Summary
Growing evidence has indicated that genetic factors contribute to the etiology of seizure disorders. Most epilepsies are multifactorial, involving a combination of additive and epistatic genetic variables. However, the genetic factors underlying epilepsy have remained unclear, partially due to epilepsy being a clinically and genetically heterogeneous syndrome. Similar to the human situation, genetic background also plays an important role in modulating both seizure susceptibility and its neuropathological consequences in animal models of epilepsy, which has too often been ignored or not been paid enough attention to in published studies. Genetic homogeneity within inbred strains and their general amenability to genetic manipulation have made them an ideal resource for dissecting the physiological function(s) of individual genes. However, the inbreeding that makes inbred mice so useful also results in genetic divergence between them. This genetic divergence is often unaccounted for but may be a confounding factor when comparing studies that have utilized distinct inbred strains. The purpose of this review is to discuss the effects of genetic background strain on epilepsy phenotypes of mice, to remind researchers that the background genetics of a knockout strain can have a profound influence on any observed phenotype, and outline the means by which to overcome potential genetic background effects in experimental models of epilepsy.
Keywords: inbred strains, epilepsy, gene targeting, flanking genes, background strain, genetics
1. Introduction
Epilepsy is one of the most prevalent neurological disorders affecting people of all ages (Chang and Lowenstein, 2003; Meldrum, 1992). Although epilepsy affects between 1-3% of the population and many cases are familial, only a few epilepsy genes have been mapped; likely due to the genetic complexities that underlie the most common epilepsies (Gurnett and Hedera, 2007; Ottman, 1997; Scheffer and Berkovic, 2003). Moreover, the heterogeneity of the epilepsies, epileptic seizures, and differences in individual patient susceptibility render the identification of genetic loci responsible for differential susceptibility to both seizures and its consequences challenging in humans. Although the importance of genetic factors in determining susceptibility to seizure induction and its neuropathological consequences is generally accepted, the specific genes responsible for this susceptibility are largely unknown.
Most traits and diseases are genetically complex, resulting from combinations of several genes or from interactions between genetic and environmental factors (Lander and Schork, 1994; Moore and Nagle, 2000). Epileptic disorders are no exception to this rule in that they are quantitative traits, resulting from variation in multiple genes with small to moderate effect (reviewed in Rees, 2010; Steinlein, 2010). Inter-individual variation in epilepsy susceptibility suggests that some subpopulations are at increased risk to the detrimental effects of seizures, and it has become clear that genetic background is an important susceptibility factor.
Mouse models offer an attractive strategy for investigating complex neurological disorders, such as epilepsy. The majority of genetic studies, especially those involving disease, have employed mice, not only because their genomes are so similar to that of humans, but also because of their availability, ease of handling, high reproductive rates, and availability of extensive sequence data for inbred strains (http://www.informatics.jax.org/). In addition, inbred mouse strains demonstrate significant strain differences (i.e. genetic heterogeneity) in susceptibility to a variety of seizure induction agents as well as the consequences of seizures, thus providing a starting point for dissecting genetic influences involved in modulating complex traits in humans. Similar to the complexity observed in humans, the determinants of this differential susceptibility in murine populations are probably multigenic, and still remain to be determined. Thus, the opportunity to study discrete epilepsy mutations on a diverse choice of strain backgrounds, to develop better models, and identify the impact of genetic modifiers on seizure severity, incidence and its consequences is now available. This review aims to update on the issue of genetic background as a potential confound in studies of epilepsy and to discuss genetic factors contributing to variance in results from different laboratories.
2. Mouse models of epilepsy: genetic heterogeneity
Rodent models of chronic epilepsy based on chemoconvulsant-induced status epilepticus capture many of the key features of acquired human epilepsy. Animals injected with chemoconvulsants, such as kainic acid or pilocarpine, undergo repetitive limbic seizures and status epilepticus for several hours followed by a variable latent period, lasting between days and several weeks, preceding a chronic phase characterized by spontaneous seizure activity (Hellier et al., 1998; Leite et al., 2002). In particular, many of the features of the neuropathology of medically intractable temporal lobe epilepsy in human tissue, such as neuronal loss and sprouting of recurrent axons (Babb et al., 1991; De Lanerolle et al., 1998; Houser et al., 1990; Mathern et al., 1995) are similar to those changes found in rodents subjected to status epilepticus induced by systemic kainic acid or pilocarpine (Ben-Ari, 1985; Buckmaster and Dudek, 1997; Gorter et al., 2001; McNamara et al., 1992; Turski et al., 1983; Williams et al., 2004). However, while inbred mouse strains have played a critical role in biomedical research due to their genetic homogeneity within inbred strains, genetic divergence between inbred mouse strains can serve as a confounding factor when comparing studies that have utilized different inbred strains. In particular, even in the absence of engineered mutations, different inbred strains can vary drastically in their susceptibility to clinically relevant diseases.
