Abstract
Recombinant human surfactant protein D (SP-D) expressed in Escherichia coli, consisting of the head and neck regions of the native molecule, bound to all strains of Streptococcus pneumoniae that were tested, but the extent of binding varied between strains of differing capsular serotypes. The recombinant protein expressed in the yeast Pichia pastoris did not bind. Full-length native SP-D aggregated pneumococci in a calcium-dependent manner that was inhibited by maltose acting as a competitive sugar. The ability of SP-D to modulate the uptake and killing of pneumococci by human neutrophils was also addressed. Neither recombinant truncated SP-D nor native full-length SP-D enhanced the killing of pneumococci by human neutrophils. Aggregation of pneumococci varied not only between strains of the same multilocus sequence type and different serotypes but also between strains of the same serotype. However, use of recombinant strains in which the serotype had been changed showed that the degree of aggregation was influenced by the capsular type. Indeed, a 19F serotype strain which was not aggregated by SP-D did exhibit aggregation when the original isogenic strain was capsule switched to capsular serotype 3. However, although our results show that SP-D is capable of aggregating most pneumococci, no correlation between the degree of aggregation and the capsule or multilocus sequence type of the pneumococcus was clearly apparent. Therefore, although the capsule serotype is not the only determinant of aggregation by SP-D, the data presented here indicate that it does have a role to play.
Streptococcus pneumoniae is carried asymptomatically in the nasopharynx in humans (2) but has long been known as an eminent cause of morbidity and mortality and as a major invasive pathogen leading to bronchopneumonia, septicemia, and meningitis (6, 25).
In the early stages after infection, natural pulmonary defense mechanisms are required for efficient resistance and clearance of the pneumococcus. Recent studies have drawn attention to the important role of lung surfactant protein D (SP-D) in natural innate immunity to microbial invasion in the respiratory tract and also its ability to modulate the adaptive immune response (5, 8, 14, 17, 18, 32).
Although SP-D has been found associated with mucosal surfaces throughout the body, it is found mostly in the fluid lining the epithelium of the alveoli, where it is produced by alveolar type II cells and nonciliated Clara cells of the lung tissue (4). As a member of the collectin family, SP-D has some basic structural characteristics in common with lung surfactant protein A, mannose binding lectin, bovine conglutinin, collectin 43, and the recently discovered ficolins (24). Predominantly, SP-D is secreted as dodecamers consisting of four collagenous trimers cross-linked by disulfide bonds to create a cruciform structure. Each trimer of the molecule consists of three polypeptide chains, and each subunit consists of four domains, a short amino-terminal end, a collagen-like region followed by a short α-helical neck region, and a C-type carbohydrate recognition domain (CRD) (4, 14).
A number of pulmonary pathogens have been reported to be aggregated by SP-D in vitro (10, 21, 26). Previous studies have shown that SP-D can interact with microorganisms via binding of the CRD to carbohydrate structures on the surface of the pathogens, leading to different effector mechanisms, such as aggregation or modulation of the microbial uptake by immune cells, thus probably affecting the outcome of the pathogen invasion (15, 19). Treatment of mice with SP-D or recombinant SP-D (rSP-D) substantially increased their survival rate in a model of invasive pulmonary aspergillosis (23). However, after infection with Haemophilus influenzae or group B streptococcus, SP-D knockout mice showed increased inflammation but no decrease in bacterial killing in lung tissue (13).
A previous study reported that highly multimerized SP-D molecules bound to serotype 4, 19, and 23 S. pneumoniae, inducing their aggregation and enhancing their uptake by neutrophils (12). Assuming that aggregation of S. pneumoniae would have an important role in limiting the spread of the pathogen through the lung tissue, in the present study we further studied the interaction between SP-D and pneumococci using a more comprehensive number of strains and serotypes of S. pneumoniae than have been previously studied. Carbohydrate moieties have been implicated in the binding of SP-D by bacteria (8, 28), but although Hartshorn et al. (12) examined the binding of collectins to a limited number of pathogenic strains of pneumococci, no investigation has studied whether the capsule type is a determinant of S. pneumoniae aggregation by SP-D. Different strains of pneumococci with the same capsule type or the same multilocus sequence type were assessed for their binding and aggregation by SP-D. Moreover, encapsulated and nonencapsulated strains of S. pneumoniae were used to investigate whether SP-D enhanced killing of different strains of pneumococci by neutrophils.
MATERIALS AND METHODS
Pneumococcal strains.
