Abstract
Human exposure to nanoparticles is inevitable from natural and anthropogenic sources. Titanium dioxide (TiO2) nanoparticles are increasingly being used in pharmaceutical and cosmetic products. Previous studies revealed that TiO2 levels were significantly increased in tissues (e.g., lymph nodes) after mice were injected with nanosized TiO2. To identify early response lymph node proteins to TiO2 nanoparticles, groups of mice were intradermally injected with a low dose of DeGussa P25 TiO2 nanoparticles or vehicle alone. The proteomes of lymph nodes at 24 h were quantitatively analyzed using trypsin-catalyzed 16O/18O labeling in conjunction with two-dimensional liquid chromatography separation and tandem mass spectrometry (2DLC-MS/MS). A total of 33 proteins were significantly changed (over 1.3-fold, p<0.05) in the mice treated with TiO2 nanoparticles, which accounted for approximately 1% of the total proteins identified. The differentially expressed proteins mainly involve the immune response (e.g., inflammation), lipid and fatty acid metabolism, mRNA processing, and nucleosome assembly. Regulation of functionally distinct classes of proteins could be mediated by estrogen receptor (ESR1), PPARγ, and c-Myc signalings, etc. The differentially expressed proteins identified in this experiment could represent early response proteins to TiO2 nanoparticle treatment in mouse lymph nodes.
Keywords: proteomics, mass spectrometry, TiO2 nanoparticle, mouse lymph node, 16O/18O labeling, LC-MS/MS
1. Introduction
Nanoparticles are typically defined as particles with at least one size domain between 1 and 100 nanometers. Engineered nanoparticles have emerged as a class of new materials for more than a decade. The unique characteristics of some materials in this size domain and various applications of nanomaterials are attractive for commercial development. Studies have indicated that nanomaterials are used in many daily consumed products such as beer and baby drinks despite the limited safety information that has been acquired (1). So far, more than 1300 nanotechnology-based consumer products, produced by ∼580 companies located in 30 countries have been reported (http://www.nanotechproject.org/inventories/consumer/). Several pharmaceutical companies already obtained approval from the U.S. Food and Drug Administration (FDA) for medical applications of nanotechnology-based drug-delivery agents, biosensors, and imaging contrast agents (2, 3). While the types of nanoparticles and applications continue to grow, concerns are mounting for the health risk of human exposure to nanomaterials (4-9). Therefore, in vivo studies to characterize biological responses to nanomaterial exposure are in growing need (10).
Among the commercially available nanomaterials, titanium dioxide (TiO2) nanoparticles are increasingly being used in personal care, paint, cosmetic products and food additives (http://www.nanotechproject.org/inventories/consumer/). Possible approaches of human intake of TiO2 nanoparticles include airborne exposure, inhalation, ingestion, skin uptake, and medical injection of engineered nanomaterials (11, 12). The uptake of nanomaterials in tissues will be dependent on the site of interaction of the nanomaterials with the organism. Intravenous injection of TiO2 nanoparticles in mice/rats resulted in elevated TiO2 levels in blood and solid tissues (13) with the highest TiO2 levels found in liver, followed by blood, spleen, lung, and kidney (13, 14) one day after the treatment. In contrast, intradermally injected nanoparticles (e.g., cadmium selenide quantum dots) were taken up by lymph nodes and translocated to the blood stream through lymphatic pathways (15, 16). Studies also indicated that lymphatic transport of nanoparticles (e.g., polypropylene sulfide) was size-dependent with more efficiency for smaller (25 nm) than larger (100 nm) particles (17). Skin penetration is also possible for some nanomaterials. Recent studies demonstrated that carboxylated quantum dots applied topically could penetrate the skin of SKH-1 mice (18), and PEG-coated quantum dots penetrated dermabraded mouse skin but not intact mouse skin (19). Recent studies have demonstrated that intact porcine skin is refractory to TiO2 penetration (20), even with repeated administration over a 4 weeks period (21); however, neither of these studies were able to rule out the possible penetration of TiO2 though damaged porcine skin and presentation to the dendritic or Langerhans cells of the skin.
The biological effects of TiO2 nanoparticle exposure and the mechanisms behind the response are not well understood. Field studies found airborne anatase TiO2 nanoparticles induced cytotoxicity response in human beings during the manufacturing process (22). Pulmonary toxicity of TiO2 nanoparticles has been examined using mouse models (23-26), and the lung inflammatory and cytotoxicity response were likely related to the particle size (24, 26), but some studies suggested no such relationships (23). To elucidate the mechanisms involved in TiO2 nanoparticle induced toxicity, a few studies have been reported. For example, in vivo studies indicated that TiO2 nanoparticles bound tightly to DNA in the TiO2 exposed mouse liver (27), could induce genotoxicity (28) and spleen injury (29). Exposure of lymphocytes to TiO2 nanoparticles significantly increased micronucleus formation and DNA breakage, elevation of p53 level, activation of DNA damage checkpoint kinases, and generation of reactive oxygen species (ROS) (30). An in vitro study also suggested that TiO2 nanoparticles could disrupt the function of proteins such as lysozyme activity inhibition (31) and Aβ fibrillation promotion by shortening the nucleation process (32). The interactions between nanoparticles and proteins (33-35) could be associated with nanoparticle size and surface properties. In addition, nonporous TiO2 nanoparticles could slow the kinetics of chemical messenger secretion without altering the number of molecules released from the mast cell granules (36). The prospective response of immune system to TiO2 nanoparticles are also under investigation (2, 37).
Omics techniques have been employed for the characterization of global molecular changes associated with nanoparticle exposure to biological systems. Microarray analysis of gene expression in zebrafish embryos exposed to TiO2 nanoparticles demonstrated that different sizes of TiO2 nanoparticles had varying effects on the expression of genes involved in the immune response, tumor necrosis factor, and endocytosis (38). Metabolomic analysis of urine samples from rats intragastrically administrated TiO2 nanoparticles suggested that they could disturb energy and amino acid metabolisms and the gut microflora environment (39). Using 2-dimensional differential gel electrophoresis (2D-DIGE) and MALDI-MS, Yang et al. identified 16 differentially expressed proteins as a result of SiO2 nanoparticle exposure to HaCaT cells (40). Nevertheless, characterization of in vivo biological effects of nanomaterials has not been fully explored and proteomic analysis of nanomaterial exposure is in its infancy. Information from global quantitative proteome analysis would enhance our understanding of nanomaterial-induced biological effects. In recent years, enzyme-catalyzed 18O labeling (41) combined with multidimensional LC separation and tandem mass spectrometry (LC-MS/MS) has been successfully developed and applied to global quantitative proteome analysis(42). In this study, the technique was applied to the measurement of proteome changes in mouse lymph nodes upon intradermal injection of commercial TiO2 nanoparticles. By comparing the lymph node proteomes of the TiO2 nanoparticle-treated mice with that of the control group, we found interesting information related to the biological consequence of TiO2 exposure to lymph nodes at the proteome level.
2. Materials and Methods
2.1. Chemicals and reagents
Ammonium bicarbonate (NH4HCO3), ammonium formate (NH4HCO2), Cremophor, guanidine hydrochloride (Gdn·HCl), iodoacetamide, and Tris were purchased from Sigma-Aldrich (St. Louis, MO). Formic acid and trifluoroacetic acid (TFA) were from Fluka (Milwaukee, WI). HPLC grade acetonitrile (CH3CN) and water were obtained from Fisher Scientific (Fair Lawn, NJ). Tris(2-carboxyethyl)phosphine hydrochloride (TCEP·HCl), Excellulose desalting columns (5K MWCO) and Bicinchonic acid (BCA) protein assay reagent kit were purchased from Pierce (Rockford, IL). Chemicon total protein extraction buffer containing protease inhibitors cocktail was from Millipore (Billerica, MA). Ethyl alcohol was obtained from Pharmco-AAPER (Hopkinsville, KY).
2.2. TiO2 nanoparticles
“Aeroxide” P25 TiO2 (Baker and Collinson, Inc., Detroit, MI) nanoparticles were chosen for this study. Quantitative elemental analysis by ICP/AES for titanium indicated a purity of greater than 99%. X-Ray diffraction confirmed that the titanium dioxide consisted of a mixture of 86% anatase and 14% rutile phases. Transmission electron microscopy found the minimum particle size was 14.2 nm and the maximum particle size was 64.6 nm. The arithmetic mean was 27.5 ± 9.8 nm. The morphology of these particles was generally round, but they were irregular in shape (that is, some sharp edges were present and some particles were elongated). These results agreed well with the manufacturer's reported value of 21 nm. Surface area measurements (BET analysis) indicate that the internal surface area attributed to pores is between 2.2 and 3.3 m2/g depending on whether the adsorption or desorption isotherm is used. The ideal surface area for smooth particles was calculated to be 50.5 m2/g compared to an actual measured value of 50.9 ± 0.2 m2/g by BET. This indicates that λ =1.01 or approximately 1% of the measured surface area is due to surface roughness. The determined BET value agreed with the manufacturer's reported value of 49 m2/g.
2.3. Animal Treatment
Isolator reared, Helicobacter-free female Crl: SKH-1 (hr/hr) hairless mice were obtained from Charles River (Portage, MI) at 5 weeks of age. The mice were housed for 2 weeks in the National Center for Toxicological Research (Jefferson, AR) Quarantine Facility and acclimated for an additional week prior to use. The treatment of the mice was approved by the Institutional Animal Care and Use Committee at this American Association for Laboratory Animal Science accredited facility. At 9 weeks of age, groups of six mice were weighed (average body weight of 23 g) and anesthetized intraperitoneally with sodium pentobarbital (25 mg/kg body weight). Anesthetized mice were injected intradermally in both the left and right dorsal flanks with either 5 μl of 1:1:8 ethyl alcohol:cremophor:water vehicle or suspensions of nanoscale TiO2 (1 mg/ml, DeGussa P25) in this vehicle using a Hamilton gas-tight syringe (Hamilton Company, Reno, NV) equipped with a 3/8-in., 26-gauge needle with a 30° (intradermal) bevel. The mice were euthanized using gaseous CO2 24 hrs post injection and both the right and left brachial, axillary and inguinal lymph nodes were collected and immediately frozen in liquid nitrogen at necropsy.