For example, susceptibility to chemically or electrically induced seizures has been studied extensively in genotypic and phenotypic diverse mouse strains (Ferraro et al., 1995; Frankel et al., 2001; Kosobud and Crabbe, 1990; McKhann et al., 2003; McLin and Steward, 2006; Müller et al., 2009; Winawer et al., 2007). It has previously been established that genetic background can considerably affect the seizure thresholds of mice. Loci responsible for strain differences in susceptibility to seizures, triggered by chemoconvulsants, such as kainic acid, or non-pharmacological manipulations, have been mapped to specific mouse chromosomes (Ferraro et al., 1997, 2001, 2004, 2007a,b, 2010, 2011; Todorova et al., 2006). The identified genomic loci map to chromosomes 1, 2, 4, 5, 7, 10, 11, 12, 15 and 18 with the highest density on chromosomes 1, 5, 7, and 10 where loci from several different crosses coincide. Congenic strains have been used to confirm and fine-map some of these seizure susceptibility loci, as well (Ferraro et al., 2007a, 2010).
Furthermore, just as in human studies (Gambardella et al., 2009) genetic background has been implicated in the genesis of hippocampal sclerosis in inbred mouse strains. Because of their diverse genetic background, inbred strains of mice manifest strikingly different patterns of background pathology. A specific phenotype can even vary in one mouse strain or disappear in another strain. Moreover, genetic background may be related to the severity of seizure-induced damage. Inbred strains of mice differ in their tendency to develop cell death after chemically-induced seizure induction with some standard inbred mouse strains being relatively resistant to hilar and pyramidal cell loss in spite of similar seizure severity (McKhann et al., 2003; McLin and Steward, 2006; Schauwecker and Steward, 1997; Schauwecker, 2003; Schauwecker et al., 2004; Shuttleworth and Connor, 2001). However, the genetic basis of variation in seizure-induced cell death remains unclear.
There are significant differences in the amount of hippocampal cell death after seizures between strains and also different patterns of neurodegeneration in affected brain areas. Therefore, hippocampal cell death after seizures and the related molecular mechanisms might not be a general phenomenon, but depend on a complicated interaction between the genetic background and the protocol of seizure induction. Furthermore, while little is known about the distinct factors that cause strain differences in hippocampal vulnerability following seizure induction, modifying genes and quantitative trait loci (QTL) are now recognized as exerting a major influence on regulating the susceptibility to seizure-induced cell death (Kong et al., 2008; Lorenzana et al., 2007; Schauwecker et al., 2004; Schauwecker, 2011). Nevertheless, the genetic basis of variation in seizure-induced cell death remains unclear.
3. Mouse models of epilepsy: gene deletions
Genetic factors play a key role in the etiology of epilepsy (reviewed in Anderman, 1982; Bate and Gardiner, 1999; Elmslie and Gardiner, 1995; Gardiner and Lehesjoki, 2000; Serratosa, 1999; Steinlein, 1999). In the quest for better animal models of epilepsy, genetically manipulated mice have been generated by homologous recombination to examine the effect of gene mutation on epilepsy phenotype. Genetic background plays a critical role in knockout mouse phenotypes (Sanford et al., 2001), including epilepsy, which has too often been ignored or not been paid enough attention to in the published studies. The vast majority of mouse models in use to study genetic influences on seizures and seizure-induced cell death are reverse genetic models in which a single gene is identified for study based on its known function or known involvement in human epilepsy and then it is manipulated in a mouse. Results from a number of studies (Ferraro et al., 2010; Mohajeri et al., 2004; Schauwecker, 2000; Yagi et al., 2009; Yu et al., 2007) clearly demonstrate that the genetic backgrounds of targeted mutants used in epilepsy research can affect susceptibility to seizures (Table 1) and its neuropathological consequences (Table 2) and must be carefully controlled.
Table 1.