Clinical isolates of S. pneumoniae from different countries were kindly provided by Mark Enright of the Wellcome Trust Centre for the Epidemiology of Infectious Disease, Oxford University, and now based in the University of Bath, Bath, United Kingdom. The serotype 3 strains M393, M110, and M290; the serotype 18C strains M9, M241, and M208; the serotype 19A strain GM70; the serotype 23F strain M41; the serotype 14A strain M322; and the serotype 9V strain M15 were used. Molecular characterization by multilocus sequence typing (MLST) was available for these isolates (9); the MLST of other strains was determined by the Scottish Meningococcus and Pneumococcus Reference Laboratory, Glasgow, United Kingdom. Strains of S. pneumoniae also were made by switching the capsule type (29): the serotype 19F strain G54, the parent of the mutant Ps3221, in which the capsule was switched to serotype 3, and the serotype 3 strain HB565 and mutant strain Ps3205, in which the capsule was switched from type 2 (Strain D39) to type 3. Other encapsulated pneumococcal strains, 1N, 2, 3, 8, 14, and 18C, and the nonencapsulated strain Rx1, obtained from the National Collection of Tissue Cultures, London, United Kingdom, were used. All pneumococcal strains were confirmed by Gram staining, catalase, optochin sensitivity, and α-hemolysis on blood agar plates.
Bacterial preparation.
For use in all assays, S. pneumoniae cells were grown overnight in brain heart infusion (Oxoid, Basingstoke, United Kingdom). The suspension was centrifuged at 1,000 × g for 15 min, washed, and then resuspended in 2 ml of Tris-buffered saline, pH 7.4 (TBS).
For binding assays, pneumococci were killed with 0.5% (vol/vol) formalin in 0.9% (wt/vol) NaCl for 1 h with shaking. The bacteria were subsequently washed three times in TBS and resuspended in carbonate-bicarbonate coating buffer, pH 9.6, before being allowed to adhere to flat-bottom enzyme-linked immunosorbent assay (ELISA) plate wells at 4°C overnight.
Human SP-D, rSP-D, and antiserum preparation.
Native human SP-D was purified by affinity chromatography from bronchoalveolar lavage fluid from a patient with alveolar proteinosis as described previously (21). Recombinant forms of the carbohydrate recognition modules of SP-D (rSP-D), consisting of three CRD head molecules held together with the α-helical neck region of the molecule, were overexpressed in both Escherichia coli and Pichia pastoris and purified (21). An anti-human SP-D antibody was raised in a rabbit against the purified yeast recombinant head-neck fragment of SP-D. The immunoglobulin G fraction was purified from rabbit serum by sodium sulfate precipitation followed by protein A chromatography, as previously described (22). ELISA and Western blotting confirmed the specificity of the antiserum for native human SP-D.
Binding of SP-D to S. pneumoniae.
Binding of SP-D to pneumococci was tested by ELISA, as described before (12). Pneumococci were allowed to adhere to ELISA plates at 4°C overnight, and the mean protein concentrations per well were determined (8 to 11 μg per well). The plates were washed with 0.05% (vol/vol) TBS-Tween 20 after each subsequent step. After being coated, the plates were blocked with 3% (wt/vol) bovine serum albumin in TBS for 1 h at 37°C. Recombinant human SP-D in TBS containing 15 mM calcium chloride, polyclonal rabbit anti-human SP-D (1 in 5,000), and goat anti-rabbit immunoglobulin G antiserum conjugated to horseradish peroxidase (Sigma-Aldrich, Poole, United Kingdom) (1 in 2,000) were added sequentially with washing between the steps. All incubation steps were carried out for 2 h at 37°C. Binding of rSP-D to the plates was detected with TMB peroxidase substrate (Bio-Rad). Reactions were developed for 15 min at 37°C and were stopped by adding 50 μl of 1 M H2SO4. Absorbance was measured with an ELISA plate reader at 450 nm.
Aggregation assay.
In a final volume of 50 μl, 30 μl of pneumococcal suspension was incubated with SP-D (10 μg/ml) for 90 min at 37°C in the presence and absence of calcium (5 mM) or in the presence of EDTA (10 mM) or maltose (100 mM). Samples were put onto slides and examined by phase-contrast microscopy (magnification, ×500). The complete surface area of each suspension on each slide was examined for aggregated bacteria. An average of 10 fields of view were counted per slide, and the average area of aggregated bacteria (in square micrometers) and the number of clumps per field of view were determined. All experimental measurements, including scoring, were performed by two observers (R.J. and A.K.) individually blinded to experimental conditions.
Killing of pneumococci by neutrophils.
Neutrophils were isolated from whole blood of healthy adult donors by a one-step isolation procedure using Polymorphprep (Nycomed Pharma, Birmingham, United Kingdom) and were suspended in RPMI 1640 until required. Prior to assay, the cells were centrifuged and resuspended in Hanks balanced salt solution (1.2 mM Ca2+; Gibco, Paisley, United Kingdom) at a final concentration of 107 cells/ml. The viability of neutrophils was checked at the beginning and end of the experiments by trypan blue exclusion. More than 98% of the neutrophils were viable and were used within 2 h of isolation. Autologous serum (obtained from volunteers who had received pneumococcal vaccine) stored at −70°C was used as an opsonin source in these experiments.
