors that may contribute to S. suis virulence, we identified an extracellular nuclease, SsnA. There is no definitive way to differentiate between virulent and nonvirulent strains of bacteria without experimental infection of swine; in general, however, bacteria isolated from an internal organ of a diseased swine were probably able to persist there as a result of inherent virulence characteristics. We have identified a significant association between isolation from an internal organ and expression of nuclease; furthermore, the designated type strains for S. suis serotypes 1 to 9 have each been shown to display nuclease-positive phenotypes. In addition, anti-rSsnAΔ immune serum was capable of inhibiting DNase activity in whole bacterial cells, and convalescent porcine serum strongly recognized rSsnAΔ in Western blots, proving that nuclease is produced in vivo.
Other streptococcal (14) and staphylococcal (10) extracellular DNases have been implicated as virulence factors. Among the reasons for such an association was the observation that nuclease activity is directed against DNA, which is essential for the functioning of the target cell. Furthermore, production of anti-DNase antibodies was noted as a common feature during GAS (3), S. agalactiae (group B Streptococcus [GBS]) (15), and staphylococcal (10) infections, which correlates with the observations in this study. If there is an association between nucleases and virulence, it may be indirect, in that the enzyme would provide oligonucleotides to serve as a nutrient source (47). Alternatively, it is conceivable that SsnA could degrade DNA in the surrounding microenvironment and affect the ability of other bacteria to be transformed (e.g., Campylobacter in the tonsillar area).
In the process of screening the SX332 pGh9:ISS1 insertion library, we identified a single nuclease-deficient mutant, with a single ISS1 insertion locus, out of ca. 10,000 individual colonies. Assuming random vector integration, this suggests the presence of a single nuclease-encoding gene within the SX332 chromosome. The site of integration of pMF1015i into ssnA was immediately downstream of the sequence encoding the first 377 aa of the mature protein. Despite containing the predicted DNA-binding domain (between aa 257 and 277), the first 377 aa would appear not to be sufficient to produce a functional enzyme. This is consistent with the observed regions of homology between SsnA and nucleases of other bacterial species (Fig. 2), secreted or otherwise, where, in most cases, extensive homology was observed downstream of approximately the first 300 amino acid residues of SsnA, suggesting that this “conserved” region is essential for enzymatic activity. With regard to substrate specificity, SsnA appears to be specific for both single- and double-stranded, linear DNA and is therefore similar to other exonucleases.
Identically sized triplicate bands displaying nuclease activity were observed in concentrated culture supernatants of the wild type and the cell wall-anchoring mutant in 5s DNA SDS-PA gels (data not shown); however, in equivalent gels, no activity was observed for the mutant strains, in which ssnA was disrupted downstream of bp 2076 in SX435 and bp 2681 in SX437, encoding the first 211 and 412 amino acid residues of SsnA in each respective mutant. Since plasmid integration into ssnA of SX435 and SX437 was sufficient to abolish nuclease activity, it is most likely that the lower-molecular-mass forms of the ca. 80-kDa band in SX332 and SX436 supernatants derived from proteolytic cleavage of the high-molecular-mass form. The ca. 80-kDa band also likely derived from proteolytic cleavage of intact, cell wall-located SsnA, since it was smaller than the moieties observed in mutanolysin extracts. Whether the observed cleavage is a real phenomenon or is nonspecific proteolysis by an S. suis protease as a result of culturing under artificial conditions is currently unknown. The fact that equivalent band sizes were observed in gels of SX332 and SX436 suggests that proteolytic cleavage occurred more than 100 aa upstream of the cell wall-anchoring motif to create the truncated ca. 80-kDa SsnA.
Although protein amounts were normalized between samples, SX436 nuclease activity from culture supernatants, analyzed in native 5s DNA gels, was less than SX332 activity in equivalent samples. Since SsnA of SX436 lacks the putative cell wall sorting motif and 100 aa of the upstream region, we hypothesize that the protein is unable to be cell wall anchored; however, in this case, the apparent nuclease activity of SX436 likely derived from extraction of small amounts of SsnA in transit across the cell membrane. Unfortunately, we were unable to show any nuclease activity in cell wall extracts analyzed on denaturing PA gels; the reason for this is unknown and led to the requirement for use of native gels, which in turn complicated the issue of accurate size determination of nuclease bands in different samples.