Reverse genetic animal models that modulate KA-induced seizure susceptibility
| Genetic background (parental strains) | Gene mutated | Transgenic/Knockout | Genotypes examined | KA-induced seizure susceptibility | Reference |
|---|---|---|---|---|---|
| CD-1 | BDNF | Knockout | +/- versus +/+ | Decreased | (Barton and Shannon, 2005) |
| C57BL/6 × SJL | TNF-α | Transgenic | Tg versus wildtype | Decreased | (Balosso et al., 2005) |
| 129S6/SvEv | NPY | Knockout | -/- versus +/+ | Increased | (Shannon and Yang, 2004) |
| 129/Sv, C57BL/6 | En2 | Knockout | -/- versus +/+ | Increased | (Tripathi et al., 2009) |
| C57BL/6, 129/SvJ | NET | Knockout | -/- versus +/+ | Decreased | (Kaminski et al., 2005) |
| C57BL/6 | GalNAcT | Knockout | -/- versus +/+ | Increased | (Wu et al., 2005) |
| C57BL/6, C3H | GSHPx-1 | Transgenic | Tg versus wildtype | Increased | (Boonplueang et al., 2005) |
| C57BL/6 | Cav2.3 | Knockout | -/- versus +/+ | Decreased | (Weiergräber et al., 2007) |
| C57BL/6 | Prnp | Knockout | -/- versus +/+ | Increased | (Rangel et al., 2007) |
| C57BL/6 × 129Sv/Ev | NR1 | Knockout | -/- versus +/+ | Increased | (Duncan et al., 2010) |
| C57BL/6 | p53 | Knockout | -/- versus +/+ | Increased | (Engel et al., 2010) |
| C57BL/6 | BACE1 | Knockout | -/- versus +/+ | Increased | (Hitt et al., 2010) |
| C57BL/6 | Mag | Knockout | -/- | Increased | (Lopez et al., 2011) |
Table 2.
Reverse genetic animal models that modulate KA-induced cell death
| Genetic background (parental strains) | Gene mutated | Transgenic/Knockout | Genotypes examined | KA-induced cell death | Reference |
|---|---|---|---|---|---|
| C57BL/6 × SJL | TNF-α | Transgenic | Tg versus wildtype | Decreased | (Balosso et al., 2005) |
| 129/Sv, C57BL/6 | En2 | Knockout | -/- versus +/+ | Increased | (Tripathi et al., 2009) |
| C57BL/6, C3H | GSHPx-1 | Transgenic | Tg versus wildtype | Increased | (Boonplueang et al., 2005) |
| C57BL/6 | Prnp | Knockout | -/- versus +/+ | Increased | (Rangel et al., 2007) |
| C57BL/6 | p53 | Knockout | -/- versus +/+ | Decreased | (Engel et al., 2010) |
| 129/SvJae | TIMP1 | Knockout | -/- versus +/+ | Decreased | (Jourquin et al., 2005) |
| C57BL/10, CBA | Hsp27 | Transgenic | Tg versus wildtype | Decreased | (Akbar et al., 2003) |
| UNKNOWN | IGF2 | Knockout | -/- versus +/+ | Decreased | (Dikkes et al., 2007) |
| FVB/N | NPN2 | Knockout | -/- versus +/+ | Increased | (Gant et al., 2009) |
| C57BL/6 | TNFR | Knockout | -/- versus +/+ | Increased | (Lu et al., 2008) |
| C57BL/6 | BACE1 | Knockout | -/- versus +/+ | Increased | (Hitt et al., 2010) |
| C57BL/6 | Pro-dynorphin | Knockout | -/- versus +/+ | Increased | (Schunk et al., 2010) |
| C57BL/6 | Mag | Knockout | -/- | Increased | (Lopez et al., 2011) |
| UNKNOWN | Zip-1/Zip-3 | Knockout | -/- versus +/+ | Decreased | (Qian et al., 2011) |
As strain background can significantly influence susceptibility to either seizures or their pathological consequences, results from genetically modified mice may be difficult to interpret due to the influence of modifier genes and differences in the homogeneity of the background strain. As an example, in Tables 1 and 2, nearly 40% of the reverse genetic animal models shown have varying strain contributions from any one individual strain. Thus, some mutations are likely to have different consequences in different genetic backgrounds. Studies performed over the last 10 years have clearly illustrated that phenotypes caused by specific genetic modifications are strongly influenced by genes unlinked to the target locus. As an example of this phenomenon, Bruno4Ff mutant mice have several different kinds of seizures, depending upon genotype, age, and strain background (Yang et al., 2007). Furthermore, susceptibility to pentylenetetrazole seizures in mice differs when the knockout loci are present on C57BL/6, 129/Sv, or FVB/N backgrounds (Ferraro et al., 1998). The effect of strain background on the severity of epilepsy caused by a sodium channel mutation (Scn2aQ54) has also been reported, suggesting that the strain SJL/J carries alleles at one or more modifier loci that increases the incidence and severity of seizures caused by the mutation (Bergren et al., 2005).