Killing of pneumococci by human neutrophils was estimated by measuring the decrease in numbers of viable bacteria. Encapsulated S. pneumoniae type 2 (strain D39) and nonencapsulated strain RX1 were first opsonized by incubation at 37°C for 30 min with 20% (vol/vol) autologous serum (collected from a volunteer who had received pneumococcal polysaccharide vaccine), native SP-D (10 μg/ml), or rSP-D (20 μg/ml). Neutrophils (5 × 106) were incubated with 5 × 105 live bacteria/ml in a final volume of 250 μl in Hanks balanced salt solution at 37°C under gentle rotation. Samples were taken at 0, 30, 60, and 120 min; serially diluted in water; and plated onto blood agar plates (BAB; Oxoid) to determine the number of viable bacteria.
Statistical analysis.
Data analysis was done by analysis of variance, followed by the Bonferroni test. Statistical significance was considered to be a P value of ≤0.05.
RESULTS
Binding of rSP-D to S. pneumoniae.
A solid-phase bacterial ELISA revealed that recombinant human SP-D (5 to 10 μg/ml), overexpressed in E. coli, bound to all strains of S. pneumoniae used in this study in the presence of calcium, irrespective of the serotype (Table 1). There was no significant difference in the extent of binding between strains of the same serotype (P ≥ 0.05), but there were differences between strains of different serotypes.
TABLE 1.
Binding of rSP-D to different strains of S. pneumoniaea
| Strain | Serotype | MLST | Binding at rSP-D concn (μg/ml) of:
|
||
|---|---|---|---|---|---|
| 0 | 5 | 10 | |||
| M393 | 3 | 260 | 0.18 ± 0.01 | 0.30 ± 0.02 | 0.34 ± 0.01 |
| M110 | 3 | 180 | 0.19 ± 0.01 | 0.27 ± 0.01 | 0.37 ± 0.04 |
| M290 | 3 | 49 | 0.13 ± 0.01 | 0.22 ± 0.02 | 0.25 ± 0.01 |
| M9 | 18C | 102 | 0.20 ± 0.02 | 0.40 ± 0.02 | 0.47 ± 0.02 |
| M241 | 18C | 118 | 0.21 ± 0.01 | 0.34 ± 0.03 | 0.46 ± 0.05 |
| M208 | 18C | 119 | 0.21 ± 0.02 | 0.31 ± 0.02 | 0.38 ± 0.07 |
| GM70 | 19A | 81 | 0.13 ± 0.09 | 0.22 ± 0.02 | 0.26 ± 0.01 |
| M41 | 23F | 81 | 0.17 ± 0.01 | 0.29 ± 0.06 | 0.32 ± 0.01 |
| M322 | 14A | 156 | 0.12 ± 0.08 | 0.19 ± 0.01 | 0.25 ± 0.01 |
| M15 | 9V | 156 | 0.10 ± 0.04 | 0.17 ± 0.01 | 0.25 ± 0.007 |
Binding was assessed by bacterial solid-phase ELISA. Experiments were done in 5 mM calcium. The values are means ± standard errors of the mean of optical densities at 450 nm from three or four replicate experiments. In all cases, the optical density was significantly increased compared with the control value (in the absence of rSP-D); P < 0.05.
Strain M9 (serotype 18C) bound rSP-D to a greater extent than all the other serotypes used (P ≤ 0.001), but the two other 18C strains tested were not significantly different in terms of SP-D binding from the other serotypes. Two pneumococcal strains of the same MLST showed differences in the extent of binding: strain M41 (serotype 23 F) bound rSP-D (5 μg/ml) to a greater extent than GM70 (serotype 19A) (P ≤ 0.05). However, two strains of different serotypes but of the same MLST, M322 and M15, had similar binding to rSP-D (P ≥ 0.05). Interestingly, recombinant SP-D synthesized in yeast did not bind any of the pneumococcal strains shown in Table 1. This was unexpected, as the same protein had previously been crystallized and the structure of SP-D had been solved by members of the laboratory. Nonspecific binding of rSP-D to microtiter wells without bacteria was not observed. At rSP-D concentrations of >10 μg/ml, no further significant increases in binding of rSP-D to pneumococci was observed (data not shown).
Aggregation of S. pneumoniae by full-length SP-D.
To investigate the physical effect resulting from rSP-D binding to pneumococci, different serotypes were incubated with multimerized full-length SP-D (10 μg/ml). As shown in Table 2, several serotypes of S. pneumoniae were aggregated by full-length SP-D. There was no aggregation in the presence of 10 mM EDTA, emphasizing that it took place in a calcium-dependent manner (data not shown). No aggregation of any strain was detected when recombinant SP-D was used (data not shown), indicating that full-length multimeric SP-D molecules are required to aggregate S. pneumoniae. Also, no clumping or aggregation of pneumococci was observed in the absence of SP-D.
TABLE 2.