In Western blots of denatured, mutanolysin-extracted SX436 proteins, an extremely faint band, corresponding to the expected size of truncated SsnA with intact signal peptide, was observed. Studies of gram-positive protein secretion in Bacillus have shown that the signal peptide of exported proteins is looped through the cell membrane, while the remainder of the protein is secreted; subsequently, the signal peptide is cleaved by a signal peptidase (40). Assuming that the same process exists in S. suis, prior to cleavage of the membrane-associated signal peptide of the truncated mutant, SsnA would be transiently attached to the cell until the signal peptide was cleaved. Therefore, the identification in mutanolysin extracts of unprocessed, truncated SsnA is encouraging. Taken as a whole, our results confirm that ssnA is the nuclease-encoding gene and strongly suggest that SsnA appears to be associated with the cell wall, most likely through linkage to the peptidoglycan, directed by the cell wall sorting signal. This observation has important implications, since, to our knowledge, this is the first description of a cell wall-anchored DNase in streptococci. A previous study suggested the presence of a single-strand-preferential DNase associated with cell wall and membrane fractions of Bacillus subtilis (6); however, despite the availability of the entire B. subtilis genome sequence (accession no. NC_000964) (23), no homologue of SsnA has been identified. This, however, does not preclude the possibility that a similar DNase may be present in other B. subtilis strains.
Our evidence suggests that SsnA is the only protein displaying DNase activity in S. suis, at least under the conditions examined in this study. This is in contrast to observations of other streptococcal species, including GAS and GBS (for a review, see reference 13). Furthermore, with an apparent molecular mass of 108.5 kDa for the mature protein, SsnA is significantly larger than those of other streptococci. SsnA does show significant homology to the product of an uncharacterized conserved ORF in GAS, which does not correspond to any of the four currently described GAS extracellular nucleases (45, 46). Interestingly, like ssnA, the GAS ORF also appears to encode a C-terminal cell wall sorting motif, although it lacks a predicted secretion signal peptide.
In conclusion, the results obtained in this study have confirmed the presence of a secreted DNase in S. suis which appears to be cell wall anchored. Furthermore, they suggest that SsnA may contribute to the virulence of S. suis; however, it should be emphasized that further work is required to confirm this observation, in order to avoid the confusion that currently exists with the association of MRP, EF, and SLY with virulence. While we have reported a significant correlation between nuclease phenotype and virulence, we acknowledge that a small proportion of apparently virulent isolates apparently did not produce nuclease; it is possible that for some strains, SsnA is expressed only in vivo. Alternatively, nothing is currently known of the regulation of ssnA, and it may well be that the supposedly nuclease-negative strains only appeared to be so under the screening conditions imposed in this study or that they produced levels of nuclease too low to be detected by the screening method employed (the TBA overlay used in this study is capable of detecting as little as 10 ng of nuclease [28]). By way of comparison, essentially all strains of GAS produce nuclease(s) (13). Furthermore, nearly all GBS strains were found to produce nuclease(s) (15), despite previous observations that the majority of strains did not; the difference in prevalence was due to the sensitivities of the nuclease detection assays. The possession of a functional nuclease enzyme(s) would appear to be important in other streptococci, and it would be surprising if the same was not also true of S. suis. Therefore, it may be the amount of nuclease produced, rather than the presence or absence of nuclease, which contributes to virulence. Regardless, if SsnA is involved in virulence, it is unlikely that it alone will predetermine the pathogenic capacity of S. suis. Rather, the presence or absence of other virulence factors will likely affect its importance, and only by continuing to identify and characterize novel virulence determinants will a fuller understanding of S. suis disease pathogenesis be obtained.
Acknowledgments
We thank Emmanuelle Maguin (INRA, Paris France) for generously providing the pG+host 9 and pGh9:ISS1 plasmids and the E. coli TG1-dev strain and Keith Stephenson (University of Leeds, Leeds, United Kingdom) for helpful discussion.
This work was funded by the Canadian Research Network on Bacterial Pathogens of Swine, the Natural Science and Engineering Research Council of Canada (NSERC), and Canadian swine producers.
Editor: V. J. DiRita
Footnotes
Published as number 328 in the Vaccine & Infectious Disease Organization journal series.