In addition to the modulating effects of genetic background on seizure susceptibility, there is an important variability in the susceptibility of different mouse strains, in response to systemic injections of chemoconvulsants such as kainate, to display the characteristic pattern of hippocampal neurodegeneration observed in rats. Thus, while a number of studies have reported neuroprotective effects against seizure-induced cell death in response to a targeted mutation, it has been difficult to discern whether any putative reductions in neuronal injury result from the targeted mutation or the background genotype (see Table 2). For example, while others have reported that deletion of the p53 gene in animals of a mixed genetic background (129/Sv × C57BL/6) conferred protection against KA-induced degeneration (Morrison et al., 1996), previous studies in our lab have indicated that irrespective of the genetic background, a p53 mutation did not influence KA-induced cell death even on those genetic backgrounds susceptible to excitotoxic cell death (Schauwecker, 2002). Thus, the difference in susceptibility to KA-induced cell death was attributable to a difference in the progenitor strains used to establish the F2 background upon which the knockout existed. These results illustrate that the effect of genetic background has important implications for studies assessing the potential neuroprotective effects of particular genes on seizure-induced cell death.
4. Gene deletions: The flanking gene effect
While mouse models of epilepsy can facilitate genetic dissection through the availability of sophisticated transgenic and knockdown techniques to explore gene function, several caveats exist with regard to the design of these experiments. For example, most researchers incorrectly assume that even when a knockout mice is extensively backcrossed to an inbred strain for 10 or more generations (99.6% ‘pure’ inbred strain), that any phenotypic effects observed are due to a single-gene ablation. Unfortunately, this is not the case as despite one's best efforts, some percentage of embryonic stem (ES) cell-derived genetic material (typically 129-derived) will remain in the genome; this indeterminate region of ES-derived genetic material is often referred to as the ‘flanking region’ (as reviewed by Crusio, 2004). This is because even when extensive backcrossing has occurred, recombination is generally random throughout the genome and thus, a knockout/congenic does not have a 100% pure strain background. It is possible that these strains can still contain discrete regions of residual ES-derived genetic material that may act in isolation or likely produce epistatic interactions with the targeted gene of interest. As one generally does not know which genes are linked to the targeted gene, any phenotypic effects observed could be the result of the ablated gene, flanking genes, and/or an epistatic interaction between the ablated gene and the flanking gene(s).
5. Mixed background: The use of hybrid strains
A further complicating factor resulting in phenotypic variability in penetrance and/or expressivity is nearly always due to the knockout allele being present on a mixed or hybrid genetic background. In such cases, targeted mutants are generated by micro-injecting 129 strain embryonic stem cells, which have undergone homologous recombination to knock out the targeted gene, into C57BL/6 (B6) blastocysts, which then develop into B6/129 chimeras. Sometimes researchers maintain the targeted mutation on this mixed B6/129 background by intercrossing the mutants to create F2 mice. However, it can be difficult to discern whether the particular phenotypic effects that a knockout mouse exhibits are the result of particular gene effects when the genetic context originated from a particular embryonic stem (ES) cell line or the result of an inbred mouse strain. Moreover, as the 129 mouse strain encompasses a large number of genetically (Simpson et al., 1997) and behaviorally (Balogh et al., 1999; Cook et al., 2002; Montkowski et al., 1997) distinct substrains, phenotype characterization of the genetically modified mouse can be difficult to assess due to the presence of anatomical, genetic, and behavioral abnormalities. For example, each targeted epilepsy mutant on a B6:129 mixed background maintained by intercrossing inherits a unique proportion of seizure-resistant and seizure-susceptible alleles that may affect its epilepsy susceptibility. This is due to the fact that most studies compare homozygous mutant, heterozygous mutant, and wild-type littermates of an F2 population to determine phenotypical changes resulting from the null mutation. However, it is important to note that the genetic background composition of these hybrid mice varies among littermates because of gene segregation from the hybrid (F1) parents. The result is that the alleles of genes that surround the targeted locus in a hybrid background can be derived from one strain in homozygous mutant mice, yet be of another strain in wild-type mice. Thus, even when one attempts to ‘backcross’ the mice to a genetically well-defined background, backcrossed mice keep holding the 129-strain genes close to the targeted locus reflecting the original ES cell strain. The possibility that subtle phenotypical changes such as behavior differences may be caused by the surrounding 129 genomes other than the targeted one has been raised extensively (Gerlai et al., 2001).