Aggregation of different serotypes of S. pneumoniae by SP-Da
| Serotype | Absence of maltose
|
Presence of maltose (100 mM)
|
||
|---|---|---|---|---|
| No. of clumps/ field of view | Avg area of aggregated bacteria (μm2) | No. of clumps/ field of view | Avg area of aggregated bacteria (μm2) | |
| 1N | 12.9 ± 0.75b | 96.68 ± 0.83b | 11.8 ± 0.99 | 36.35 ± 0.42d |
| 3 | 4.33 ± 0.28 | 27.64 ± 0.67 | 0.8 ± 0.22d | 12.6 ± 0.41d |
| 14 | 4 ± 0.66 | 25.48 ± 0.78 | 1.3 ± 0.25d | 11.93 ± 0.38d |
| 8 | 2.17 ± 0.29 | 17.78 ± 0.45 | 1 ± 0.21d | 16.68 ± 0.11d |
| 2 (D39) | 1.11 ± 0.35c | 128.62 ± 0.87c | No clumps | No aggregates |
| RX1 | 4 ± 0.25 | 108 ± 2.6 | 1.7 ± 0.26 | 14.44 ± 0.6d |
| E. coli Y1088 (5 μg/ml) | 3.9 ± 0.28 | 97.2 ± 1.11 | 2.73 ± 0.33 | 21.99 ± 0.51d |
Pneumococci were incubated with SP-D (10 μg/ml) for 90 min in the presence of calcium (5 mM) and with or without 100 mM maltose before examination by phase-contrast microscopy (×500). The average area of aggregated bacteria was measured, and the number of clumps per field of view was determined. A substantial difference in the extent of aggregation was detected when maltose was used as a competitive sugar. The data are the means ± standard errors of the mean of four or five experiments. No clumping or aggregation of pneumococci was observed in the absence of SP-D (data not shown).
P < 0.05, significantly higher than the values of all other strains.
P < 0.05, significantly lower than the values of all other strains.
P < 0.05, significantly different from the control value in the absence of maltose.
The average area of aggregated bacteria and the number of clumps per field of view differed among the serotypes. For instance, four clumps of aggregated bacteria per field of view were detected for serotype 14, significantly different (P ≤ 0.001) from serotype 2, for which only one clump was detected; the average area of aggregated bacteria was significantly greater with serotype 1N than with serotype 8 (P ≤ 0.001). The supposition that SP-D binds to carbohydrate structures on the surface of the pneumococcus was tested by using maltose (100 mM) as a competitive sugar. Significant inhibition of aggregation was observed with all strains (P ≤ 0.05), as shown in Table 2.
To further explore the mechanism of aggregation, strains of S. pneumoniae of different capsule types and MLSTs were utilized. As shown in Table 3, SP-D induced aggregation of several, but not all, strains of S. pneumoniae, and the extent of aggregation differed among the aggregated strains. Pneumococcal strains of the same serotype showed differences in the amount of aggregation by SP-D. For instance, the serotype 3 strain M110 was strongly aggregated by SP-D, while the serotype 3 strain M393 showed no aggregation (Fig. 1 and Table 3). The same was also true for 18C strains. However, pneumococcal strains with the same multilocus sequence type but different capsules were also differently aggregated by SP-D. For example, strains GM70 and M41 are both multilocus sequence type 81, but only the latter strain was aggregated by the surfactant protein.
TABLE 3.
Aggregation of different strains and serotypes of S. pneumoniae by SP-Da
| Strain | Serotype | MLST | No. of clumps/ field of view | Avg area of aggregated bacteria (μm2) |
|---|---|---|---|---|
| M393 | 3 | 260 | No clumps | No aggregates |
| M110 | 3 | 180 | 3.2 ± 0.38 | 179 ± 0.2 |
| M290 | 3 | 49 | 4.3 ± 0.53 | 124 ± 0.5 |
| M9 | 18C | 102 | 5.2 ± 0.50b | 156 ± 0.4 |
| M241 | 18C | 113 | No clumps | No aggregates |
| M208 | 18C | 119 | No clumps | No aggregates |
| GM70 | 19A | 81 | No clumps | No aggregates |
| M41 | 23F | 81 | 4.2 ± 0.37 | 64 ± 0.1 |
| M322 | 14A | 156 | 0.7 ± 0.21c | 6.1 ± 0.2 |
| M15 | 9V | 156 | No clumps | No aggregates |
| HB565 | 3 | 378 | 3.1 ± 0.20 | 88 ± 0.6 |
| Ps3205 | 3 | NDd | No clumps | No aggregates |
| D39 | 2 | 128 | 1.11 ± 0.35 | 128.62 ± 0.87 |
| Ps3221 | 3 | ND | 2.2 ± 0.29 | 14.2 ± 0.3 |
| G54 | 19F | 63 | No clumps | No aggregates |
Pneumococci were incubated with SP-D (10 μg/ml) for 90 min in the presence of calcium (5 mM) and were examined by phase-contrast microscopy (×500). The average area of agglutinated bacteria was measured and was significantly different (P < 0.05) among all aggregated strains. The number of clumps per field of view was determined for aggregated strains. The results are means ± standard errors of the mean of three to five experiments.
P < 0.05, significantly higher than other strains.
P < 0.05, significantly lower than other strains.
ND, not determined.