REFERENCES
- 1.Allen, A. G., S. Bolitho, H. Lindsay, S. Khan, C. Bryant, P. Norton, P. Ward, J. Leigh, J. Morgan, H. Riches, S. Eastty, and D. Maskell. 2001. Generation and characterization of a defined mutant of Streptococcus suis lacking suilysin. Infect. Immun. 69:2732-2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alouf, J. E., and C. Geoffroy. 1991. The family of antigenically-related cholesterol-binding (′sulphydryl-activated') cytolytic toxins, p. 147-186. In J. E. Alouf and J. H. Freer (ed.), Sourcebook of bacterial proteins. Academic Press, Inc., New York, N.Y.
- 3.Ayoub, E. M., and L. W. Wannamaker. 1962. Evaluation of the streptococcal deoxyribonuclease B and diphosphopyridine nucleotidase antibody tests in acute rheumatic fever and acute glomerulonephritis. Pediatrics 29:527-538. [Google Scholar]
- 4.Bendjennat, M., A. Blanchard, M. Loutfi, L. Montagnier, and E. Bahraoui. 1999. Role of Mycoplasma penetrans endonuclease P40 as a potential pathogenic determinant. Infect. Immun. 67:4456-4462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berthelot-Herault, F., H. Morvan, A. M. Keribin, M. Gottschalk, and M. Kobisch. 2000. Production of muraminidase-released protein (MRP), extracellular factor (EF) and suilysin by field isolates of Streptococcus suis capsular types 2, 1/2, 9, 7 and 3 isolated from swine in France. Vet. Res. 31:473-479. [DOI] [PubMed] [Google Scholar]
- 6.Birnboim, H. C. 1966. Cellular site in Bacillus subtilis of a nuclease which preferentially degrades single-stranded nucleic acids. J. Bacteriol. 91:1004-1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Biswas, I., A. Gruss, S. D. Ehrlich, and E. Maguin. 1993. High-efficiency gene inactivation and replacement system for gram-positive bacteria. J. Bacteriol. 175:3628-3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Charland, N., J. Harel, M. Kobisch, S. Lacasse, and M. Gottschalk. 1998. Streptococcus suis serotype 2 mutants deficient in capsular expression. Microbiology 144:325-332. [DOI] [PubMed] [Google Scholar]
- 9.Chau, P. Y., C. Y. Huang, and R. Kay. 1983. Streptococcus suis meningitis. an important underdiagnosed disease in Hong Kong. Med. J. Aust. 1:414-416. [PubMed] [Google Scholar]
- 10.Chesbro, W., and R. Walker. 1972. Detection of staphylococcal nuclease elaborated during lethal infections of mice. Infect. Immun. 6:1028-1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Greeff, A., H. Buys, R. Verhaar, J. Dijkstra, L. van Alphen, and H. E. Smith. 2002. Contribution of fibronectin-binding protein to pathogenesis of Streptococcus suis serotype 2. Infect. Immun. 70:1319-1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dodd, I. B., and J. B. Egan. 1990. Improved detection of helix-turn-helix DNA-binding motifs in protein sequences. Nucleic Acids Res. 18:5019-5026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ferreira, B. T., L. C. Benchetrit, A. C. De Castro, T. G. Batista, and L. Barrucand. 1992. Extracellular deoxyribonucleases of streptococci: a comparison of their occurrence and levels of production among beta-hemolytic strains of various serological groups. Zentbl. Bakteriol. 277:493-503. [DOI] [PubMed] [Google Scholar]
- 14.Ferretti, J. J., T. T. Huang, W. L. Hynes, H. Malke, D. Simon, A. Suvorov, and C. E. Yu. 1991. Extracellular product genes of group A streptococci, p. 201-205. In G. M. Dunny, P. P. Cleary, and L. L. McKay (ed.), Genetics and molecular biology of streptococci, lactococci, and enterococci. American Society for Microbiology, Washington, D.C.
