Abstract
Ticks are mites specialized in acquiring blood from vertebrates as their sole source of food and are important disease vectors to humans and animals. Among the specializations required for this peculiar diet, ticks evolved a sophisticated salivary potion that can disarm their host’s hemostasis, inflammation, and immune reactions. Previous transcriptome analysis of tick salivary proteins has revealed many new protein families indicative of fast evolution, possibly due to host immune pressure. The hard ticks (family Ixodidae) are further divided into two basal groups, of which the Metastriata have 11 genera. While salivary transcriptomes and proteomes have been described for some of these genera, no tick of the genus Hyalomma has been studied so far. The analysis of 2,084 expressed sequence tags (EST) from a salivary gland cDNA library allowed an exploration of the proteome of this tick species by matching peptide ions derived from MS/MS experiments to this data set. We additionally compared these MS/MS derived peptide sequences against the proteins from the bovine host, finding many host proteins in the salivary glands of this tick. This annotated data set can assist the discovery of new targets for anti-tick vaccines as well as help to identify pharmacologically active proteins.
Keywords: Tick, hematophagy, salivary glands, sialome
Introduction
Ticks are specialized mites, divided into two large families, the Argasidae (soft ticks) and Ixodidae (hard ticks), and the monotypic Nuttalliellidae [1]. Soft ticks take relatively fast meals on their hosts, usually lasting less than one hour, while hard ticks stay attached for days or weeks to their hosts. The Ixodidae are further subdivided into the basal Prostriata group, with the single genus Ixodes, and the Metastriata, with 11 recognized genera organized into 4 subfamilies [2].
Among the several adaptations to blood feeding, ticks evolved a complex saliva consisting of a mixture of pharmacologically active components that affects their host’s hemostasis, inflammation, and immunity and also contains antimicrobial factors [3–8]. Perhaps due to their host’s immune response, which could neutralize such activities, these salivary proteins appear to evolve quickly, as indicated by the discovery of unique protein families among different tick genera and large sequence diversity within protein families that are common to all ticks, such as the lipocalin or Kunitz superfamilies [3]. Gene duplications are also common, leading to the existence of many multigene families within individual tick species, as exemplified by the Kunitz, lipocalin, basic tail, and ixodegrin families [3, 9].
In the past 8 years, salivary transcriptomes, or sialomes (from the Greek sialo = saliva), have been described from several tick species, including the soft ticks Argas monolakensis [10, 11], Ornithodoros parkeri [12] and Ornithodoros coriaceus [13]; the prostriates Ixodes scapularis [14, 15], Ixodes pacificus [16], and Ixodes ricinus [17]; the metastriates Amblyomma americanum [18], Amblyomma cajennense [19], and Amblyomma variegatum [20] belonging to the metastriate Amblyomminae subfamily; and Dermacentor andersoni [21] and Rhipicephalus appendiculattus [22], members of the larger Rhipicephalinae subfamily. Within this last subfamily, the genera Anomalohimalayia, Cosmiomma, Hyalomma, Margaropus, Nosomma, and Rhipicentor remain unexplored.
To investigate the diversity of the sialome of a member of the Hyalomma genus, we analyzed the sialotranscriptome and sialoproteome of adult female Hyalomma marginatum rufipes, which is a common three-host tick found in Africa and Europe, and also a competent vector of Crimean Congo fever [23–27]. Immature stages H. m. rufipes feed on small vertebrates including mammals but mostly birds, while adults feed on large mammals, including cattle, from where our samples were obtained [28–32].
Material and Methods
Ticks and SG preparation
Ticks were removed from zebu cows located on Point G in Bamako, Mali, in December 2008. The SGs were dissected by one of us (JMA) and transferred to RNAlater (Ambion, Austin, Texas, USA). The vials were kept at 4°C for 24 hours then stored at 30°C until use. Tick carcasses were saved and analyzed by Dr. Dmitry A. Apanaskevich (Assistant Curator, U.S. National Tick Collection, Institute of Arthropodology and Parasitology, Georgia Southern University, Statesboro, Georgia, USA). They were all identified to be adult female specimens of H. m. rufipes Koch, 1844.
cDNA library construction and sequencing
H. m rufipes mRNA from one pair of SGs was isolated using the Micro-FastTrack mRNA isolation kit (Invitrogen, San Diego, California, USA). The PCR-based cDNA library was made following the instructions for the SMART cDNA library construction kit (Clontech, Palo Alto, California, USA). This system utilizes oligoribonucleotide (SMART IV) to attach an identical sequence at the 5′ end of each reverse-transcribed cDNA strand. This sequence is then utilized in subsequent PCR reactions and restriction digests.
First-strand synthesis was carried out using PowerScript reverse transcriptase at 42°C for 1 hour in the presence of the SMART IV and CDS III (3′) primers. Second-strand synthesis was performed using a long distance (LD) PCR-based protocol, using Advantage™ Taq polymerase (Clontech) mix in the presence of the 5′ PCR primer and the CDS III (3′) primer. The cDNA synthesis procedure resulted in the creation of SfiI A and B restriction enzyme sites at the ends of the PCR products that are used for cloning into the phage vector. PCR conditions were as follows: 95°C for 20 sec; 24 cycles of 95°C for 5 sec., 68°C for 6 min. A small portion of the cDNA obtained by PCR was analyzed on a 1.1% agarose gel to check quality and range of cDNA synthesized. Double-stranded cDNA was immediately treated with proteinase K (0.8 μg/ml) at 45°C for 20 min, and the enzyme was removed by ultrafiltration though a Microcon YM-100 centrifugal filter device (Amicon Inc., Beverly, California, USA). The cleaned, double-stranded cDNA was then digested with SfiI at 50°C for 2 hours, followed by size fractionation on a ChromaSpin–400 column (Clontech). The profile of the fractions was checked on a 1.1% agarose gel, and fractions containing cDNAs of more than 400 bp were pooled and concentrated using a Microcon YM-100.
The cDNA mixture was ligated into the λ TriplEx2 vector (Clontech), and the resulting ligation mixture was packaged using the GigaPack® III Plus packaging extract (Stratagene, La Jolla, California, USA) according to the manufacturer’s instructions. The packaged library was plated by infecting log-phase XL1-Blue Escherichia coli cells (Clontech). The percentage of recombinant clones was determined by blue-white selection screening on LB/MgSO4 plates containing X-gal/IPTG. Recombinants were also determined by PCR, using vector primers (5′ λ TriplEx2 sequencing primer and 3′ λ TriplEx2 sequencing) flanking the inserted cDNA, with subsequent visualization of the products on a 1.1% agarose/EtBr gel.
The H. m. rufipes SG cDNA library was plated on LB/MgSO4 plates containing X-gal/IPTG to an average of 250 plaques per 150-mm Petri plate. Recombinant (white) plaques were randomly selected and transferred to 96-well MICROTEST ™ U-bottom plates (BD BioSciences, Franklin Lakes, New Jersey, USA) containing 100 μl of SM buffer [0.1 M NaCl; 0.01 M MgSO4; 7 H2O; 0.035 M Tris-HCl (pH 7.5); 0.01% gelatin] per well. The plates were covered and placed on a gyrating shaker for 30 min at room temperature. The phage suspension was either immediately used for PCR or stored at 4°C for future use.
To amplify the cDNA using a PCR reaction, 4 μl of the phage sample was used as a template. The primers were sequences from the λ TriplEx2 vector and named pTEx2 5seq (5′-TCC GAG ATC TGG ACG AGC-3′) and pTEx2 3LD (5′-ATA CGA CTC ACT ATA GGG CGA ATT GGC-3′), positioned at the 5′ and the 3′ end of the cDNA insert, respectively. The reaction was carried out in 96-well flexible PCR plates (Fisher Scientific, Pittsburgh, Pennsylvania, USA) using the TaKaRa EX Taq polymerase (TAKARA Mirus Bio, Madison, Wisconsin, USA), on a Perkin Elmer GeneAmp® PCR system 9700 (Perkin Elmer Corp., Foster City, California, USA). The PCR conditions were: one hold of 95°C for 3 min; 25 cycles of 95°C for 1 min, 61°C for 30 sec; 72°C for 6 min. Approximately 200–250 ng of each PCR product was transferred to Thermo-Fast 96-well PCR plates (ABgene Corp., Epsom, Surrey, UK) and frozen at −20°C. Samples were shipped on dry ice to the Rocky Mountain Laboratories Genomics Unit with primer and template combined together in an ABI 96-well Optical Reaction Plate (P/N 4306737) following the manufacturer’s recommended concentrations. Sequencing reactions were set up as recommended by Applied Biosystems BigDye® Terminator v3.1 cycle sequencing kit by adding 1 μl ABI BigDye® Terminator ready reaction mix v3.1 (P/N 4336921), 3 μl 5× ABI sequencing buffer (P/N 4336699), and 2 μl of water for a final volume of 10 μl. Cycle sequencing was performed at 96°C for 10 sec, 50°C for 5 sec, 60°C for 4 min for 27 cycles on either a Bio-Rad Tetrad 2 (Bio-Rad Laboratories, Hercules, California, USA) or ABI 9700 (Applied Biosystems, Inc., Foster City, California, USA) thermal cycler. Fluorescently labeled extension products were purified following Applied Biosystems BigDye® XTerminator™ purification protocol and subsequently processed on an ABI 3730xL DNA Analyzer (Applied Biosystems, Inc.). The AB1 file generated for each sample from the 3730xL DNA analyzer was provided to researchers in Rockville, Maryland, USA, through a secure network drive for all subsequent downstream sequencing analysis. In addition to the sequencing of the cDNA clones, primer extension experiments were performed in selected clones to further extend sequence coverage.
Bioinformatics tools used
ESTs were trimmed of primer and vector sequences. The BLAST suite of programs [33], CAP3 assembler [34], and ClustalW [35] software were used to compare, assemble, and align sequences, respectively. For functional annotation of the transcripts, we used blastx [33] to compare the nucleotide sequences with the NR protein database of the NCBI and to the Gene Ontology database [36]. The program reverse position-specific BLAST (RPS-BLAST) [33] was used to search for conserved protein domains in the Pfam [37], SMART [38], Kog [39], and conserved domains databases [40]. We have also compared the transcripts with other subsets of mitochondrial and rRNA nucleotide sequences downloaded from NCBI and to several organism proteomes downloaded from NCBI, ENSEMBL, or VectorBase and to the assembled EST salivary database described before [3], and found in http://exon.niaid.nih.gov/transcriptome/tickreview/Sup-Table-1.xls from where the fasta set can also be recovered at http://exon.niaid.nih.gov/transcriptome/tick_review/tick_proteins_fasta.zip. Segments of the three-frame translations of the EST (because the libraries were unidirectional, six-frame translations were not used) starting with a methionine found in the first 300 predicted amino acids, or the predicted protein translation in the case of complete coding sequences, were submitted to the SignalP server [41] to help identify translation products that could be secreted. O-glycosylation sites on the proteins were predicted with the program NetOGlyc [42]. Functional annotation of the transcripts was based on all the comparisons above.
For sequence comparisons and phylogenetic analysis, we retrieved tick sequences from GenBank, and we have also deducted protein sequences from ESTs deposited in Dbest, as described and made accessible in a previous review article [3]. Phylogenetic analysis and statistical neighbour-joining bootstrap tests of the phylogenies were done with the Mega package [43] after sequence alignment performed by Clustal [44]. Codon volatility was calculated as previously described [45].
Proteomic characterization using one-dimensional gel electrophoresis and tandem mass spectrometry (MS)
The soluble protein fraction from salivary gland homogenates from H. marginatum corresponding to approximately 200 μg of protein was brought up in reducing Laemmli gel-loading buffer. The sample was boiled for 10 min and applied to two lanes (~50 and ~150 μg in each lane) and resolved on a NuPAGE 4–12% Bis-Tris precast gel. The separated proteins were visualized by staining with SimplyBlue (Invitrogen). The gel was sliced into 20 individual sections that were destained and digested overnight with trypsin at 37°C. Peptides were extracted and desalted using ZipTips (Millipore, Bedford, MA) and resuspended in 0.1% TFA prior to S analysis.