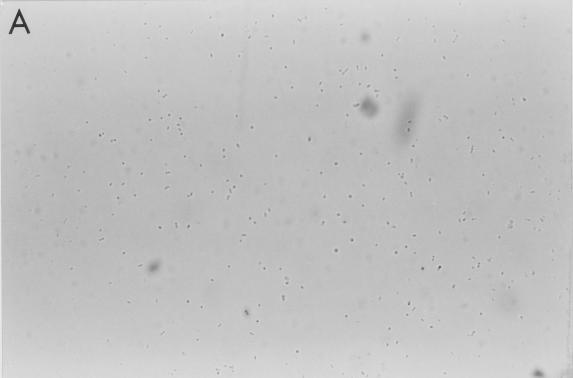
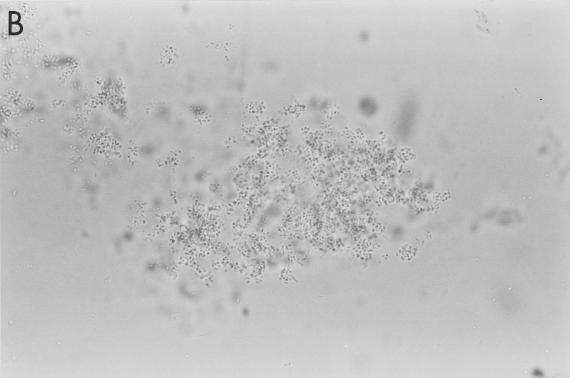
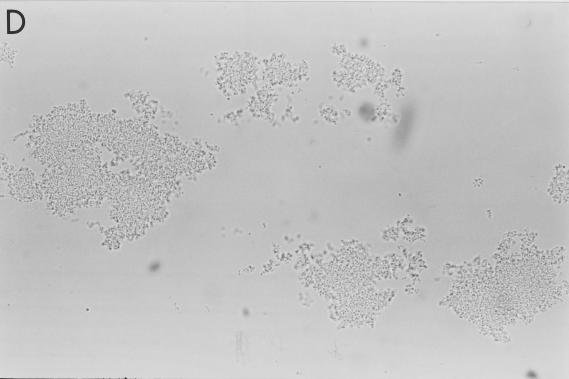
FIG. 1.
Aggregation of S. pneumoniae by SP-D. Strains M110 (A and B) and M9 (C and D) were incubated with 0 and 10 μg of full-length SP-D molecules/ml, respectively, for 90 min in the presence of calcium (5 mM) and were examined by phase-contrast microscopy (magnification, ×500). The results are representative of at least four similar experiments.
Aggregation by SP-D was also tested by using strains of S. pneumoniae whose serotypes had been switched in the laboratory. Strain HB565, serotype 3, was strongly aggregated by SP-D, whereas strain Ps3205, in which the capsule had been switched to type 3 from type 2 (the type 2 was D39, which showed aggregation) was not aggregated by SP-D. Furthermore, strain Ps3221, in which the capsule had been switched to type 3 from 19F, showed aggregation by SP-D, while strain G54, the type 19F parent of PS3221, did not show any aggregation.
Killing of S. pneumoniae by neutrophils in the presence of SP-D and rSP-D.
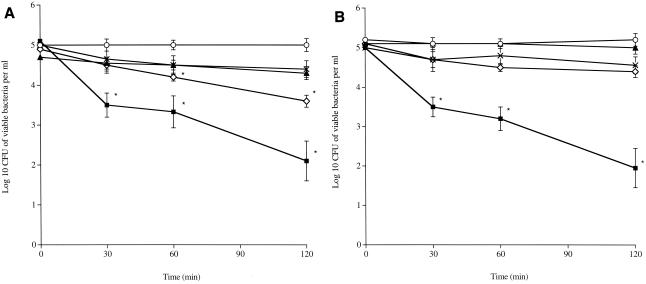
We further investigated the interaction between S. pneumoniae and SP-D by testing if SP-D could act as an opsonin for pneumococci. Experiments were done with encapsulated serotype 2 and nonencapsulated Rx1 S. pneumoniae and human neutrophils in the presence or absence of SP-D (10 μg/ml), rSP-D (20 μg/ml), or autologous sera of vaccinated donors. Data showed that killing by neutrophils was enhanced in the presence of serum, with an ∼3-log-unit decrease (P ≤ 0.01 compared to time zero) in CFU for both pneumococcal strains over 120 min of incubation with neutrophils (Fig. 2). In contrast, in the presence of SP-D or rSP-D and neutrophils, the numbers of pneumococci remained constant over the 120 min for both strains (P ≥ 0.05). Remarkably, in the absence of any opsonin, the number of Rx1 nonencaspulated pneumococci showed a significant decrease at 60 min (P < 0.01) compared to time zero. There was no change in the number of pneumococci when they were incubated with serum in the absence of neutrophils (Fig. 2).
FIG. 2.