- 15.Ferrieri, P., E. D. Gray, and L. W. Wannamaker. 1980. Biochemical and immunological characterization of the extracellular nucleases of group B streptococci. J. Exp. Med. 151:56-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fischetti, V. A., V. Pancholi, and O. Schneewind. 1990. Conservation of a hexapeptide sequence in the anchor region of surface proteins from gram-positive cocci. Mol. Microbiol. 4:1603-1605. [DOI] [PubMed] [Google Scholar]
- 17.Framson, P. E., A. Nittayajarn, J. Merry, P. Youngman, and C. E. Rubens. 1997. New genetic techniques for group B streptococci: high-efficiency transformation, maintenance of temperature-sensitive pWV01 plasmids, and mutagenesis with Tn917. Appl. Environ. Microbiol. 63:3539-3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Froehlich, B. J., and J. R. Scott. 1991. A single-copy promoter-cloning vector for use in Escherichia coli. Gene 108:99-101. [DOI] [PubMed] [Google Scholar]
- 19.Gottschalk, M., R. Higgins, M. Jacques, K. R. Mittal, and J. Henrichsen. 1989. Description of 14 new capsular types of Streptococcus suis. J. Clin. Microbiol. 27:2633-2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gottschalk, M., A. Lebrun, H. Wisselink, J. D. Dubreuil, H. Smith, and U. Vecht. 1998. Production of virulence-related proteins by Canadian strains of Streptococcus suis capsular type 2. Can. J. Vet. Res. 62:75-79. [PMC free article] [PubMed] [Google Scholar]
- 21.Higgins, R., and M. Gottschalk. 1999. Streptococcal diseases, p. 563-570. In A. D. Leman, B. E. Straw, W. L. Mengeling, S. D'Allaire, and D. J. Taylor (ed.), Diseases of swine, 8th ed. Iowa State University Press, Ames, Iowa.
- 22.Kling, D. E., L. C. Madoff, and J. L. Michel. 1999. Subcellular fractionation of group B Streptococcus. BioTechniques 27:24-28. [DOI] [PubMed] [Google Scholar]
- 23.Kunst, F., N. Ogasawara, I. Moszer, A. M. Albertini, G. Alloni, V. Azevedo, M. G. Bertero, P. Bessieres, A. Bolotin, S. Borchert, R. Borriss, L. Boursier, A. Brans, M. Braun, S. C. Brignell, S. Bron, S. Brouillet, C. V. Bruschi, B. Caldwell, V. Capuano, N. M. Carter, S. K. Choi, J. J. Codani, I. F. Connerton, A. Danchin, et al. 1997. The complete genome sequence of the Gram-positive bacterium Bacillus subtilis. Nature 390:249-256. [DOI] [PubMed] [Google Scholar]
- 24.Lun, S., J. Perez-Casal, W. Connor, and P. J. Willson. 2003. Role of suilysin in pathogenesis of Streptococcus suis capsular serotype 2. Microb. Pathog. 34:27-37. [DOI] [PubMed] [Google Scholar]
- 25.Luque, I., C. Tarradas, R. Astorga, A. Perea, H. J. Wisselink, and U. Vecht. 1999. The presence of muramidase released protein and extracellular factor protein in various serotypes of Streptococcus suis isolated from diseased and healthy pigs in Spain. Res. Vet. Sci. 66:69-72. [DOI] [PubMed] [Google Scholar]
- 26.Maguin, E., H. Prevost, S. D. Ehrlich, and A. Gruss. 1996. Efficient insertional mutagenesis in lactococci and other gram-positive bacteria. J. Bacteriol. 178:931-935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Perch, B., K. B. Pedersen, and J. Henrichsen. 1983. Serology of capsulated streptococci pathogenic for pigs: six new serotypes of Streptococcus suis. J. Clin. Microbiol. 17:993-996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Poquet, I., S. D. Ehrlich, and A. Gruss. 1998. An export-specific reporter designed for gram-positive bacteria: application to Lactococcus lactis. J. Bacteriol. 180:1904-1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rosenthal, A. L., and S. A. Lacks. 1977. Nuclease detection in SDS-polyacrylamide gel electrophoresis. Anal. Biochem. 80:76-90. [DOI] [PubMed] [Google Scholar]
- 30.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 31.Sanford, S. E. 1987. Gross and histopathological findings in unusual lesions caused by Streptococcus suis in pigs. I. Cardiac lesions. Can. J. Vet. Res. 51:481-485. [PMC free article] [PubMed] [Google Scholar]
- 32.Sanford, S. E., and M. E. Tilker. 1982. Streptococcus suis type II-associated diseases in swine: observations of a one-year study. J. Am. Vet. Med. Assoc. 181:673-676. [PubMed] [Google Scholar]
- 33.Shortle, D. 1983. A genetic system for analysis of staphylococcal nuclease. Gene 22:181-189. [DOI] [PubMed] [Google Scholar]
- 34.Smith, H. E., M. Damman, J. van der Velde, F. Wagenaar, H. J. Wisselink, N. Stockhofe-Zurwieden, and M. A. Smits. 1999. Identification and characterization of the cps locus of Streptococcus suis serotype 2: the capsule protects against phagocytosis and is an important virulence factor. Infect. Immun. 67:1750-1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smith, H. E., U. Vecht, A. L. Gielkens, and M. A. Smits. 1992. Cloning and nucleotide sequence of the gene encoding the 136-kilodalton surface protein (muramidase-released protein) of Streptococcus suis type 2. Infect. Immun. 60:2361-2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smith, H. E., U. Vecht, H. J. Wisselink, N. Stockhofe-Zurwieden, Y. Biermann, and M. A. Smits. 1996. Mutants of Streptococcus suis types 1 and 2 impaired in expression of muramidase-released protein and extracellular protein induce disease in newborn germfree pigs. Infect. Immun. 64:4409-4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Spellerberg, B., B. Pohl, G. Haase, S. Martin, J. Weber-Heynemann, and R. Lutticken. 1999. Identification of genetic determinants for the hemolytic activity of Streptococcus agalactiae by ISS1 transposition. J. Bacteriol. 181:3212-3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Staats, J. J., B. L. Plattner, G. C. Stewart, and M. M. Changappa. 1999. Presence of the Streptococcus suis suilysin gene and expression of MRP and EF correlates with high virulence in Streptococcus suis type 2 isolates. Vet. Microbiol. 70:201-211. [DOI] [PubMed] [Google Scholar]
- 39.Strangmann, E., H. Froleke, and K. P. Kohse. 2002. Septic shock caused by Streptococcus suis: case report and investigation of a risk group. Int. J. Hyg. Environ. Health 205:385-392. [DOI] [PubMed] [Google Scholar]
- 40.Tjalsma, H., A. Bolhuis, J. D. Jongbloed, S. Bron, and J. M. van Dijl. 2000. Signal peptide-dependent protein transport in Bacillus subtilis: a genome-based survey of the secretome. Microbiol. Mol. Biol. Rev. 64:515-547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vecht, U., H. J. Wisselink, M. L. Jellema, and H. E. Smith. 1991. Identification of two proteins associated with virulence of Streptococcus suis type 2. Infect. Immun. 59:3156-3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vecht, U., H. J. Wisselink, N. Stockhofe-Zurwieden, and H. E. Smith. 1996. Characterization of virulence of the Streptococcus suis serotype 2 reference strain Henrichsen S 735 in newborn gnotobiotic pigs. Vet. Microbiol. 51:125-136. [DOI] [PubMed] [Google Scholar]
- 43.Vecht, U., H. J. Wisselink, J. E. van Dijk, and H. E. Smith. 1992. Virulence of Streptococcus suis type 2 strains in newborn germfree pigs depends on phenotype. Infect. Immun. 60:550-556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Walsh, B., A. E. Williams, and J. Satsangi. 1992. Streptococcus suis type 2: pathogenesis and clinical disease. Rev. Med. Microbiol. 3:65-71. [Google Scholar]
- 45.Wannamaker, L. W. 1958. The differentiation of three distinct deoxyribonucleases of group A streptococci. J. Exp. Med. 107:797-812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wannamaker, L. W., B. Hayes, and W. Yasminch. 1967. Streptococcal nucleases. II. Characterisation of DNase D. J. Exp. Med. 126:497-508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wilson, A. T. 1945. Nucleic acid derivatives as growth factors for certain group A hemolytic streptococci. Proc. Soc. Exp. Biol. Med. 58:249-257. [Google Scholar]
- 48.Wisselink, H. J., H. E. Smith, N. Stockhofe-Zurwieden, K. Peperkamp, and U. Vecht. 2000. Distribution of capsular types and production of muramidase-released protein (MRP) and extracellular factor (EF) of Streptococcus suis strains isolated from diseased pigs in seven European countries. Vet. Microbiol. 74:237-248. [DOI] [PubMed] [Google Scholar]