Nanoflow reversed-phase liquid chromatography tandem MS (RPLS-MS/MS) was performed using an Agilent 1100 nanoflow LC system (Agilent technologies, Palo Alto, CA) coupled online with a linear ion-trap (LIT) mass spectrometer (LTQ, ThermoElectron, San José, CA). NanoRPLC columns were slurry-packed in-house with 5 μm, 300-Å pore size C-18 phage (Jupiter, Phenomenex, CA) in a 75-μm i.d. × 10-cm fused silica capillary (Polymicro Technologies, Phoenix, AZ) with a flame-pulled tip. After sample injection, the column was washed for 30 min with 98% mobile phase A (0.1% formic acid in water) at 0.5 μL/min, and peptides were eluted using a linear gradient of 2% mobile phase B (0.1% formic acid in acetonitrile) to 42% mobile phase B in 40 min at 0.25 μL/min, then to 98% B for an additional 10 min. The LIT-mass spectrometer was operated in a data-dependent MS/MS mode in which each full MS scan was followed by seven MS/MS scans where the seven most abundant molecular ions were dynamically selected for collision-induced dissociation (CID) using a normalized collision energy of 35%. Dynamic exclusion was applied to minimize repeated selection of peptides previously selected for CID.
Tandem mass spectra were searched using SEQUEST on a 20-node Beowulf cluster against the H. marginatum rufipes described in this paper and the Bos taurus proteome (downloaded from ftp://ftp.ncbi.nih.gov/genomes/Bos_taurus/protein/) with methionine oxidation included as dynamic modification. Only tryptic peptides with up to two missed cleavage sites meeting a specific SEQUEST scoring criteria [delta correlation (ΔCn) ≥ 0.08 and charge-state-dependent cross correlation (Xcorr) ≥ 1.9 for [M + H]1+, ≥ 2.2 for [M + 2H]2+, and 3.5 for [M + 3H]3+] were considered as legitimate identifications. The peptides identified by MS were converted to Prosite block format [46] by a program written in Visual Basic. This database was used to search matches in the Fasta-formatted database of salivary proteins, using the program Seedtop, which is part of the Blast package. The result of the Seedtop search is piped into the hyperlinked spreadsheet to produce a text file as shown in supplemental table S2. Notice that the ID lines indicate, for example, 18_73, which means that one match was found for fragment number 73 from gel band 18. Because the same tryptic fragment can be found in many gel bands, another program was written to count the number of fragments for each gel band, displaying a summarized result in an Excel table. The summary in this form of 11 →18 | 12 →18 | 13→2 | indicates that 18 fragments were found in Fraction 11, while 18 and 2 peptides were found in fractions 12 and 13, respectively. Furthermore, this summary included protein identification only when two or more peptide matches to the protein were obtained from the same gel slice.
Results and Discussion
Overview of the assembled salivary expressed sequence tag (EST) set
A total of 2,084 cDNA clones were used to assemble a database (Additional file 1 [Supplemental Table S1]) to yield 1,167 clusters of related sequences, 993 of which contained only one EST. The 1,167 clusters were compared, using the programs blastx, blastn, or RPS-BLAST [33], to the nonredundant (NR) protein database of the National Center of Biological Information (NCBI), National Library of Medicine, NIH, to a gene ontology database [36], to the conserved domains database of the NCBI [40], and to a custom-prepared subset of the NCBI nucleotide database containing either mitochondrial or rRNA sequences.
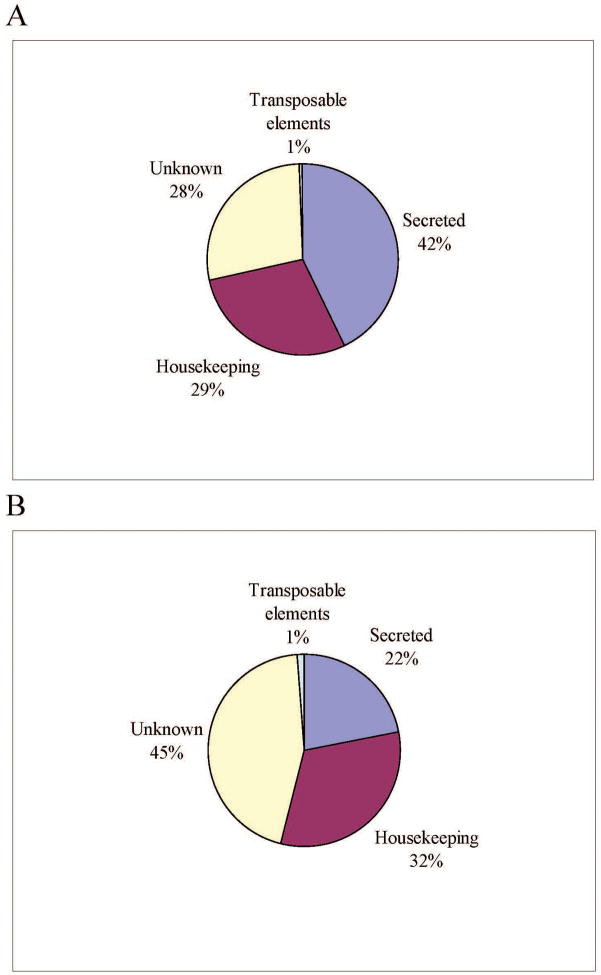
Manual annotation of the contigs resulted in four broad categories of expressed genes (Table 1 and Figure 1). The putatively housekeeping (H) category contained 29% of the sequences and had on average 1.59 sequences per cluster, and the secreted (S) category had 42% of the ESTs with an average of 3.51 ESTs/clusters, while 28% of the ESTs, mostly singletons, were not classifiable, constituting the Unknown (U) group. The transcripts of the U class could represent novel proteins or derive from the less conserved 3′ or 5′ untranslated regions of genes, as was indicated for the sialotranscriptome of Anopheles gambiae [47]. Sequences deriving from transposable elements (TE) accounted for the remaining sequences, mostly singletons. TE-related sequences may indicate either the presence of active transposition in the tick, or more likely, the expression of sequences suppressing transposition. Low-level expression of TE sequences have been a relatively common finding in previous sialotranscriptomes.
Table 1.
Main classes of transcripts found in the cDNA library of Hyalomma marginatum rufipes
| Class | Number of Contigs | Number of ESTs | ESTs/Contigs |
|---|---|---|---|
| Secreted | 255 | 894 | 3.51 |
| Housekeeping | 374 | 594 | 1.59 |
| Unknown | 526 | 583 | 1.11 |
| Transposable elements | 12 | 13 | 1.08 |
| Total | 1167 | 2084 |
Figure 1. Distribution of expressed sequence tags (ESTs) and assembled contigs.
Distribution of ESTs (A) and assembled contigs (B) obtained from a cDNA library from the salivary glands of adult female Hyalomma marginatum rufipes ticks.
Housekeeping (H) genes
The 594 ESTs attributed to H-class genes expressed in the salivary glands (SGs) of H. m. rufipes were further characterized into 20 subgroups according to function (Table 2 and Additional file 1 [Supplemental Table S1]). Transcripts associated with the protein synthesis machinery represented 44% of all transcripts associated within the H class, an expected result given the secretory nature of the organ. Energy metabolism accounted for the second most abundant H class, with 10.8% of the transcripts. Another 10.3% of the transcripts were classified as either “hypothetical conserved” or “conserved secreted” proteins. These represent highly conserved proteins of unknown function, presumably associated with cellular function but still uncharacterized. This functional distribution is typical of previous sialotranscriptomes.
Table 2.
Housekeeping classes of transcripts found in the cDNA library of Hyalomma marginatum rufipes
| Class | Number of Contigs | Number of ESTs | ESTs/Contig | Percent |
|---|---|---|---|---|
| Protein synthesis machinery | 123 | 262 | 2.13 | 44.11 |
| Metabolism, energy | 50 | 64 | 1.28 | 10.77 |
| Unknown, conserved | 48 | 61 | 1.27 | 10.27 |
| Signal transduction | 16 | 44 | 2.75 | 7.41 |
| Protein modification machinery | 32 | 40 | 1.25 | 6.73 |
| Transcription machinery | 29 | 36 | 1.24 | 6.06 |
| Protein export machinery | 17 | 17 | 1.00 | 2.86 |
| Cytoskeletal | 10 | 17 | 1.70 | 2.86 |
| Nuclear regulation | 10 | 11 | 1.10 | 1.85 |
| Proteasome machinery | 9 | 10 | 1.11 | 1.68 |
| Oxidant metabolism/detoxification | 5 | 5 | 1.00 | 0.84 |
| Metabolism, amino acid | 5 | 5 | 1.00 | 0.84 |
| Transporters/storage | 4 | 4 | 1.00 | 0.67 |
| Metabolism, carbohydrate | 4 | 4 | 1.00 | 0.67 |
| Metabolism, nucleotide | 3 | 4 | 1.33 | 0.67 |
| Metabolism, lipid | 4 | 4 | 1.00 | 0.67 |
| Transcription factor | 1 | 2 | 2.00 | 0.34 |
| Metabolism, intermediate | 2 | 2 | 1.00 | 0.34 |
| Extracellular matrix/cell adhesion | 1 | 1 | 1.00 | 0.17 |
| Immunity | 1 | 1 | 1.00 | 0.17 |
| Total | 374 | 594 |
Possibly secreted (S) class of expressed genes
A total of 894 ESTs contributed to 255 contigs and singletons associated with putative H. m. rufipes salivary-secreted components (Table 3 and Additional file 1). These include previously known gene families [48] such as metalloproteases, lipocalins, protease inhibitor domain-containing peptides, immunomodulators, antimicrobial peptides, basic tail, and glycine-rich peptides. Several other deducted sequences code for proteins that have some sequence similarity to other known proteins or to proteins not previously described in tick sialotranscriptomes.
Table 3.
Secreted classes of transcripts found in the cDNA library of Hyalomma marginatum rufipes
| Subclass | Number of ESTs |
|---|---|
| Protease inhibitors | |
| Kazal domain | 2 |
| Kunitz domain | 15 |
| Madainin family | 32 |
| TIL domain | 2 |
| Basic tail family found in argasids and ixodids | 11 |
| Lipocalins | 18 |
| RGD domain family (may not contain RGD) | 1 |
| Glycine-rich proteins | |
| Large GGY protein | 73 |
| Cuticle Ala-rich family | 1 |
| Metastriate spider silk-like | 56 |
| Glycine-rich protein group GYG | 3 |
| Glycine-rich similar to RNA binding protein | 3 |
| Other glycine-rich proteins | 81 |
| Mucins | 12 |
| Putative proline-rich polypeptides | 4 |
| 28-kDa metastriate family | 1 |
| 8.9-kDa family | 6 |
| Immunity-related products | 27 |
| DAP-36 fragment | 1 |
| Dermacentor 9-kDa family | 400 |
| Enzymes | |
| Metalloproteases | 3 |
| Ribonucleases | 2 |
| Metastriate insulin growth factor binding protein | 1 |
| Conserved secreted protein | 1 |
| Similar to previously orphan proteins | 23 |
| Other possibly secreted | 115 |
| Total | 894 |
Detailed analysis of the sialome of H. m. rufipes
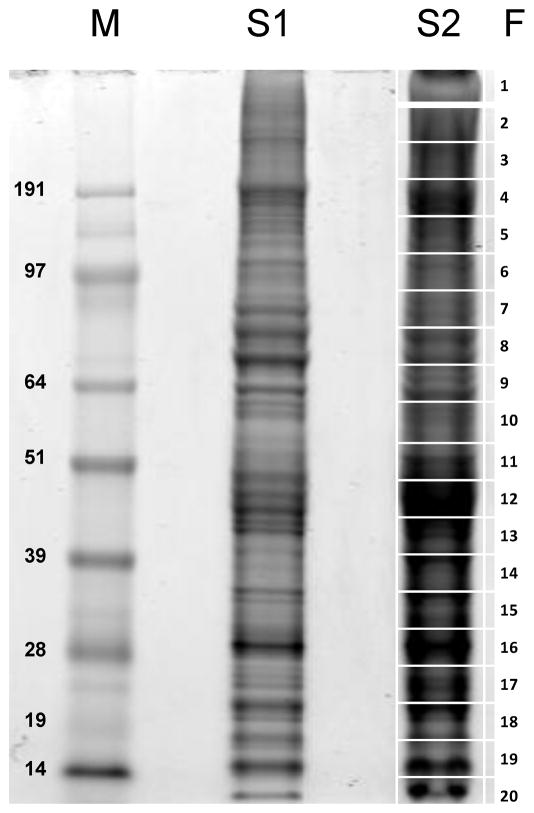
From the sequenced cDNAs, a total of 249 protein sequences were derived, 101 of which code for putative secreted products (Additional file 2 [Supplemental Table S2]). This set of 101 proteins includes 74 that are presumably full length, while the remaining 27 are truncated. With this transcriptome-derived protein database, we characterized the tick salivary proteome via analysis of salivary gland homogenates fractionated by electrophoresis on SDS-polyacrylamide gels, bands of which were tryptic digested, fractionated by reversed phase chromatography, followed by in line electro spray into a mass spectrometer for tandem mass spectrometry analysis (Figure 2). Follows a description of the protein families deducted from the transcriptome analysis, with information of the proteomic experiment as summarized in Table 4 and supplemental files 1 and 2.