Neutrophil killing of nonencapsulated S. pneumoniae strain RX1 (A) and encapsulated strain D39 type 2 (B). Pneumococci (5 × 105) were incubated either with neutrophils alone (5 × 106) (⋄) or with neutrophils in the presence of 20% (vol/vol) autologous serum (▪), in the presence of SP-D (10 μg/ml) (▴), or in the presence of rSP-D (20 μg/ml) (×), or with serum alone (○) for 120 min. Samples were taken at 0, 30, 60, and 120 min to determine the number of viable bacteria. The results are the means ± standard errors of the mean of four independent experiments. *, P < 0.05 (significantly different) compared to time zero.
DISCUSSION
Although SP-D was first identified as a pulmonary surfactant with innate immune functions, it has also been found in numerous tissue sites in humans, where it has been proposed to bind and aggregate a number of diverse bacterial, fungal, and viral pathogens. However, SP-D is expressed primarily in the tracheal-bronchial tract, supporting the concept that it plays a major role as a defense molecule in and around the respiratory tract. Our understanding of the recognition pattern molecules on the surfaces of both gram-negative and gram-positive bacteria is becoming clearer, with many of the potential targets for binding by collectins containing carbohydrate moieties. In this study, we wished to see if SP-D could be an effective host defense molecule, first by binding and aggregating different strains and serotypes of S. pneumoniae and, second, by determining if SP-D was an effective opsonin for killing of S. pneumoniae by human neutrophils.
Our results show that SP-D is capable of binding and aggregating most pneumococci in the presence of calcium. However, different pneumococcal strains and serotypes were not aggregated to the same extent. As with Klebsiella pneumoniae (27), pneumococcal aggregation by SP-D was inhibited by maltose as a competitive sugar, and consistent with previous studies (16, 21), aggregation was inhibited with EDTA. These results indicate that SP-D, via its calcium-dependent C-type lectin domains, binds to carbohydrate structures present on the pneumococcal surface.
Full-length SP-D aggregated pneumococcal strains, whereas rSP-D, lacking the collagen domain, did not, probably because the long, rigid-structure collagen domain of SP-D is essential and allows better linking and interactions among the aggregated bacteria (4, 20). This is partly due to the oligomerization of full-length SP-D into multimers of trimers via N-terminal intra- and interchain disulfide cross-linking, which enhances the avidity of the CRD for carbohydrate ligands on the surfaces of different individual bacteria (11). Truncated recombinant forms of human SP-D generated in E. coli and lacking the collagen domain have been used successfully in previous studies to agglutinate E. coli Y1088 (7), and truncated and chimeric forms of SP-D have been expressed in CHO-K1 cells and proved to be effective at agglutinating Salmonella enterica and Streptococcocus pneumoniae, as well as Influenza A virus (11). These studies, together with our present work, emphasize how the production of recombinant trimers of SP-D in various expression systems can influence the binding and recognition properties of SP-D for various pathogens. The different ability of SP-Dneck+CRD to aggregate selected microorganisms cannot be accounted for by protein misfolding, since the crystal structure for both yeast- and E. coli-expressed forms of SP-D have confirmed their structural integrity (5, 30). However other factors may influence the ability of recombinant trimers of SP-D to bind to specific bacterial strains. For example, SP-D has been demonstrated to bind to nonterminal sugar residues in polysaccharide (1). Such interaction would presumably require precise orientation of the polysaccharide and protein. With the constraints of the collagen stalk removed from the rSP-D fragment, its ability to bind to the surfaces of some bacteria might be affected. However the bioactivity of both yeast- and E. coli-expressed rSP-D and their potential as therapeutic compounds for eradicating lung infections and for allergy suppression in vivo are encouraging, despite the differences in activity observed between native and rSP-D, as recently highlighted by the work of Clark and Reid (3). Although full-length SP-D aggregated many pneumococcal strains, this study showed no correlation between the degree of aggregation by SP-D and the capsule or the MLST of the pneumococcus. While we can conclude from this that the capsule serotype is not the sole determinant of aggregation of S. pneumoniae by SP-D, the data for the isogenic mutants in which the capsule has been changed show that capsule is still a factor. Overall, it seems that a combination of factors, specific for each pneumococcal strain, determines the interaction of SP-D and pneumococci. Using a solid-phase-based binding assay, van de Wetering and colleagues (31) showed that two major cell wall components of gram-positive bacteria, lipoteichoic acid of Bacillus subtilis and peptidoglycan of Staphylococcus aureus, were bound by SP-D in a calcium-dependent and carbohydrate-inhibitable manner. Thus, it is possible to suggest that the combined extent of binding of SP-D to the pneumococcal lipoteichoic acid, peptidoglycan, and capsular polysaccharide determine the interaction.
Despite this interaction, the presence of SP-D did not lead to enhanced killing of pneumococci by neutrophils. This is in contrast to a previous study that reported that SP-D did promote the binding of S. pneumoniae strains 4, 19, and 23 to neutrophils (12). An explanation for the apparent differences seen in this study and the study of Hartshorn and colleagues (12) is the assay system. In our study, the measure was pneumococcus killing, which required not only the binding of bacteria to neutrophils but also active uptake and killing. Hartshorn and colleagues (12) assayed only binding via fluorescence, although the authors did argue that by eye they could see differences in the intensities of fluorescence of internal compared to external bacteria.