6. Complications in interpretation of findings from other laboratories
A number of studies have investigated and described epilepsy in rodents, but it is difficult to compare these studies with each other due to experimental differences including background strain effects, epistatic interactions between the background strain and the mutation, developmental effects of a null mutation, method of seizure induction, and differences in experimental endpoints. Recent studies have raised concerns about the use of mice with an inappropriately controlled genetic background, which sometimes leads to distorted experimental outcomes (Crusio, 2004; Gerlai, 2001; Linder, 2001, 2006; Mohajeri et al., 2004).
6.1 Genetic background
Converging observations indicate that discrepancies in a number of different pathological and physiological features among different knock-out lines could be the result of genetic background. First and foremost, it is likely that they are due to a strain effect - especially if analyses are conducted using F2 mice on a different mixed genetic background. Secondly, genetic background can significantly affect both susceptibility to seizure induction, as well as the severity of seizure-induced damage, as discussed above. For example, the DBA/2J strain has been shown to be somewhat seizure sensitive (Ferraro et al., 1995, 1997) irrespective of the mode of seizure induction, and the C57BL/6 strain has previously been shown to be resistant to neuronal death induced by kainate administration (Schauwecker and Steward, 1997; Schauwecker et al., 2004). Thus, apart from differences in experimental procedures, it is likely that disparate results are due to the use of different genetic backgrounds.
6.2 Residual background genome
Discrepancies between laboratories using the same null mutant strain could be the result of genetic influences that subsequently affect the background strain. For example, some authors note that animals were backcrossed for 4 or 5 generations, which is substantially reduced from the requisite 10 generations considered a ‘full’ backcross. Thus, each time the mouse with the knockout is crossed with a mouse of a constant genetic background, the average percentage of the genetic material of the offspring that is derived from that constant background increases. One of the disadvantages of inadequate backcrossing of mice is that sections of the genome from the non-recurrent parents are often still present and can have unwanted traits associated with them. Importantly, even mice that have been made congenic by extensive backcrossing to one strain, such as C57BL/6, can retain small regions of 129-derived material in areas elsewhere in the genome that are resistant to genetic recombination (Eisener-Dorman et al., 2009). Thus, it is possible that subtle genetic variation can interact with the targeted mutation in an epistatic fashion. As a result, the genetic background of these mutants can be an unpredictable determinant of the phenotypic outcome.
Interactions between a mutation and genetic background may be especially relevant to the study of a candidate gene. While it has been reported that genetic factors can contribute to the risk for epilepsy disorders, no single gene accounts for more than a small proportion of the overall risk (reviewed in Rees, 2010; Steinlein, 2010). This is likely due to the understanding that both genetic and environmental factors contribute to susceptibility, however it remains unclear how their interaction influences risk. Therefore, traits such as susceptibility to seizure-induced cell death and seizure susceptibility are likely to result from epistatic gene-gene interactions and gene-environment interactions. As a result, any functional interactions between a given mouse mutation and its effect on seizure susceptibility and genetic background may be relevant to understanding the polygenic nature of epilepsy and/or seizure disorders.
6.3 Substrains
Diversity within substrains, especially as is prevalent within the 129/Sv strain, can become an issue when trying to take into account ‘flanking gene’ issues, as discussed earlier. There is a tremendous amount of genetic diversity among the 129/Sv substrains of mice and as such, 129 substrains have been classified according to their common parental lineages (http://www.informatics.jax.org/mgihome/nomen/strain_129.shtml): P (the original parent strain); S (strains derived from a congenic strain outcrossed to introduce the steel mutation); and T (derived from the 129 congenic that originally carried a teratoma mutation). However, the origin and history of some mutations maintained on a 129 background is unknown.