2.4. Lymph node proteome extraction and trypsin digestion
The right brachial, axillary and inguinal lymph nodes from each mouse were combined and lysed. After adding 100 µL of Chemicon total protein extraction buffer, the lymph nodes were ground using a 1 mL Dounce tissue grinder (Wheaton Science International, Millville, NJ). The resulting lysates were centrifuged at 11,000 × g for 20 min at 4 °C and the supernatants were collected. Protein concentrations were measured by the BCA protein assay. Equal amounts of protein from three TiO2 nanoparticle treated mice were combined, as was the protein from the 3 control mice. The extracted proteome samples from both the treatment and control groups were desalted using Excellulose desalting columns and lyophilized. Protein samples were re-dissolved with 6 M Gdn·HCl in 50 mM Tris-HCl, pH 8.3. The samples were reduced by adding TCEP·HCl to the final concentration of 10 mM and boiling in a water bath for 10 min. Proteins were further alkylated in 50 mM iodoacetamide with an incubation at 37 °C in dark for 2 hrs. After alkylation, each sample was buffer-exchanged to 25 mM NH4HCO3 (pH 8.3) through an Excellulose desalting column (5K MWCO, Pierce, Rockford, IL) that was pre-equilibrated with 25 mM NH4HCO3 (pH 8.3). The samples were digested with trypsin (Promega, Madison, WI) at 37 °C for 16 hrs using a protein to enzyme ratio of 50:1 (w/w). The tryptic peptides were further cleaned by Alltech Extract-Clean SPE C18 HC column (Grace, Deerfield, IL). Samples were lyophilized and stored at -80 °C.
2.5. Trypsin-catalyzed 16O/18O labeling
Peptide C-terminal 16O/18O labeling was performed as described previously (43). For 18O labeling, 50 µg of proteome tryptic peptides from the TiO2 nanoparticle treated mouse group were dissolved in 17% CH3CN in 18O-enriched water (97%, Sigma-Aldrich, St. Louis, MO). Sequencing grade modified trypsin (Promega, Madison, WI) dissolved in 18O-water was added to the samples at a ratio of 30:1 (w/w, protein-to-trypsin), and the mixture was incubated at 37 °C for 16 hrs. The reactions were quenched by boiling the samples for 10 min in a water bath and then cooling down to room temperature, followed by addition of 0.2% TFA. The sample was immediately lyophilized to dryness. The identical procedure was carried out in parallel for 16O labeling of the same amounts of peptides from the control group in which the regular 16O-water was used instead of 18O-water. In addition, reverse labeling was performed as well in which the sample from TiO2 nanoparticle treated mouse group was labeled using 16O-water while the control sample was labeled using 18O-water. The 16O- and 18O-labeled samples stored separately at -80 °C for further analysis.
2.6. Strong cation exchange liquid chromatographic (SCXLC) fractionation
Prior to SCXLC fractionation, each pair of 16O-labeled and 18O-labeled samples were dissolved in 25% CH3CN/0.1% TFA and combined (i.e., 16O-control/18O-treatment pair or 18O-control/16O-treatment pair). A Dionex UltiMate 3000 Nano and Cap LC system (Dionex Softron GmbH, Germering, Germany) was used to deliver mobile phase A (25% CH3CN in water) and mobile phase B (25% CH3CN/0.5 M NH4HCO2, pH 3.0). The 16O/18O-labeled peptides were loaded onto a 1 mm × 150 mm Polysulfoethyl A column (PolyLC Inc., Columbia, MD) and eluted at a flow rate of 50 μL/min, using the following NH4HCO2/CH3CN multistep gradient: 3% mobile phase B for 5 min, followed a linear increase to 10% B for 18 min, a linear increase to 45% B for 26 min, then a linear increase to 100% B for 1 min, and maintained at 100% B for 10 min. The separation was monitored using a laser induced fluorescence detector equipped with 266 nm diode pumped solid state pulsed laser (ZETALIF Discovery, Picometrics, Toulouse, France) to detect native fluorescence at an emission wavelength of 340 nm. Thirty one fractions were collected at two-minute intervals. The fractions were lyophilized and stored at -80 °C for LC-MS/MS analysis
2.7. Nanoflow reversed-phase LC-MS/MS analysis
Nanoflow RPLC separation of peptides was conducted using a 9 cm long × 75 µm inner diameter (i.d.) fused silica capillary electrospray ionization (ESI) column which was coupled online to an Orbitrap mass spectrometer (LTQ-Orbitrap XL, Thermo Electron, San Jose, CA) for MS/MS analysis of each SCXLC fraction. The ESI column was slurry packed with 5 μm, 300 Å pore size Jupiter C18 RP particles (Phenomenex, Torrence, CA) against a 9 cm × 75 µm i.d. fused-silica capillary (Polymicro Technologies, Phoenix, AZ) with a flame-pulled fine i.d. (i.e., 5-7 μm) tip. Mobile phases A (0.1% formic acid in water) and B (0.1% formic acid in CH3CN) were delivered by a Dionex UltiMate 3000 Nano and Cap LC system (Dionex Softron GmbH, Germering, Germany). Peptides were loaded in 30 min while the column was maintained with 2% solvent B at a flow rate of 1 µL/min, and then separated using a step gradient of 2%-42% solvent B for 65 min and 42%-98% solvent B for 15 min at a flow rate of ∼250 nL/min. Following the MS survey scan with a resolution of 6×104 and a mass range of m/z 300-1800 in the Orbitrap analyzer, data-dependent MS/MS scans were acquired in the linear ion trap analyzer in which the 7 most intense peptide molecular ions in the MS scan were sequentially and dynamically selected for subsequent collision-induced dissociation (CID) using a normalized collision energy of 35%. Dynamic exclusion was enabled with duration of 1 min to prevent repeated acquisition of MS/MS spectra of the same peptide for which the MS/MS spectrum had been acquired in the previous scan. Electrospray voltage was set at 1.6 kV, and the voltage and temperature for the ion source capillary were 47 V and 160 °C, respectively
2.8. Peptide identification
The raw MS/MS data were searched using the SEQUEST cluster running under BioWorks (Rev. 3.3.1 SP1) (Thermo Electron, San Jose, CA) against a mouse IPI proteome database (version 3.78, containing 54,928 protein sequence entries) downloaded from the European Bioinformatics Institute (EBI) (http://www.ebi.ac.uk). Reversed protein sequences of all the protein entries were added to the same database for an estimation of false identification rate. Peptide mass tolerance of 10 ppm and fragment ion tolerance of 1 Da were set with tryptic specificity allowing two missed cleavages. SEQUEST criteria were Xcorr ≥ 1.7 for [M+H]1+ ions, ≥ 2.5 for [M+2H]2+ ions and ≥ 3.2 for [M+3H]3+ ions, and P ≤ 0.01 for identification of fully tryptic peptides. A dynamic 4.0085 Da modification on the C-terminus was also set in a single search to identify both 18O-labeled peptides and peptides with normal C-terminus. In addition, dynamic oxidation of Met by the addition of one oxygen (+15.9949 Da) and Cys carboxyamidomethylation (+57.0215 Da) were included. These criteria were applied to filter the peptide identifications from both forward and reversed protein sequences.
2.9. Quantitation, normalization and statistical analysis
The identified peptides were quantified using the BioWorks' PepQuan module (Thermo Electron, San Jose, CA), which calculated the relative abundance (e.g., ratios of 18O/16O, H/L) of peptides based on the areas of their extracted ion chromatograms (XIC) using a minimum intensity threshold of 100 counts, mass tolerance of 0.03 Da and smoothing point of 5. After logarithmic transformation, the abundance ratio distribution of all the peptides in each dataset was plotted to fit with normal distribution and nonlinear regression was performed according to the method described previously (44). The mean ratio was used to normalize the abundance ratios of the dataset. When multiple peptides were identified from the same protein, an average ratio was calculated. Student's t-test was performed for the proteins in each quantitative dataset and p value was calculated.
2.10. Pathway and network analysis
To investigate the pathways and networks involving the lymph node proteins differentially expressed between the control and TiO2 nanoparticle treated mice, the MetaCore (GeneGo, St. Joseph, MI) program was used to build protein interaction networks. MetaCore is an integrated software suite for functional analysis of experimental data and it contains curated protein interaction networks on the basis of manually curated database of human, mouse, rat protein-protein, protein-DNA, protein-RNA and protein-compound interactions. The significantly changed proteins from our experiments and the proteins from the MetaCore database were used to generate networks using the shortest paths algorithm (maximum 2 steps in the path) and pre-filters as lymphocyte and mouse (M. musculus).
3. Results
3.1. Forward and reverse trypsin-catalyzed 16O/18O labeling strategy for confident quantitation
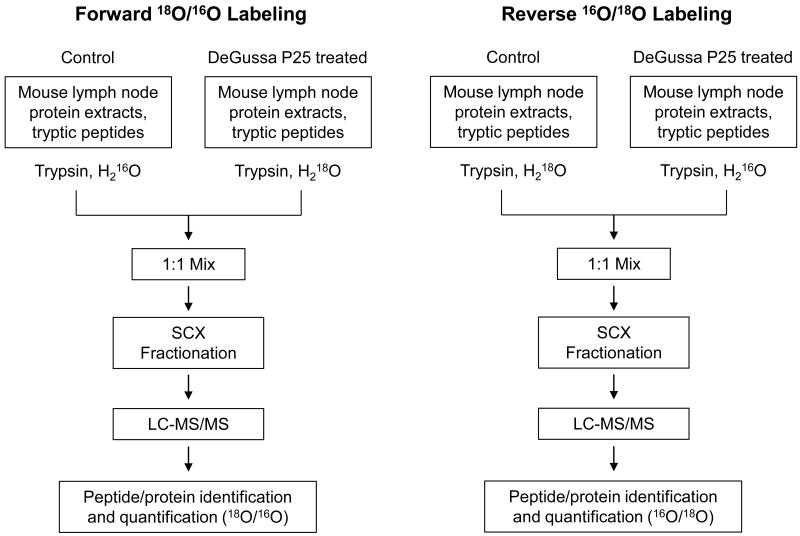
Enzyme-catalyzed oxygen exchange has been used for stable isotope labeling in quantitative proteomics. The labeling is performed typically by incubation of peptides with trypsin in the presence of 18O-coded water (45). The two oxygen atoms in the C-terminal carboxylate group of a peptide are exchanged with two 18O atoms from the water, resulting in an addition of 4.0085 Da to the peptide. This labeling technique has been applied to many quantitative proteomic studies for relative quantitation of protein expression. However, oxygen back-exchange during sample processing and analysis and/or incomplete labeling especially for proteome samples have been reported (46). The labeling technique has been improved to achieve complete labeling and prevent oxygen back-exchange (42, 47, 48). These improvements include decoupling of 18O labeling from the protein digestion step (42, 47), 18O labeling performed in a buffer containing 20% methanol which enhances trypsin activity (42), and inhibition of post-labeling trypsin activity by thermal deactivation to prevent oxygen back-exchange in 18O-labeled samples (47, 48). To overcome the described potential issues and eliminate protein quantitation errors, we carried out both forward and reverse 16O/18O labeling in parallel for the same pair of control and TiO2 nanoparticle treated mouse lymph node proteomes. The quantitative proteomic approach employing this labeling strategy is illustrated in Figure 1. The protein samples of lymph nodes from the control and TiO2 nanoparticle treated mice were firstly digested with trypsin and desalted. Fifty micrograms of peptides from the TiO2 nanoparticle treated mouse lymph nodes were labeled with 18O and mixed in equal amounts with 16O-labeled control sample to form the forward labeling mixture as defined here. In reverse labeling, the control sample was 18O-labeled while the treated sample was 16O-labeled. The resulting two mixtures were individually separated using SCX liquid chromatography into 31 fractions, and each SCX fraction was analyzed by nanoflow RPLC-MS/MS.