Clearly, even if SP-D does not directly promote the killing of S. pneumoniae by neutrophils, SP-D's ability to bind to and agglutinate pneumococci may still be important in vivo, as it may enhance clearance via the mucociliary system and retard the spread of the pathogen through the respiratory tract, thus allowing other arms of both the innate and adaptive immune systems to come into play.
Acknowledgments
The Wellcome Trust supported the work in Leicester. R.J. was in receipt of a studentship from The Lebanese University.
The assistance of Mark Enright, by providing strains, and of Matthew Diggle, by determining MLSTs, is gratefully acknowledged.
Editor: A. D. O'Brien
REFERENCES
- 1.Allen, M., A. Laederach, P. Reilly, and R. Mason. 2001. Polysaccharide recognition by surfactant protein D: novel interactions of a C-type lectin with nonterminal glucosyl residues. Biochemistry 40:7789-7798. [DOI] [PubMed] [Google Scholar]
- 2.Austrian, R. 1986. Some aspects of pneumococcal carrier state. J. Antimicrob. Chemother. 18:35-45. [DOI] [PubMed] [Google Scholar]
- 3.Clark, H., and K. B. M. Reid. 2002. Structural requirements for SP-D function in vitro and in vivo: therapeutic potential of recombinant SP-D. Immunobiology 205:619-631. [DOI] [PubMed] [Google Scholar]
- 4.Crouch, E., A. Persson, D. Chang, and J. Heuser. 1994. Molecular structure of pulmonary surfactant D (SP-D). J Biol. Chem. 269:17311-17319. [PubMed] [Google Scholar]
- 5.Crouch, E. C., K. Hartshorn, and I. Ofek. 2000. Collectins and pulmonary innate immunity. Immunol. Rev. 173:52-65. [DOI] [PubMed] [Google Scholar]
- 6.Cundell, D. R., and E. I. Toumanen. 1994. Identification of carbohydrate receptor specificity of Streptococcus pneumoniae for pulmonary and vascular cells. Microb. Pathog. 17:361-374. [DOI] [PubMed] [Google Scholar]
- 7.Eda, S., Y. Suzuki, T. Kawai, K. Ohtani, T. Kase, Y. Fujinaga, T. Sakamoto, T. Kurimura, and N. Wakamiya. 1997. Structure of a truncated human surfactant protein D is less effective in agglutinating bacteria than the native structure and fails to inhibit haemogglutination by influenza A virus. Biochem. J. 15:393-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eggleton, P., and K. B. M. Reid. 1999. Lung surfactant involved in innate immunity. Curr. Opin. Immunol. 11:28-33. [DOI] [PubMed] [Google Scholar]
- 9.Enright, M. C., and B. G. Spratt. 1998. A multilocus sequence typing scheme for Streptococcus pneumoniae: identification of clones associated with serious invasive disease. Microbiology 144:3049-3060. [DOI] [PubMed] [Google Scholar]
- 10.Ferguson, J. S., D. R. Voelker, J. A. Ufnar, A. J. Dawson, and L. S. Schlesinger. 2002. Surfactant protein D inhibition of human macrophage uptake of Mycobacterium tuberculosis is independent of bacterial agglutination. J. Immunol. 168:1309-1314. [DOI] [PubMed] [Google Scholar]
- 11.Hartshorn, K., D. Chang, K. Rust, M. White, J. Heuser, and E. Crouch. 1996. Interactions of recombinant human pulmonary surfactant protein D and SP-D multimers with influenza A. Am. J. Physiol. 271:L753-L762. [DOI] [PubMed] [Google Scholar]
- 12.Hartshorn, K. L., E. Crouch, M. R. White, M. L. Colamussi, A. Kakkanatt, B. A. I. Tauber, V. Shepherd, and K. Sastry. 1998. Pulmonary surfactant proteins A and D enhance neutrophil uptake of bacteria. Am. J. Physiol. 274:L958-L969. [DOI] [PubMed] [Google Scholar]
- 13.Hartshorn, K. L., M. R. White, and E. C. Crouch. 2002. Contributions of the N- and C-terminal domains of surfactant protein D to the binding, aggregation, and phagocytic uptake of bacteria. Infect. Immun. 70:6129-6139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Holmskov, U. 1999. Lung surfactant proteins (Sp-A and SP-D) in non-adaptive host responses to infection. J. Leukoc. Biol. 66:747-752. [DOI] [PubMed] [Google Scholar]
- 15.Kishore, U., J. Y. Wang, H. J. Hoppe, and K. B. M. Reid. 1996. The α-helical neck region of human lung surfactant protein D is essential for the binding of the carbohydrate recognition domains to lipopolysaccharides and phospholipids. Biochem. J. 318:505-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuan, S. F., K. Rust, and E. Crouch. 1992. Interactions of surfactant protein D with bacterial lipopolysaccharides: surfactant protein D is an Escherichia coli binding protein in bronchoalveolar lavage. J. Clin. Investig. 90:97-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lawson, P. R., and K. B. M. Reid. 2000. The roles of surfactant proteins A and D in innate immunity. Immunol. Rev. 173:66-78. [DOI] [PubMed] [Google Scholar]