As well, this has become an even thornier issue recently with the use of different substrains of C57BL/6 mice. C57BL/6J mice were developed at The Jackson Laboratory, while C57BL/6N mice were developed at The National Institutes of Health, and C57BL/6C mice were derived from C57BL/6N mice at The National Cancer Institute. C57BL/6NCrl mice are obtained from Charles River Laboratories, which originally obtained their substrain from the NIH. While others have reported behavioral and substrain polymorphism differences among these C57BL/6 substrains (Matsuo et al., 2010; Tsang et al., 2005; Zurita et al., 2010), it is imperative that Investigators state explicitly which substrain was used. Moreover, it is also important to reduce the use of incomplete informal abbreviations to designate strain names. For example, B6 is a common abbreviation used for C57BL/6 mice, but it is unclear which substrain of C57BL/6 mouse was examined. This is also applicable to the genetic background of mutants, as it is possible that genetic variation between and among substrains could interact with the targeted mutation to modify gene:gene interactions.
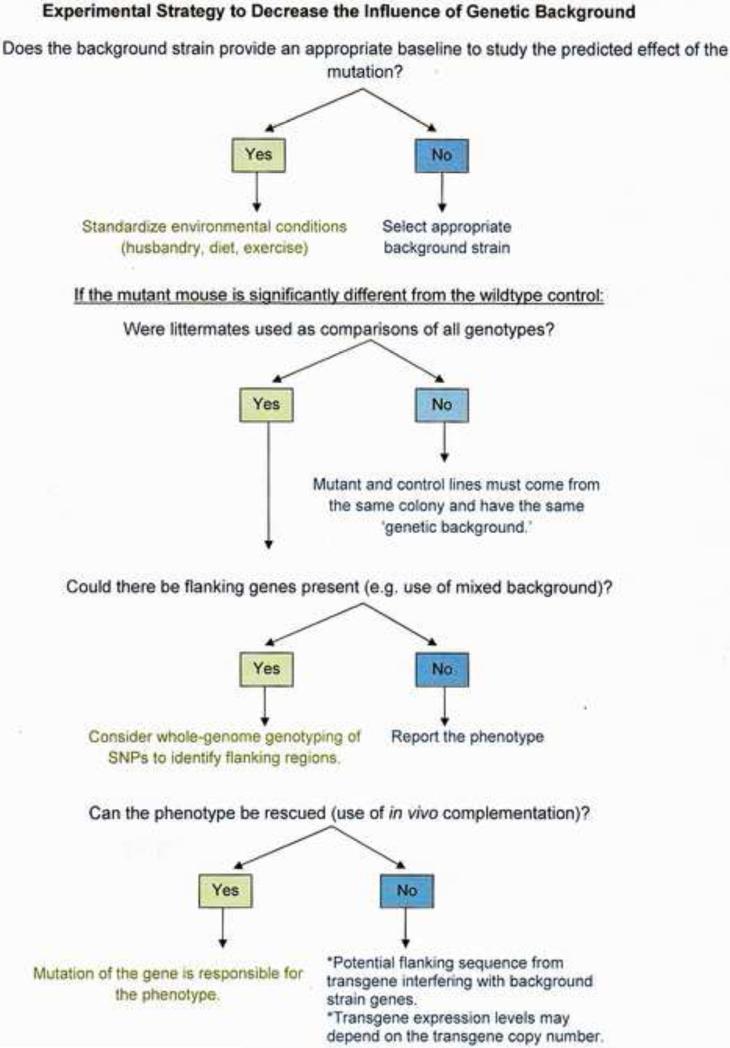
7. Experimental solutions to resolve the issue of genetic background (Figure 1)
Figure 1.
Experimental Strategy to Decrease the Influence of Genetic Background
7.1 Controlling for genetic background
Even if one does not use a mixed background, it is important to take into consideration phenotypic differences for studying mutants as they could affect the mutant phenotype, exaggerating or masking seizure-induced cell death or other defects. In particular, the genetic background onto which a gene-targeted allele is placed can cause considerable variation that can present itself as completely different phenotypes, as variations in penetrance of phenotype, or as variable expressivity of phenotype. Thus, compared to mutants on a mixed genetic background, those on a pure genetic background will give a more definitive answer to the function of the targeted gene because such mutants and their controls will differ only with respect to the targeted gene. As well, while it is imperative to employ appropriate breeding strategies to control for potentially confounding genetic background issues, the mutation of interest should be studied in two or more genetic backgrounds to determine if the same phenotype is observed in each background. This would enable one to determine if there are any interactions between the genetic background and the presence or absence of the gene product of interest.