Figure 2. 1D gel electrophoresis of Hyalomma marginatum rufipes salivary gland homogenates.
The numbers at the left indicate the MW in kDA of the protein standards shown in the left lane labelled M. The right gel lane (S2) shows the separation of ~ 150 μg salivary gland proteins. The grid at the right (1–20) represents the gel slices submitted for tryptic digest and MS/MS identification. S1 depicts a lane with less protein (50 μg) than S2.
Table 4.
Tick proteins identified in the polyacrylamide gel shown in Figure 2
| Sequence name | Description | Fraction Number --> Number of ions | Coverage in aa residues (1) | Percent coverage (1) |
|---|---|---|---|---|
| HEX-706 | Ubiquitin | 1 -> 12| 2 -> 12| 3 -> 12| | 144 | 52.4 |
| HEX-879 | 40s ribosomal protein S27 | 1 -> 4| 2 -> 4| | 38 | 45.2 |
| HEX-1118 | ubiquitin/ribosomal protein S27a fusion protein | 1 -> 6| 2 -> 6| 10 -> 6| 17 -> 6| | 54 | 37.5 |
| HEX-1165 | proteasome (prosome, macropain) 26S subunit, non-ATPase, 1 | 6 -> 18| | 292 | 109.0 |
| HEX-1043 | putative cement protein RIM36 | 8 -> 10| 6 -> 8| 18 -> 8| | 248 | 306.2 |
| HEX-1057 | hypothetical glycine rich secreted cement protein | 8 -> 26| 9 -> 20| 17 -> 16| | 510 | 314.8 |
| HEX-235 | putative cement protein | 8 -> 32| 9 -> 24| 10 -> 16| | 750 | 230.8 |
| HEX-1097 | apoptosis-promoting RNA-binding protein TIA-1/TIAR | 9 -> 18| 8 -> 10| | 258 | 80.9 |
| HEX-1107 | Mitochondrial chaperonin, Cpn60/Hsp60p | 10 -> 8| 11 -> 4| | 104 | 46.2 |
| HEX-153 | protein disulfide isomerase | 12 -> 10| 13 -> 4| | 154 | 46.2 |
| HEX-18 | aldehyde dehydrogenase, putative | 12 -> 18| 11 -> 4| | 220 | 80.9 |
| HEX-779 | Alpha tubulin | 12 -> 52| 13 -> 26| 11 -> 16| | 776 | 236.6 |
| HEX-911 | putative mitochondrial processing peptidase beta-subunit | 13 -> 14| 12 -> 8| 16 -> 4| | 182 | 152.9 |
| HEX-296 | 40S ribosomal protein SA (P40)/Laminin receptor 1 | 13 -> 14| 15 -> 6| 14 -> 4| | 206 | 86.2 |
| HEX-1089 | ribosomal protein L3, putative | 13 -> 8| 15 -> 4| | 150 | 56.8 |
| HEX-750 | hypothetical glycine rich secreted protein | 15 -> 2| | 42 | 33.6 |
| HEX-1117 | hypothetical glycine rich secreted protein | 15 -> 2| | 42 | 22.0 |
| HEX-11 | mitochondrial malate dehydrogenase | 15 -> 20| 16 -> 8| 17 -> 4| | 264 | 201.5 |
| HEX-910 | 60s ribosomal protein L6 | 15 -> 24| 16 -> 8| | 318 | 152.9 |
| HEX-350 | guanine nucleotide-binding protein | 15 -> 32| 16 -> 14| 17 -> 4| | 448 | 182.9 |
| HEX-702 | 60S acidic ribosomal protein P0 | 15 -> 6| | 144 | 133.3 |
| HEX-115 | RNA-binding protein musashi/mRNA cleavage and polyadenylation factor I complex, subunit HRP1 | 15 -> 8| 14 -> 6| | 102 | 49.0 |
| HEX-497 | Tick lipocalin superfamily member - Subgroup B | 16 -> 14| | 172 | 98.9 |
| HEX-180 | Ribosomal protein L8 | 16 -> 14| 17 -> 2| | 202 | 120.2 |
| HEX-186 | 60s ribosomal protein L23 | 16 -> 18| 17 -> 8| | 280 | 142.9 |
| HEX-1021 | ADP/ATP translocase, putative | 16 -> 18| 2 -> 14| 3 -> 12| | 296 | 216.1 |
| HEX-215 | 40S ribosomal protein S2/30S ribosomal protein S5 | 16 -> 20| 7 -> 2| | 342 | 133.6 |
| HEX-743 | 40S ribosomal protein S6 | 16 -> 22| | 250 | 151.5 |
| HEX-549 | 40S ribosomal protein S3 | 16 -> 26| 15 -> 6| 17 -> 6| | 324 | 180.0 |
| HEX-550 | hypothetical secreted protein with basic head | 16 -> 3| 1 -> 2| | 36 | 40.9 |
| HEX-1143 | putative cement protein | 16 -> 6| | 48 | 15.4 |
| HEX-986 | cathepsin L-like cysteine proteinase B | 16 -> 6| | 86 | 56.6 |
| HEX-240 | putative cement protein | 16 -> 7| | 105 | 52.2 |
| HEX-737 | eukaryotic translation initiation factor 3 subunit 5 epsilon-like | 16 -> 8| | 128 | 77.6 |
| HEX-1155 | ribosomal protein L7-like | 16 -> 8| | 96 | 90.6 |
| HEX-225 | Multifunctional chaperone (14-3-3 family) | 17 -> 14| 16 -> 10| 18 -> 4| | 140 | 69.0 |
| HEX-866 | 60S ribosomal protein L13 | 17 -> 14| 19 -> 4| | 150 | 76.5 |
| HEX-619 | 60s ribosomal protein L10 | 17 -> 16| 18 -> 6| | 166 | 102.5 |
| HEX-870 | 40S ribosomal protein S8 | 17 -> 20| 18 -> 4| 19 -> 4| | 264 | 126.9 |
| HEX-892 | glutathione S-transferase | 17 -> 67| 18 -> 54| 19 -> 6| | 886 | 579.1 |
| HEX-423 | RNA binding motif-containing protein, putative | 17 -> 8| | 118 | 100.0 |
| HEX-975 | Mitochondrial F1F0-ATP synthase, subunit OSCP/ATP5 | 18 -> 14| | 204 | 167.2 |
| HEX-710 | 60S ribosomal protein L14 | 18 -> 16| | 162 | 114.1 |
| HEX-1007 | hypothetical secreted protein with 3 TIL domains | 18 -> 2| | 20 | 7.2 |
| HEX-739 | NADH-ubiquinone oxidoreductase, NUFS7/PSST/20 kDa subunit | 18 -> 24| | 372 | 204.4 |
| HEX-1158 | 40S ribosomal protein S7 | 18 -> 26| 17 -> 12| 19 -> 4| | 385 | 233.3 |
| HEX-455 | Ribosomal protein S4 | 18 -> 27| 19 -> 22| 17 -> 18| | 265 | 175.5 |
| HEX-980 | histamine release factor | 18 -> 4| | 40 | 26.5 |
| HEX-366 | proteasome (prosome, macropain) subunit, beta type 2 | 18 -> 6| | 94 | 62.3 |
| HEX-1069 | Gly-Tyr rich salivary protein | 18 -> 6| | 82 | 38.5 |
| HEX-1036 | ribosomal protein L35a | 18 -> 8| 19 -> 8| | 64 | 49.6 |
| HEX-896 | lysozyme | 19 -> 10| 20 -> 5| | 144 | 142.6 |
| HEX-766 | ribosomal protein S19, putative | 19 -> 10| 20 -> 6| | 104 | 86.0 |
| HEX-68 | ribosomal protein L15 | 19 -> 12| | 122 | 62.2 |
| HEX-881 | 40S ribosomal protein S14, putative | 19 -> 12| 20 -> 8| | 176 | 144.3 |
| HEX-1090 | ribosomal protein S17 | 19 -> 14| | 264 | 266.7 |
| HEX-608 | ribosomal protein L28, putative | 19 -> 14| 18 -> 6| | 142 | 103.6 |
| HEX-751 | 60S ribosomal protein L27, putative | 19 -> 16| 18 -> 4| | 176 | 128.5 |
| HEX-473 | ribosomal protein L31 | 19 -> 16| 20 -> 8| | 306 | 248.8 |
| HEX-909 | translation initiation factor 5A | 19 -> 18| | 212 | 146.2 |
| HEX-356 | 60S ribosomal protein L32 | 19 -> 34| 18 -> 4| | 408 | 304.5 |
| HEX-1038 | hypothetical secreted peptide precursor | 19 -> 4| 20 -> 4| | 42 | 49.4 |
| HEX-244 | ribosomal protein L27A, putative | 19 -> 6| | 62 | 52.1 |
| HEX-1139 | translation machinery-associated protein, putative | 19 -> 6| 18 -> 4| | 78 | 42.9 |
| HEX-392 | ribosomal protein S16 | 19 -> 8| 20 -> 8| | 78 | 70.9 |
| HEX-122 | histone H4, putative | 20 -> 31| 19 -> 4| | 322 | 322.0 |
| HEX-182 | ribosomal protein S12 | 20 -> 4| | 60 | 48.4 |
| HEX-247 | 60s acidic ribosomal protein P1 | 20 -> 4| | 48 | 39.3 |
| HEX-600 | cytochrome B5, putative | 20 -> 4| | 50 | 58.8 |
| HEX-867 | cytochrome C oxidase subunit VIc | 20 -> 4| | 56 | 77.8 |
| HEX-44 | putative superoxide dismutase Cu-Zn | 20 -> 6| 19 -> 4| | 84 | 54.5 |
| HEX-650 | cytochrome c oxidase polypeptide IV | 20 -> 8| 19 -> 6| | 98 | 55.1 |
On fraction with higher number of ions only. Includes redundant ions. Coverage may be greater than 100%.
Putative secreted proteins with presumed or experimentally validated function
Metalloprotease domains
Transcripts coding for metalloproteases have been commonly found in tick sialotranscriptomes [3, 10, 49], and these may be associated with fibrinogenolytic activity as previously found in I. scapularis [50]. HEX-267 is a truncated sequence coding for the carboxy terminal region of a metalloprotease sequence from Haemaphysalis found in GenBank, with only 25 % similarity over 228 residues but with 68% identity to a homologue deducted from ESTs from Rhipicephalus microplus. It also displays the CDD domain for arthropod metalloproteases.
HEX-920 codes for a 5′ truncated endonuclease that may or may not be secreted in saliva. Although not found in Ixodes sialotranscriptomes, these types of transcripts have been found in transcriptomes of Rhipicephalus and Amblyomma, all with a signal peptide indicative of secretion [3]. DNAse activity has not been described in tick saliva but is present in saliva of mosquitoes of the Culex genus [51], where it may work in concert with hyaluronidases to decrease the viscosity of the extracellular matrix and help the formation of the feeding lesion. Endonuclease transcripts are also commonly found in sand fly and tsetse sialotranscriptomes [52, 53].
Protease inhibitor-containing domains
A total of 62 ESTs from the sialotranscriptome of H. m. rufipes code for proteins containing signatures of proteins previously associated with a protease inhibitory function, which are either ubiquitous or particular to ticks. A more detailed analysis of these transcripts follows.
Kunitz domain containing proteins
The Kunitz domain acquired its name from the Kunitz pancreatic trypsin inhibitor, also known as aprotinin, and later found to be ubiquitous [54]. Tick sialotranscriptomes [3], as well as those of the hematophagous flies of the genera Culicoides [55, 56] and Simulium [57], abound with transcripts coding for members of this family. Proteins containing single or multiple Kunitz domain were described in ticks, where Ixolaris, a double Kunitz protein, and Pentalaris, containing five domains, have been functionally characterized [58–62]. The Kunitz fold can also perform functions beyond protease inhibition, such as ion channel inhibition [63–66]; indeed, recently a modified Kunitz domain peptide from R. appendiculatus [67] was shown to activate maxiK channels in an in vitro system, suggesting a vasodilator function. Fifteen ESTs were found in the H. m. rufipes sialotranscriptome coding for members of the Kunitz family, allowing the deduction of two full-length coding sequences (CDS), one (Hex-1093) from a single and the other (Hex-13) from a double Kunitz family. Both polypeptides have less than 50% identity to their closest matches to the NR and to the assembled dataset described in Francischetti, et al. [3].
TIL domain containing proteins
The TIL (for trypsin inhibitor-like) domain typically contain ten cysteines forming five disulphide bonds and is found in many protease inhibitors. It belongs to the family I8 of the MEROPS database [68]. These polypeptides may also exert antimicrobial function [69]. Members of this family have been found ubiquitously in blood-feeding insect and tick sialomes, but very few have been characterized. A tick hemolymph anti microbial peptide (AMP) was previously reported to be a member of this family [70]. More recently, tick proteins containing TIL domains were characterized from R. microplus as subtilisin inhibitors with antimicrobial activity and expressed in various tick organs, including the SGs [71]. Hex-1007 is an interesting member of this family, as having 3 TIL domains in tandem starting at positions 91, 147, and 209. It has over 58% identity to proteins deducted from tick sialome ESTs and proteins from Amblyomma deposited on GenBank. Two ions matching the Hex-1007 sequence were obtained by MS/MS from gel fraction 18 (Fig 2, table 4). This region of the gel is near the 19 kDa marker, a smaller MW than the predicted 28 kDa for this protein. However, it is common for proteins containing many disulfide bonds to appear more compact and thus move faster when submitted to electrophoresis. Alternatively, this protein may be processed into shorter peptides.
Hex-55 is a shorter peptide that actually does not show a typical TIL domain, yet produces weak matches to proteins that are typical members of the family.
Basic tail protein family
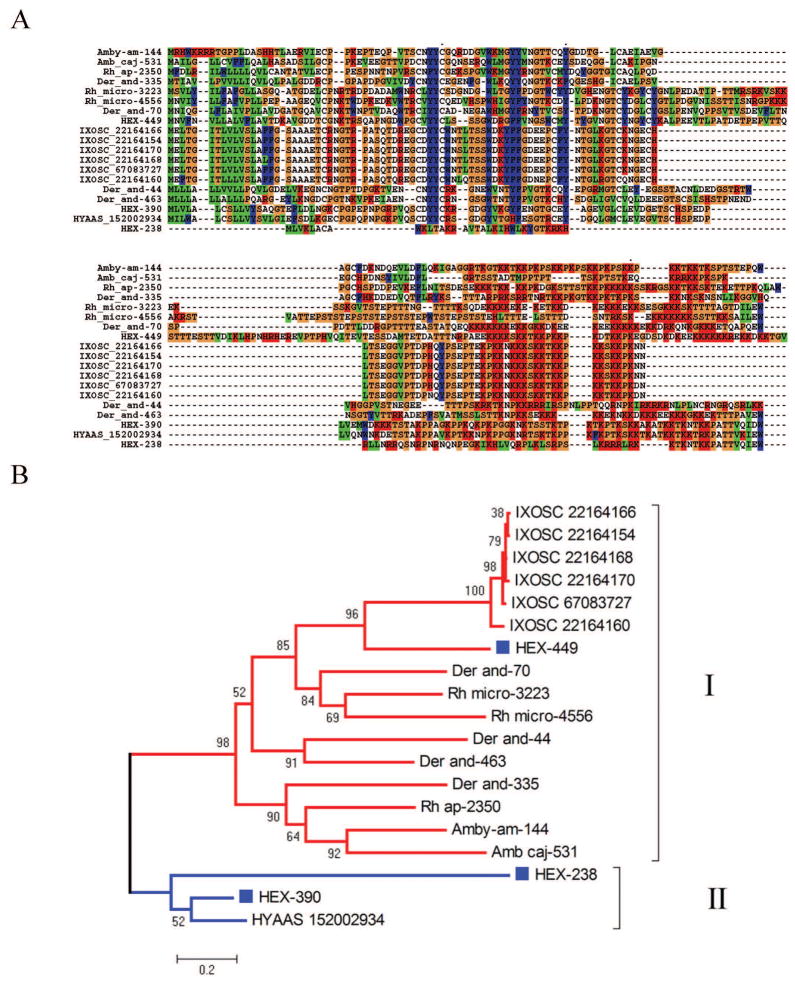
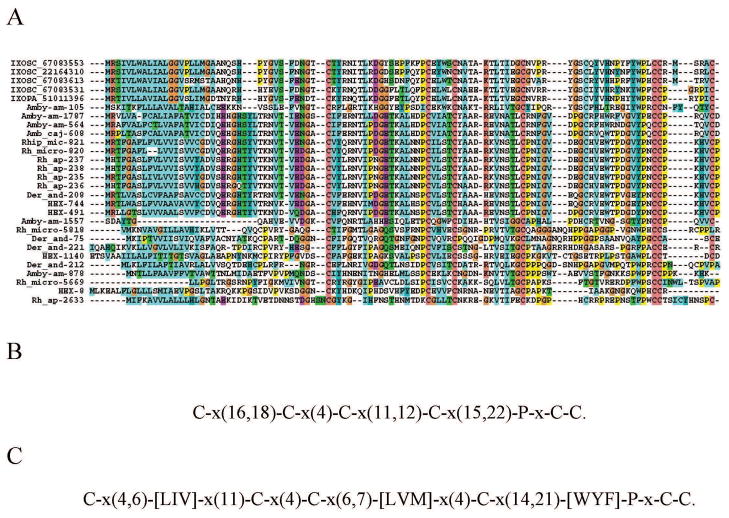
This protein family was so named due to a stretch of lysine residues in the carboxy terminus region of an expanded family of salivary proteins found in the I. scapularis sialome [14, 15]. They are unique to ticks [3] and can be identified by the PFAM domain PF07771, although many members are so divergent that they do not register it. Many members also have the conserved block C-x(13,21)-Y-Y-C-x(16,19)-C. Some members of this family have been characterized in I. scapularis as anti-clotting [72], thus their inclusion in this section. Hex-449 is a typical member of the family, having the characteristic PFAM domain and the YF-YF block. Its closest known relative is a protein reconstructed from D. andersoni ESTs, to which it has 37% identity and 50% similarity. Hex-390 presents 72% identity to a salivary protein from Hyalomma asiaticum named P18, and to basic tail proteins from Ixodes and Ornithodoros. It does not have the characteristic PFAM domain, but has the YF-YF signature, as well as a poly lysine tail. Hex-238 appears to be a very divergent member, presenting weak similarity to the P18 protein when compared to the NR database. Alignment of the H. m. rufipes sequences with those of other ticks suggests that Hex-238 may be a truncated member of the family or a protein resulting from a missing exon (Figure 3A). The phylogenetic tree shows that HEX-449 is a canonical basic tail protein within clade I as shown in Figure 3B. This clade has strong bootstrap support, indicating a common origin for this protein family in metastriates and prostriates. Clade II, however, does not group with the remaining proteins and does not have strong bootstrap support, suggesting it may derive from a different ancestor, or more likely have evolved beyond recognition of a common ancestor. Future inclusion of novel members of this family may lead to the merging of these clades.
Figure 3. The basic tail proteins of Hyalomma marginatum rufipes and other ticks.
A Clustal alignment. B Phylogenetic tree deducted from the alignment in A after 10,000 bootstraps. The number on the branches indicates percentage bootstrap support. The Hyalomma marginatum rufipes sequences are indicated by HEX and a square mark. The names initiating by IXOSC are from Ixodes scapularis, and the numbers are NCBI accession codes. HYAAS indicate a protein from H. asiaticum followed by its NCBI accession code. The remaining proteins derive from deducted expressed sequence tags available at DBEST and were described in a previous review [3]. The bar at the bottom indicates 20% amino acid divergence. For more detail, see text.
Madanin thrombin inhibitor family
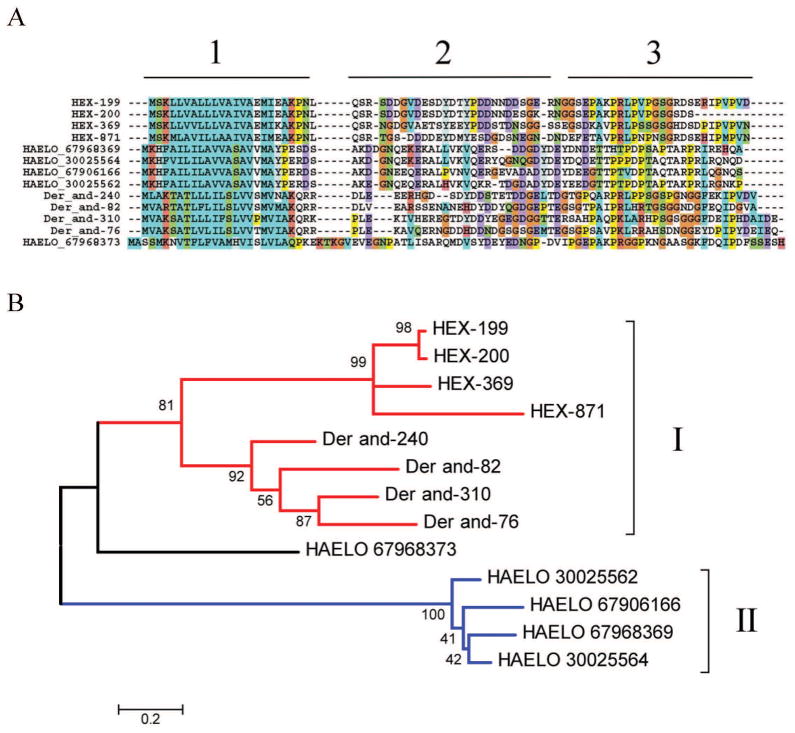
Madanins was the name given for related small polypeptides (~6 kDa) isolated from the tick Haemaphysalis longicornis that possess anti-thrombin activity [73]. Later another peptide, named chimadanin, was isolated from the same tick [74]. Additionally, variegin[75] was isolated from A. variegatum as a novel anti-thrombin peptide. Recently, these peptides were suggested to be part of a exclusive metastriate superfamily [3]. The transcriptome of H. m. rufipes indicates the presence of at least four genes that are possibly polymorphic. Alignment of the Ha. longicornis sequences with those of H. m. rufipes and deducted sequences from Dermacentor indicates three regions on these peptides: the signal peptide region, a second region with predominantly negatively charged peptides, and a third proline/serine/threonine-enriched region (marked as 1, 2 and 3 on Figure 4A). The phylogram provides strong bootstrap support for a common origin between H. m. rufipes and D. andersoni sequences, both members of the Rhipicephalinae subfamily (marked with I in Figure 4A), while the Haemaphysalis madanins are so divergent as to constitute a separate clade (marked II in Figure 4B), with the chimadanin (HAELO 67968373) possibly being a link between the two clades. Additional sequencing within the Rhipicephalinae and Haemaphysaline subfamilies may uncover more detailed phylogenetic relationships of these proteins. Notice that these mature peptides are small, with near 60 amino acids, and contain no cysteines, making them relatively straightforward for direct chemical synthesis. The anti-thrombin function of these peptides in Hyalomma and Dermacentor remains to be confirmed.
Figure 4. The Madanin family of metastriate ticks.
A Clustal alignment. B Phylogenetic tree deducted from the alignment in A after 10,000 bootstraps. The number on the branches indicates percentage bootstrap support. The Hyalomma marginatum rufipes sequences are indicated by HEX. The names initiating by HAELO are from Haemaphysalis longicornis, and the numbers are NCBI accession codes. The remaining proteins derive from deducted expressed sequence tags from Dermacentor andersoni available at DBEST and were described in a previous review [3]. The bar at the bottom indicates 20% amino acid divergence. For more detail, see text.
Lipocalin family
The lipocalin family is extremely diverse in ticks, where it serves multiple functions, as chelators of agonists (kratagonists) of hemostasis and inflammation, and other unrelated functions, such as anti-complement [3]. A previous review characterized 301 tick salivary lipocalins into 10 major groups [3]. The sialotranscriptome of H. m. rufipes yielded 18 ESTs that are similar to previously described tick lipocalins (Table 3). From these ESTs, five lipocalins can be derived. Two of these lipocalins, HEX-614, which is 22% identical and 41% similar to an A. americanum salivary protein, and less so to other tick lipocalins and HEX-938, which is most probably a splice variant of the same gene coding for HEX-614, are very divergent, forming a clade of their own with the Amblyomma proteins. HEX-133 produces a best match to another Amblyomma protein, previously classified as the Metastriate specific group III [3]. HEX-497 matches an R. microplus lipocalin with only 34% identity and belongs to Group I, subgroup B of lipocalins. HEX-497 appears abundantly expressed as indicated by the finding of 14 MS/MS ions producing a coverage of 99 % of the protein found in gel band 16, near the 28 kDa marker. Finally, HEX-43 matches a Ha. longicornis protein at 29% identity, belonging to the Group IIa of lipocalins. The specific functions of any of these lipocalins remain to be identified.
Glycine-rich family
Glycine-rich protein is a generic name encompassing a diverse group of proteins, including short and long proteins. Some of these have many GY repeats that are found in small antimicrobial peptides [76] but may be also found in cuticle proteins, where the tyrosine residue may be involved in crosslinking reactions. Very long glycine-rich proteins are found in metastriate ticks, are similar to spider-silk proteins, and may function as cement proteins to attach the tick mouthparts to their hosts [77, 78]. A total of 217 ESTs from the H. m. rufipes sialotranscriptome was classified as possibly coding for glycine-rich proteins (Table 3), from which 21 CDS were derived. Some of these coding sequences derive from abundantly expressed transcripts such as HEX-1069, a protein containing GY repeats deriving from 30 ESTs, and a homologue of the protein annotated as cement protein 64P-BA1 from R. appendiculatus. HEX-1069, as well as HEX-1143 and HEX-20 were identified by MS/MS in the gel shown in Fig 2 with 6 to 7 ions each at gel fractions 18 (HEX-1069) and 16 (HEX-1143 and HEX-20). HEX-235 is also abundantly expressed, with 42 ESTs, and is similar to the R. appendiculatus protein annotated as putative cement protein RIM36 and found to produce strong antibody response in cattle [79]. It was identified in the proteome experiment (Fig 2 and table 4) at fraction 8, a region of the gel between the markers for 97 and 64 kDa. The glycine rich proteins HEX-1057 and HEX-1043 were also identified in the same gel fraction, and those coded by HEX-750 and HEX-1117 were found in band 15, between the 28 and 39 kDa markers.
Mucins
Mucins are serine- and/or threonine-rich proteins, usually of low complexity, and having the motifs for being linked to N-acetyl-galactosamine residues [42]. Some of these invertebrate mucins also contain chitin-binding domains, suggesting they may coat the feeding channels of blood-sucking arthropods. Transcripts coding for mucins are commonly found in sialotranscriptomes of blood-sucking arthropods. HEX-930 and HEX-264 are probable alleles, deriving from four and three ESTs, respectively. They have a chitin-binding domain and are 50% identical to a Haemaphysalis protein annotated as a mucin.
HEX-826 represents the sequence of a threonine-rich protein with seven predicted galactosylation sites, a mature MW of 7.8 kDa, and is similar to a D. andersoni protein deducted from salivary ESTs but not to other protein in the NR database.
Immunity-related products
Twenty-seven ESTs in the H. m. rufipes sialotranscriptome code for proteins assigned to an immunity function (Table 3). Coding sequence for a peptidoglycan recognition protein, an ixoderin/ficolin, also involved in microbial pattern recognition and possibly associated with the activation of the invertebrate complement system [80], and a typical lysozyme were deduced from these ESTs. This lysozyme (coded by HEX-896) was identified in Fig 2 gel fraction 19 in a region of the gel consistent with its expected MW.
Putative secreted proteins with unknown function
8.9-kDa family
We previously characterized 60 tick salivary proteins as members of the uniquely Ixodidae protein family named 8.9-kDa family, of unknown function. The H. m. rufipes sialotranscriptome provides evidence for five members of this family. Alignment of these proteins with their relatives allows for detection of a conserved framework of cysteines, including a double Cys-Cys in their carboxy terminals and a few other conserved residues (Figure 5) indicative of a fast divergence of these proteins from a common ancestor. The 8.9 kDa protein coded by HEX-1038 was identified in gel fractions 18, 19 and 20 (Fig 2 and table 4).
Figure 5. The 8.9-kDa family of Ixodidae.
A Clustal alignment. B Conserved cysteine and proline framework. C Conserved framework including conserved amino acid substitutions. The Hyalomma marginatum rufipes sequences are indicated by HEX. The names initiating by IXOSC are from Ixodes scapularis, and the numbers are NCBI accession codes. The remaining proteins derive from deducted expressed sequence tags available at DBEST and were described in a previous review [3]. For more detail, see text.
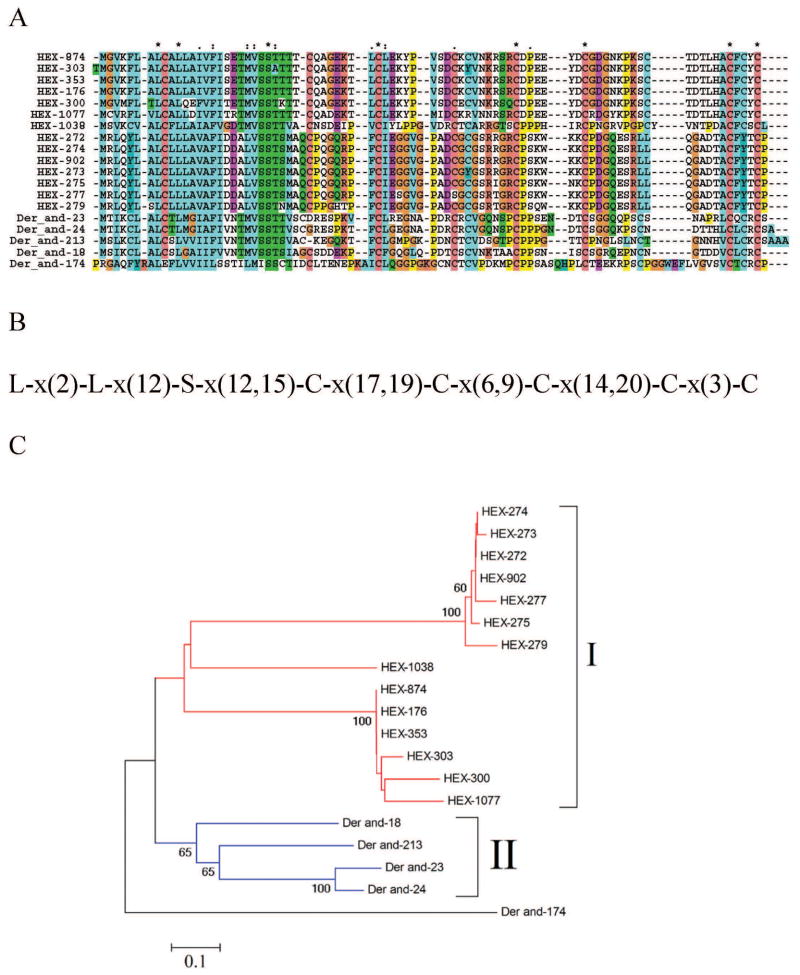
Dermacentor-Hyalomma specific 9-kDa family
We have previously characterized a D. andersoni-specific family, based on five protein sequences, named Dermacentor-specific 9-kDa expansion, due to their inability to significantly match any protein in the NR database, but being related among themselves, indicating gene duplications in D. andersoni followed by fast divergence. Somewhat surprisingly, the sialotranscriptome of H. m. rufipes produced 400 ESTs coding for members of this unique family, from which 14 coding sequences were derived. This is the most abundantly expressed family in H. m. rufipes. The coding sequences match various D. andersoni proteins from 36 to 49% identity, over nearly 100% of their lengths. These 14 proteins are possibly the product of at least 4 genes, two of which may be polymorphic, one producing the proteins HEX-902, HEX-272, HEX-275, HEX-277, HEX-274, and HEX-273, and the other producing proteins HEX-874, HEX-353, HEX-300, HEX-303, and HEX-176, as these proteins are within 10% identity from each other. The remaining proteins, HEX-1077 and HEX-1038, appear to derive from different genes. Alignment of the Hyalomma with the Dermacentor proteins shows a framework of conserved cysteine residues as well as two leucines and a serine residue. Phylogenetic analysis reveals two main mono-specific clades (marked I and II on Figure 6C) consisting of Hyalomma and Dermacentor proteins.
Figure 6. The 9-kDa family of Dermacentor and Hyalomma.
A Clustal alignment. B Conserved cysteine, leucine, and serine framework. C Phylogram derived from the alignment in A. The numbers on the branches represent percent bootstrap support (less than 50% are omitted). The Dermacentor sequences derive from deducted expressed sequence tags available at DBEST and were described in a previous review [3]. The bar at the bottom indicates 20% amino acid divergence. For more detail, see text.
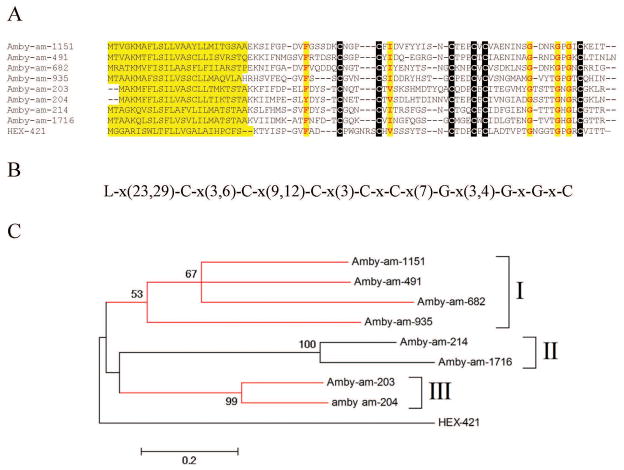
Hyalomma proteins similar to previously described orphan tick proteins
We have previously catalogued 917 tick proteins within 19 protein families as orphans, because they did not produce significant matches to proteins outside their own original genus [3]. The sialotranscriptome of H. m. rufipes contains transcripts that allow us to “de-orphanize” a few of these families, as follows: HEX-434 is similar to a R. microplus protein, while HEX-421 is similar to a monospecific family within A. americanum, within which they share a common framework of six cysteines, three glycines, and two additional sites with hydrophobic amino acids (Figure 7). Finally, a group of Hyalomma proteins with a common polylysine stretch in their mid-region, thus named “basic belly” proteins, matches an Ornithodoros protein. The basic belly protein coded by HEX-550 was identified in the gel band 16 (Fig 2 and table 4), near the 28 kDa marker. However, HEX-550 has a predicted MW of 7.8 kDa. Although many of the basic belly proteins have a signal peptide, the deducted polylysine stretch is coded by a polyA region, suggesting that these CDS could be artifacts derived from a 3′ untranslated region.
Figure 7. The AmbHy9 kDa family of Amblyomma and Hyalomma.
A Clustal alignment showing the signal peptide region and a conserved amino acid framework. B Conserved cysteine, glycine, and hydrophobic amino acids framework. C Phylogram derived from the alignment in A showing three Amblyomma americanum clades (I–III). The numbers on the branches represent percent bootstrap support (less than 50% are omitted). The Amblyomma sequences derive from deducted expressed sequence tags from available at DBEST and were described in a previous review [3]. The bar at the bottom indicates 20% amino acid divergence. For more detail, see text.
Orphan Hyalomma proteins
Additional file 2 presents 22 protein sequences coding for secreted products without any significant similarities to known proteins. Most of these putative polypeptides are small, and their CDS could derive from the 3′ region of truncated transporters that produce intramembrane helices that are interpreted as signal peptides.
Housekeeping proteins and transposable elements
The EST set acquired in this study allowed for the description of 144 coding sequences associated with housekeeping functions, including a set of conserved hypothetical proteins that might be related to protein synthesis or protein modification. Many of these products were identified in various electrophoresis gel bands (Fig 2 and table 4), including various ribosomal proteins, products associated with protein modification such as glutathione S-transferases, and proteins associated with energy metabolism. Strong signal for tubulin was also found in fraction 12 and neighbouring fractions. Two class I transposon sequence fragments were also extracted from the dataset (Additional file 2).
Bovine proteins identified in salivary gland homogenates
We and others have previously reported that host proteins appear in tick saliva [15, 81]. Indeed, there are tick lipocalins postulated to be carriers of host immunoglobulins from the tick hemolymph to tick saliva [81–84], with a possible role of detoxifying host proteins that may cross from the midgut to the hemolymph. We have previously identified host albumin, hemoglobin and immunoglobulin chains in the saliva of Ixodes scapularis. This study was done near 10 years ago, with no host mammalian proteomes available, and using low sensitivity Edman degradation of proteins [15]. Presently we searched for host proteins in the salivary gland homogenates of H. marginatum rufipes, by supplying the predicted proteome of Bos taurus to the Sequest program that searches the MS/MS generated ions against a target database (Table 5 and Supplemental file S3). To properly analyze the bovine proteome, we organized the proteins in a hyperlinked spreadsheet which was blasted against the available predicted proteins of the tick Ixodes scapularis (downloaded from http://iscapularis.vectorbase.org/Ixodes_scapularis/Info/Index), the only tick genome known, and, to facilitate protein annotation, against the SWISSPROT protein database and the Gene Ontology database. MS/MS derived peptides originating from the study reported in Fig 2 were mapped to this spreadsheet as indicated in the methods section. We thus obtained matches to 425 bovine proteins (supplemental file S3, worksheet named Bos matches). However, many of these matches are to very conserved proteins, such as histones, tubulins or ribosomal proteins, that are 100% or nearly so conserved with tick proteins. These matches could derive from tick as well as bovine proteins. We conservatively excluded from the bovine set those proteins producing more than 50% identity to tick proteins, as well as all myosins, obtaining a list of 77 bovine proteins (Supplemental file S3, worksheet 2). Several of these 77 proteins were related proteins by being either splice variants, or closely related gene families, such as hemoglobin. We thus removed these redundancies to produce table 4, with 22 bovine proteins that appear in the tick salivary gland proteome. The table is ordered by the fraction number shown in fig 2, from higher to lower MW. The predicted mature masses of the proteins (Table 4) are in accordance with the gel order, except for complement C3 and fibrinogen; C3 appears on fraction 10, between the markers for 64 and 51 kDa, and incompatible with the C3 predicted mass of 185 kDa, indicating C3 cleavage; further C3 fragments appear on fractions 14 and 17. Fibrinogen appears most abundantly covered on fraction 14, under the 39 kDa marker, while the mature protein has a predicted mass of 53 kDa, indicating fibrinogen cleavage. Notice that the list of bovine proteins includes abundant ions for serum albumin, hemoglobin and immunoglobulin chains, as well as for alpha-2-macroglobulin. Proteins abundant in red cells such as band 3 anion transport protein and carbonic anhydrase were also found, as well as leukocyte-derived products azurocidin and the antimicrobial cathelicidin.
Table 5.
Bovine proteins identified in the polyacrylamide gel shown in Figure 2
| NCBI accession | Mature MW | Description | Fraction --> Number of Ions | Coverage in aa residues (1) | Percent protein coverage (1) |
|---|---|---|---|---|---|
| gi|157954061 | 165 | alpha-2-macroglobulin | Fr-2 -> 16| Fr-3 -> 15| Fr-7 -> 14| | 279 | 18.5 |
| gi|30794360 | 104 | band 3 anion transport protein | Fr-2 -> 4| Fr-3 -> 4| Fr-5 -> 2| | 47 | 5.1 |
| gi|297462813 | 119 | ceruloplasmin | Fr-6 -> 2| | 33 | 3.1 |
| gi|76677897 | 138 | complement factor H precursor | Fr-6 -> 2| | 33 | 2.7 |
| gi|76627249 | 103 | glucosidase, alpha; neutral C | Fr-7 -> 2| | 24 | 2.6 |
| gi|297462666 | 80 | primary amine oxidase | Fr-8 -> 3| | 48 | 6.4 |
| gi|27806815 | 88 | plasminogen precursor | Fr-9 -> 4| | 55 | 6.8 |
| gi|114326282 | 75 | serotransferrin precursor | Fr-9 -> 8| Fr-10 -> 5| | 124 | 17.6 |
| gi|99028969 | 185 | complement C3 | Fr-10 -> 9| Fr-14 -> 6| Fr-17 -> 3| | 155 | 9.3 |
| gi|30794280 | 67 | serum albumin precursor | Fr-11 -> 16| Fr-10 -> 14| Fr-12 -> 3| | 219 | 36.1 |
| gi|119914040 | 31 | Serpin A3-3 | Fr-11 -> 2| | 31 | 10.4 |
| gi|218931172 | 53 | fibrinogen beta chain | Fr-14 -> 10| Fr-13 -> 5| Fr-12 -> 4| | 138 | 27.9 |
| gi|297464089 | 44 | serpin peptidase inhibitor, clade B like | Fr-14 -> 2| | 23 | 5.9 |
| gi|297477165 | 24 | azurocidin 1 preproprotein-like | Fr-16 -> 2| | 32 | 13.0 |
| gi|30466252 | 29 | carbonic anhydrase 2 | Fr-16 -> 5| | 75 | 28.8 |
| gi|139948632 | 22 | immunoglobulin lambda-like | Fr-17 -> 16| Fr-18 -> 8| Fr-16 -> 5| | 254 | 108.1 |
| gi|297460000 | 23 | Ig kappa chain | Fr-17 -> 5| | 97 | 41.3 |
| gi|297461641 | 22 | immunoglobulin lambda-like complement component 4 binding | Fr-17 -> 8| Fr-18 -> 2| Fr-16 -> 2| | 131 | 55.5 |
| gi|76677514 | 18 | protein | Fr-18 -> 3| | 42 | 21.5 |
| gi|27808640 | 18 | peptidoglycan recognition protein | Fr-19 -> 4| | 57 | 30.0 |
| gi|27819608 | 16 | hemoglobin subunit beta | Fr-20 -> 37| Fr-19 -> 7| Fr-18 -> 5| | 500 | 344.8 |
| gi|27807341 | 14 | cathelicidin-1 precursor | Fr-20 -> 4| Fr-19 -> 2| | 64 | 41.3 |
On fraction with higher number of ions only; redundant ions are considered; coverage may be higher than 100%.
The appearance of host proteins on tick salivary gland homogenates could be considered an artefact of contamination during dissection, possibly from the tick gut. However, our samples were carefully collected and no EST produced matches to bovine sequences, as could happen in the case the SG were contaminated with bovine blood. Host Ig in tick hemolymph and saliva were also previously characterized in detailed studies [81]. As indicated before [15], it is interesting to speculate whether these host proteins, while passing through the tick salivary glands, may be submitted to the tick protein glycosylation machinery, although no significant increase in mass for any product was found. Incorporation of these tick epitopes into self molecules may be a strategy for tick suppression of host immunity against carbohydrate antigens.
Overview of proteomics results
One hundred and fifteen contigs were identified by the proteomic data. The distribution of the matched proteins among functional classes, considering only those that obtained at least two ion matches in one gel slice (supplemental file S1, worksheet named “proteome analysis” and Table 6) shows members of the protein synthesis machinery as the most abundantly detected, followed by secreted proteins, protein modification machinery and energy metabolism; these classes account for over 90% of the identified proteins. No correlation was found between the transcript abundance (measured by their number of EST’s) and the number of matching MS/MS ions for each contig (R=0.13) (Supplemental data S1, worksheet “proteome analysis”).
Table 6.
Functional classification of tick transcripts identified by MS/MS
| Class | Number of Contigs | Number of ions | Ions/contig |
|---|---|---|---|
| Protein synthesis machinery | 61 | 835 | 13.69 |
| Secreted | 23 | 182 | 8.00 |
| Protein modification machinery | 12 | 289 | 24.08 |
| Metabolism, energy | 10 | 126 | 12.60 |
| Proteasome machinery | 3 | 46 | 15.33 |
| Transcription machinery | 3 | 34 | 11.33 |
| Nuclear regulation | 2 | 62 | 31.00 |
| Cytoskeletal | 2 | 56 | 28.00 |
| Oxidant metabolism / detoxification | 2 | 48 | 24.00 |
| Signal transduction | 1 | 32 | 32.00 |
| Metabolism, carbohydrate | 1 | 4 | 4.00 |
| Total | 115 | 1676 |
Conclusions
Several protein families previously found in tick salivary transcriptomes were identified in H. m. rufipes, such as the Kunitz, basic tail, madanin, lipocalin, glycine-rich, mucins, immunity-related, and 8.9-kDa family, as well as protein families previously found only in the metastriate Dermacentor genus, such as the 9-kDa family. Most of these proteins have no known function. Many orphan proteins were found that do not match known proteins, but have signal peptides indicative of secretion, suggesting these are Hyalomma-specific proteins. This annotated dataset can assist in the discovery of new targets for anti-tick vaccines, as well as help to identify pharmacologically active proteins. In this current study, this annotated transcript data was used to identify salivary protein expression in a proteomic experiment. We additionally identified bovine host proteins in salivary homogenates reinforcing the idea that host proteins are recycled back to the host after ingestion.
Supplementary Material
Hyperlinked Excel spreadsheet containing annotated assembled ESTs (Supplementary Table 1): http://exon.niaid.nih.gov/transcriptome/Hmrufipes/Hmr-S1.xls
Hyperlinked Excel spreadsheet containing annotated coding sequences (Supplementary Table 2): http://exon.niaid.nih.gov/transcriptome/Hmrufipes/Hmr-S2.xls
Hypelinked Excel spreasheet containing annotated bovine proteins (Supplementary Table 3): http://exon.niaid.nih.gov/transcriptome/Hmrufipes/Hmr-S3.xls
Acknowledgments
This work was supported by the Intramural Research Program of the Division of Intramural Research, National Institute of Allergy and Infectious Diseases, National Institutes of Health. We thank NIAID intramural editor Brenda Rae Marshall for assistance. We are grateful for the support of the Research Technologies Section, Rocky Mountain Laboratories, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Hamilton, MT 59840, USA, for DNA sequencing.
Because all authors are government employees and this is a government work, the work is in the public domain in the United States. Notwithstanding any other agreements, the NIH reserves the right to provide the work to PubMedCentral for display and use by the public, and PubMedCentral may tag or modify the work consistent with its customary practices. You can establish rights outside of the U.S. subject to a government use license.
Abbreviations
- AMP
antimicrobial peptide
- CDS
coding sequence
- EST
expressed sequence tag
- H class
housekeeping
- NR
non-redundant
- NCBI
National Center of Biological Information
- S class
secreted
- SG
salivary gland
- TIL
trypsin inhibitor-like
- TE
transposable element
- U class
unknown function
Footnotes
Authors’ contributions
IMBF and JMA helped with data analysis, sample preparation, and manuscript editing. NM helped with tick collection and manuscript editing.. VMP helped with DNA sequencing. JMCR did most of the data analysis and wrote the bulk of the manuscript. All authors read and approved the final manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Nava S, Guglielmone AA, Mangold AJ. An overview of systematics and evolution of ticks. Front Biosci. 2009;14:2857–77. doi: 10.2741/3418. [DOI] [PubMed] [Google Scholar]
- 2.Barker SC, Murrell A. Systematics and evolution of ticks with a list of valid genus and species names. Parasitology. 2004;129 (Suppl):S15–36. doi: 10.1017/s0031182004005207. [DOI] [PubMed] [Google Scholar]
- 3.Francischetti IMB, Sá-Nunes A, Mans BJ, Santos IM, Ribeiro JMC. The role of saliva in tick feeding. Frontiers in Biosciences. 2009;14:2051–88. doi: 10.2741/3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hovius JW, Levi M, Fikrig E. Salivating for knowledge: potential pharmacological agents in tick saliva. PLoS medicine. 2008;5:e43. doi: 10.1371/journal.pmed.0050043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Steen NA, Barker SC, Alewood PF. Proteins in the saliva of the Ixodida (ticks): pharmacological features and biological significance. Toxicon. 2006;47:1–20. doi: 10.1016/j.toxicon.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 6.Brossard M, Wikel SK. Tick immunobiology. Parasitology. 2004;129 (Suppl):S161–76. doi: 10.1017/s0031182004004834. [DOI] [PubMed] [Google Scholar]
- 7.Valenzuela JG. Exploring tick saliva: from biochemistry to ‘sialomes’ and functional genomics. Parasitology. 2004;129 (Suppl):S83–94. doi: 10.1017/s0031182004005189. [DOI] [PubMed] [Google Scholar]
- 8.Francischetti IM. Platelet aggregation inhibitors from hematophagous animals. Toxicon. 2010;56:1130–44. doi: 10.1016/j.toxicon.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mans BJ, Louw AI, Neitz AW. Evolution of hematophagy in ticks: common origins for blood coagulation and platelet aggregation inhibitors from soft ticks of the genus Ornithodoros. Mol Biol Evol. 2002;19:1695–705. doi: 10.1093/oxfordjournals.molbev.a003992. [DOI] [PubMed] [Google Scholar]
- 10.Mans BJ, Andersen JF, Francischetti IM, Valenzuela JG, Schwan TG, Pham VM, et al. Comparative sialomics between hard and soft ticks: Implications for the evolution of blood-feeding behavior. Insect biochemistry and molecular biology. 2008;38:42–58. doi: 10.1016/j.ibmb.2007.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mans BJ, Andersen JF, Schwan TG, Ribeiro JM. Characterization of anti-hemostatic factors in the argasid, Argas monolakensis: Implications for the evolution of blood-feeding in the soft tick family. Insect biochemistry and molecular biology. 2008;38:22–41. doi: 10.1016/j.ibmb.2007.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Francischetti IM, Mans BJ, Meng Z, Gudderra N, Veenstra TD, Pham VM, et al. An insight into the sialome of the soft tick, Ornithodorus parkeri. Insect biochemistry and molecular biology. 2008;38:1–21. doi: 10.1016/j.ibmb.2007.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Francischetti IM, Meng Z, Mans BJ, Gudderra N, Hall M, Veenstra TD, et al. An insight into the salivary transcriptome and proteome of the soft tick and vector of epizootic bovine abortion, Ornithodoros coriaceus. Journal of proteomics. 2008;71:493–512. doi: 10.1016/j.jprot.2008.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ribeiro JM, Alarcon-Chaidez F, Francischetti IM, Mans BJ, Mather TN, Valenzuela JG, et al. An annotated catalog of salivary gland transcripts from Ixodes scapularis ticks. Insect Biochem Mol Biol. 2006;36:111–29. doi: 10.1016/j.ibmb.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 15.Valenzuela JG, Francischetti IMB, Pham VM, Garfield MK, Mather TN, Ribeiro JMC. Exploring the sialome of the tick, Ixodes scapularis. J Exp Biol. 2002;205:2843–64. doi: 10.1242/jeb.205.18.2843. [DOI] [PubMed] [Google Scholar]
- 16.Francischetti IM, My Pham V, Mans BJ, Andersen JF, Mather TN, Lane RS, et al. The transcriptome of the salivary glands of the female western black-legged tick Ixodes pacificus (Acari: Ixodidae) Insect biochemistry and molecular biology. 2005;35:1142–61. doi: 10.1016/j.ibmb.2005.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chmelar J, Anderson JM, Mu J, Jochim RC, Valenzuela JG, Kopecky J. Insight into the sialome of the castor bean tick, Ixodes ricinus. BMC genomics. 2008;9:233. doi: 10.1186/1471-2164-9-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aljamali MN, Hern L, Kupfer D, Downard S, So S, Roe BA, et al. Transcriptome analysis of the salivary glands of the female tick Amblyomma americanum (Acari: Ixodidae) Insect molecular biology. 2009;18:129–54. doi: 10.1111/j.1365-2583.2009.00863.x. [DOI] [PubMed] [Google Scholar]
- 19.Batista IF, Chudzinski-Tavassi AM, Faria F, Simons SM, Barros-Batestti DM, Labruna MB, et al. Expressed sequence tags (ESTs) from the salivary glands of the tick Amblyomma cajennense (Acari: Ixodidae) Toxicon. 2008;51:823–34. doi: 10.1016/j.toxicon.2007.12.011. [DOI] [PubMed] [Google Scholar]
- 20.Nene V, Lee D, Quackenbush J, Skilton R, Mwaura S, Gardner MJ, et al. AvGI, an index of genes transcribed in the salivary glands of the ixodid tick Amblyomma variegatum. International journal for parasitology. 2002;32:1447–56. doi: 10.1016/s0020-7519(02)00159-5. [DOI] [PubMed] [Google Scholar]
- 21.Alarcon-Chaidez FJ, Sun J, Wikel SK. Transcriptome analysis of the salivary glands of Dermacentor andersoni Stiles (Acari: Ixodidae) Insect biochemistry and molecular biology. 2007;37:48–71. doi: 10.1016/j.ibmb.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 22.Nene V, Lee D, Kang’a S, Skilton R, Shah T, de Villiers E, et al. Genes transcribed in the salivary glands of female Rhipicephalus appendiculatus ticks infected with Theileria parva. Insect Biochem Mol Biol. 2004;34:1117–28. doi: 10.1016/j.ibmb.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 23.Zeller HG, Cornet JP, Diop A, Camicas JL. Crimean-Congo hemorrhagic fever in ticks (Acari: Ixodidae) and ruminants: field observations of an epizootic in Bandia, Senegal (1989–1992) Journal of medical entomology. 1997;34:511–6. doi: 10.1093/jmedent/34.5.511. [DOI] [PubMed] [Google Scholar]
- 24.Zeller HG, Cornet JP, Camicas JL. Experimental transmission of Crimean-Congo hemorrhagic fever virus by west African wild ground-feeding birds to Hyalomma marginatum rufipes ticks. The American journal of tropical medicine and hygiene. 1994;50:676–81. doi: 10.4269/ajtmh.1994.50.676. [DOI] [PubMed] [Google Scholar]
- 25.Zeller HG, Cornet JP, Camicas JL. Crimean-Congo haemorrhagic fever virus infection in birds: field investigations in Senegal. Research in virology. 1994;145:105–9. doi: 10.1016/s0923-2516(07)80012-4. [DOI] [PubMed] [Google Scholar]
- 26.Shepherd AJ, Swanepoel R, Shepherd SP, Leman PA, Mathee O. Viraemic transmission of Crimean-Congo haemorrhagic fever virus to ticks. Epidemiology and infection. 1991;106:373–82. doi: 10.1017/s0950268800048524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Linthicum KJ, Bailey CL. Ecology of Crimean-Congo hemorrhagic fever. In: Mather DESaTN., editor. Ecological dynamics of tick-borne zoonoses. New York: Oxford University Press; 1994. pp. 392–427. [Google Scholar]
- 28.Horak IG, Golezardy H, Uys AC. Ticks associated with the three largest wild ruminant species in southern Africa. The Onderstepoort journal of veterinary research. 2007;74:231–42. doi: 10.4102/ojvr.v74i3.126. [DOI] [PubMed] [Google Scholar]
- 29.Horak IG, MacIvor KM. The scrub hare, a reliable indicator of the presence of Hyalomma ticks in the Cape Province. Journal of the South African Veterinary Association. 1987;58:15–9. [PubMed] [Google Scholar]
- 30.Salih DA, Julla, Hassan SM, El Hussein AM, Jongejan F. Reliminary survey of ticks (Acari: Ixodidae) on cattle in Central Equatoria State, Southern Sudan. The Onderstepoort journal of veterinary research. 2008;75:47–53. doi: 10.4102/ojvr.v75i1.87. [DOI] [PubMed] [Google Scholar]
- 31.Wanzala W, Okanga S. Ticks (Acari: Ixodidae) associated with wildlife and vegetation of Haller park along the Kenyan coastline. Journal of medical entomology. 2006;43:789–94. doi: 10.1603/0022-2585(2006)43[789:taiaww]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 32.Ruiz-Fons F, Fernandez-de-Mera IG, Acevedo P, Hofle U, Vicente J, de la Fuente J, et al. Ixodid ticks parasitizing Iberian red deer (Cervus elaphus hispanicus) and European wild boar (Sus scrofa) from Spain: geographical and temporal distribution. Veterinary parasitology. 2006;140:133–42. doi: 10.1016/j.vetpar.2006.03.033. [DOI] [PubMed] [Google Scholar]
- 33.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang X, Madan A. CAP3: A DNA sequence assembly program. Genome Res. 1999;9:868–77. doi: 10.1101/gr.9.9.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–82. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–9. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bateman A, Birney E, Durbin R, Eddy SR, Howe KL, Sonnhammer EL. The Pfam protein families database. Nucleic Acids Res. 2000;28:263–6. doi: 10.1093/nar/28.1.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Letunic I, Goodstadt L, Dickens NJ, Doerks T, Schultz J, Mott R, et al. Recent improvements to the SMART domain-based sequence annotation resource. Nucleic Acids Res. 2002;30:242–4. doi: 10.1093/nar/30.1.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tatusov RL, Fedorova ND, Jackson JD, Jacobs AR, Kiryutin B, Koonin EV, et al. The COG database: an updated version includes eukaryotes. BMC bioinformatics. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marchler-Bauer A, Panchenko AR, Shoemaker BA, Thiessen PA, Geer LY, Bryant SH. CDD: a database of conserved domain alignments with links to domain three-dimensional structure. Nucleic Acids Res. 2002;30:281–3. doi: 10.1093/nar/30.1.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nielsen H, Engelbrecht J, Brunak S, von Heijne G. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- 42.Julenius K, Molgaard A, Gupta R, Brunak S. Prediction, conservation analysis, and structural characterization of mammalian mucin-type O-glycosylation sites. Glycobiology. 2005;15:153–64. doi: 10.1093/glycob/cwh151. [DOI] [PubMed] [Google Scholar]
- 43.Kumar S, Tamura K, Nei M. MEGA3: Integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Brief Bioinform. 2004;5:150–63. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- 44.Jeanmougin F, Thompson JD, Gouy M, Higgins DG, Gibson TJ. Multiple sequence alignment with Clustal X. Trends Biochem Sci. 1998;23:403–5. doi: 10.1016/s0968-0004(98)01285-7. [DOI] [PubMed] [Google Scholar]
- 45.Friedman R, Hughes AL. Codon volatility as an indicator of positive selection: data from eukaryotic genome comparisons. Molecular biology and evolution. 2005;22:542–6. doi: 10.1093/molbev/msi038. [DOI] [PubMed] [Google Scholar]
- 46.Hulo N, Bairoch A, Bulliard V, Cerutti L, De Castro E, Langendijk-Genevaux PS, et al. The PROSITE database. Nucleic acids research. 2006;34:D227–30. doi: 10.1093/nar/gkj063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Arca B, Lombardo F, Valenzuela JG, Francischetti IM, Marinotti O, Coluzzi M, et al. An updated catalogue of salivary gland transcripts in the adult female mosquito, Anopheles gambiae. J Exp Biol. 2005;208:3971–86. doi: 10.1242/jeb.01849. [DOI] [PubMed] [Google Scholar]
- 48.Francischetti IMB, Sa-Nunes A, Mans B, Santos IM, Ribeiro JMC. The role of saliva in tick feeding. In: delaFuente J, Kocan KM, Sonenshine DE, editors. Molecular biology of ticks and the tick-pathogen interface: Frontiers in Bioscience. 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Harnnoi T, Sakaguchi T, Nishikawa Y, Xuan X, Fujisaki K. Molecular characterization and comparative study of 6 salivary gland metalloproteases from the hard tick, Haemaphysalis longicornis. Comparative biochemistry and physiology. 2007;147:93–101. doi: 10.1016/j.cbpb.2006.12.008. [DOI] [PubMed] [Google Scholar]
- 50.Francischetti IM, Mather TN, Ribeiro JM. Cloning of a salivary gland metalloprotease and characterization of gelatinase and fibrin(ogen)lytic activities in the saliva of the Lyme disease tick vector Ixodes scapularis. Biochemical and biophysical research communications. 2003;305:869–75. doi: 10.1016/s0006-291x(03)00857-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Calvo E, Ribeiro JM. A novel secreted endonuclease from Culex quinquefasciatus salivary glands. J Exp Biol. 2006;209:2651–9. doi: 10.1242/jeb.02267. [DOI] [PubMed] [Google Scholar]
- 52.Valenzuela JG, Garfield M, Rowton ED, Pham VM. Identification of the most abundant secreted proteins from the salivary glands of the sand fly Lutzomyia longipalpis, vector of Leishmania chagasi. J Exp Biol. 2004;207:3717–29. doi: 10.1242/jeb.01185. [DOI] [PubMed] [Google Scholar]
- 53.Haddow JD, Poulis B, Haines LR, Gooding RH, Aksoy S, Pearson TW. Identification of major soluble salivary gland proteins in teneral Glossina morsitans morsitans. Insect biochemistry and molecular biology. 2002;32:1045–53. doi: 10.1016/s0965-1748(02)00042-5. [DOI] [PubMed] [Google Scholar]
- 54.Ascenzi P, Bocedi A, Bolognesi M, Spallarossa A, Coletta M, De Cristofaro R, et al. The bovine basic pancreatic trypsin inhibitor (Kunitz inhibitor): a milestone protein. Current protein & peptide science. 2003;4:231–51. doi: 10.2174/1389203033487180. [DOI] [PubMed] [Google Scholar]
- 55.Wilson AD, Heesom KJ, Mawby WJ, Mellor PS, Russell CL. Identification of abundant proteins and potential allergens in Culicoides nubeculosus salivary glands. Veterinary immunology and immunopathology. 2008;122:94–103. doi: 10.1016/j.vetimm.2007.10.011. [DOI] [PubMed] [Google Scholar]
- 56.Campbell CL, Vandyke KA, Letchworth GJ, Drolet BS, Hanekamp T, Wilson WC. Midgut and salivary gland transcriptomes of the arbovirus vector Culicoides sonorensis (Diptera: Ceratopogonidae) Insect Mol Biol. 2005;14:121–36. doi: 10.1111/j.1365-2583.2004.00537.x. [DOI] [PubMed] [Google Scholar]
- 57.Andersen JF, Pham VM, Meng Z, Champagne DE, Ribeiro JM. Insight into the Sialome of the Black Fly, Simulium vittatum. Journal of proteome research. 2009;8:1474–88. doi: 10.1021/pr8008429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Monteiro RQ, Rezaie AR, Bae JS, Calvo E, Andersen JF, Francischetti IM. Ixolaris binding to factor X reveals a precursor state of factor Xa heparin-binding exosite. Protein Sci. 2008;17:146–53. doi: 10.1110/ps.073016308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nazareth RA, Tomaz LS, Ortiz-Costa S, Atella GC, Ribeiro JM, Francischetti IM, et al. Antithrombotic properties of Ixolaris, a potent inhibitor of the extrinsic pathway of the coagulation cascade. Thrombosis and haemostasis. 2006;96:7–13. doi: 10.1160/TH06-02-0105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Monteiro RQ, Rezaie AR, Ribeiro JM, Francischetti IM. Ixolaris: a factor Xa heparin-binding exosite inhibitor. The Biochemical journal. 2005;387:871–7. doi: 10.1042/BJ20041738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Francischetti IM, Mather TN, Ribeiro JM. Penthalaris, a novel recombinant five-Kunitz tissue factor pathway inhibitor (TFPI) from the salivary gland of the tick vector of Lyme disease, Ixodes scapularis. Thrombosis and haemostasis. 2004;91:886–98. doi: 10.1160/TH03-11-0715. [DOI] [PubMed] [Google Scholar]
- 62.Monteiro RQ, Rezaie AR, Ribeiro JM, Francischetti IM. Ixolaris: A factor Xa heparin binding exosite inhibitor. Biochem J. 2004 doi: 10.1042/BJ20041738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lucchesi KJ, Moczydlowski E. On the interaction of bovine pancreatic trypsin inhibitor with maxi Ca(2+)-activated K+ channels. A model system for analysis of peptide-induced subconductance states. The Journal of general physiology. 1991;97:1295–319. doi: 10.1085/jgp.97.6.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kwong PD, McDonald NQ, Sigler PB, Hendrickson WA. Structure of beta 2-bungarotoxin: potassium channel binding by Kunitz modules and targeted phospholipase action. Structure. 1995;3:1109–19. doi: 10.1016/s0969-2126(01)00246-5. [DOI] [PubMed] [Google Scholar]
- 65.Harvey AL. Recent studies on dendrotoxins and potassium ion channels. General pharmacology. 1997;28:7–12. doi: 10.1016/s0306-3623(96)00173-5. [DOI] [PubMed] [Google Scholar]
- 66.Castaneda O, Harvey AL. Discovery and characterization of cnidarian peptide toxins that affect neuronal potassium ion channels. Toxicon. 2009 doi: 10.1016/j.toxicon.2009.02.032. [DOI] [PubMed] [Google Scholar]
- 67.Paesen GC, Siebold C, Dallas ML, Peers C, Harlos K, Nuttall PA, et al. An ion-channel modulator from the saliva of the brown ear tick has a highly modified Kunitz/BPTI structure. Journal of molecular biology. 2009;389:734–47. doi: 10.1016/j.jmb.2009.04.045. [DOI] [PubMed] [Google Scholar]
- 68.Rawlings ND, Morton FR, Barrett AJ. MEROPS: the peptidase database. Nucleic acids research. 2006;34:D270–2. doi: 10.1093/nar/gkj089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li WF, Ma GX, Zhou XX. Apidaecin-type peptides: biodiversity, structure-function relationships and mode of action. Peptides. 2006;27:2350–9. doi: 10.1016/j.peptides.2006.03.016. [DOI] [PubMed] [Google Scholar]
- 70.Fogaca AC, Almeida IC, Eberlin MN, Tanaka AS, Bulet P, Daffre S. Ixodidin, a novel antimicrobial peptide from the hemocytes of the cattle tick Boophilus microplus with inhibitory activity against serine proteinases. Peptides. 2005;27:667–74. doi: 10.1016/j.peptides.2005.07.013. [DOI] [PubMed] [Google Scholar]
- 71.Sasaki SD, de Lima CA, Lovato DV, Juliano MA, Torquato RJ, Tanaka AS. BmSI-7, a novel subtilisin inhibitor from Boophilus microplus, with activity toward Pr1 proteases from the fungus Metarhizium anisopliae. Experimental parasitology. 2008;118:214–20. doi: 10.1016/j.exppara.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 72.Narasimhan S, Koski RA, Beaulieu B, Anderson JF, Ramamoorthi N, Kantor F, et al. A novel family of anticoagulants from the saliva of Ixodes scapularis. Insect molecular biology. 2002;11:641–50. doi: 10.1046/j.1365-2583.2002.00375.x. [DOI] [PubMed] [Google Scholar]
- 73.Iwanaga S, Okada M, Isawa H, Morita A, Yuda M, Chinzei Y. Identification and characterization of novel salivary thrombin inhibitors from the ixodidae tick, Haemaphysalis longicornis. European journal of biochemistry / FEBS. 2003;270:1926–34. doi: 10.1046/j.1432-1033.2003.03560.x. [DOI] [PubMed] [Google Scholar]
- 74.Nakajima C, Imamura S, Konnai S, Yamada S, Nishikado H, Ohashi K, et al. A novel gene encoding a thrombin inhibitory protein in a cDNA library from Haemaphysalis longicornis salivary gland. The Journal of veterinary medical science / the Japanese Society of Veterinary Science. 2006;68:447–52. doi: 10.1292/jvms.68.447. [DOI] [PubMed] [Google Scholar]
- 75.Koh CY, Kazimirova M, Trimnell A, Takac P, Labuda M, Nuttall P, et al. Variegin, a novel class of fast and tight-binding thrombin inhibitor from the tropical bont tick. The Journal of biological chemistry. 2007 doi: 10.1074/jbc.M705600200. [DOI] [PubMed] [Google Scholar]
- 76.Couillault C, Pujol N, Reboul J, Sabatier L, Guichou JF, Kohara Y, et al. TLR-independent control of innate immunity in Caenorhabditis elegans by the TIR domain adaptor protein TIR-1, an ortholog of human SARM. Nat Immunol. 2004;5:488–94. doi: 10.1038/ni1060. [DOI] [PubMed] [Google Scholar]
- 77.Sauer JR, McSwain JL, Bowman AS, Essenberg RC. Tick salivary gland physiology. Annual review of entomology. 1995;40:245–67. doi: 10.1146/annurev.en.40.010195.001333. [DOI] [PubMed] [Google Scholar]
- 78.Maruyama SR, Anatriello E, Anderson JM, Ribeiro JM, Brandao LG, Valenzuela JG, et al. The expression of genes coding for distinct types of glycine-rich proteins varies according to the biology of three metastriate ticks, Rhipicephalus (Boophilus) microplus, Rhipicephalus sanguineus and Amblyomma cajennense. BMC genomics. 11:363. doi: 10.1186/1471-2164-11-363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bishop R, Lambson B, Wells C, Pandit P, Osaso J, Nkonge C, et al. A cement protein of the tick Rhipicephalus appendiculatus, located in the secretory e cell granules of the type III salivary gland acini, induces strong antibody responses in cattle. International journal for parasitology. 2002;32:833–42. doi: 10.1016/s0020-7519(02)00027-9. [DOI] [PubMed] [Google Scholar]
- 80.Fujita T, Matsushita M, Endo Y. The lectin-complement pathway--its role in innate immunity and evolution. Immunological reviews. 2004;198:185–202. doi: 10.1111/j.0105-2896.2004.0123.x. [DOI] [PubMed] [Google Scholar]
- 81.Wang H, Nuttall PA. Excretion of host immunoglobulin in tick saliva and detection of IgG-binding proteins in tick haemolymph and salivary glands. Parasitology. 1994;109 (Pt 4):525–30. doi: 10.1017/s0031182000080781. [DOI] [PubMed] [Google Scholar]
- 82.Wang H, Nuttall PA. Immunoglobulin G binding proteins in male Rhipicephalus appendiculatus ticks. Parasite Immunol. 1995;17:517–24. doi: 10.1111/j.1365-3024.1995.tb00882.x. [DOI] [PubMed] [Google Scholar]
- 83.Wang H, Nuttall PA. Immunoglobulin-G binding proteins in the ixodid ticks, Rhipicephalus appendiculatus, Amblyomma variegatum and Ixodes hexagonus. Parasitology. 1995;111:161–5. doi: 10.1017/s0031182000064908. [DOI] [PubMed] [Google Scholar]
- 84.Wang H, Nuttall PA. Immunoglobulin-binding proteins in ticks: new target for vaccine development against a blood-feeding parasite. Cell Mol Life Sci. 1999;56:286–95. doi: 10.1007/s000180050430. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Hyperlinked Excel spreadsheet containing annotated assembled ESTs (Supplementary Table 1): http://exon.niaid.nih.gov/transcriptome/Hmrufipes/Hmr-S1.xls
Hyperlinked Excel spreadsheet containing annotated coding sequences (Supplementary Table 2): http://exon.niaid.nih.gov/transcriptome/Hmrufipes/Hmr-S2.xls
Hypelinked Excel spreasheet containing annotated bovine proteins (Supplementary Table 3): http://exon.niaid.nih.gov/transcriptome/Hmrufipes/Hmr-S3.xls