- 18.LeVine, A. M., J. A. Whitsett, J. A. Gwozdz, T. R. Richardson, J. H. Fisher, M. S. Burhans, and T. R. Korfhagen. 2000. Distinct effects of surfactant protein A and D deficiency during bacterial infection on the lung. J. Immunol. 165:3934-3940. [DOI] [PubMed] [Google Scholar]
- 19.Lim, B. L., J. Y. Wang, U. Holmskov, H. J. Hoppe, and K. B. M. Reid. 1994. Expression of the carbohydrate recognition domains of lung surfactant protein D and demonstration of its binding to lipopolysaccharides of gram-negative bacteria. Biochem. Biophys. Res. Commun. 202:1674-1680. [DOI] [PubMed] [Google Scholar]
- 20.Lu, J., H. Wiedehann, U. Holmskov, S. Thiel, R. Timpl, and K. B. M. Reid. 1993. Structural similarities between lung surfactant protein D and conglutinin, two distinct C-type lectins containing collagen-like sequences. Eur. J. Biochem. 215:793-799. [DOI] [PubMed] [Google Scholar]
- 21.Madan, T., P. Eggleton, U. Kishore, P. Strong, S. S. Aggrawal, P. U. Sarma, and K. B. M. Reid. 1997. Binding of pulmonary surfactant Sp-A and SP-D to Aspergillus fumigatus conidia enhances phagocytosis and killing by human neutrophils and alveolar macrophages. Infect. Immun. 65:3171-3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Madan, T., U. Kishore, A. Shah, P. Eggleton, P. Strong, J. Y. Wang, S. S. Aggrawal, P. U. Sarma, and K. B. M. Reid. 1997. Lung surfactant proteins A and D can inhibit specific IgE binding to the allergens of Aspergillus fumigatus and block allergen-induced histamine release from human basophils. Clin. Exp. Immunol. 110:241-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Madan, T., U. Kishore, M. Singh, P. Strong, E. M. Hussain, K. B. M. Reid, and U. Sarma. 2001. Protective role of lung surfactant protein D in a murine model of invasive pulmonary aspergillosis. Infect. Immun. 69:2728-2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matsushita, M., and T. Fujita. 2002. The role of ficolins in innate immunity. Immunobiology 205:490-497. [DOI] [PubMed] [Google Scholar]
- 25.Mitchell, T. J., J. E. Alexander, P. J. Morgan, and P. W. Andrew. 1997. Molecular analysis of virulence factors of Streptococcus pneumoniae. J. Appl. Bacteriol. Symp. Ser. 26:625-715. [PubMed] [Google Scholar]
- 26.Murray, E., W. Khamri, M. M. Walker, P. Eggleton, A. P. Moran, J. A. Ferris, S. Knapp, K. N. Karim, M. Worku, P. Strong, K. B. M. Reid, and M. R. Thursz. 2002. Expression of surfactant protein D in the human gastric mucosa and during Helicobacter pylori infection. Infect. Immun. 70:1481-1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ofek, I., A. Mesika, N. Kalina, Y. Keisari, R. Podschun, H. Sahly, D. Chang, D. McGragor, and E. Crouch. 2001. Surfactant protein D enhances phagocytosis and killing of unencapsulated phase variants of Klebsiella pneumoniae. Infect. Immun. 69:24-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Palaniyar, N., J. Nadesalingam, and K. B. M. Reid. 2002. Pulmonary innate immune proteins and receptors that interact with gram-positive bacterial ligands. Immunobiology 205:575-594. [DOI] [PubMed] [Google Scholar]
- 29.Pozzi, G., L. Masala, F. Iannelli, R. Manganelli, L. S. Havarstein, L. Piccoli, D. Simon, and D. A. Morrison. 1996. Competence for genetic transformation in encapsulated strains of Streptococcus pneumoniae: two allelic variants of the peptide pheromone. J. Bacteriol. 178:6087-6090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shrive, A. K., H. A. Tharia, P. Strong, U. Kishore, I. Burns, P. J. Rizkallah, K. B. Reid, and T. J. Greenhough. 2003. High-resolution structural insights into ligand binding and immune cell recognition by human lung surfactant protein D. J. Mol. Biol. 331:509-523. [DOI] [PubMed] [Google Scholar]
- 31.van de Wetering, J. K., H. van Eijk, L. M. van Golde, T. Hartung, J. A. van Strijp, and J. J. Batenburg. 2001. Characteristics of surfactant protein A and D binding to lipoteichoic acid and peptidoglycan, two major cell wall components of gram-positive bacteria. J. Infect. Dis. 184:1143-1151. [DOI] [PubMed] [Google Scholar]
- 32.Wright, J. R. 1997. Immunomodulatory functions of surfactant. Physiol. Rev. 77:931-962. [DOI] [PubMed] [Google Scholar]