Figure 1.
Schematic flowcharts of forward and reverse trypsin-catalyzed 16O/18O labeling combined with SCX fractionation and LC-MS/MS for comparative analysis of proteome changes in lymph nodes from DeGussa P25 TiO2 nanoparticle-treated vs control mice.
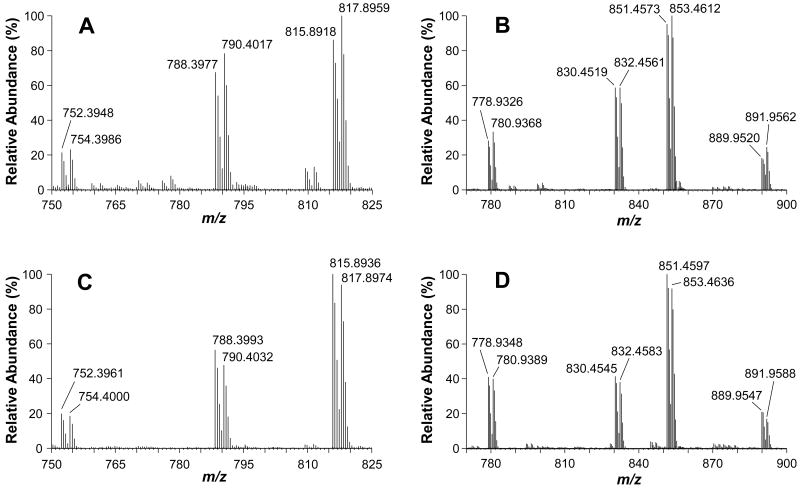
Careful examination of MS data from both forward and reverse labeled samples suggest that 18O labeling for a vast majority of peptides was generally complete and no obvious oxygen back-exchange was observed during the experiment. Although a systematic evaluation has not been done on the factors that might contribute to oxygen back-exchange, the samples were handled in a way to minimize if not eliminate this problem. Even when samples were kept at 5 °C for 2 days in the HPLC sample tray, there was no obvious oxygen back-exchange observed (data not shown). An example illustrating 18O labeling efficiencies in both forward and reverse approaches is shown in Figure 2. Three and four pairs of relatively abundant MS peaks with LC elution time of approximately 25 min and 38 min, respectively, were detected in the defined mass ranges from a fraction of forward 18O-labeled sample as shown in Figure 2A and 2B. The corresponding peaks of the same peptides from the reverse labeling are shown in Figure 2C and 2D, respectively. For these peptide pairs, the abundance ratios of 16O/18O are generally 1:1, showing no significant changes in expression. A careful comparison of the 18O-labeled (heavy) peptides (the TiO2 nanoparticle treatment) at m/z 754.3986, 790.4017 and 817.8959 with their 16O-countparts (light, the control) reveals that these 18O-peptides are 8.4%, 15.5% and 15.7% higher in abundance, respectively (Figure 2A). In the reverse labeling, the same peptides from the TiO2 nanoparticle treated sample are 16O-labeled. As shown in Figure 2C, the 16O-labeled peptides at m/z 752.3961, 788.3993 and 815.8936 are 7.2%, 18.3%, 6.6% more abundant than the 18O-countparts. Similar results are observed for the peaks in Figure 2B and 2D.
Figure 2.
MS spectra showing complete trypin-catalyzed 18O labeling. Three and four major pairs of peptides at LC elution time of approximately 25 min (A) and 38 min (B), respectively, were detected in the defined mass ranges from a fraction of forward 18O-labeled sample. The corresponding peaks of the same peptides from the reverse labeling are shown in C and D, respectively.
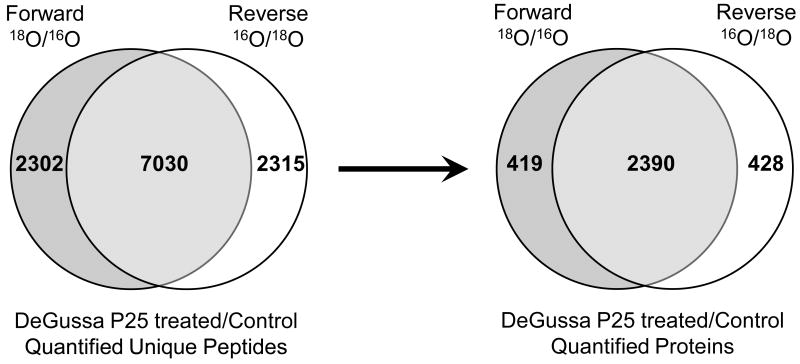
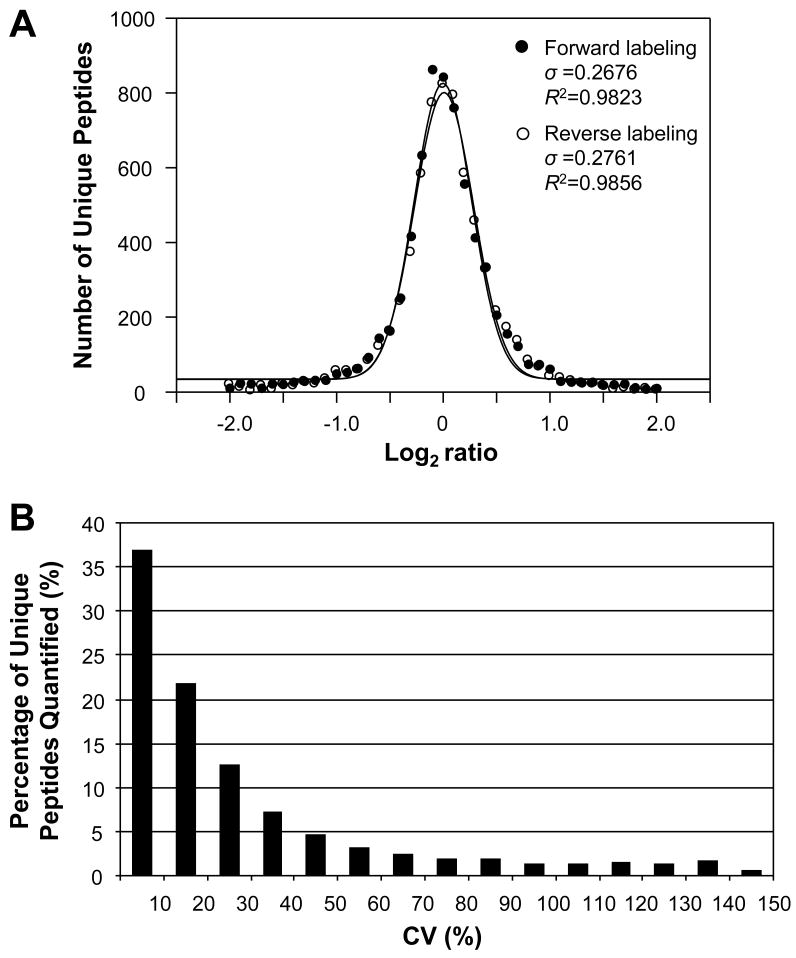
To examine the overall efficiency of forward and reverse 18O labeling and compare the quantitation variance at the whole proteome level, statistical distribution was plotted for the 7030 common unique peptides (Figure 4) identified and quantified from both forward and reverse 18O labeling experiments. A normal distribution was obtained in terms of the number of unique peptides plotted within binned log2 ratio for both labeling approaches (Figure 3A). These distributions were quite similar, indicating 18O labeling was highly reproducible. Nonlinear regression analyses indicated that both datasets had similar coefficients of determination (R2=0.9823 for forward labeling, and R2=0.9856 for reverse labeling). The standard deviation (σ) of the two datasets was close as well with the values of 0.2676 for forward labeling and 0.2761 for reverse labeling. The standard deviation values were back-calculated to an approximate ratio of 1.2 for both labeling approaches. It should be noted that the real abundance change in the peptide ratio datasets were implied in the calculated abundance ratio, the actual standard deviation owing to incomplete 18O labeling would be less. The above analyses indicate that a vast majority of the peptides have a ratio of or close to 1:1, as illustrated in Figure 3A as well. The results from this study are similar to or better than some previously published large-scale 18O labeling datasets (49).
Figure 4.
Venn diagrams showing the number of unique peptides (left) and proteins (right) as well as their overlaps identified from forward and reverse trypsin-catalyzed 16O/18O labeling analyses.
Figure 3.
A) Normal distribution of the 7030 common unique peptides quantified from both forward and reverse 18O labeling experiments within binned base 2 logarithms of peptide abundance ratios. An average ratio was calculated if a peptide was identified and quantified multiple times. B) Plot of relative number of peptides (%) versus binned coefficient of variation (CV) of abundance ratios for the 7030 common unique peptides quantified from both forward and reverse 18O labeling experiments. The CV of a peptide was calculated for its ratios obtained from forward and reverse 18O labeling analyses, indicating the ratio variance between forward and reverse labeling.
Since forward and reverse 18O labeling approaches were employed in this study, another way to evaluate the overall labeling efficiency is to calculate the quantitation variance between forward and reverse labeling analyses for the same peptides. Theoretically, the coefficient of variation (CV, %) of the abundance ratios of the same peptide from forward and reverse 18O labeling should be zero if the labeling is complete, i.e., the peptide has exactly the same ratio. Calculation of CV for the 7030 common unique peptides (Figure 4) quantified from both forward and reverse 18O labeling experiments revealed that approximately 37%, 22%, and 13% of the peptides had CVs of <10%, 10-20%, and 20-30%, respectively (Figure 3B). Therefore, a total of 72% of the peptides had CV values less than 30%. Nonetheless, it should be pointed out that the actual number could be more than 72% since the ratio variance between forward and reverse labeling was originated not only from incomplete labeling but also from other factors such as errors in peptide peak integration, etc. Taken together, these data suggest that the overall 18O labeling efficiency in this study is high at the proteome level and consistent quantitation can be achieved between forward and reverse 18O labeling. For a limited number of peptides with incomplete labeling, the abundance ratios of the same peptide from forward and reverse labeling are inconsistent. By comparing the quantitative data from forward and reverse labeling approaches, quantitation error resulted from incomplete labeling can be eliminated, thus confident results can be obtained.
3.2. Quantitative proteomic analysis of mouse lymph nodes upon DeGussa P25 TiO2 nanoparticle treatment
To identify protein expression alterations in mouse lymph nodes associated with DeGussa P25 TiO2 nanoparticle treatment, both forward and reverse trypsin-catalyzed 18O-labelings were employed as described previously to achieve reliable and confident protein quantitation. A total of 25,069 and 23,144 peptides, corresponding to 9,332 and 9,345 unique peptide sequences regardless of 16O/18O labeling, were identified and quantified from forward and reverse labeling experiments respectively using the Xcorr criteria defined in the methods and p≤0.01 (Figure 4). The false discovery rate (FDR) was estimated to be <0.1% at the peptide level for these peptide datasets as a result of a reversed protein sequence database search. Approximately 75% of these unique peptides overlapped between forward and reverse labeling experiments. From these indentified peptides, a total of 2,809 and 2,818 proteins were identified and quantified from the forward and reverse labeling, respectively, with 2,390 proteins identified commonly (i.e., ∼85% overlap) from these two labeling approaches (Figure 4).
An evaluation of quantitation errors and statistic analysis was performed for the identification of reliable protein expression changes as a result of TiO2 nanoparticle treatment. The CV (%) of the abundance ratio was calculated for each protein with multiple identifications. The average CV of all the proteins with multiple IDs within each of the two labeling experiments was approximately 30%. In addition, Student's t-test was performed for each protein with multiple peptide identifications to examine whether the protein abundance change as a result of TiO2 nanoparticle treatment was statistically significant or not as compared to the control. By using p<0.05 from this t-test, proteins with single peptide identifications or with high quantitative standard deviation were excluded from further consideration. Based on the CV data and application of the t-test, we set 1.3-fold protein abundance changes (the abundance ratio of treatment to control was ≥1.3 or ≤0.77) with statistical significance p<0.05 to define differentially expressed proteins, which must also be consistently observed from both forward and reverse labeling experiments. As a result, 19 lymph node proteins were up-regulated and 14 were down-regulated in the mice treated with TiO2 nanoparticles compared to control animals. The quantitative data along with subcellular localization of these significantly changed proteins are summarized in Table 1. The detailed peptide identification information such as individual peptide fold change, Xcorr, DeltaCn and so forth were listed in Supplementary Table 1.
Table 1.
Proteins showing statistically significant change in their expression (≥1.3-fold change and p<0.05) in mouse lymph nodes at 24 hours after DeGussa P25 TiO2 nanoparticles treatment as revealed from trypsin-catalyzed 16O/18O labeling analysis.
| Forward 18O/16O Label (P25/Control) |
Reverse 16O/18O Label (P25/Control) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Accession | Gene | Description | Subcellular Localization |
Total Peptide Count |
Fold Change |
p value | Total Peptide Count |
Fold Change |
p value | Biological Function |
| IPI00118173.1 | Aoc3 | Isoform 1 of Membrane primary amine oxidase | Plasma Membrane | 17 | 1.38 | 4.35E-05 | 11 | 1.63 | 1.29E-03 | Amine metabolism, cell adhesion, inflammatory response |
| IPI00122475.2 | Camp | Cathelin-related antimicrobial peptide (LL37) | Secreted | 10 | -2.43 | 1.99E-20 | 8 | -2.84 | 3.32E-12 | Antimicrobial activity |
| IPI00317340.1 | Ltf | Lactotransferrin | Secreted | 4 | -3.50 | 8.29E-03 | 3 | -2.81 | 3.35E-02 | Antimicrobial activity, iron transport |
| IPI00230768.5 | S100a8 | Protein S100-A8 (Calgranulin A) | Cytoplasm | 4 | -2.38 | 4.38E-05 | 6 | -5.46 | 4.29E-09 | Immune response, antimicrobial, chemotaxis |
| IPI00222556.5 | S100a9 | Protein S100-A9 (Calgranulin B) | Cytoplasm | 2 | -4.01 | 4.96E-02 | 2 | -4.06 | 1.07E-02 | Inflammation, antimicrobial, chemotaxis |
| IPI00157508.2 | Chi3l3 | Chitinase-3-like protein 3 | Secreted | 9 | -2.55 | 8.38E-07 | 15 | -1.67 | 7.98E-10 | Inflammatory response |
|
| ||||||||||
| IPI00223783.2 | Plin1 | Isoform 1 of Perilipin-1 | Cytoplasm | 12 | 1.70 | 2.52E-03 | 9 | 1.71 | 5.33E-03 | Lipid metabolism |
| IPI00132874.4 | Mgll | Monoglyceride lipase | Plasma Membrane | 5 | 1.35 | 5.34E-08 | 12 | 1.43 | 2.93E-06 | Lipid metabolism, fatty acid biosynthesis |
| IPI00454049.4 | Echs1 | Enoyl-CoA hydratase, mitochondrial | Cytoplasm | 11 | 1.38 | 5.88E-05 | 9 | 1.38 | 1.67E-03 | Lipid metabolism, fatty acid metabolism |
| IPI00134961.1 | Acadm | Medium-chain specific acyl-CoA dehydrogenase, mitochondrial | Cytoplasm | 8 | 1.55 | 1.76E-05 | 10 | 1.67 | 3.93E-15 | Lipid metabolism, fatty acid metabolism |
| IPI00126248.3 | Acly | ATP-citrate synthase, mitochondrial | Cytoplasm | 72 | 1.38 | 6.34E-13 | 57 | 1.39 | 1.72E-11 | Lipid synthesis, carbohydrate metabolism |
| IPI00421241.5 | Acacb | Acetyl-Coenzyme A carboxylase beta | Cytoplasm | 8 | 1.60 | 1.23E-08 | 8 | 1.59 | 2.36E-03 | Lipid synthesis, fatty acid biosynthesis |
| IPI00114710.3 | Pcx | Pyruvate carboxylase, mitochondrial (PYC) | Cytoplasm | 53 | 1.57 | 2.03E-15 | 36 | 1.53 | 6.70E-18 | Lipid synthesis, gluconeogenesis |
| IPI00116705.5 | Fabp4 | Fatty acid-binding protein, adipocyte (A-FABP) | Cytoplasm | 37 | 1.42 | 3.56E-10 | 24 | 1.32 | 3.29E-02 | Lipid transport |
|
| ||||||||||
| IPI00421085.1 | Cpsf6 | Cleavage and polyadenylation specificity factor subunit 6 | Nucleus | 9 | -1.59 | 4.30E-03 | 13 | -1.38 | 2.22E-02 | mRNA processing |
| IPI00122418.1 | Luc7l3 | Isoform 2 of Luc7-like protein 3 | Nucleus | 3 | -1.31 | 3.02E-04 | 4 | -1.41 | 6.52E-05 | mRNA processing, apoptosis, stress response |
| IPI00129430.1 | Sfpq (PSF) | Splicing factor, proline- and glutamine-rich | Nucleus | 26 | -1.79 | 1.80E-12 | 33 | -1.98 | 1.30E-09 | mRNA processing, DNA repair, transcription regulation |
| IPI00129323.1 | Srsf3 (Sfrs3) | Isoform Long of Serine/arginine-rich splicing factor 3 | Nucleus | 4 | -2.90 | 4.46E-02 | 9 | -1.71 | 6.17E-06 | mRNA processing, mRNA splicing |
| IPI00113746.3 | U2af2 | Splicing factor U2AF 65 kDa subunit (U2AF-65) | Nucleus | 9 | -1.67 | 1.27E-05 | 7 | -1.70 | 1.13E-08 | mRNA processing, mRNA splicing |
| IPI00121135.5 | Srsf2 | Serine/arginine-rich splicing factor 2 | Nucleus | 15 | -1.47 | 5.61E-10 | 25 | -1.40 | 7.91E-36 | mRNA processing, mRNA splicing |
| IPI00468896.7 | Ddx46 | Isoform 1 of Probable ATP-dependent RNA helicase DDX46 | Nucleus | 12 | -1.38 | 1.57E-07 | 15 | -1.81 | 1.74E-10 | mRNA processing, mRNA splicing |
|
| ||||||||||
| IPI00329998.3 | Hist1h4a | Histone H4 | Nucleus | 52 | 1.46 | 2.03E-32 | 38 | 1.62 | 2.73E-14 | Nucleosome assembly |
| IPI00114642.4 | Hist1h2bn; Hist1h2bj;Hi st1h2bl;Hist 1h2bf | Histone H2B type 1-F/J/L | Nucleus | 18 | 1.33 | 6.23E-09 | 22 | 1.43 | 2.51E-16 | Nucleosome assembly |
| IPI00153400.2 | H2afj | Histone H2A.J | Nucleus | 12 | 1.54 | 1.63E-10 | 9 | 1.62 | 3.40E-02 | Nucleosome assembly |
| IPI00223713.5 | Hist1h1c | Histone H1.2 | Nucleus | 14 | 1.73 | 2.90E-09 | 12 | 2.29 | 1.85E-04 | Nucleosome assembly, nucleosome positioning |
|
| ||||||||||
| IPI00474959.2 | Rcn2 | Reticulocalbin 2 | Cytoplasm | 2 | -2.53 | 6.57E-04 | 2 | -1.66 | 2.55E-04 | Calcium ion binding |
| IPI00113886.1 | Lmnb2 | Isoform B3 of Lamin-B2 | Nucleus | 9 | 1.48 | 3.55E-04 | 4 | 1.55 | 5.68E-04 | Component of nuclear membrane, structural molecule activity |
| IPI00420261.5 | Hmgb1 (Hmg1) | High mobility group protein B1 (Amphoterin) | Nucleus | 34 | 1.35 | 1.67E-10 | 36 | 1.41 | 8.49E-10 | DNA repair, DNA ligation, DNA recombination, apoptosis |
| IPI00109044.8 | 2900073G1 5Rik | Myosin light chain, regulatory B-like | Cytoplasm | 20 | 1.45 | 5.90E-15 | 14 | 1.43 | 4.37E-09 | Motor activity |
| IPI00114894.1 | Myh11 | Isoform 1 of Myosin-11 | Cytoplasm | 22 | 1.46 | 2.15E-05 | 15 | 2.16 | 2.14E-03 | Motor activity |
| IPI00113659.4 | Ifi30 | Interferon gamma inducible protein 30 (IP-30) | Cytoplasm | 4 | -1.30 | 4.62E-03 | 9 | -1.32 | 3.17E-05 | Oxidoreductase activity |
| IPI00352984.4 | Xdh | Xanthine dehydrogenase/oxidase | Cytoplasm | 15 | 1.48 | 4.32E-07 | 11 | 1.45 | 3.16E-08 | Oxidoreductase activity |
| IPI00116222.1 | Hibadh | 3-hydroxyisobutyrate dehydrogenase, mitochondrial | Cytoplasm | 18 | 1.46 | 1.92E-03 | 13 | 1.38 | 4.09E-09 | Valine metabolism, pentose-phosphate shunt |
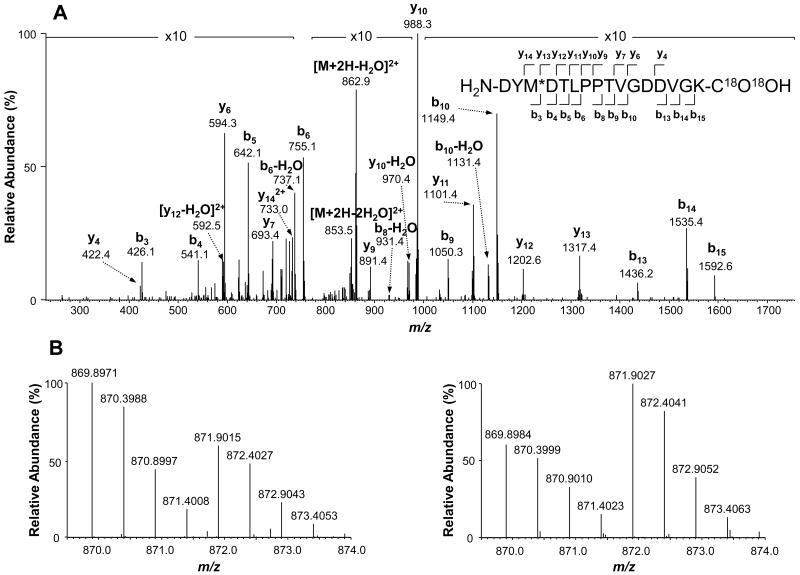
For the peptides from the 33 differentially expressed proteins, almost half of them (49%) are methionine (Met)-containing peptides or the peptides with missed cleavage sites. Jorge et al. discussed that peptides containing missed cleavage sites or oxidized Met residues do not reliably reflect correct protein concentrations (49); however, Bonzon-Kulichenko et al. argued that partial digestions and Met oxidation do not affect protein quantification and that variances at the scan, peptide, and protein levels are stable and reproducible(50). The results from this experiment are in agreement with Bonzon-Kulichenko's observation in that a vast majority of the peptides containing Met residues or from partial digestion had consistent fold changes between the forward and reverse 18O labeling experiments and the abundance changes of these peptides were in agreement with other regular peptides within the same protein. Approximately 4% of Met-containing or partially digested peptides had inconsistent fold changes between the forward and reverse 18O labeling analyses. The results from this study along with those from other laboratories suggest that quantification reliability of Met-containing or partially digested peptides depends on the experimental procedures and conditions used for sample preparation.
Among the differentially expressed proteins, most of them are cytoplasmic and nuclear proteins while there are only three secreted and two plasma membrane proteins. Biological function annotation indicates that most altered proteins are involved in immune response (e.g., inflammation) and antimicrobial activity, lipid and fatty acid metabolism, mRNA processing, and nucleosome assembly. Proteins associated with immune response and antimicrobial activity include protein S100-A8, S100-A9, chitinase-3-like protein 3, amine oxidase, cathelin-related antimicrobial peptide, and lactotransferrin. These proteins were down-regulated upon TiO2 nanoparticle treatment except for amine oxidase. Another class of down-regulated proteins are related to mRNA processing (e.g., splicing), encompassing cleavage and polyadenylation specificity factor subunit 6 (CPSF6), Luc7-like protein 3, proline- and glutamine-rich splicing factor, serine/arginine-rich splicing factors 2 and 3, splicing factor U2AF 65 kDa subunit, and probable ATP-dependent RNA helicase DDX46. As an example, Figure 5A shows an MS/MS spectrum of the 18O-labeled peptide DYM*DTLPPTVGDDVGK from protein CPSF6. The MS spectra of 16O/18O-labled peptide pairs indicate that the abundance ratios are consistent between the forward (18O/16O=0.63) and reverse (16O/18O=0.70) labeling (Figure 5B), suggesting protein CPSF6 was down-regulated in the mouse lymph nodes due to DeGussa P25 TiO2 nanoparticle treatment. In contrast, proteins associated with lipid and fatty acid metabolism were up-regulated, which include perilipin-1, monoglyceride lipase, enoyl-CoA hydratase, medium-chain specific acyl-CoA dehydrogenase, ATP-citrate synthase, acetyl-Coenzyme A carboxylase beta, pyruvate carboxylase, and adipocyte fatty acid-binding protein. Four of them are mitochondrial proteins. In addition, histone H1, H2A, H2B and H4, which are responsible for nucleosome assembly, were up-regulated.
Figure 5.
Identification and quantitative comparison of the peptide DYM*DTLPPTVGDDVGK from cleavage and polyadenylation specificity factor subunit 6 (CPSF6). (A) MS/MS spectrum of the 18O-labeled version of this peptide. (B) MS spectra of the doubly charged peptide pairs showing the relative abundance of this peptide as a result of forward (left) and reverse (right) 18O-labeling. Consistent abundance ratios between the forward (18O/16O=0.63) and reverse (16O/18O=0.70) labeling indicate down-regulation of CPSF6 in the mouse lymph nodes due to DeGussa P25 TiO2 nanoparticle treatment. * indicates the oxidized methionine residue in the peptide.
3.3. Pathway and network analysis of differentially expressed proteins
The 33 differentially expressed proteins (Table 1) were subjected to molecular network analysis by MetaCore program (GeneGo, Inc, St Joseph, MI) using the shortest paths algorithm (maximum 2 steps in the path for interaction) and pre-filters as lymphocyte and M musculus. Figure 6 shows the most significant network resolved, which includes 15 differentially expressed proteins and the 10 proteins in the MetaCore database. One of the main hubs in the network is the ESR1 (estrogen receptor) whose transcriptional activity is regulated by multiple signaling pathways (thick cyan lines) (51). Lactoferrin is the only differentially expressed protein interacting directly with ESR1. Another hub is c-Myc, which directly interacts with 5 differentially expressed proteins (ACADM, HMGB1, IP-30, PSF, and PYC). Interestingly, PPARγ is a hub that mainly connects up-regulated proteins (A-FABP, Perilipin, and PYC) involved in lipid metabolism except for SFRS3. It has been reported that U2AF-65 physically interacts with CPSF6 and increases its activity (52). Our results indicated that both U2AF-65 and CPSF6 were down-regulated after TiO2 nanoparticle treatment. It is also worth noting that calgranulin A and B interact with each other and increase their activities in normal circumstances (53), but this study showed that they were down-regulated. An analysis of this network revealed that the top five biological processes involved by those differentially expressed proteins are cellular responses to stimulus, stress, organic substance, multi-organism process and chemical stimulus (Table 2).
Figure 6.
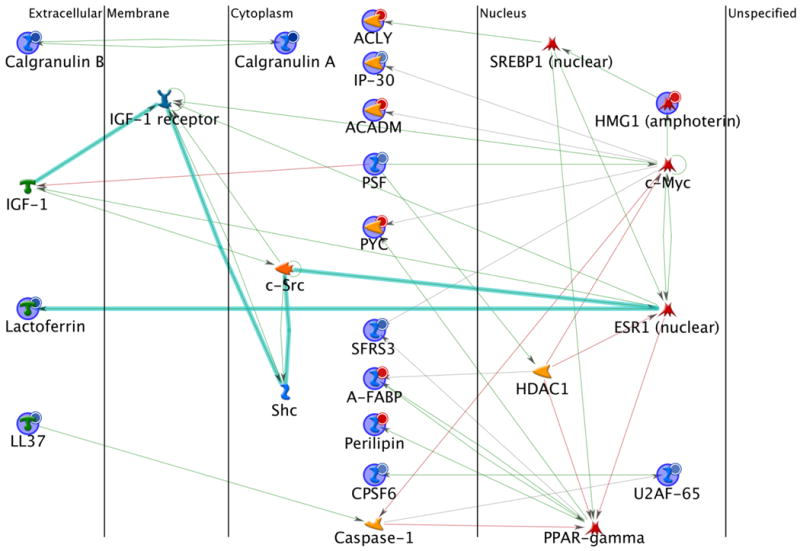
Protein interaction network of differentially expressed lymph node proteins between the DeGussa P25 TiO2 nanoparticles treated and the control mice. Network analysis was performed using the MetaCore program via the shortest paths algorithm for all the 33 differentially expressed proteins. As a result, 15 proteins were included in this map. The lines between the gene (protein) symbols show different effects between up- and down-streams: inhibition (pink lines), activation (green lines), unspecified (grey lines) and fragments of canonical pathways (thick cyan lines). Up-regulated proteins are marked with red circles and down-regulated with blue circles.
Table 2.
Top five Gene Ontology (GO) processes involved by the differentially expressed proteins. GO processes were ranked by the number of genes linked to the corresponding GO process.
| Gene Ontology (GO) Process | Number of Genes | Gene | p value |
|---|---|---|---|
| Response to stimulus | 20 | S100a8, S100a9, Ltf, Camp, Chi3l3, Xdh, Acacb, Aoc3, Mgll, Ifi30, Acadm, Sfpq, Pcx, Srsf3, Fabp4, Plin1, Luc7l3, Hmgb1, U2af2, Hist1h2bn/Hist1h2bj/Hist1h2bl/Hist1h2bf | 9.21E-07 |
| Response to stress | 17 | S100a8, S100a9, Ltf, Camp, Chi3l3, Aoc3, Mgll, Acadm, Sfpq, Pcx, Srsf3, Fabp4, Plin1, Luc7l3, Hmgb1, U2af2, Hist1h2bn/Hist1h2bj/Hist1h2bl/Hist1h2bf | 5.94E-08 |
| Response to organic substance | 14 | S100a8, S100a9, Ltf, Camp, Acacb, Ifi30, Acadm, Sfpq, Pcx, Srsf3, Fabp4, Plin1, Hmgb1, U2af2 | 3.26E-07 |
| Multi-organism process | 13 | S100a8, S100a9, Ltf, Camp, Ifi30, Acadm, Sfpq, Pcx, Srsf3, Fabp4, Hmgb1, U2af2, Hist1h2bn/Hist1h2bj/Hist1h2bl/Hist1h2bf | 4.66E-07 |
| Cellular response to chemical stimulus | 12 | S100a8, S100a9, Ltf, Camp, Ifi30, Acadm, Sfpq, Pcx, Srsf3, Fabp4, Plin1, Hmgb1 | 5.52E-07 |
4. Discussion
In this study, 2390 proteins were commonly identified and quantified from forward and reverse trypsin-catalyzed 16O/18O labelings of the proteins extracted from the control and TiO2 nanoparticle-treated mouse lymph nodes, using combined SCX fractionation and nanoflow LC-MS/MS experiments. Compared to early studies employing two-dimensional gel electrophoresis (2-DE) combined with mass spectrometry (54-58), protein coverage for mouse lymph nodes was significantly increased in this study.
The TiO2 nanoparticle employed here is DeGussa P25. The average particle size is ∼28 nm and the specific surface area is ∼51 m2/g. Early studies showed that nanoparticles traffic to the draining lymph nodes in a size-dependent manner, where micron-sized particles required dendritic cells to transport them from the injection site to lymph nodes. In contrast, small nanoparticles (20-200 nm) and virus-like particles (30 nm) were transported to lymph nodes through free drainage, and it would take two or more hours for nanoparticles of 30 nm or less to travel to lymph nodes (59). The migrated nanoparticles could remain in the lymph nodes for days. A recent study of organic nanoparticles and inorganic/organic hybrid nanoparticles also reported the rapid transportation of nanoparticles with hydrodynamic diameter (HD) less than 34 nm and a noncationic surface charge from the lung to mediastinal lymph nodes (60). In order to determine the impact of the TiO2 nanoparticles on the lymph node proteome, we selected the 24 hr time-point to allow sufficient time for them to be to present in the lymph nodes.
In previous in vivo studies using a high dose of TiO2 nanoparticles, liver DNA cleavage (27), genotoxicity (28) and spleen injury (29) were found. Trouiller et al. (28) discovered DeGussa P25 TiO2 nanoparticles induced 8-hydroxy-2′-deoxyguanosine, gamma-H2AX foci, micronuclei, and DNA deletions after treating mice daily with in their drinking water containing DeGussa P25 TiO2 nanoparticles up to 0.6 mg/mL for 5 days (100 mg/kg body weight daily). Li et al. (27) found intraperitoneal injection of anatase TiO2 nanoparticles (∼ 5 nm) at a daily dose of 150 mg/kg body weight for 14 days could cause liver DNA cleavage and hepatocyte apoptosis in mice. Li et al. (29) also found spleen injuries after the same high dose treatment of mice for 45 days. In comparison, our study introduced DeGussa P25 TiO2 nanoparticles into mice at a low dose (∼0.2 mg/kg body weight) through intradermal injection and their effects were evaluated after 24 hours. The low dose TiO2 nanoparticle treatment allows the early assessment of protein expression changes in the mouse lymph nodes exposed to TiO2 nanoparticles before the apparent pathological damages are observed. Results from this study indicate this treatment affected only 33 proteins (up- or down-regulated using a filter of ≥1.3 fold and p<0.05), which occupied only ∼1% of the total number of proteins quantified. This subset of proteins could represent early response proteins to the perturbation with exogenous TiO2 nanoparticles.
A set of proteins associated with the immune response and antimicrobial activity were down-regulated (i.e., CAMP, LTF, S00A8, S00A9, and CHI3L3) except for amine oxidase (AOC3) which was up-regulated as a result of TiO2 nanoparticle treatment. Network analysis indicated that ESR1 interacts with lactotransferrin (LTF) (Figure 6), an important iron-binding protein that declined about three folds. Lactotransferrin (i.e., lactoferrin) is present in milk and other body secretary fluids with an antimicrobial activity. In the non-specific immune system, lactoferrin performs multiple functions such as iron homeostasis regulation, host defense against microbial infections, anti-inflammation activity, regulation of cellular growth and differentiation, and protection against cancer development and metastasis. Studies have suggested that nuclear hormone receptor ESR1 can bind the LTF promoter and activate its expression (51). However ESR1 was not identified directly in this experiment. Related proteins in the identified network are calgranulin A (S100A8) and B (S100A9). Both proteins participate in the inflammatory amphoterin signaling network. Calgranulin A and B proteins are two Ca2+-binding proteins of the S100 family. The two proteins can form a heterodimer and leukocyte L1 antigen complex, present in the serum and interstitial fluid in several infectious and/or inflammatory disorders, and may play an important role in leukocyte trafficking (53). Calgranulin A/B dimer was reported to be membrane-associated and present in acute inflammation but absent in chronic inflammation (61). Down-regulation of calgranulin A and B and other immune responsive proteins after TiO2 nanoparticle treatment may suggest potential early signs of low level/chronic inflammation or the immediate immune response of lymph nodes at the molecular level as a result of low dose treatment.
A class of lipid and fatty acid metabolism-related proteins was up-regulated as described early. Protein network analysis indicated that PPARγ is one of the major hubs in the significantly altered network. PPARγ protein, albeit not identified and quantified, is a regulator of adipocyte differentiation. The genes activated by PPARγ stimulate lipid uptake and adipogenesis by adipocytes (62). In the identified network (Figure 6), downstream proteins of PPARγ include lipid droplet-associated protein perilipin-1, adipocyte-type fatty acid-binding protein (A-FABP) and pyruvate carboxylase (PYC). These proteins are associated with lipid/fatty acid metabolism and were all up-regulated upon TiO2 nanoparticle treatment. PPARγ also interacts with serine/arginine-rich splicing factor 3 (SFRS3) gene promoter but the binding effect was not clear (63). However, SFRS3 is associated with c-Myc as well. It may be related to mRNA processing. In our study, SFRS3 was down-regulated upon TiO2 nanoparticles treatment. Up-regulated lipid metabolism proteins that are directly or indirectly associated with c-Myc include PYC, medium-chain specific acyl-CoA dehydrogenase (ACADM) and ATP-citrate synthase (ACLY). Currently, it is not clear what effects of up-regulation of lipid metabolism proteins are on the functions of mouse lymph nodes, and further studies are needed.
TiO2 nanoparticle treatment also resulted in expression changes of other groups of proteins such as mRNA processing proteins and histone isoforms. Seven proteins associated with mRNA processing/splicing (i.e., CPSF6, DDX46, SFPQ, SRSF2, SRSF3, and U2AF-65) were down-regulated in TiO2 nanoparticle-treated mice. There are more than one hundred fifty proteins involved in mRNA processing. Down-regulation of seven of them may not suggest the slowdown of mRNA processing. Likewise, transcription factor c-Myc is believed to regulate the expression of ∼15% of all the genes including those involved in cell division, cell growth, and apoptosis (64). In this study, however, only a few proteins whose abundance changed because of treatment were identified to be regulated by c-Myc and these proteins are mainly related to mRNA processing or lipid metabolism although c-Myc is one of the major hubs in the significantly altered protein network. Interestingly, histone isoforms H1, H2A, H2B and H4 were up-regulated upon TiO2 nanoparticle treatment. However, histone H3 was identified with only one unique peptide from the forward 18O-labeled sample, and the abundance change was less than 1.3-fold. Since the TiO2 nanoparticle treatment was in low doses and analysis was conducted at an early time point, the differentially expressed proteins identified in the current experiment could represent early response proteins to this type of perturbation. With extended treatment with a low dose of this nanoparticle, further protein alterations could be observed or, conversely, the lymph nodes might adapt. When high doses of TiO2 nanoparticles are employed for relative long time (days to weeks) treatment, more proteins might be expected to be changed in expression and be more apparently associated with pathological changes as observed in the previous studies (27-29).
5. Conclusions
Alterations in protein expression levels in mouse lymph nodes in response to low dose and short treatment time with DeGussa P25 TiO2 nanoparticles were analyzed using trypsin-catalyzed 16O/18O labeling in conjunction with two-dimensional LC separation and tandem mass spectrometry. Forward and reverse 16O/18O labeling resulted in quantification of 2809 and 2818 proteins, respectively, with a total of 2390 proteins commonly quantified from both labeling approaches. While 18O labeling was generally complete, more confident quantification could be achieved by comparing the consistency of protein abundance ratios of forward and reverse 16O/18O labeling. A total of 19 lymph node proteins were up-regulated and 14 were down-regulated over 1.3 fold with p<0.05, in the mice treated with TiO2 nanoparticles. This accounted for approximately 1% (33 proteins) of the total proteins identified from mouse lymph nodes. Biological function annotation indicates that the abundance changed proteins mainly involve in immune response (e.g., inflammation) and antimicrobial activity, lipid and fatty acid metabolism, mRNA processing, and nucleosome assembly. Protein network analysis indicates that the main regulators of protein expression could be estrogen receptor (ESR1), PPARγ, and c-Myc signalings. The differentially expressed proteins identified in this experiment could represent early response proteins to TiO2 nanoparticle treatment in mouse lymph nodes although their functions need to be further explored in relation to this type of perturbation.
Supplementary Material
Identified unique peptides for the differentially expressed proteins (≥1.3-fold change and p<0.05) in mouse lymph nodes at 24 hours after DeGussa P25 TiO2 nanoparticles treatment as revealed from trypsin-catalyzed 16O/18O labeling analysis. The symbols of *and # in the peptides stand for methionine oxidation (addition of one oxygen) and cysteine carboxyamidomethylation, respectively. The peptides labeled with either 16O or 18O but having the same sequence and modifications were considered as a single unique peptide and its identification times were counted and expression ratios were averaged. For a single unique peptide, the maximum SEQUEST cross-correlation score (XC) is presented along with its charge state (z), identification probability (P), final score (Sf), and DeltaCn. The p value for proteins represents the confidence level for protein expression/fold changes.
Acknowledgments
We are grateful to Dr. Donna Mendrick for critical reading and comments on this manuscript. This study was supported in part with funds from National Center for Toxicological Research, U.S. Food and Drug Administration (NCTR/FDA) and through an interagency agreement between the FDA and the National Toxicology Program at NIEHS (FDA 224-07-0007, NIH Y1ES1027). The views presented in this article do not necessarily reflect those of the U. S. Food and Drug Administration.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Schultz WB, Barclay L. A hard pill to swallow: Barriers to effective FDA regulation of nanotechnology-based dietary supplements. Woodrow Wilson International Center for Scholars; Washington, DC: 2009. pp. 1–28. [Google Scholar]
- 2.Dobrovolskaia MA, McNeil SE. Immunological properties of engineered nanomaterials. Nat Nanotechnol. 2007;2:469–478. doi: 10.1038/nnano.2007.223. [DOI] [PubMed] [Google Scholar]
- 3.Lammers T, Hennink WE, Storm G. Tumour-targeted nanomedicines: principles and practice. Br J Cancer. 2008;99:392–397. doi: 10.1038/sj.bjc.6604483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Handy RD, Shaw BJ. Toxic effects of nanoparticles and nanomaterials: implications for public health, risk assessment and the public perception of nanotechnology. Health Risk Soc. 2007;9:125–144. [Google Scholar]
- 5.Maynard AD, Aitken RJ, Butz T, Colvin V, Donaldson K, Oberdorster G, Philbert MA, Ryan J, Seaton A, Stone V, Tinkle SS, Tran L, Walker NJ, Warheit DB. Safe handling of nanotechnology. Nature. 2006;444:267–269. doi: 10.1038/444267a. [DOI] [PubMed] [Google Scholar]
- 6.Sundaram SK, Weber TJ. Special issue: on nanotoxicity. Int J Nanotechnol. 2008;5:1–2. [Google Scholar]
- 7.Sweet L, Strohm B. Nanotechnology - life-cycle risk management. Hum Ecol Risk Assess. 2006;12:528–551. [Google Scholar]
- 8.Landsiedel R, Ma-Hock L, Kroll A, Hahn D, Schnekenburger J, Wiench K, Wohlleben W. Testing metal-oxide nanomaterials for human safety. Adv Mater. 2010;22:2601–2627. doi: 10.1002/adma.200902658. [DOI] [PubMed] [Google Scholar]
- 9.Xu LG, Liu Y, Bai R, Chen CY. Applications and toxicological issues surrounding nanotechnology in the food industry. Pure Appl Chem. 2010;82:349–372. [Google Scholar]
- 10.Fischer HC, Chan WC. Nanotoxicity: the growing need for in vivo study. Curr Opin Biotech. 2007;18:565–571. doi: 10.1016/j.copbio.2007.11.008. [DOI] [PubMed] [Google Scholar]
- 11.Oberdorster G, Oberdorster E, Oberdorster J. Nanotoxicology: an emerging discipline evolving from studies of ultrafine particles. Environ Health Persp. 2005;113:823–839. doi: 10.1289/ehp.7339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Holgate ST. Exposure, uptake, distribution and toxicity of nanomaterials in humans. J Biomed Nanotechnol. 2010;6:1–19. doi: 10.1166/jbn.2010.1098. [DOI] [PubMed] [Google Scholar]
- 13.Sugibayashi K, Todo H, Kimura E. Safety evaluation of titanium dioxide nanoparticles by their absorption and elimination profiles. J Toxicol Sci. 2008;33:293–298. doi: 10.2131/jts.33.293. [DOI] [PubMed] [Google Scholar]
- 14.Fabian E, Landsiedel R, Ma-Hock L, Wiench K, Wohlleben W, van Ravenzwaay B. Tissue distribution and toxicity of intravenously administered titanium dioxide nanoparticles in rats. Arch Toxicol. 2008;82:151–157. doi: 10.1007/s00204-007-0253-y. [DOI] [PubMed] [Google Scholar]
- 15.Kim S, Lim YT, Soltesz EG, De Grand AM, Lee J, Nakayama A, Parker JA, Mihaljevic T, Laurence RG, Dor DM, Cohn LH, Bawendi MG, Frangioni JV. Near-infrared fluorescent type II quantum dots for sentinel lymph node mapping. Nat Biotechnol. 2004;22:93–97. doi: 10.1038/nbt920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gopee NV, Roberts DW, Webb P, Cozart CR, Siitonen PH, Warbritton AR, Yu WW, Colvin VL, Walker NJ, Howard PC. Migration of intradermally injected quantum dots to sentinel organs in mice. Toxicol Sci. 2007;98:249–257. doi: 10.1093/toxsci/kfm074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reddy ST, van der Vlies AJ, Simeoni E, Angeli V, Randolph GJ, O'Neil CP, Lee LK, Swartz MA, Hubbell JA. Exploiting lymphatic transport and complement activation in nanoparticle vaccines. Nat Biotechnol. 2007;25:1159–1164. doi: 10.1038/nbt1332. [DOI] [PubMed] [Google Scholar]
- 18.Mortensen LJ, Oberdorster G, Pentland AP, DeLouise LA. In vivo skin penetration of quantum dot nanoparticles in the murine model: The effect of UVR. Nano Lett. 2008;8:2779–2787. doi: 10.1021/nl801323y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gopee NV, Roberts DW, Webb P, Cozart CR, Siitonen PH, Latendresse JR, Warbitton AR, Yu WW, Colvin VL, Walker NJ, Howard PC. Quantitative Determination of Skin Penetration of PEG-Coated CdSe Quantum Dots in Dermabraded but not Intact SKH-1 Hairless Mouse Skin. Toxicol Sci. 2009;111:37–48. doi: 10.1093/toxsci/kfp139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gontier E, Ynsa MD, Biro T, Hunyadi J, Kiss B, Gaspar K, Pinheiro T, Silva JN, Filipe P, Stachura J, Dabros W, Reinert T, Butz T, Moretto P, Surleve-Bazeille JE. Is there penetration of titania nanoparticles in sunscreens through skin? A comparative electron and ion microscopy study. Nanotoxicology. 2008;2:218–231. [Google Scholar]
- 21.Sadrieh N, Wokovich AM, Gopee NV, Zheng JW, Haines D, Parmiter D, Siitonen PH, Cozart CR, Patri AK, McNeil SE, Howard PC, Doub WH, Buhse LF. Lack of significant dermal penetration of titanium dioxide from sunscreen formulations containing nano- and submicron-size TiO2 particles. Toxicol Sci. 2010;115:156–166. doi: 10.1093/toxsci/kfq041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liao CM, Chiang YH, Chio CP. Assessing the airborne titanium dioxide nanoparticle-related exposure hazard at workplace. J Hazard Mater. 2009;162:57–65. doi: 10.1016/j.jhazmat.2008.05.020. [DOI] [PubMed] [Google Scholar]
- 23.Warheit DB, Webb TR, Sayes CM, Colvin VL, Reed KL. Pulmonary instillation studies with nanoscale TiO2 rods and dots in rats: toxicity is not dependent upon particle size and surface area. Toxicol Sci. 2006;91:227–236. doi: 10.1093/toxsci/kfj140. [DOI] [PubMed] [Google Scholar]
- 24.Warheit DB, Webb TR, Reed KL, Frerichs S, Sayes CM. Pulmonary toxicity study in rats with three forms of ultrafine-TiO2 particles: differential responses related to surface properties. Toxicology. 2007;230:90–104. doi: 10.1016/j.tox.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 25.Lee K, Yang YS, Kwon SJ, Lee JS, Choi SJ, Seo HS, Kang MS, Lee BC, Kim SN, Yang HS, Han YA, Ryu HJ, Heo JD, Cho KH, Song CW. Lung injury study by 15 days inhalation exposure of titanium dioxide nanoparticles in rats. Toxicol Lett. 2009;189:S186–S186. [Google Scholar]
- 26.Grassian VH, Adamcakova-Dodd A, Pettibone JM, O'Shaughnessy PT, Thorne PS. Inflammatory response of mice to manufactured titanium dioxide nanoparticles: Comparison of size effects through different exposure routes. Nanotoxicology. 2007;1:211–226. [Google Scholar]
- 27.Li N, Ma LL, Wang J, Zheng L, Liu J, Duan YM, Liu HT, Zhao XY, Wang SS, Wang H, Hong FS, Xie YN. Interaction between nano-anatase TiO2 and liver DNA from mice in vivo. Nanoscale Res Lett. 2010;5:108–115. doi: 10.1007/s11671-009-9451-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Trouiller B, Reliene R, Westbrook A, Solaimani P, Schiestl RH. Titanium dioxide nanoparticles induce DNA damage and genetic instability in vivo in mice. Cancer Res. 2009;69:8784–8789. doi: 10.1158/0008-5472.CAN-09-2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li N, Duan YM, Hong MM, Zheng L, Fei M, Zhao XY, Wang J, Cui YL, Liu HT, Cai JW, Gong SJ, Wang H, Hong FS. Spleen injury and apoptotic pathway in mice caused by titanium dioxide nanoparticules. Toxicol Lett. 2010;195:161–168. doi: 10.1016/j.toxlet.2010.03.1116. [DOI] [PubMed] [Google Scholar]
- 30.Kang SJ, Kim BM, Lee YJ, Chung HW. Titanium dioxide nanoparticles trigger p53-mediated damage response in peripheral blood lymphocytes. Environ Mol Mutagen. 2008;49:399–405. doi: 10.1002/em.20399. [DOI] [PubMed] [Google Scholar]
- 31.Xu Z, Liu XW, Ma YS, Gao HW. Interaction of nano-TiO2 with lysozyme: insights into the enzyme toxicity of nanosized particles. Environ Sci Pollut Res. 2010;17:798–806. doi: 10.1007/s11356-009-0153-1. [DOI] [PubMed] [Google Scholar]
- 32.Wu WH, Sun X, Yu YP, Hu J, Zhao L, Liu Q, Zhao YF, Li YM. TiO2 nanoparticles promote beta-amyloid fibrillation in vitro. Biochem Biophys Res Commun. 2008;373:315–318. doi: 10.1016/j.bbrc.2008.06.035. [DOI] [PubMed] [Google Scholar]
- 33.Lundqvist M, Stigler J, Elia G, Lynch I, Cedervall T, Dawson KA. Nanoparticle size and surface properties determine the protein corona with possible implications for biological impacts. Proc Natl Acad Sci USA. 2008;105:14265–14270. doi: 10.1073/pnas.0805135105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gessner A, Waicz R, Lieske A, Paulke B, Mader K, Muller RH. Nanoparticles with decreasing surface hydrophobicities: influence on plasma protein adsorption. Int J Pharm. 2000;196:245–249. doi: 10.1016/s0378-5173(99)00432-9. [DOI] [PubMed] [Google Scholar]
- 35.Asuri P, Bale SS, Karajanagi SS, Kane RS. The protein-nanomaterial interface. Curr Opin Biotechnol. 2006;17:562–568. doi: 10.1016/j.copbio.2006.09.002. [DOI] [PubMed] [Google Scholar]
- 36.Maurer-Jones MA, Lin YS, Haynes CL. Functional assessment of metal oxide nanoparticle toxicity in immune cells. ACS Nano. 2010;4:3363–3373. doi: 10.1021/nn9018834. [DOI] [PubMed] [Google Scholar]
- 37.Izhaky D, Pecht I. What else can the immune system recognize? Proc Natl Acad Sci USA. 1998;95:11509–11510. doi: 10.1073/pnas.95.20.11509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yeo MK, Kim HE. Gene expression in zebrafish embryos following exposure to TiO2 nanoparticles. Mol Cell Toxicol. 2010;6:97–104. [Google Scholar]
- 39.Bu Q, Yan GY, Deng PC, Peng F, Lin HJ, Xu YZ, Cao ZX, Zhou T, Xue AQ, Wang YL, Cen XB, Zhao YL. NMR-based metabonomic study of the sub-acute toxicity of titanium dioxide nanoparticles in rats after oral administration. Nanotechnology. 2010;21 doi: 10.1088/0957-4484/21/12/125105. [DOI] [PubMed] [Google Scholar]
- 40.Yang XF, Liu JJ, He HW, Zhou L, Gong CM, Wang XM, Yang LQ, Yuan JH, Huang HY, He LH, Zhang B, Zhuang ZX. SiO2 nanoparticles induce cytotoxicity and protein expression alteration in HaCaT cells. Part Fibre Toxicol. 2010;7 doi: 10.1186/1743-8977-7-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schnolzer M, Jedrzejewski P, Lehmann WD. Protease-catalyzed incorporation of O-18 into peptide fragments and its application for protein sequencing by electrospray and matrix-assisted laser desorption/ionization mass spectrometry. Electrophoresis. 1996;17:945–953. doi: 10.1002/elps.1150170517. [DOI] [PubMed] [Google Scholar]
- 42.Blonder J, Hale ML, Chan KC, Yu LR, Lucas DA, Conrads TP, Zhou M, Popoff MR, Issaq HJ, Stiles BG, Veenstra TD. Quantitative profiling of the detergent-resistant membrane proteome of Iota-b toxin induced Vero cells. Journal of Proteome Research. 2005;4:523–531. doi: 10.1021/pr049790s. [DOI] [PubMed] [Google Scholar]
- 43.Yu LR, Zhu ZY, Chan KC, Issaq HJ, Dimitrov DS, Veenstra TD. Improved titanium dioxide enrichment of phosphopeptides from HeLa cells and high confident phosphopeptide identification by cross-validation of MS/MS and MS/MS/MS spectra. J Proteome Res. 2007;6:4150–4162. doi: 10.1021/pr070152u. [DOI] [PubMed] [Google Scholar]
- 44.Yu LR, Chan KC, Tahara H, Lucas DA, Chatterjee K, Issaq HJ, Veenstra TD. Quantitative proteomic analysis of human breast epithelial cells with differential telomere length. Biochemical and Biophysical Research Communications. 2007;356:942–947. doi: 10.1016/j.bbrc.2007.03.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fenselau C, Yao XD. O-18(2)-Labeling in Quantitative Proteomic Strategies: A Status Report. Journal of Proteome Research. 2009;8:2140–2143. doi: 10.1021/pr8009879. [DOI] [PubMed] [Google Scholar]
- 46.Stewart II, Thomson T, Figeys D. O-18 labeling: A tool for proteomics. Rapid Commun Mass Sp. 2001;15:2456–2465. doi: 10.1002/rcm.525. [DOI] [PubMed] [Google Scholar]
- 47.Storms HF, van der Heijden R, Tjaden UR, van der Greef J. Considerations for proteolytic labeling-optimization of O-18 incorporation and prohibition of back-exchange. Rapid Commun Mass Sp. 2006;20:3491–3497. doi: 10.1002/rcm.2738. [DOI] [PubMed] [Google Scholar]
- 48.Petritis BO, Qian WJ, Camp DG, Smith RD. A simple procedure for effective quenching of trypsin activity and prevention of O-18-labeling back-exchange. J Proteome Res. 2009;8:2157–2163. doi: 10.1021/pr800971w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jorge I, Navarro P, Martinez-Acedo P, Nunez E, Serrano H, Alfranca A, Redondo JM, Vazquez J. Statistical model to analyze quantitative proteomics data obtained by 18O/16O labeling and linear Ion trap mass spectrometry: application to the study of vascular endothelial growth factor-induced angiogenesis in endothelial cells. Mol Cell Proteomics. 2009;8:1130–1149. doi: 10.1074/mcp.M800260-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bonzon-Kulichenko E, Perez-Hernandez D, Nunez E, Martinez-Acedo P, Navarro P, Trevisan-Herraz M, Ramos MD, Sierra S, Martinez-Martinez S, Ruiz-Meana M, Miro-Casas E, Garcia-Durado D, Redondo JM, Burgos JS, Vazquez J. A robust method for quantitative high-throughput analysis of proteomes by 18O Labeling. Mol Cell Proteomics. 2011;10:1–14. doi: 10.1074/mcp.M1110.003335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Carroll JS, Meyer CA, Song J, Li W, Geistlinger TR, Eeckhoute J, Brodsky AS, Keeton EK, Fertuck KC, Hall GF, Wang QB, Bekiranov S, Sementchenko V, Fox EA, Silver PA, Gingeras TR, Liu XS, Brown M. Genome-wide analysis of estrogen receptor binding sites. Nat Genet. 2006;38:1289–1297. doi: 10.1038/ng1901. [DOI] [PubMed] [Google Scholar]
- 52.Millevoi S, Loulergue C, Dettwiler S, Karaa SZ, Keller W, Antoniou M, Vagner S. An interaction between U2AF 65 and CF I(m) links the splicing and 3′ end processing machineries. EMBO J. 2006;25:4854–4856. doi: 10.1038/sj.emboj.7601331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kerkhoff C, Klempt M, Sorg C. Novel insights into structure and function of MRP8 (S100A8) and MRP14 (S100A9) BBA-Mol Cell Res. 1998;1448:200–211. doi: 10.1016/s0167-4889(98)00144-x. [DOI] [PubMed] [Google Scholar]
- 54.Naranjo V, Villar M, Martin-Hernando MP, Vidal D, Hofle U, Gortazar C, Kocan KM, Vazquez J, de la Fuente J. Proteomic and transcriptomic analyses of differential stress/inflammatory responses in mandibular lymph nodes and oropharyngeal tonsils of European wild boars naturally infected with Mycobacterium bovis. Proteomics. 2007;7:220–231. doi: 10.1002/pmic.200600527. [DOI] [PubMed] [Google Scholar]
- 55.Kimura Y, Yokoyama R, Ishizu Y, Nishigaki T, Murahashi Y, Hijikata A, Kitamura H, Ohara O. Construction of quantitative proteome reference maps of mouse spleen and lymph node based on two-dimensional gel electrophoresis. Proteomics. 2006;6:3833–3844. doi: 10.1002/pmic.200500586. [DOI] [PubMed] [Google Scholar]
- 56.Leak LV, Liotta LA, Krutzsch H, Jones M, Fusaroa VA, Ross SJ, Zhaos YM, Petricoin EF. Proteomic analysis of lymph. Proteomics. 2004;4:753–765. doi: 10.1002/pmic.200300573. [DOI] [PubMed] [Google Scholar]
- 57.Shriver C, Sullivan A, Somiari S, Russell S, Heckman C, Hooke J, Somiari RI. Proteomics analysis of breast tumors and lymph nodes by 2-dimensional differential in-gel electrophoresis. Ann Surg Oncol. 2003;10:S15–S15. [Google Scholar]
- 58.Antonucci F, Chilosi M, Santacatterina M, Herbert B, Righetti PG. Proteomics and immunomapping of reactive lymph-node and lymphoma. Electrophoresis. 2002;23:356–362. doi: 10.1002/1522-2683(200202)23:2<356::AID-ELPS356>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 59.Manolova V, Flace A, Bauer M, Schwarz K, Saudan P, Bachmann MF. Nanoparticles target distinct dendritic cell populations according to their size. Eur J Immunol. 2008;38:1404–1413. doi: 10.1002/eji.200737984. [DOI] [PubMed] [Google Scholar]
- 60.Choi HS, Ashitate Y, Lee JH, Kim SH, Matsui A, Insin N, Bawendi MG, Semmler-Behnke M, Frangioni JV, Tsuda A. Rapid translocation of nanoparticles from the lung airspaces to the body. Nat Biotech. 2010;28:1300–1303. doi: 10.1038/nbt.1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bhardwaj RS, Zotz C, Zwadloklarwasser G, Roth J, Goebeler M, Mahnke K, Falk M, Meinardushager G, Sorg C. The calcium-binding proteins MRP8 and MRP14 form a membrane-associated heterodimer in a subset of monocytes/macrophages present in acute but absent in chronic inflammatory lesions. Eur J Immunol. 1992;22:1891–1897. doi: 10.1002/eji.1830220732. [DOI] [PubMed] [Google Scholar]
- 62.Rosen ED, Spiegelman BM. PPAR gamma: a nuclear regulator of metabolism, differentiation, and cell growth. J Biol Chem. 2001;276:37731–37734. doi: 10.1074/jbc.R100034200. [DOI] [PubMed] [Google Scholar]
- 63.Nielsen R, Pedersen TA, Hagenbeek D, Moulos P, Siersbaek R, Megens E, Denissov S, Borgesen M, Francoijs KJ, Mandrup S, Stunnenberg HG. Genome-wide profiling of PPAR gamma:RXR and RNA polymerase II occupancy reveals temporal activation of distinct metabolic pathways and changes in RXR dimer composition during adipogenesis. Gene Dev. 2008;22:2953–2967. doi: 10.1101/gad.501108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gearhart J, Pashos EE, Prasad MK. Pluripotency redux - Advances in stem-cell research. New Engl J Med. 2007;357:1469–1472. doi: 10.1056/NEJMp078126. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Identified unique peptides for the differentially expressed proteins (≥1.3-fold change and p<0.05) in mouse lymph nodes at 24 hours after DeGussa P25 TiO2 nanoparticles treatment as revealed from trypsin-catalyzed 16O/18O labeling analysis. The symbols of *and # in the peptides stand for methionine oxidation (addition of one oxygen) and cysteine carboxyamidomethylation, respectively. The peptides labeled with either 16O or 18O but having the same sequence and modifications were considered as a single unique peptide and its identification times were counted and expression ratios were averaged. For a single unique peptide, the maximum SEQUEST cross-correlation score (XC) is presented along with its charge state (z), identification probability (P), final score (Sf), and DeltaCn. The p value for proteins represents the confidence level for protein expression/fold changes.