7.2 Constructing targeted mutants
When one is considering the use of a targeted mutation, however, one must take into account that any variation observed for a given phenotype may not necessarily be the result of the loss of gene function alone. Strategies that may be considered for this purpose are described in Figure 1. For example, when constructing targeted mutations, one should try to construct genetically altered strains that are coisogenic to controls (Linder 2006; Silver, 1995) and on well-defined ES cell lines and well-defined genetic backgrounds. Breeding of mutant and control animals should be accomplished through the same line (e.g. same parents), otherwise different recombinant inbred lines will be established in which large chromosomal pieces containing a multitude of genes will differ. Thus, littermates should be utilized in all experiments such that genetic variance will not be a contributing factor in phenotype modification.
As powerful as it may be, the genetic knockout approach also has its intrinsic limitations that may confound the correct interpretation of the phenotypic analysis of knockout mice. For example, it can be a faulty strategy to continuously inbreed and maintain separate +/+, +/-, and -/- lines, as this will result in genetic drift and potentially the creation of phenotypic line differences that are unrelated to the mutant line. Such extraneous mutated genes (“hitchhiker” or “passenger) may become incorporated into the genome of future generations creating serious artifacts for phenotyping. As a result, random segregation events occurring over many generations will generate different alleles for background genes and could potentially influence the phenotype of the mutation. When these extraneous mutated genes are transmitted to future generations, they can introduce false positives or false negatives into the interpretation of the phenotype of the mutation.
Placing a mutation on several different genetic backgrounds allows the study of different aspects of gene function that might not be apparent on a single genetic background. However, if the use of a single inbred strain for genetic manipulation is not possible, then congenic mice should be created. Congenic strains are created by transferring a short segment of the chromosome around a marker gene of interest from one strain into an inbred genetic background by repeated backcrossing and selection (Weil et al., 1997). However, it is important to note that the use of congenic strains does not completely eliminate the potential confound of “passenger” genes. For example, the persistence of phenotypic differences between homozygous null mutants and wild type animals may be due to the proximity of the “passenger” and targeted genes on the chromosomal segment and difficulties in separating the effects between the two genes. For example, it has been estimated that hundreds of ES cell donor genes can flank a target mutation locus even after 12 generations of backcrossing (Bolivar et al., 2001; Crusio, 1996; Gerlai, 1996; Wolfer and Lipp, 2000; Wolfer et al., 2002). If polymorphisms in the flanking genes exist between the ES cell donor strain and the congenic strain, there may be phenotypic differences that exist between the homozygous null mutants (-/-) and the wild type controls (+/+) that are inaccurately attributed to the mutation (Bolivar et al., 2001; Gerlai, 1996; Wolfer et al., 2002). Lastly, to definitively demonstrate that null mutation of a specific gene is responsible for phenotype modification, one could perform a ‘rescue’ experiment, in which the knockout phenotype is rescue by in vivo complementation, typically bacterial artificial chromosome (BAC) transgenesis. This process entails the design of a BAC construct that contains the wild-type gene of interest, microinjection of the resulting construct into embryos, and then implantation of the embryos into pseudo pregnant females. Transgenic mice are identified by genotyping for the BAC transgene and these animals are then crossed to knockout mice to generate mice that carry the BAC transgene on the knockout background. Transgene rescue of mouse gene function by BAC-encoded transgenes have been reported for several loci (Ferraro et al., 2007b; Kibar et al., 2003; Means et al., 2001). One of the advantages of this approach is that the BAC transgene is present during development and should theoretically show an expression pattern similar to the endogenous gene. Such rescue strategies support the interpretation that the original deficit in the knockout mouse was due to the gene mutation and not to some other nonspecific effect.
While the development of BAC transgenic mice has helped overcome the limitations of traditional transgenic approaches, BAC transgenic mice are privy to caveats and potential confounds in interpretation as well. For example, like conventional transgenics, it is possible that essential mouse sequences at the site of the transgene integration may be disrupted. As well, BAC transgenes can integrate into more than one genomic location (Chandler et al., 2007), as the precise site of integration of a BAC transgene cannot be well controlled. Similarly, because BAC transgenes are much larger constructs as compared to conventional transgenes, they are more susceptible to fragmentation (Chandler et al., 2007; Deal et al., 2006). Nevertheless, BAC transgenics are a potentially feasible way to complement the loss of mouse gene function and can complement gene modification strategies currently in use.
7.3 Flanking gene effects
One viable solution used to eliminate flanking gene effects on a mutant gene of interest is to create a coisogenic strain by using an ES cell line and a background strain all derived from the same inbred strain. The creation of targeted mutant mice with C57BL/6J ES cells is one such alternative. Some have suggested that the solution to the flanking gene problem is to apply speed-congenic techniques to rapidly repeatedly backcross mice to a preferred inbred background, followed by genotyping for the absence or presence of the mutation. However, even though this technique is highly effective at reducing the size of the flanking region, a small flanking region will still exist that could exceed more than 300 genes (Crusio, 2004; Wolfer et al., 2002). However, if one is utilizing already created ‘null mutants’ to assess a particular phenotype, to assess the possibility of ‘flanking gene’ effects, one could broadly scan the genetic background for regions of variability by whole-genome single nucleotide polymorphism (SNP) genotyping, using the vast number of known single-nucleotide polymorphisms. Even though it may be difficult to identify each and every 129-derived locus, it may help identify regions of variability that contain one or more genes influencing the observed phenotype As well, the use of a polymorphic marker for the targeted genomic region that can permit rapid exclusion of flanking allele effects is equally useful. Lastly, all Investigators should be explicit as possible about which inbred strains (and/or substrains) are under study, as well as their gender and age. For targeted mutants, the genetic background should also be explicitly stated, including either the percentage of recipient genome and/or the number of backcrosses performed in order to determine whether flanking (“passenger”) gene effects may be substantial.
8. Conclusions
In summary, this brief review has stressed potential pitfalls and artifacts that can complicate the interpretation of experimental results. It is important to consider the genetic background of mouse models used in epilepsy research as well as the map position of the gene to be targeted. The availability of numerous inbred strains represents a useful tool for the genetic dissection of the basis of differential susceptibility to seizures and seizure-induced cell death. Knowledge of which genes maintain the balance between susceptibility and resistance may help to develop pharmacogenetic strategies tailored to individual genomes in order to improve our therapeutic protocols. As well, the ease by which the mouse genome can be experimentally manipulated makes the mouse invaluable for dissecting the pathogenetic and pathophysiologic mechanisms underlying the epilepsies in humans. However, it is important to not only recognize the benefits of knockout technology, but also its potential caveats. Due to the means by which many knockout mice are created, phenotypes originally attributed to the knockout gene under study may actually be attributed to tightly-linked loci immediately flanking the ablated gene, or even to unlinked loci located throughout the genetic background of the knockout strain. Thus, the importance of genetic heterogeneity in the investigation of the genetic basis underlying the penetrance of specific phenotypes cannot be underestimated. It is important to keep in mind that the construction and propagation of any mutant mouse must address not only the single gene mutation, but also the genetic background.
Acknowledgements
This writing was supported by NIH Grant NS38696.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akbar MT, Lundberg AMC, Liu K, Vidyadaran S, Wells KE, Dolatshad H, Wynn S, Wells DJ, Latchman DS, de Belleroche J. The neuroprotective effects of heat shock protein 27 overexpression in transgenic animals against kainate-induced seizures and hippocampal cell death. J Biol Chem. 2003;278:19956–19965. doi: 10.1074/jbc.M207073200. [DOI] [PubMed] [Google Scholar]
- Anderman E. Multifactorial inheritance of generalized and focal epilepsy. In: Anderson VE, Hauser WA, Penry JK, Sing CF, editors. Genetic Basis of the Epilepsies. Raven Press; New York: 1982. pp. 355–374. [Google Scholar]
- Babb TL, Kupfer WR, Pretorius JK, Crandall PH, Levesque MF. Synaptic reorganization by mossy fibers in human epileptic fascia dentate. Neuroscience. 1991;42:351–363. doi: 10.1016/0306-4522(91)90380-7. [DOI] [PubMed] [Google Scholar]
- Balogh SA, McDowell CS, Stavnezer AJ, Denenberg VH. A behavioral and neuroanatomical assessment of an inbred substrain of 129 mice with behavioral comparisons to C57BL/6 mice. Brain Res. 1999;836:38–48. doi: 10.1016/s0006-8993(99)01586-3. [DOI] [PubMed] [Google Scholar]
- Balosso S, Ravizza T, Perego C, Peschon J, Campbell IL, De Simoni MG, Vezzani A. Tumor necrosis factor-α inhibits seizures in mice via p75 receptors. Ann Neurol. 2005;57:804–812. doi: 10.1002/ana.20480. [DOI] [PubMed] [Google Scholar]
- Barton ME, Shannon HE. Behavioral and convulsant effects of the (S) enantiomer of the group I metabotropic glutamate receptor agonist 3,5-DHPG in mice. