Abstract
The Saccharomyces cerevisiae DEAD-box protein Mss116p is a general RNA chaperone that functions in splicing mitochondrial group I and group II introns. Recent X-ray crystal structures of Mss116p in complex with ATP analogs and single-stranded RNA show that the helicase core induces a bend in the bound RNA, as in other DEAD-box proteins, while a C-terminal extension induces a second bend, resulting in RNA crimping. Here, we illuminate these structures by using high-throughput genetic selections, unigenic evolution, and analyses of in vivo splicing activity to comprehensively identify functionally important regions and permissible amino acid substitutions throughout Mss116p. The functionally important regions include those containing conserved sequence motifs involved in ATP and RNA binding or interdomain interactions, as well as previously unidentified regions, including surface loops that may function in protein-protein interactions. The genetic selections recapitulate major features of the conserved helicase motifs seen in other DEAD-box proteins, but also show surprising variations, including multiple novel variants of motif III (SAT). Patterns of amino acid substitutions indicate that the RNA bend induced by the helicase core depends upon ionic and hydrogen-bonding interactions with the bound RNA; identify a subset of critically interacting residues; and indicate that the bend induced by the C-terminal extension results primarily from a steric block. Finally, we identified two conserved regions, one the previously noted post-II region in the helicase core and the other in the C-terminal extension, which may help displace or sequester the opposite RNA strand during RNA unwinding.
Keywords: Catalytic RNA, RNA chaperone, ribozyme, RNA helicase, RNA-protein interaction
Introduction
DEAD-box proteins are a large, ubiquitous family of ATP-dependent RNA helicases that mediate RNA and RNP structural rearrangements in a variety of cellular processes, including translation, RNA splicing, ribosome assembly, RNA degradation, and nuclear transport.1–5 Unlike processive RNA helicases, DEAD-box proteins unwind RNA duplexes by local strand separation, enabling them to elicit RNA and RNP conformational changes without globally unfolding RNA structure.6 All DEAD-box proteins contain a conserved helicase core consisting of two tandem RecA-like domains with a series of 13 conserved motifs that function in RNA or ATP binding or interdomain interactions.7 In many DEAD-box and related proteins, the helicase core is flanked by additional N- and/or C-terminal domains, and in some cases, these additional domains specialize the proteins for specific functions by contributing to substrate binding or providing additional enzymatic activities.1,2
X-ray crystal structures of DEAD-box proteins in ternary complexes with single-stranded RNA and non-hydrolyzable ATP analogs have provided some insight into their RNA-unwinding mechanism.8–14 In the absence of substrates, the helicase core is in an open conformation in which its two RecA-like domains are separated and can move relative to each other via a flexible linker. The binding of ATP and RNA leads to a compact closed conformation in which the two core domains interact extensively. In the closed conformation, the conserved motifs are brought together at or near the interface between the domains and contribute to ATP- and RNA-binding sites on opposite sides of the core. The RNA-binding site, a cleft formed at the domain interface, binds a short segment of an RNA strand and uses a wedge α-helix containing the conserved motif Ic (motifs are named according to Fairman-Williams et al.7) to bend the strand in a manner that would disrupt base pairing with an opposite strand. Biochemical experiments show that ATP hydrolysis is not required for strand separation, but is required for release of the bound RNA strand, enabling recycling of the enzyme for another round of RNA unwinding.15,16 Major unanswered questions include how DEAD-box proteins bind double-stranded RNA; the identification of protein regions that may contribute to displacement or sequestration of the strand opposite that bound by the helicase core; the mechanism by which ATP binding and hydrolysis are coupled to RNA binding and release; the nature of the conformational changes that occur during different steps of RNA unwinding; the function of ancillary domains; and the roles of partner proteins.
The related DEAD-box proteins Mss116p of S. cerevisiae and CYT-19 of Neurospora crassa have emerged as important model systems for studying DEAD-box protein mechanisms. These proteins function as general RNA chaperones in the splicing of mitochondrial (mt) group I and group II introns, other mt RNA processing reactions, and translational activation.17–19 Biochemical studies show that Mss116p and CYT-19 bind group I and group II intron RNAs non-specifically and use their ATP-dependent RNA-unwinding activity to resolve stable, inactive structures (“kinetic traps”) that limit the rate of RNA folding.18–21 The splicing of some introns may require this basic activity as well as additional activities, such as strand annealing or non-specific RNA binding.22–24 Although Mss116p and CYT-19 can by themselves promote the splicing of some introns in vitro, they function in vivo in concert with other proteins, such as intron-encoded maturases and host-encoded splicing factors, that stabilize the active RNA structure.18,19,21 Additionally, Mss116p was found recently to interact with and affect the activity of the mt RNA polymerase, positioning it to influence the folding of nascent RNAs.25
Mss116p and CYT-19 belong to subfamilies of DEAD-box proteins in which the helicase core is followed by a distinctive, largely α-helical C-terminal extension (CTE), and an unstructured basic tail (C-tail; Fig. 1(a)).26 Preceding the helicase core is an N-terminal extension (NTE), which is larger in Mss116p than in CYT-19 (52 and 11 amino acid residues, respectively). In both Mss116p and CYT-19, the CTE is required for activity of the helicase core, with mutations within the CTE destabilizing and inactivating the protein.13,26 The C-tail is not required for ATP-dependent RNA unwinding, but contributes to the non-specific binding of RNA substrates.26–28
Fig. 1.
Schematic of Mss116p and S. cerevisiae genetic assay used for unigenic evolution analysis and genetic selections. (a) Mss116p schematic showing protein domains and conserved sequence motifs named according to Fairman-Williams et al.7 MT, mt import sequence (black stripes); NTE, N-terminal extension; CTE, C-terminal extension (orange); C-tail, basic C-terminal tail (blue). (b) Genetic assay. Mss116p is expressed from CEN plasmid pHRH197B in an mss116Δ strain derived from S. cerevisiae wild-type 161-U7/1+2+, which contains 12 mt group I and group II introns. The region encompassing the NTE and helicase core that was analyzed by unigenic evolution in this work is shown in gray. B and X are BsrGI and XbaI sites introduced via silent mutations for use in library constructions (see Materials and Methods). For unigenic evolution analysis, functional Mss116p variants that support RNA splicing were selected from the mutant library by plating on growth medium containing glycerol (YPG) at 30°C (Gly+ phenotype) and checked for cold- and high-temperature sensitivity by replating on the same medium at 24 and 37°C. The mutagenized region of the MSS116 gene was amplified by colony PCR and sequenced to identify the mutations.
Recently, we obtained high-resolution (1.9–2.1 Å) X-ray crystal structures of Mss116p, which show the entire helicase core and CTE in ternary complexes with a single-stranded RNA oligonucleotide (U10 RNA) and a series of ATP analogs.13 The construct used for crystallography, denoted Mss116p/Δ598–664, was deleted for the C-tail, and the NTE was present but not visible, suggesting flexibility. The structures showed that the helicase core of Mss116p binds ATP and RNA similarly to other DEAD-box proteins and that the CTE is an extension of the RNA-binding side of helicase core domain 2. The CTE interacts extensively with domain 2, explaining why mutations within the CTE destabilize and inactivate the protein.13 Surprisingly, Mss116p was seen to induce two bends in the bound RNA, one by using the motif Ic wedge helix in the helicase core as in other DEAD-box proteins, and the other by using a second wedge helix in the CTE, resulting in RNA crimping.
To complement the crystal structures, we used small angle X-ray scattering (SAXS) to obtain solution structures of full-length Mss116p and CYT-19 and deletion mutants in the presence and absence of substrates.28 This SAXS analysis provided information about conformational changes upon binding of substrates and flexible regions, which could not be visualized by X-ray crystallography. The SAXS solution structures for Mss116p showed that the NTE emerges from core domain 1 away from the region that binds RNA, while the C-tail emerges from domain 2 in position to interact with RNA regions neighboring that bound to the core. The C-tails of Mss116p and CYT-19 appear to be largely unstructured, and the SAXS analysis showed them to be flexibly attached, enabling them to move over a wide arc to interact with 5′- or 3′-extensions of oligonucleotides bound to the core. These findings support models in which the C-tail serves as a flexible tether that binds non-specifically to different locations on large RNAs and enables the core to sample neighboring regions and different orientations for RNA unwinding.26–28
In addition to this structural information, an important advantage of the Mss116p experimental system is the availability of facile yeast genetic assays for its splicing and translational activation activities, making it possible to readily correlate biochemical activities in vitro with physiological functions in vivo.19 Previously, we used high-throughput unigenic evolution analysis with genetic assays for Mss116p’s RNA splicing activity to identify functionally important regions of the CTE and basic tail.26 Here, we extended this approach to the NTE and helicase core and interpret the combined results for the entire protein in light of the recently determined X-ray crystal and SAXS solution structures. Our results provide an overview of functionally important regions and permissible amino acid substitutions throughout Mss116p, provide new insights into the contribution of the helicase core and CTE to RNA unwinding, and identify protein regions potentially involved in displacing the strand opposite that bound by the core.
Results and Discussion
Unigenic evolution analysis
To systematically identify functionally important features of Mss116p’s NTE and helicase core, we employed a high-throughput method termed unigenic evolution.29 This method involves using a genetic assay to isolate a collection of functional variants from a protein library containing random PCR-induced mutations and then statistically analyzing ratios of missense to silent mutations across a sliding window to assess the degree of constraint on different protein regions. The degree of constraint is expressed by a parameter termed the mutability (M) value, with negative and positive M values indicating hypomutable and hypermutable regions, respectively (see Materials and Methods).
For the unigenic evolution analysis of Mss116p, we used a genetic assay in which an mss116Δ strain (mss116Δ/1+2+), containing twelve mt group I and II introns, is complemented by Mss116p expressed from a low copy number, centromere-containing (CEN) plasmid (pHRH197B: Fig. 1(b)).19 The twelve group I and group II introns in mss116Δ/1+2+ are dependent upon Mss116p for efficient splicing and are located in the genes encoding cytochrome oxidase subunit 1 (COX1) and cytochrome b (COB), which are required for respiratory growth on non-fermentable carbon sources, such as glycerol. Thus, the activity or lack of activity of Mss116p variants can be scored readily by growth or lack of growth on glycerol-containing medium (Gly+ and Gly− phenotypes, respectively).
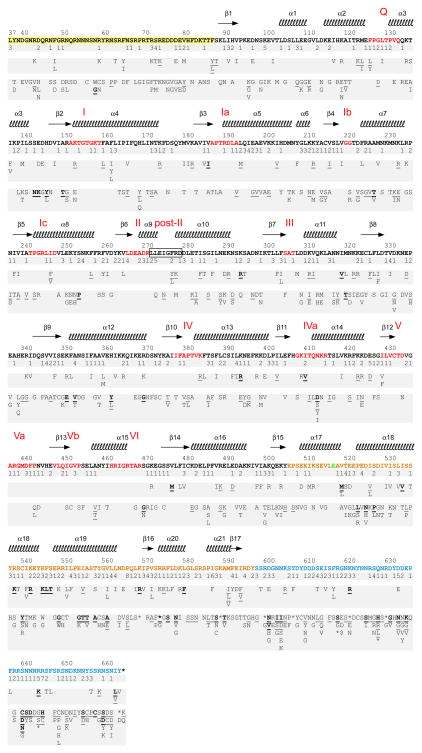
To analyze the function of the NTE and helicase core, we constructed a library of Mss116p variants containing random PCR-induced mutations in these regions (amino acid residues 37–516) in the CEN plasmid pHRH197B (Fig. 1(b); see Materials and Methods). The library was transformed into S. cerevisiae strain mss116Δ/1+2+, and the transformants were plated on glycerol medium at 30°C to select Gly+ colonies that express functional Mss116p variants. After picking colonies and replating to confirm their Gly+ phenotype, the mutagenized region of the complementing plasmids was amplified by colony PCR and sequenced to identify mutations. In total, we isolated 178 functional Mss116p variants containing 969 mutations of which 612 were missense and 357 were silent. In the previous unigenic evolution analysis of the CTE and basic tail (amino acid residues 518–664), we used the same method to isolate 111 functional Mss116p variants containing 703 mutations of which 468 were missense and 235 were silent.26 Figure 2 summarizes the mutations that were detected in both unigenic evolution analyses. Below, we focus first on the NTE and helicase core and then discuss the CTE and C-tail.
Fig. 2.
Summary of mutations in functional Mss116p variants that promote mt group I and group II intron splicing. Variants with mutations in the NTE and helicase core (amino acid residues 37–516) are from this work, and variants in the CTE and basic tail (amino acid residues 518–664) are from Mohr et al.26 The amino acid sequence of wild-type Mss116p is shown, with the NTE highlighted in yellow, conserved helicase motifs in red, CTE in orange, and C-tail in blue. E517 (green) overlaps the XbaI site used to clone the mutated fragments and was not mutagenized. Secondary structure elements from the Mss116p crystal structure13 are shown above the wild-type Mss116p sequence, and mutations found in the unigenic evolution analyses are summarized below in three tiers. The top tier indicates the number of silent mutations; the middle tier shows conservative amino acid substitutions; and the bottom tier shows non-conservative amino acid substitutions. The frequency of occurrence of an amino acid substitution is indicated as follows: plain, once; underlined, twice; bold and underlined, three or more times. Stars denote stop codons.
Mutability of the NTE and helicase core
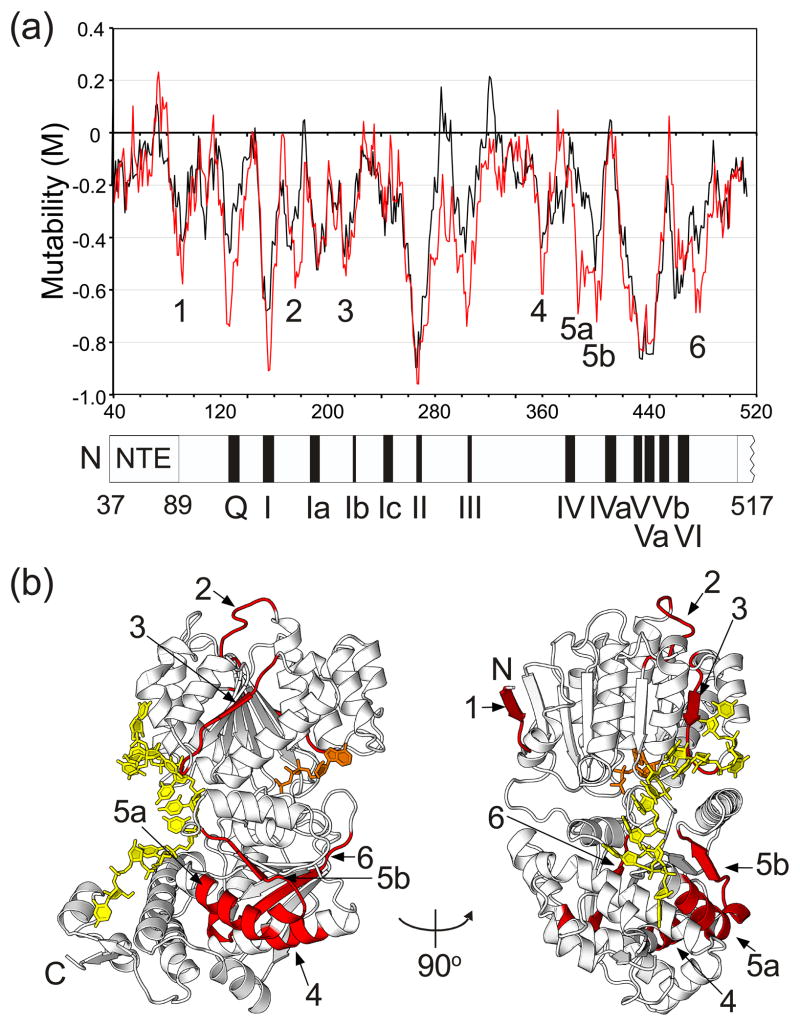
Figure 3(a) shows a mutability plot for the NTE and helicase core based on statistical analysis of the 969 mutations in these regions obtained in the unigenic analysis. The plot shows M values as a function of amino acid position calculated across an 11-amino acid sliding window. This relatively small sliding window was used here to assess the hypomutability of the short conserved sequence motifs and to identify specific structural features for more detailed analysis. A disadvantage is that the window is not sufficiently large to establish statistical significance in all cases. However, as longer windows average adjacent hyper- and hypomutable regions to give broad hypomutable peaks throughout the helicase core, the shorter window was deemed more useful for identification of functionally important features in an initial survey. The plot in black shows M values calculated by the conventional method in which all missense mutations are treated equally, and the plot in red shows M values calculated by a modified method in which strictly defined conservative substitutions30 are treated as silent (see Figure 3 legend for details). The hypomutability in some regions appears more pronounced when conservative substitutions are treated as silent. We found no functional variants with premature termination codons that would result in truncated proteins.
Fig. 3.
Mutability plot and crystal structure of Mss116p showing hypomutable regions of the NTE and helicase core identified by unigenic evolution. (a) Mutability plot. The plot shows M values as a function of Mss116p position at the center of an 11-amino acid residue sliding window, calculated as described in Materials and Methods. The plot in black was calculated with all missense mutations treated equally, and the plot in red was calculated with conservative changes (R=K, D=E, L=I=V=F, F=Y, S=T)30 treated as silent. Negative mutability values indicate hypomutable regions, and positive mutability values indicate hypermutable regions. For this plot, hypomutable peak regions are defined as those having M values ≤ −0.4. (b) Mss116p crystal structure (PDB ID 3I5X) showing the location of hypomutable peak regions 1–6 (red) from the mutability plot. Region 1, residues 89–95; region 2, residues 175–182; region 3, residues 213–221; region 4, residues 360–365; region 5a, residues 387–396; region 5b, residues 398–407; region 6, residues 473–484. U10 RNA (yellow) and AMP-PNP (orange) are shown in stick representation.
The mutability plots show that the NTE (amino acid residues 37–88) is not significantly hypomutable, except possibly near its boundary with the helicase core, suggesting that the NTE contributes minimally to Mss116p function in vivo. Consistent with this finding, an Mss116p mutant lacking the NTE (Mss116p/ΔNTE) functioned as efficiently as the wild-type protein in supporting glycerol growth of the Δmss116/1+2+ strain (Fig. S1) and almost as efficiently as the wild-type protein in splicing the mt group II intron aI5γ in vitro (second-order rate constants determined from protein concentration dependencies were 1.6 × 106 and 1.0 × 106 M−1 min−1 for wild-type Mss116p and Mss116p/ΔNTE, respectively; Fig. S2). The NTE was present in the protein but not visible in the previous crystal structures of Mss116p/Δ598–664 ternary complexes,13 and crystal structures of Mss116p constructs deleted for the NTE by itself or together with the C-tail were essentially identical to that obtained previously for Mss116p/Δ598–664 (root-mean-square deviations of 0.14 and 0.18 Å, respectively, over 508 Cα atoms; Fig. S3; Table S1). Together, these findings indicate that the NTE is a flexible attachment that contributes minimally to Mss116p’s splicing function and does not affect folding of the remainder of the protein. Additionally, the crystal structure of Mss116p/ΔNTE in which the C-tail is present but not visible provides further evidence that the C-tail is flexibly attached (Fig. S3).
By contrast to the NTE, the mutability plot shows that the helicase core (amino acid residues 89–505) contains regions of hypomutability throughout. Most of the markedly hypomutable regions (M values ≤ −0.4 in this plot) correspond to or overlap conserved DEAD-box protein motifs, including Q, I, Ia, Ib, II, III, V, Va, Vb, and VI, and conversely, most of the DEAD-box protein motifs are within such hypomutable regions. The exceptions are motif Ic (discussed below) and motifs IV and IVa, which have multiple variable positions that tolerate nonconservative substitutions (Fig. 4).
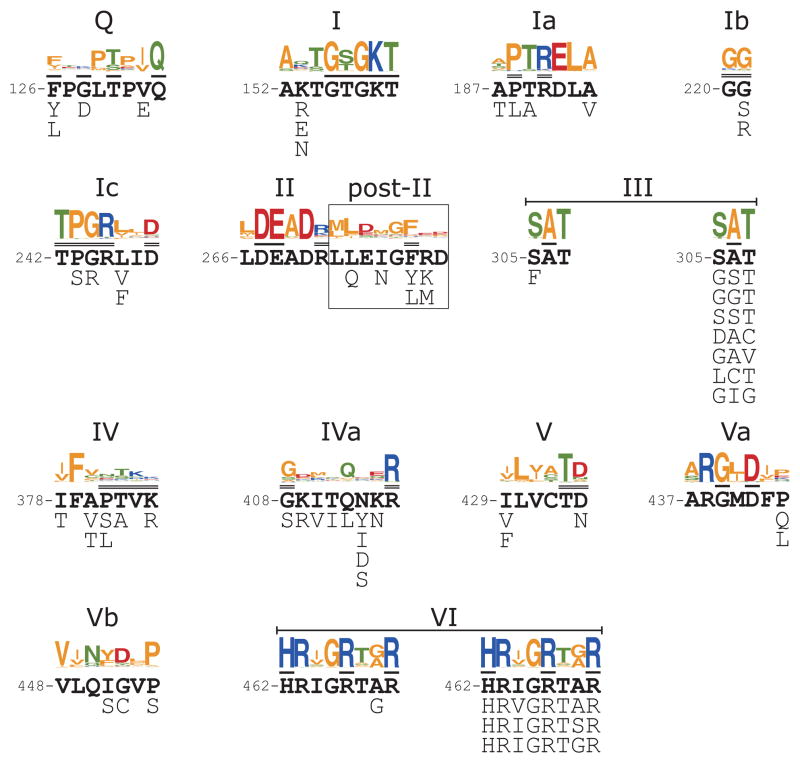
Fig. 4.
Functional amino acid substitutions in conserved DEAD-box protein motifs. The figure shows amino acid substitutions found within the DEAD-box protein motifs in functional Mss116p variants selected in the unigenic evolution analysis supplemented in three cases (motifs II, III, and VI) by additional selections in which the coding sequence for the motif was randomized (motifs II and III) or doped with 70% of the wild-type nucleotide residue and 10% of each mutant nucleotide residue at each position (motif VI). For each conserved motif, the top shows the motif sequence generated by WebLogo60 from a multiple alignment of E. coli, S. cerevisiae, and human DEAD-box protein sequences (67 sequences from supplemental Figure 1 of ref.7). The wild-type Mss116p sequence is shown in bold below the WebLogo, with residues contacting ATP or ssRNA (U10 RNA) in the Mss116p crystal structure overlined once or twice, respectively. The amino acid substitutions found in functional Mss116p variants are shown below the wild-type Mss116p sequence. For motifs III, and VI, the functional amino acid substitutions from the unigenic evolution analysis are shown below to the left, and functional sequences from the supplementary selections are shown below to the right. In the supplementary selection for motif III, the GGT variant was found in two independent isolates with different DNA sequences. In the supplementary selection for motif VI, the HRIGRTgR variant was found in the unigenic evolution and in two independent isolates in the supplementary selection. Amino acid residues: acidic, red; basic, blue; non-polar, orange; polar, green.
Five other hypomutable peak regions do not correspond to conserved DEAD-box protein motifs (peaks 1–4 and a double peak 5a/b; minimum M values at V93, S177, C215, E361/H362, and P402/E405, respectively), and an additional hypomutable peak (peak 6; S476/F479) overlaps motif VI but is not centered on the motif. These six peaks, whose locations are shown on the structure of Mss116p in Figure 3(b), potentially identify additional functionally important regions that do not contain known conserved motifs. Hypomutable peaks 1 and 2 correspond to surface loops in core domain 1, while hypomutable peaks 3–6 include both surface regions and internal elements that could be necessary to maintain Mss116p’s structure. The hypomutable surface loops in peaks 1 and 2 are distant from the ATP- and RNA-binding sites, with no obvious structural basis for their hypomutability, and they could be required for oligomerization or interaction with another protein (e.g., the mt RNA polymerase; see25). Other hypomutable peaks with surface regions that could be involved in protein-protein interactions are peak 3, which is located in domain 1 close to peak 2, and peak 5a/b, which is located in domain 2 roughly on the same side of the protein as peaks 2 and 3. Notably, hypomutable peak 5a/b in domain 2 corresponds to a region of DEAD-box protein eIF4A that interacts with eIF4G31 and was suggested previously to be a region commonly used by DEAD-box proteins for protein-protein interactions.32
Variations in conserved DEAD-box protein motifs
Figure 4 summarizes functional substitutions in the 13 conserved DEAD-box protein motifs of Mss116p found in the unigenic evolution analysis. In each case, the motif in Mss116p is shown in bold, the consensus from a collection of E. coli, S. cerevisiae, and human DEAD-box proteins is shown above in WebLogo format,7 and the functional variations found in the unigenic evolution analysis are summarized below (or below left for motifs III and VI in which additional functional variants were isolated in supplementary selections described below). In most cases, the non-synonymous changes found within the motifs correspond to variable positions in the consensus that either do not contact or make main-chain contacts with substrates. However, some variations found in functional Mss116p variants in the unigenic evolution analysis are surprising and noteworthy.
In the Q motif, which functions in the specific binding of ATP, F126 forms a platform that stacks with the adenine base.13 Other DEAD-box proteins usually have an F, Y, or W at this position, consistent with this pi-electron stacking function, and stacking of the adenine base with each of these three residues has been seen in DEAD-box protein structures.7,8,10,12–14,33,34 Such stacking must not be essential, however, because the unigenic evolution yielded two functional variants in which F126 is replaced by L, which lacks pi-electrons and cannot stack similarly with an adenine base, and one of these variants (F126L R234K V360L) grows at or near wild-type rates when plated on glycerol-containing medium (YPG) at 24, 30, and 37°C (not shown). These findings indicate that hydrophobic interaction of the side chain with the adenine base can provide adequate function, and indeed, some DEAD-box proteins naturally have a hydrophobic residue (V, M, I, L) at this position.7,33,35
Motifs Ib (GG) and Ic (TPGRxxD) interact with the bound ssRNA at the conserved bending point in the helicase core.8–14,34 In motif Ib, the unigenic evolution analysis gave functional variants in which G221 is replaced with an S or R residue. The amide N atom of G221 H-bonds with a phosphoryl oxygen of ssRNA, and the mutation to an S or R is likely functional because this main-chain interaction is preserved while the side chain is exposed to the solvent. Although a G at this position is conserved in other DEAD-box proteins, some have substitutions of N, Q, A, S, or T (see Fig. S1 of ref. 7).
In motif Ic, the unigenic evolution analysis gave functional variants with the nonconservative substitutions P243S and G244R. Although the P in motif Ic is conserved strongly in DEAD-box proteins, some have a T at this position (see Fig. S1 of ref. 7). The G244 residue in motif Ic, which is also strongly conserved in DEAD-box proteins, serves two main purposes: (i) its amino group H-bonds to the 2′ oxygen of U6 of the ssRNA, and (ii) it is the point of the wedge that bends the ssRNA. The larger R side chain at this position would clash with the bound RNA, perhaps further exaggerating the bend.
Motif II (DEAD) and motif VI (HRxGRxxR) form key parts of the ATPase active site and are highly conserved in DEAD-box proteins.7 In motif II, we found no functional variants either in the unigenic analysis or in a supplementary selection in which the codons for the four amino acid residues were randomized. In motif VI, the unigenic evolution analysis and a supplementary selection yielded only three different variant sequences (HRvGRTAR, HRIGRTsR, and HRIGRTgR (where the lowercase letter indicates the changed amino acid), and all of these changes occurred at variable positions in the consensus. Thus, as in other DEAD-box proteins, motifs II and VI are highly constrained in Mss116p.
Novel variations in motif III (SAT)
Motif III (SAT) has been analyzed extensively in other DEAD-box proteins (36 and refs. therein). It is involved in a series of interactions that extend across the interface between domains 1 and 2, including H-bonds between the two alcohol side chains and residues in motifs II and VI and an H-bond via a water between the alanine amino group and ATP, and these interactions are thought to help couple ATP and RNA binding. The latter suggestion was based on findings that mutations within motif III of eIF4a inhibit RNA-unwinding, but retain high RNA-dependent ATPase activity.37 A recent, detailed study of motif III in the yeast DEAD-box protein Ded1p suggests that mutations in motif III inhibit RNA unwinding by weakening the strength of ATP-dependent ssRNA binding, which results in a decreased kcat for ATP hydrolysis.36
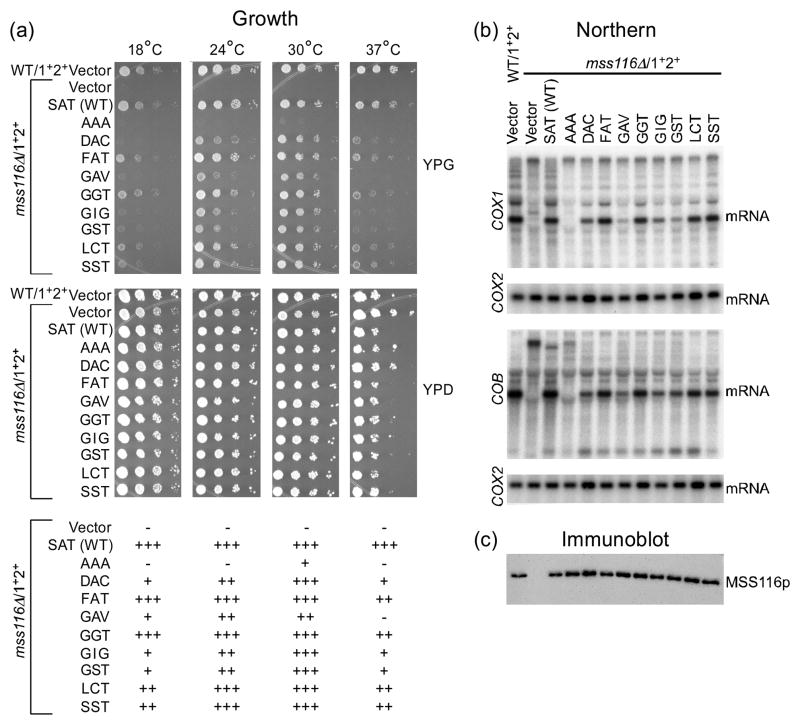
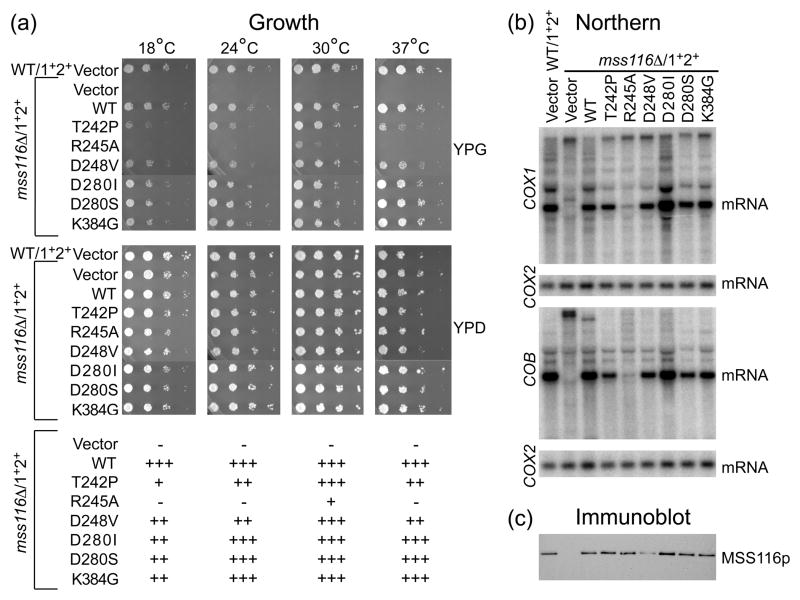
The unigenic evolution analysis of Mss116p yielded a single functional variant with a nonconservative substitution (FAT), which was found previously in a cyanobacterial DEAD-box protein (CrhC), but not tested for function.38 However, a more saturating supplementary selection in which the three motif III codons in Mss116p were randomized simultaneously yielded seven additional variants (DAC, GAV, GGT, GIG, GST, LCT and SST). Figure 5(a) shows complementation assays in which the eight Mss116p motif III variants were expressed from a CEN plasmid in the mss116Δ/1+2+ strain and tested for their ability to support growth on glycerol at different temperatures. All eight of these motif III variants functioned relatively efficiently in supporting glycerol growth of the Δmss116/1+2+ strain at 30°C, although they were temperature sensitive to different extents at 37°C. Most were also cold sensitive at 18°C and 24°C, except for FAT and GGT, which functioned similarly to wild-type Mss116p at low temperatures. By contrast, a previously studied Mss116p motif III mutant (AAA)24, included in the assays for comparison, was more strongly impaired: it gave only barely detectable glycerol growth at 30°C and was Gly− at other temperatures (Fig. 5(a); note the AAA mutant was better able to support glycerol growth in strains with smaller numbers of mt introns.24)
Fig. 5.
Analysis of Mss116p motif III mutants. The strains analyzed were wild-type 161-U7/1+2+ and mss116Δ/1+2+ containing the CEN plasmid vector pRS416 or CEN plasmids expressing wild-type Mss116p or the indicated motif III mutants. (a) Growth test. S. cerevisiae strains were grown in YNBD medium lacking uracil to select for the CEN plasmid, and serial dilutions were plated on medium containing glycerol (YPG) or glucose (YPD), and incubated at 18, 24, 30, or 37°C. Scoring of glycerol growth phenotypes is shown below: +++, 90–100% wild type; ++, 50–89% wild type; +, <50% wild-type; and −, no growth. (b) Northern hybridization. The blots show whole-cell RNAs from the indicated strains separated in a 1.5% agarose gel and hybridized with 32P-labeled oligonucleotide probes complementary to COX1 exon 6 or COB exon 6. The blots were then stripped and rehybridized with a probe for the intronless COX2 gene to assess equal loading. (c) Immunoblot. The blot shows TCA-precipitated proteins from the same strains separated in a 0.1% SDS/4–12% polyacrylamide gradient gel and probed with anti-Mss116p antibody. After imaging, the blot was stripped of antibodies and stained with AuroDye Forte to confirm equal loading (not shown).
The ability of the motif III variants to promote splicing of mt group I and II introns in vivo was analyzed by Northern hybridizations (Fig. 5(b)). In these experiments, the strains were grown at 30°C on the non-repressing fermentable sugar raffinose, enabling us to examine RNA splicing in mutants that are strongly defective in Mss116p function. Blots of whole cell RNAs from the different strains were hybridized with exon probes for the COX1 or COB gene, then stripped and rehybridized with a probe for COX2, a gene which lacks introns, to assess equal loading. The Northern blots showed that those SAT mutants that support growth on glycerol promote splicing of COX1 or COB introns to different degrees, but in all cases efficiently enough to produce distinct bands corresponding to COX1 and COB mRNAs, as expected from their Gly+ phenotype. By contrast, the AAA mutant produced undetectable amounts of the mature mRNAs, as expected from its severely impaired glycerol-growth phenotype in the multi-intron strain. Among the new motif III mutants FAT, GGT, LCT, and SST were most efficient in promoting splicing; DAC and GIG were moderately efficient; and GAV was least efficient. Curiously, GST appeared moderately efficient in splicing COB introns, but less efficient in splicing COX1 introns. Immunoblots showed that all of the variant proteins were expressed from the CEN plasmid at levels comparable to that of the wild-type protein (Fig. 5(c)).
The degree of variation in motif III mutants in the Mss116p variants is greater than that seen or suspected possible from studies of other DEAD-box proteins, including Ded1p where similar codon randomization was done separately for the first and third positions.36 The Mss116p variants that are fully or moderately functional in vivo at 30°C violate the consensus sequence alcohol-small amino acid-alcohol at each of the three positions, with the moderately functional variant GIG violating this consensus at all three positions. The FAT variant, which we find to be almost fully functional in Mss116p in vivo, places a large phenylalanine side chain at a position in a tight interface where it would not easily fit in the crystal structure. The pattern of amino acid substitutions in the mutants indicate that: (i) the putative H-bonds between the alcohols of S305 and T307 and residues in motifs II and VI in the crystal structure are not essential for activity; (ii) A306 and T307 can be replaced with residues having similarly sized nonpolar or uncharged polar side chains; and (iii) S305 can tolerate substitutions with larger side chains (F, D, L, H, M, Q). The degree of variation seen in the Mss116p motif III mutants may reflect that all three codons were randomized simultaneously or that Mss116p is less dependent upon motif III interactions than are other DEAD-box proteins due to its higher RNA-binding affinity.26,36 Nevertheless, motif III interactions are still required for Mss116p activity, as the AAA mutation strongly impairs Mss116p function in the multi-intron strain (see above), presumably due primarily to loss of side-chain interactions. Further biochemical and structural analyses of motif III mutants in Mss116p and other DEAD-box proteins will be needed to address these issues.
Analysis of helicase core residues that bind ssRNA
The crystal structure of Mss116p identified a series of amino acid residues in the helicase core domains 1 and 2 and the CTE that contact U10 RNA and are thus potentially important for Mss116p function.13 The helicase core contacts residues U3–U10, but U9 and U10 have weak electron density, indicating mobility and uncertainty in their positions. The core contacts with U3–U8 are made primarily through residues in the conserved motifs and are part of an RNA-binding tract spanning core domains 1 and 2, similar to that seen in other DEAD-box protein structures.13 However, structures of Mss116p and other DEAD-box proteins provide only a static picture of these contacts and little insight into their relative contributions to RNA binding.
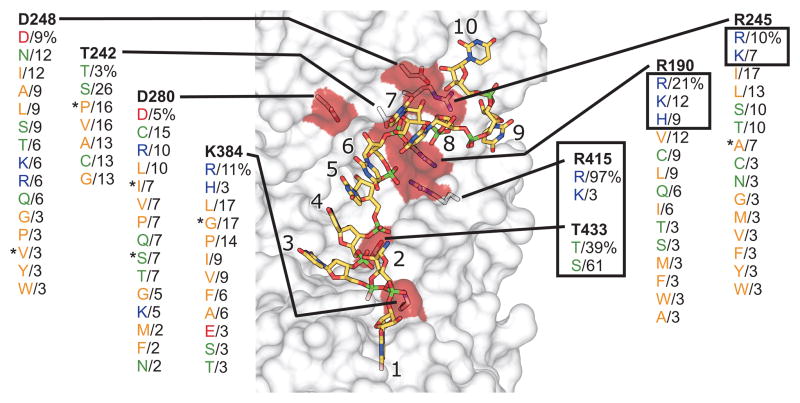
Figure 6 shows a view of Mss116p’s RNA-binding tract highlighting the eight residues whose side chains make ionic or H-bond contacts (directly or through a water molecule) with U10 RNA. In the unigenic evolution analysis, seven of these residues (R190, T242, R245, D248, D280, R415, T433) were invariant, and the remaining residue (K384) showed only the conservative substitution to R. By contrast, a number of residues that make only main-chain hydrophobic and/or H-bond contacts to U10 RNA (P188, G221, P243, G244, P381, T382, and G408) showed non-conservative substitutions that could retain the original contacts.
Fig. 6.
Genetic selection for functional substitutions of RNA-binding tract residues that make side-chain contacts with bound RNA. The figure shows the surface of the RNA-binding cleft of Mss116p (gray transparent) highlighting the eight amino acid residues whose side chains make ionic contacts or H-bond directly or through a water molecule to U10 RNA in the Mss116p crystal structure (red surface and stick representation; C, yellow; N, blue; O, red; P, green; PDB ID 3I5X).13 The codon for each residue was randomized and functional Mss116p variants were selected by growth on glycerol at 30°C. The figure summarizes functional amino acid substitutions found in 30–40 randomly selected functional variants for each position, with the percentage of variants containing the substitution shown beneath the wild-type residue (bold). The U10 RNA contacts for R190, R245, R415, T242, and T433 are described in the text. The remaining three residues (D248, D280, and K384) were not conserved in the selections and were replaced by a variety of non-synonymous residues whose side chains cannot make the same contacts. D248 is part of motif Ic and is near the point of the α8 wedge helix, where its carboxyl group H-bonds with the 2′-OH of U7. It is conserved in other DEAD-box proteins (see Fig. 4), but its ready replacement by non-synonymous residues in functional variants indicates that the steric block rather than the side-chain interaction at this position is critical for Mss116p function. D280 is part of the post-II region, where its carboxyl group H-bonds through a water molecule with the 2′-OH of U6. It is not conserved in other DEAD-box proteins and its replacement by non-synonymous residues indicates that the indirect H-bond is not critical for Mss116p function. Finally, K384 is part of motif IV, where its basic side chain makes an ionic contact with the 5′ phosphate of U3. It is not conserved in other DEAD-box proteins, and its ready replacement by non-synonymous amino acid residues indicates that this ionic contact is not critical. Asterisks indicate variants analyzed further in Figure 7.
Because the unigenic evolution analysis was not mutationally saturating, we further assessed the conservation of the eight residues whose side chains make ionic or H-bond contacts with the RNA by carrying out additional selections. In these selections, we constructed CEN plasmid libraries in which the codon for each of these eight residues was randomized. We then transformed the libraries into the Δmss116/1+2+ strain and selected and sequenced 30–40 Gly+ colonies from each library to identify functional amino acid substitutions (see Materials and Methods). Although many of these residues are part of conserved motifs and are highly conserved or invariant in other DEAD-box proteins, these more saturating selections revealed a range of constraints on different amino acid residues.
Surprisingly, only two of the eight residues showed strong conservation. The first, R415, makes an ionic contact to the U6 phosphate immediately preceding the bend and is present with different selected codons in 97% of the variants. The remaining 3% of the variants at this position have K, which can make a similar contact. The second conserved residue, T433, H-bonds to a U5 phosphoryl oxygen and is either conserved or replaced only by S, which can make the same contact. The strong selection at these two residues indicates that their side-chain contacts are critical for Mss116p function, and we confirmed that mutants with alanine substitutions at these positions are unable to complement the splicing defects in the mss116Δ/1+2+ strain (Gly− phenotype; Fig. S4).
Two other residues, R190 and R245, which make both side- and main-chain contacts to U10 RNA, show some selection for basic residues, but were replaced in the majority of selected variants by other residues, mostly hydrophobics, indicating that their side-chain contacts are not essential. A previous study reported that the Mss116p mutant R245E does not support respiratory growth or promote COX1 or COB intron splicing and concluded that this R residue is critical for RNA binding.40 In agreement with this finding, we find that E or D are not selected at this position, but the spectrum of permissible substitutions indicates that this lack of function is likely due to the charge change from a basic to an acidic residue and not because the R side-chain contact is essential.
The remaining four residues that make side-chain contacts in the crystal structure were readily replaced by non-synonymous residues. T242, whose alcohol H-bonds to a phosphoryl oxygen of U7 immediately after the bend, shows some selection for a side-chain alcohol (T or S) but can be replaced by residues with small side chains that cannot make the H-bond. The dispensability of T242 could reflect that two other residues (R245 and G220) make main-chain H-bonds to the same phosphoryl oxygen. The remaining three residues (D248, D280, and K384) make other apparently non-critical contacts, including surprisingly the H-bonds to RNA 2′-OH groups by D248 and D280 (see legend Figure 6 for details).
To confirm the results of the selections, we tested Mss116p mutants with a sampling of selected non-synonymous substitutions at different positions for their ability to function in vivo. Growth tests showed that mutants with non-synonymous substitutions at the four least conserved positions in the selections functioned as well as wild-type Mss116p in supporting glycerol growth of the Δmss116/1+2+ strain at 30°C, although all of these mutants appeared to be at least somewhat cold sensitive and two (T242P and D248V) were also heat sensitive (Fig. 7(a)). For R245, which makes both a side-chain and main-chain contact to U10 RNA, we tested the mutant R245A, which can make the main-chain but not the side-chain contact. This mutant showed slow growth on glycerol at 30°C and little or no growth at the other temperatures, suggesting that the side-chain contact to the bound RNA significantly enhances Mss116p function even though it is not essential. Similar results were obtained for R190, which also makes both side- and main-chain contacts to U10 RNA, although in this case, the relative contribution of the side-chain contact appears somewhat less than for R245 (Fig. S4). Northern hybridizations for cells grown at 30°C showed that glycerol growth correlates with the ability to support mt RNA splicing in all cases tested (Fig. 7(b)), and immunoblots showed that the mutant proteins were expressed at levels comparable to the wild-type protein (except D248V, which is expressed at lower levels Fig. 7(c)).
Fig. 7.
Analysis of Mss116p RNA-binding tract mutants. The strains analyzed were wild-type 161-U7/1+2+ and mss116Δ/1+2+ containing the CEN plasmid vector pRS416 or CEN plasmids expressing wild-type Mss116p or the indicated RNA-binding tract mutants. (a) Growth test. Serial dilutions of cells grown in YNBD lacking uracil were plated on media containing glycerol (YPG) or glucose (YPD), and incubated at 18, 24, 30, or 37°C. Scoring of glycerol growth phenotypes is shown below: +++, 90–100% wild type; ++, 50–89%, wild type; +, <50% wild type; and −, no growth. (b) Northern hybridization. Whole-cell RNAs from the indicated strains were separated in a 1.5% agarose gel, and blots were hybridized with 32P-labeled probes for COX1, COB and COX2, as described in the legend of Figure 5. (c) Immunoblot. TCA-precipitated proteins from the same strains were separated in a 0.1% SDS/4–12% polyacrylamide gradient gel, and the blots were probed with anti-Mss116p antibody. After imaging, the blots were stripped of antibodies and stained with AuroDye Forte to confirm equal loading (not shown).
Together, the selections for residues of the RNA-binding tract of the helicase core identify two amino acid residues, R415 and T433, whose side-chain contacts are essential for Mss116p function and two additional residues, R190 and R245, whose side-chain contacts enhance Mss116p function but whose main-chain contacts are sufficient for some function. All four of these residues are conserved in most DEAD-box proteins (exceptions for some of the arginines include DDX19/Dbp5, DDX25, and their homologs12,14,34), and the X-ray crystal structures of Vasa and eIF4AIII show them making the same contacts with ssRNA as in Mss116p.8–10 The remaining four residues whose side-chains contact U10 RNA in the Mss116p crystal structure could be replaced readily by non-synonymous residues in functional variants, suggesting that they contribute less strongly to RNA binding and/or that there is a tradeoff at these positions, in which the positive effect of higher RNA-binding affinity on RNA unwinding is balanced by the negative effect of slowing dissociation of the unwound RNA strand.
Post-II region
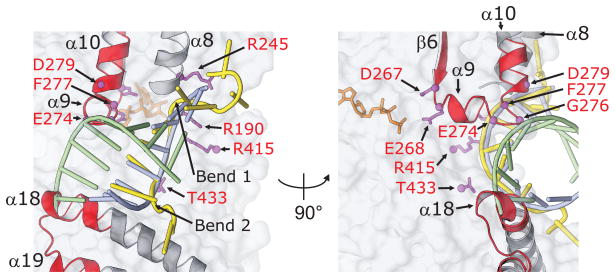
Although structures of DEAD-box proteins with bound dsRNA have not been determined, modeling suggests that the region downstream of motif II (denoted post-II), which consists of the end of α9, the following loop, and the beginning of α10, may contribute to displacement of the RNA strand opposite that which binds in the RNA-binding cleft.8,13 The unigenic evolution analysis shows that this region is part of a large hypomutable peak centered on motif II (Fig. 3(a)). Figure 8 shows a close-up of the structure of this region illustrating the clash with a modeled dsRNA. The unigenic evolution data show strong conservation of α9 and some residues in the loop between α9 and α10 that clash with the opposite RNA strand in the model (E274 and G276 are invariant, and F277, which also contributes to the binding of U10 RNA, shows only conservative substitutions; Fig. 2). These same three loop residues are conserved or replaced by synonymous residues in other DEAD-box proteins, suggesting functional importance (Fig. 4). We note that any large structural rearrangement in the post-II region resulting from the contact with the opposite RNA strand could potentially affect the positioning of the DEAD motif (D267–D270), which is located ~10–15 Å away at the other end of α9, with a consequent effect on ATP-binding or hydrolysis.
Fig. 8.
Conserved regions of Mss116p potentially involved in RNA strand separation. Close up of the RNA-binding region of Mss116p crystal structure highlighting the two wedge helices (α8 and α18) used to crimp the bound RNA strand, the post-II region (end of α9, the following loop, and the beginning of α10), and a conserved region of the CTE that modeling suggests may contribute to displacement of the complementary RNA strand (adapted from Figure 4(d) of 13). Hypomutable regions and amino acid residues conserved in unigenic evolution analyses and genetic selections are shown in red; the bound U10 RNA is shown in yellow; and an ideal A-form RNA duplex aligned to U4–U6 of the bound U10 RNA by least squares superpositioning is shown with blue and green strands. Comparison of the path of U10 RNA with the modeled duplex RNA illustrates the two bends (1 and 2) that result in crimping of the bound RNA strand (cf., yellow and blue strands). The complementary strand of the modeled duplex (green) clashes both with conserved residues in the post-II region and with the C-terminus of α18, which falls within hypomutable region 7 of the CTE (see Fig. 9).
Finally, the modeling shows that in addition to the clash with post-II, the displaced RNA strand of the modeled RNA duplex undergoes an additional clash about ~10 Å away from post-II with the conserved C-terminus of α18, the second wedge helix in the CTE that contributes to RNA crimping (Fig. 8; discussed further below).
The C-terminal extension
Mss116p belongs to a subfamily of DEAD-box proteins in which the helicase core is followed by a structured CTE and an unstructured basic tail.26 The crystal structure of Mss116p/Δ598–664 showed that the CTE is a compact module that interacts with and forms an extension of the RNA-binding side of domain 2 and functions both to stabilize the core and introduce a second bend at the 5′ end of the bound RNA, resulting in RNA crimping.13 Three α-helices (α17–19) in the CTE pack beneath the RNA-binding side of domain 2, with the remainder of the CTE packing against α18 and α19. α18 is a wedge helix that induces the second bend in the bound RNA, while α19 appears to play a critical role in positioning α18 and stabilizing the CTE.13,26 SAXS analysis and far UV-circular dichroism show that the C-tail is a largely unstructured, flexible extension that can bind non-specifically to different sites of large RNA substrates and tether the core for unwinding of neighboring duplexes.28
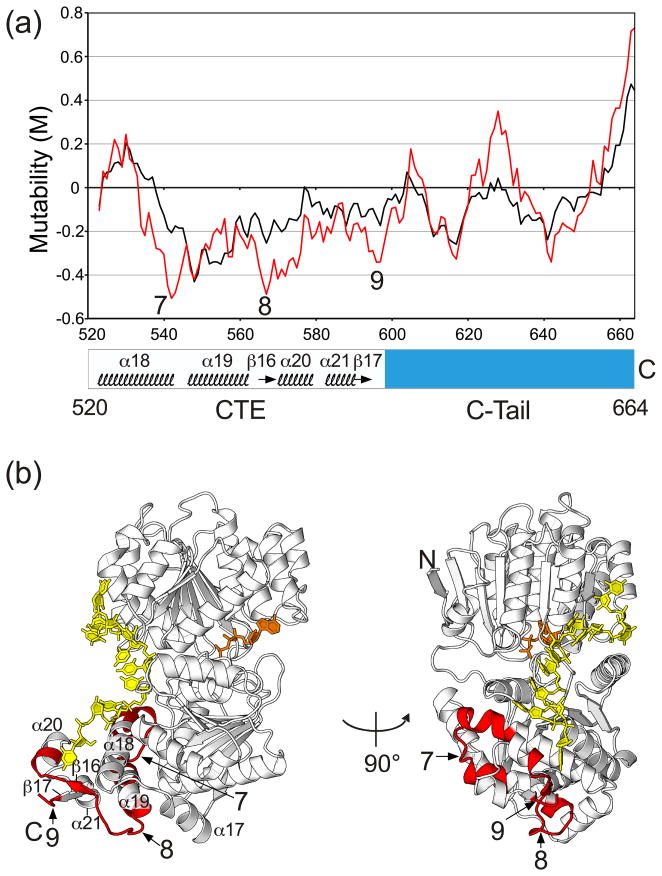
The unigenic evolution data for the CTE and C-tail that were obtained previously could now be interpreted in light of structural information from X-ray crystallography and SAXS. Figure 9(a) shows a mutability plot for the CTE and basic tail recalculated from previous data, using an 11-amino acid residue sliding window to match the window size used for the analysis of the NTE and helicase core. The mutability plot shows that the CTE has three markedly hypomutable regions (regions 7–9; M values ≤ −0.2), and these regions are highlighted on the Mss116p structure in Figure 9(b).
Fig. 9.
Mutability plot and crystal structure of Mss116p showing hypomutable regions of the CTE and basic tail identified by unigenic evolution. (a). Mutability plot. The plot shows M values as a function of Mss116p position calculated for the center of an 11-amino acid sliding window, based on previous unigenic evolution data.26 The plot in black was calculated with all missense mutations treated equally, and the plot in red was calculated with conservative changes (R=K, D=E, L=I=V=F, F=Y, S=T)30 treated as silent. Negative mutability values indicate hypomutable regions, and positive mutability values indicate hypermutable regions. In this plot, hypomutable peak regions are defined as those having M values ≤ −0.2. (b) Mss116p X-ray crystal structure with hypomutable peak regions 7–9 from the mutability plot highlighted in red (region 7, residues 537–553; region 8, residues 560–576; region 9, residues 594–598). U10 RNA (yellow) and AMP-PNP (orange) are shown in stick representation.
Based on the crystal structure and mutants analyzed previously, critical regions of the CTE were expected to include its interface with core domain 2; α18, which contacts the RNA at the site of the second bend; and α19, which is the site of inactivating point mutations in both Mss116p and CYT-19 and may be required to position α18.13,26 The interface between domain 2 and the CTE involves largely hydrophobic contacts along with several H-bonds and salt bridges.13 Consistent with a critical role for these interactions in stabilizing the core, all functional Mss116p variants contain only synonymous or functionally equivalent amino acid replacements at all of the solvent-inaccessible, hydrophobic residues at the interface in both domain 2 (I356, F357, V360, C481, F487) and the CTE (L533, Y544, I555). Further, even those hydrophobic residues in the interface that are partially solvent accessible (domain 2: F352, F385, F388, I392, V453, L457; CTE: I511, L516, A518, V519, F546, I551, L562) appear to tolerate only synonymous changes.
Hypomutable region 7 corresponds to the C-terminus of α18 (the CTE wedge helix), the loop between α18 and α19, and N-terminus of α19. It contains 17 residues (positions 537–553), ten of which are involved in interface contacts with domain 2 and seven of which are on the protein surface (Fig. 9(b)). The C-terminus of α18 corresponds to the second region identified above as possibly helping to displace the strand opposite that bound in the RNA-binding cleft, and the following loop and N-terminus of α19 may in some way contribute to this function.
The majority of α18, including residues 532–539 that contact the bound U10 RNA in the crystal structure, lies outside of the hypomutable peak region. This lower degree of conservation may reflect that the loop leading into α18 and residues 523–528 of α18 are solvent-exposed and relatively unconstrained, as long as the structure of α18 is maintained. Consistent with this hypothesis, all 57 variants with mutations in α18 are predicted to have an extended α-helix in this region by the secondary structure prediction algorithm JPRED.41,42 In the crystal structure, the middle of α18 contacts U1–U3 of the bound U10 RNA, but the path of the RNA upstream of the bend at U3 is ambiguous due to a crystal contact and may not be physiologically relevant.13 U3 at the site of the bend is involved in non-specific, hydrophobic interactions with four serines (532, 535, 536, and 539) located on three consecutive turns of α18. Consistent with the nonspecific contacts, we observed non-synonymous substitutions at three of these four positions. The remaining residue, S539, shows only the synonymous change of S to T in the unigenic evolution analysis, but CYT-19 and other DEAD-box proteins with homologous CTEs have non-synonymous residues at this position, again suggesting a non-specific interaction.13,26 Together, these findings indicate that the steric block due to the position of α18 is more important for RNA crimping than are interactions between the RNA and specific amino acid residues at the site of the bend.
The two remaining markedly hypomutable regions in the CTE, regions 8 and 9, come together on the surface of the CTE opposite that which contacts U10 RNA (Fig. 9(b)). Region 8 corresponds to β16 and the preceding surface loop, and region 9 corresponds to the end of β17. The C-terminal truncation Mss116p/Δ569–664 in which this region is deleted remained highly active in RNA splicing, but had decreased thermostability,13 suggesting a contribution to structural stabilization. The hypomutable surfaces could be involved in oligomerization, protein-protein interactions, or interactions that help position the C-tail.
The C-terminal tail
The basic C-tail (residues 597–664), which contributes to the non-specific binding of large RNA substrates, contains 13 basic residues (10 R, 3 K, and no H residues) and 11 acidic residues (9 D and 2 E residues) and has a calculated pI of 9.9. Notably, ten of the basic and acidic residues are found in clusters of two or three like residues, and this pattern is also seen in the C-tails of Mss116p and CYT-19 homologs.26 These basic and acidic clusters may interact electrostatically, accounting for the partial retraction of the C-tail suggested by the SAXS analysis.28 Mss116p’s C-tail is also rich in hydrophilic S and N residues (13 S and 15 N residues), which favor extrusion into the solvent and could contribute to RNA binding.
The mutability plot for the C-tail with an 11-amino acid residue sliding window shows two moderately hypomutable regions (M values ≤ −0.2; residues 615–618 and 641–643), which correspond to the sequences ISFR and NNN, respectively. These sequences are not conserved at the same position in the C-tails of other Mss116p homologs,26 but the alignments of the C-tail residues are uncertain, and it is possible that functionally equivalent residues are present at other positions. Surprisingly, the unigenic evolution analysis reveals several regions of the C-tail for which non-synonymous substitutions appear to be favored (mutability plot in which synonymous missense mutations are treated as silent (red) shows greater hypermutability than the plot in which all missense mutations are treated as equal (black)). The functional amino acid substitutions found in the unigenic evolution analysis show that every charged residue in the C-tail, except R639, can be replaced by an uncharged residue, consistent with a model which the C-tail binds RNA non-specifically via multiple non-specific electrostatic interactions that are individually dispensable so long as the other interactions are maintained.
Conclusions and perspectives
Here, we used high-throughput genetic selections and analysis of in vivo splicing activity to identify functionally important regions and permissible amino acid substitutions throughout the DEAD-box RNA chaperone Mss116p, and we interpret the results in the framework of recently determined X-ray crystal and SAXS solution structures for this protein. First, we carried out an unbiased selection of libraries of Mss116p containing random mutations induced by mutagenic PCR and used unigenic evolution to identify conserved (“hypomutable”) regions that may be functionally important. Then, we analyzed specific regions in greater detail by using more saturating genetic selections. In the case of motif III (SAT) and RNA-binding tract mutations, we extended analysis of growth phenotypes by analyzing RNA-splicing activity of individual variants in vivo. We thus comprehensively analyzed features identified in the crystal structures and sharpened focus for further biochemical and structural analyses.
The power of the genetic strategy used here is illustrated by the analysis of residues in the RNA-binding cleft of the helicase core that contact the bound ssRNA in the crystal structures. First, the unigenic evolution analysis showed that a number of amino acid residues whose side chains make ionic or H-bond contacts with the RNA are conserved, while residues that make main-chain contacts could in most cases be replaced by non-synonymous residues that could make the same contacts. Then, more saturating selections in which eight residues whose side-chains contact the bound RNA were randomized individually indicated a hierarchy of importance of the different interactions. Only two of these residues (R415 and T433) were strongly conserved in functional Mss116p variants, indicating that their side-chain contacts are critical for Mss116p function. Two other R-residues (R190 and R245), which make both main- and side-chain contacts with U10 RNA, were less strongly conserved, and analysis of the variants R190A and R245A, indicated that the side-chain contacts enhance Mss116p function but are not essential. Surprisingly, the remaining four residues that make side-chain contacts with U10 RNA in the crystal structure were not conserved in the selection, and we confirmed that their replacement by non-synonymous residues has little or no effect on Mss116p’s RNA-splicing function at normal expression levels in vivo. These latter residues may contribute less strongly to RNA binding, and/or there may be a tradeoff at these positions in which the ability to bind the RNA strand strongly is balanced by the need to release the bound strand following RNA unwinding. Thus, the genetic selections enabled us to rapidly evaluate the functional significance of individual RNA-protein contacts identified in the crystal structure, provided insight into the function of individual amino acid residues, and identified specific features, interactions, and amino acid substitutions that will be of interest for more detailed biochemical analysis.
In addition to the RNA-binding cleft in the helicase core, our results provide new insights into the functionality of regions of the CTE that induce the second bend in the bound RNA. Thus, we find that all residues contacting the RNA in the CTE wedge helix (α18) can be replaced by non-synonymous residues, but that despite these amino acid sequence changes, an α-helix is predicted to be maintained in all functional variants. These findings suggest that the RNA bend induced by the CTE results primarily from a steric block due to the position of α18 rather than due to ionic or H-bond interactions with specific residues, as is the case for the bend induced by the helicase core. The N-terminal portion of α19, which interacts with and potentially supports α18, is also strongly conserved, in agreement with previous findings that mutations in α19 inactivate the protein.13,18,43 In future work, mutations that disrupt or shorten these helices might be used to assess the contribution of the second bend to the efficiency of RNA unwinding.
Other noteworthy findings in our study include the identification of: (i) new functional substitutions in the conserved DEAD-box protein motifs, including replacements of the conserved F residue of the Q motif that contraindicate a strict requirement for stacking with the adenine base of ATP, and new functional combinations of amino acid residues in the SAT motif, which is involved in a network of interactions between the two core domains; (ii) conserved surface loops potentially involved in oligomerization or partner protein interactions; and (iii) two conserved regions, one the previously identified post-II region in the helicase core, and the other in the CTE that may be involved in helping to displace or sequester the RNA strand opposite that bound by the helicase core. More detailed genetic and biochemical analysis of these latter regions may provide critical insights into how DEAD-box proteins bind dsRNA and initial steps in RNA strand separation about which little is currently known.
Materials and Methods
S. cerevisiae strains and growth media
The S. cerevisiae wild-type strain 161-U7 (MATa ade1 lys1 ura3) contains eight mt group I introns (aI3α, aI4α, aI5α, aI5β, bI2, bI3, bI4, bI5 and four mt group II introns (aI1, aI2, aI5γ and bI1).19 mss116Δ/1+2+ is a derivative of 161-U7 in which the MSS116 gene was replaced by a kanr disruption cassette.19 Strains were grown in yeast peptone medium (1% yeast extract, 2% peptone) supplemented with 2% D-(+)-glucose (YPD) or 3% glycerol (YPG). For complementation tests of Mss116p variants, transformants containing CEN plasmids were selected by plating on YNBD (yeast nitrogen base minimal medium without amino acids; Becton Dickinson, Sparks, MD) supplemented with ammonium sulfate (5 g/l), adenine (20 mg/l), tryptophan (30 mg/l) and lysine (30 mg/l), with 2% D-(+)-glucose as the carbon source. For Northern and immunoblotting experiments, cells were grown in Hartwell’s complete medium lacking uracil with 2% raffinose as the carbon source.44 Solid media contained 2% bacto agar (Becton Dickinson).
Recombinant plasmids
The S. cerevisiae CEN-plasmids used to express Mss116p for genetic assays are derivatives of pHRH108.19 This plasmid contains the promoter and coding sequence of the MSS116 gene from wild-type 161-U7 cloned as a 3-kb HindIII fragment in the corresponding site of the S. cerevisae CEN plasmid vector pRS416.45 The plasmid vector carries a URA3 marker, enabling selection of strains containing it on medium lacking uracil. pHRH197 was derived from pHRH108 by adding a SacII site just downstream of the MSS116 stop codon and two silent mutations (A222G and C1546T), which destroy and create an XbaI site, respectively.43 pHRH197B, which was used for library construction for unigenic evolution analysis, was derived from pHRH197 by introducing two additional silent mutations in the MSS116 ORF, T393C and A110G, which destroy and create a BsrGI site, respectively. pHRH197-Mss116p/ΔNTE, which expresses Mss116p deleted for the NTE (residues 37–87), was derived from pHRH197 by Quikchange mutagenesis (Agilent Technologies, Santa Clara, CA).
pMAL-Mss116p is a derivative of pMal-c2x (New England Biolabs, Ipswich, MA) and uses a tac promoter to express wild-type Mss116p (beginning at codon 37 after the mt targeting sequence) with maltose-binding protein (MalE) fused to its N-terminus via a TEV protease cleavable linker.22 Variants of this plasmid, which express MalE fused to the N-termini of Mss116p/ΔNTE (deletion of the N-terminal extension; residues 37–87) or Mss116p/ΔNTE+ΔC-tail (deletion of residues 37–87 and 598–664), were created by replacing the 296-bp BamHI/BsrGI fragment of pMAL-Mss116p and pMAL-Mss116p/Δ598–664, respectively, with synthetic double-strand oligonucleotides that cleanly delete the desired amino acid sequence.
Cloning was done in E. coli DH5α (Invitrogen, Carlsbad, CA) grown in Luria-Bertani (LB) medium (0.5% yeast extract, 1% peptone, and 1% NaCl, pH 7), with 2% Difco agar for solid media, and ampicillin (100 μg/ml) and kanamycin (40 μg/ml) added as required for selections. All constructs were sequenced through the regions amplified by PCR to insure that no adventitious mutations had been introduced.
Protein expression, purification, and storage
Wild-type Mss116p, Mss116p/ΔNTE, and Mss116p/ΔNTE+ΔC-tail were expressed with a cleavable N-terminal MalE tag and purified as described.22,39,46 Proteins used for biochemical assays were dialyzed into Mss116p assay buffer (20 mM Tris-HCl (pH 7.5), 500 mM KCl, 1 mM EDTA, 1 mM DTT, and 50% glycerol), flash-frozen immediately after purification, and stored at −80°C. Proteins used for crystallization were dialyzed into Mss116p crystallization buffer (10 mM Tris-HCl (pH 7.5), 250 mM NaCl, 1 mM DTT, 50 mM arginine and glutamate, and 50% glycerol), stored on ice, and used within 1–2 weeks after purification.
Unigenic evolution analysis
For unigenic evolution analysis, we constructed a library of Mss116p variants with random mutations in the NTE and helicase core (amino acid residues 37–516) by mutagenic PCR of the MSS116 ORF in pHRH197.29,47 The PCR was done in 100 μl of reaction medium containing 0.06 pmol of pHRH197, 50 pmol each of primers MSS116-A110Gs, 5′-TCAAGAAGATTGTACAATGATG and 1543Xba(as) 5′-TCGGTCACTGCCTCTAGAAC, 0.39 mM dATP, 0.15 mM dCTP, 1.17 mM dGTP, 3.85 mM dTTP, 11 mM MgCl2, 0.5 mM MnCl2, and 5 units Taq DNA polymerase (Invitrogen). The PCR conditions were initial denaturation at 94°C for 5 min, followed by 16 cycles of 91°C for 1 min, 51°C for 1 min, and 72°C for 10 min, plus a final extension at 72°C for 15 min. The ~1.5-kb PCR product containing the mutagenized segment of the MSS116 ORF was gel-purified, digested with BsrGI and XbaI, and swapped for a stuffer DNA fragment (347-bp XbaI-BsrGI fragment containing the Ll.LtrB group II intron from pACD2x),48 which had been inserted between the XbaI and BsrGI sites of pHRH197B. The use of an intermediate plasmid containing stuffer DNA prevents contamination of the library by the wild-type MSS116 sequence from uncut pHRH197B. The mutation frequency, determined by sequencing 39 randomly chosen clones from the unselected library, was 0.78%, corresponding to ~10 amino acid substitutions per protein.
For unigenic evolution analysis, the library was transformed into mss116Δ/1+2+, and cells were plated on YPG at 30°C to select those expressing splicing-competent Mss116p variants. The initial colonies were streaked onto YPG plates, and single colonies from the streaks were patched onto YPG plates and incubated at 24°C, 30°C, and 37°C to verify the phenotype and assess temperature sensitivity. To identify mutations in functional Mss116p variants, the mutated region of Mss116p was amplified from colonies that grew at 30°C by colony PCR (http://www.imbf.ku.dk/LisbyHolmberg/colonyPCR.htm), with primers MSS116-LEADs 5′-CGCACACCTGTTCTTGCAAG and MSS116-seqXBAa 5′-TACAGGATCTGTAGGATGAG. The PCR products were purified by using Sera-Mag magnetic beads (Seradyne, Indianapolis, IN) and sequenced using primers MSS116-LEADs, MSS116–550s 5′-CAACAAGAGATTTGGCCTTG, MSS116–1100s 5′-TTGCACCAACTGTTAAATTC, and/or MSS116-KT2 5′-ACACCTGTTCTTGCAAGCAGA.
Mutational data were analyzed as described.29,47,49 Briefly, the mutability value (M) in an 11-amino acid residue sliding window was calculated by using the formula:
where fOmis is the observed frequency of missense mutations and fEmis is their expected frequency. The latter was calculated for each codon based on the probability of a single nucleotide change producing a missense mutation, correcting for the transition/transversion ratio of 3.4 for the variants present in the unselected library. Negative mutability values indicate hypomutability, with a minimum value of −1 indicating no missense mutations in a given window. Positive values indicate hypermutability and were normalized by using the formula:
so that all mutability values fall between +1 and −1.
Targeted selection for mutations in specific regions of Mss116p
Functional Mss116p variants were selected from libraries with randomized nucleotide residues at codons for motif II (DEAD), motif III (S305A306T307), each of the eight residues whose side chains contact U10 RNA (R190, T242, R245, D248, D280, K384, R415, and T433), and a doped library (70% wild-type and 10% each other nucleotide) for motif VI (H462RIGRTAR469). The mutant inserts were generated by two PCRs with Phusion Flash polymerase (New England Biolabs), one producing a 5′ fragment of the Mss116p gene without mutations and the other using a primer containing the randomized or doped nucleotide residues to produce an overlapping 3′ fragment containing the mutations. The two PCR fragments were then gel-purified and used for a second PCR to recreate a continuous DNA fragment with BsrGI and XbaI sites introduced by the outside primers. This final PCR product was digested with BsrGI and XbaI and ligated between the corresponding sites of pHRH197B. The resulting libraries were electroporated into MegaX DH10B cells (Invitrogen) for plasmid DNA isolation and sequencing. In capillary electrophoresis sequencing reactions, all libraries showed approximately equal peak heights for all four nucleotide traces at the randomized positions, and 32 randomly selected clones from each library were sequenced to confirm the expected nucleotide frequencies. The libraries were transformed into yeast 161-U7 mss116Δ/1+2+ cells, and transformants expressing functional Mss116p variants were selected on YPG plates at 30°C. Colonies were restreaked onto fresh YPG plates, and single colonies from the restreaks were used to amplify the mutated region of Mss116p by colony PCR. The PCR fragments were then purified and sequenced to identify functional substitutions at each mutated position. For phenotypic analysis, individual variants identified in the selections were reconstructed in the CEN plasmid pHRH197B via PCR with primers that introduce the mutations, and the regions subjected to PCR were sequenced to verify the absence of adventitious secondary mutations.
S. cerevisiae complementation assays
S. cerevisiae mss116Δ/1+2+ was transformed with CEN plasmids expressing wild-type or mutant Mss116p proteins and plated on YNBD medium lacking uracil to select for complementation of the strain’s ura3 mutation by the URA3 marker on the plasmid. The transformants were then grown in liquid YNBD cultures to A600 = ~1.0 (5 × 107 cells /ml), serially diluted in a microtiter plate, and stamped onto agar plates containing YPD or YPG medium. The plates were incubated at 18, 24, 30, or 37°C and photographed at different times to compare growth rates of wild-type and mutant strains.
Northern hybridizations and immunoblotting
For Northern hybridizations, S. cerevisiae strains were grown in Hartwell’s complete medium lacking uracil with 2% raffinose at 30°C to A600 = 1.0 to 2.0, and whole-cell RNA was isolated as described.50 After extraction with phenol/chloroform/isoamyl alcohol (25:24:1, by volume) and ethanol precipitation, portions of the RNA (1.2 μg) were denatured by incubating with 20% (v/v) glyoxal for 15 min at 65°C and run in a 1.5% (w/v) agarose gel. The gel was then blotted to a nylon membrane (Hybond-XL; GE Healthcare, Piscataway, NJ), which was hybridized with 5′-end-labeled DNA oligonucleotide probes (COB exon 6 probe: 5′-AAGGTACTTCTACATGGCATGCT; COX1 exon 6 probe: 5′-ATTTCATCCTGCGAAAGCATCAGGAT; COX2 exon probe: 5′-AGGTAATGATACTGCTTCGATC) and visualized by scanning with a PhosphorImager (Typhoon Trio, GE Healthcare).
For immunoblotting, whole-cell proteins were isolated by trichloroacetic acid (TCA) precipitation.26 After washing with 1 M Tris base, the protein pellets were dissolved in 150 μl of SDS-PAGE sample buffer, and portions (~60 μg) were run in a 0.1% (w/v) SDS/4–12% (w/v) polyacrylamide gradient gel (NuPage, Invitrogen). The gel was blotted to a Sequi-Blot™ PVDF membrane (Bio-Rad, Hercules, CA), using a Bio-Rad Criterion blotter apparatus, and the blot was probed with a guinea pig anti-Mss116 antibody (1:5,000 dilution),43 developed using an ECL Plus Western Blotting-kit (GE Healthcare), and imaged using a Cell Biosciences (Santa Clara, CA) HD2 imager or Kodak Biomax XAR film. To confirm equal loading of protein samples, the PVDF membrane was stripped of antibodies using Restore™ Plus Western Blot Stripping Buffer (ThermoScientific, Waltham, MA) and stained with AuroDye Forte (GE Healthcare) following the manufacturers’ directions.
RNA splicing assays
RNA splicing assays were as described.21 Briefly, 32P-labeled precursor RNA containing the aI5γ group II intron from S. cerevisiae was transcribed by using a T7 Megascript kit (Ambion, Austin, TX) from plasmid pJD20 that had been linearized by digestion with HindIII.51 Splicing reactions were done in a thermocycler at 30°C with 10 nM 32P-labeled precursor RNA and the indicated amounts of Mss116p in 50 μl of reaction medium containing 100 mM KCl, 8 mM MgCl2, 50 mM Na-MOPS, pH 7.5, 5% glycerol, and 1 mM ATP-Mg2+. Products were analyzed by electrophoresis in a denaturing 4% polyacrylamide gel, which was dried and quantified with a PhosphorImager (Typhoon Trio) using ImageQuant TL software (GE Healthcare). The data for the disappearance of precursor RNA from splicing time courses were fitted to single-exponential equations using KaleidaGraph 4.1 (Synergy Software, Reading, PA).
X-ray crystallography
Ternary complexes were formed by incubating 90 μM protein (Mss116p/ΔNTE or Mss116p/ΔNTE+ΔC-tail), 180 μM U10 ssRNA oligonucleotide (Integrated DNA Technologies, Coralville, IA), 1 mM AMP-PNP-Mg2+, 2 mM MgCl2 on the bench top for 10 min. For Mss116p/ΔNTE, sitting drops were assembled in a sealed 96-well plate using 0.5 μl of complex and 0.5 μl of Natrix condition 39 (100 mM ammonium acetate, 20 mM MgCl2 • 6H2O, 50 mM HEPES-Na pH 7.0, and 5% polyethylene glycol 8,000; Hampton Research, Aliso Viejo, CA) suspended over a 50 μl reservoir solution consisting of a 1:1 mix of Natrix condition 39 and Mss116p crystallization buffer. For Mss116p/ΔNTE+ΔC-tail, sitting drops were assembled in a sealed 24-well plate using 1 μl of complex and 1 μl of screening solution (0.1 M ammonium tartrate dibasic, pH 7.0, 10% PEG3350) suspended over a 200 μl reservoir solution consisting of a 1:1 mix of screening solution and Mss116p storage buffer. Sitting drops were stored at 22 C. Crystals were removed from the drops and flash cooled immediately in liquid N2.
Synchrotron X-ray diffraction data were collected at LS-CAT beamline 21-ID-G at the Advanced Photon Source, Argonne National Laboratory. Data collection and refinement parameters are summarized in Table S1. Diffraction intensities were indexed and scaled with HKL2000.52 Both the Mss116p/ΔNTE and Mss116p/ΔNTE+ΔC-tail ternary complexes crystallized in the same space group with essentially the same unit cell dimensions as the previously published Mss116p/ΔC-tail complex.13 To build and refine models of these complexes, the Rfree flags from the previous AMP-PNP ternary complex dataset were transferred to the new datasets using tools from CCP4,53 and the previous Mss116p/ΔC-tail protein model was then refined against the other datasets using Refmac.54 The models were completed by several cycles of manual model building in Coot55 and refinement in Refmac. TLS (translation, libration, screw) vibrational motions were used at the end of refinement with TLS groups identified by the TLSMD server.56 Model validation was performed using MolProbity.57
Structural figures and analysis
Structural figures were created using the PyMOL Molecular Graphics System, Version 1.4 (Schrödinger, LLC, New York, NY). Solvent accessibility was determined by the PISA Server.58 Models were compared to each other using LSQMAN.59
PDB accession codes
The coordinates and structure factors for ternary complexes of Mss116p/ΔNTE and Mss116p/ΔNTE+ΔC-tail with AMP-PNP and U10 RNA were deposited in the PDB and have accession codes 3SQW and 3SQX, respectively.
Supplementary Material
Acknowledgments
This work was supported by NIH grant GM037951. Use of the APS was supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under contract number DE-AC02-06CH11357. Use of LS-CAT Sector 21 was supported by the Michigan Economic Development Corporation and the Michigan Technology Tri-Corridor (grant 085P1000817). We thank Lilian Lamech for constructing pMAL-Mss116p/ NTE and pMAL-Mss116p/ NTE+ C-tail, Anna L. Mallam for protein preparations, and Anna L. Mallam and Rick Russell for comments on the manuscript.
Abbreviations used
- CEN plasmid
centromere-containing plasmid
- C-tail
C-terminal tail
- CTE
C-terminal extension
- Gly
glycerol
- M value
mutability value
- mt
mitochondrial
- NTE
N-terminal extension
- TCA
trichloroacetic acid
- YNBD
yeast nitrogen base minimal medium with glucose
- YPD and YPG
yeast peptone medium with glucose and glycerol, respectively
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Cordin O, Banroques J, Tanner NK, Linder P. The DEAD-box protein family of RNA helicases. Gene. 2006;367:17–37. doi: 10.1016/j.gene.2005.10.019. [DOI] [PubMed] [Google Scholar]
- 2.Hilbert M, Karow AR, Klostermeier D. The mechanism of ATP-dependent RNA unwinding by DEAD box proteins. Biol Chem. 2009;390:1237–1250. doi: 10.1515/BC.2009.135. [DOI] [PubMed] [Google Scholar]
- 3.Linder P. The dynamic life with DEAD-box RNA helicases. In: Jankowsky E, editor. RSC Biomolecular Sciences: RNA Helicases. Vol. 20. Royal Society of Chemistry; Cambridge, UK: 2010. pp. 32–60. [Google Scholar]
- 4.Jankowsky E. RNA helicases at work: binding and rearranging. Trends Biochem Sci. 2011;36:19–29. doi: 10.1016/j.tibs.2010.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jarmoskaite I, Russell R. DEAD-box proteins as RNA helicases and chaperones. WIREs RNA. 2011;2:135–152. doi: 10.1002/wrna.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang Q, Del Campo M, Lambowitz AM, Jankowsky E. DEAD-box proteins unwind duplexes by local strand separation. Mol Cell. 2007;28:253–263. doi: 10.1016/j.molcel.2007.08.016. [DOI] [PubMed] [Google Scholar]
- 7.Fairman-Williams ME, Guenther UP, Jankowsky E. SF1 and SF2 helicases: family matters. Curr Opin Struct Biol. 2010;20:313–324. doi: 10.1016/j.sbi.2010.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Andersen CB, Ballut L, Johansen JS, Chamieh H, Nielsen KH, Oliveira CL, Pedersen JS, Séraphin B, Le Hir H, Andersen GR. Structure of the exon junction core complex with a trapped DEAD-box ATPase bound to RNA. Science. 2006;313:1968–1972. doi: 10.1126/science.1131981. [DOI] [PubMed] [Google Scholar]
- 9.Bono F, Ebert J, Lorentzen E, Conti E. The crystal structure of the exon junction complex reveals how it maintains a stable grip on mRNA. Cell. 2006;126:713–725. doi: 10.1016/j.cell.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 10.Sengoku T, Nureki O, Nakamura A, Kobayashi S, Yokoyama S. Structural basis for RNA unwinding by the DEAD-box protein Drosophila Vasa. Cell. 2006;125:287–300. doi: 10.1016/j.cell.2006.01.054. [DOI] [PubMed] [Google Scholar]
- 11.Yang Q, Jankowsky E. The DEAD-box protein Ded1 unwinds RNA duplexes by a mode distinct from translocating helicases. Nat Struct Mol Biol. 2006;13:981–986. doi: 10.1038/nsmb1165. [DOI] [PubMed] [Google Scholar]
- 12.Collins R, Karlberg T, Lehtiö L, Schütz P, van den Berg S, Dahlgren LG, Hammarström M, Weigelt J, Schüler H. The DEXD/H-box RNA helicase DDX19 is regulated by an α-helical switch. J Biol Chem. 2009;284:10296–10300. doi: 10.1074/jbc.C900018200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Del Campo M, Lambowitz AM. Structure of the yeast DEAD box protein Mss116p reveals two wedges that crimp RNA. Mol Cell. 2009;35:598–609. doi: 10.1016/j.molcel.2009.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.von Moeller H, Basquin C, Conti E. The mRNA export protein DBP5 binds RNA and the cytoplasmic nucleoporin NUP214 in a mutually exclusive manner. Nat Struct Mol Biol. 2009;16:247–254. doi: 10.1038/nsmb.1561. [DOI] [PubMed] [Google Scholar]
- 15.Chen Y, Potratz JP, Tijerina P, Del Campo M, Lambowitz AM, Russell R. DEAD-box proteins can completely separate an RNA duplex using a single ATP. Proc Natl Acad Sci USA. 2008;105:20203–20208. doi: 10.1073/pnas.0811075106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu F, Putnam A, Jankowsky E. ATP hydrolysis is required for DEAD-box protein recycling but not for duplex unwinding. Proc Natl Acad Sci USA. 2008;105:20209–20214. doi: 10.1073/pnas.0811115106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Séraphin B, Simon M, Boulet A, Faye G. Mitochondrial splicing requires a protein from a novel helicase family. Nature. 1989;337:84–87. doi: 10.1038/337084a0. [DOI] [PubMed] [Google Scholar]
- 18.Mohr S, Stryker JM, Lambowitz AM. A DEAD-box protein functions as an ATP-dependent RNA chaperone in group I intron splicing. Cell. 2002;109:769–779. doi: 10.1016/s0092-8674(02)00771-7. [DOI] [PubMed] [Google Scholar]
- 19.Huang HR, Rowe CE, Mohr S, Jiang Y, Lambowitz AM, Perlman PS. The splicing of yeast mitochondrial group I and group II introns requires a DEAD-box protein with RNA chaperone function. Proc Natl Acad Sci USA. 2005;102:163–168. doi: 10.1073/pnas.0407896101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mohr S, Matsuura M, Perlman PS, Lambowitz AM. A DEAD-box protein alone promotes group II intron splicing and reverse splicing by acting as an RNA chaperone. Proc Natl Acad Sci USA. 2006;103:3569–3574. doi: 10.1073/pnas.0600332103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Del Campo M, Mohr S, Jiang Y, Jia H, Jankowsky E, Lambowitz AM. Unwinding by local strand separation is critical for the function of DEAD-box proteins as RNA chaperones. J Mol Biol. 2009;389:674–693. doi: 10.1016/j.jmb.2009.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Halls C, Mohr S, Del Campo M, Yang Q, Jankowsky E, Lambowitz AM. Involvement of DEAD-box proteins in group I and II intron splicing. Biochemical characterization of Mss116p, ATP-hydrolysis-dependent and -independent mechanisms, and general RNA chaperone activity. J Mol Biol. 2007;365:835–855. doi: 10.1016/j.jmb.2006.09.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Karunatilaka KS, Solem A, Pyle AM, Rueda D. Single-molecule analysis of Mss116-mediated group II intron folding. Nature. 2010;467:935–939. doi: 10.1038/nature09422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Potratz JP, Del Campo M, Wolf RZ, Lambowitz AM, Russell R. ATP-dependent roles of the DEAD-box protein Mss116p in group II intron splicing in vitro and in vivo. J Mol Biol. 2011;411:661–679. doi: 10.1016/j.jmb.2011.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Markov DA, Savkina M, Anikin M, Del Campo M, Ecker K, Lambowitz AM, De Gnore JP, McAllister WT. Identification of proteins associated with the yeast mitochondrial RNA polymerase by tandem affinity purification. Yeast. 2009;26:423–440. doi: 10.1002/yea.1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mohr G, Del Campo M, Mohr S, Yang Q, Jia H, Jankowsky E, Lambowitz AM. Function of the C-terminal domain of the DEAD-box protein Mss116p analyzed in vivo and in vitro. J Mol Biol. 2008;375:1344–1364. doi: 10.1016/j.jmb.2007.11.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grohman JK, Del Campo M, Bhaskaran H, Tijerina P, Lambowitz AM, Russell R. Probing the mechanisms of DEAD-box proteins as general RNA chaperones: the C-terminal domain of CYT-19 mediates general recognition of RNA. Biochemistry. 2007;46:3013–3022. doi: 10.1021/bi0619472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mallam AL, Jarmoskaite I, Tijerina P, Del Campo M, Seifert S, Guo L, Russell R, Lambowitz AM. Solution structures of DEAD-box RNA chaperones reveal conformational changes and nucleic acid tethering by a basic tail. Proc Natl Acad Sci USA. 2011;108:12254–12259. doi: 10.1073/pnas.1109566108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Deminoff SJ, Tornow J, Santangelo GM. Unigenic evolution: a novel genetic method localizes a putative leucine zipper that mediates dimerization of the Saccharomyces cerevisiae regulator Gcr1p. Genetics. 1995;141:1263–1274. doi: 10.1093/genetics/141.4.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Henikoff S, Henikoff JG. Amino acid substitution matrices from protein blocks. Proc Natl Acad Sci USA. 1992;89:10915–10919. doi: 10.1073/pnas.89.22.10915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schütz P, Bumann M, Oberholzer AE, Bieniossek C, Trachsel H, Altmann M, Baumann U. Crystal structure of the yeast eIF4A-eIF4G complex: an RNA-helicase controlled by protein-protein interactions. Proc Natl Acad Sci USA. 2008;105:9564–9569. doi: 10.1073/pnas.0800418105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Karow AR, Klostermeier D. A structural model for the DEAD box helicase YxiN in solution: localization of the RNA binding domain. J Mol Biol. 2010;402:629–637. doi: 10.1016/j.jmb.2010.07.049. [DOI] [PubMed] [Google Scholar]
- 33.Schütz P, Karlberg T, van den Berg S, Collins R, Lehtiö L, Högbom M, Holmberg-Schiavone L, Tempel W, Park HW, Hammarström M, Moche M, Thorsell AG, Schüler H. Comparative structural analysis of human DEAD-box RNA helicases. PLoS One. 2010;5:e12791. doi: 10.1371/journal.pone.0012791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Montpetit B, Thomsen ND, Helmke KJ, Seeliger MA, Berger JM, Weis K. A conserved mechanism of DEAD-box ATPase activation by nucleoporins and InsP6 in mRNA export. Nature. 2011;472:238–242. doi: 10.1038/nature09862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rudolph MG, Heissmann R, Wittmann JG, Klostermeier D. Crystal structure and nucleotide binding of the Thermus thermophilus RNA Helicase Hera N-terminal domain. J Mol Biol. 2006;361:731–743. doi: 10.1016/j.jmb.2006.06.065. [DOI] [PubMed] [Google Scholar]
- 36.Banroques J, Doere M, Dreyfus M, Linder P, Tanner NK. Motif III in superfamily 2 “helicases” helps convert the binding energy of ATP into a high-affinity RNA binding site in the yeast DEAD-box protein Ded1. J Mol Biol. 2010;396:949–966. doi: 10.1016/j.jmb.2009.12.025. [DOI] [PubMed] [Google Scholar]
- 37.Pause A, Sonenberg N. Mutational analysis of a DEAD box RNA helicase: the mammalian translation initiation factor eIF-4A. EMBO J. 1992;11:2643–2654. doi: 10.1002/j.1460-2075.1992.tb05330.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chamot D, Magee WC, Yu E, Owttrim GW. A cold shock-induced cyanobacterial RNA helicase. J Bacteriol. 1999;181:1728–1732. doi: 10.1128/jb.181.6.1728-1732.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Del Campo M, Tijerina P, Bhaskaran H, Mohr S, Yang Q, Jankowsky E, Russell R, Lambowitz AM. Do DEAD-box proteins promote group II intron splicing without unwinding RNA? Mol Cell. 2007;28:159–166. doi: 10.1016/j.molcel.2007.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bifano AL, Turk EM, Caprara MG. Structure-guided mutational analysis of a yeast DEAD-box protein involved in mitochondrial RNA splicing. J Mol Biol. 2010;398:429–443. doi: 10.1016/j.jmb.2010.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cuff JA, Clamp ME, Siddiqui AS, Finlay M, Barton GJ. JPred: a consensus secondary structure prediction server. Bioinformatics. 1998;14:892–893. doi: 10.1093/bioinformatics/14.10.892. [DOI] [PubMed] [Google Scholar]
- 42.Cuff JA, Barton GJ. Application of multiple sequence alignment profiles to improve protein secondary structure prediction. Proteins. 2000;40:502–511. doi: 10.1002/1097-0134(20000815)40:3<502::aid-prot170>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 43.Huang H-R. Ph D thesis. The University of Texas Southwestern Medical Center; 2004. Functional studies of intron- and nuclear-encoded splicing factors in the mitochondria of Saccharomyces cerevisiae. [Google Scholar]
- 44.Amberg DC, Burke D, Strathern JN. Methods in Yeast Genetics: a Cold Spring Harbor Laboratory Course Manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, N.Y: 2005. [Google Scholar]
- 45.Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Del Campo M, Lambowitz AM. Crystallization and preliminary X-ray diffraction of the DEAD-box protein Mss116p complexed with an RNA oligonucleotide and AMP-PNP. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2009;65:832–835. doi: 10.1107/S1744309109027225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cui X, Matsuura M, Wang Q, Ma H, Lambowitz AM. A group II intron-encoded maturase functions preferentially in cis and requires both the reverse transcriptase and X domains to promote RNA splicing. J Mol Biol. 2004;340:211–231. doi: 10.1016/j.jmb.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 48.San Filippo J, Lambowitz AM. Characterization of the C-terminal DNA-binding/DNA endonuclease region of a group II intron-encoded protein. J Mol Biol. 2002;324:933–951. doi: 10.1016/s0022-2836(02)01147-6. [DOI] [PubMed] [Google Scholar]
- 49.Behrsin CD, Brandl CJ, Litchfield DW, Shilton BH, Wahl LM. Development of an unbiased statistical method for the analysis of unigenic evolution. BMC Bioinformatics. 2006;7:150. doi: 10.1186/1471-2105-7-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schmitt ME, Brown TA, Trumpower BL. A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res. 1990;18:3091–3092. doi: 10.1093/nar/18.10.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jarrell KA, Dietrich RC, Perlman PS. Group II intron domain 5 facilitates a trans-splicing reaction. Mol Cell Biol. 1988;8:2361–2366. doi: 10.1128/mcb.8.6.2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 53.Collaborative Computational Project Number 4, C. The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 54.Murshudov GN, Skubak P, Lebedev AA, Pannu NS, Steiner RA, Nicholls RA, Winn MD, Long F, Vagin AA. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr D Biol Crystallogr. 2011;67:355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Painter J, Merritt EA. Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Crystallogr D Biol Crystallogr. 2006;62:439–450. doi: 10.1107/S0907444906005270. [DOI] [PubMed] [Google Scholar]
- 57.Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall WB, 3rd, Snoeyink J, Richardson JS, Richardson DC. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 59.Kleywegt GJ. Experimental assessment of differences between related protein crystal structures. Acta Crystallogr D Biol Crystallogr. 1999;55:1878–1884. doi: 10.1107/s0907444999010495. [DOI] [PubMed] [Google Scholar]
- 60.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.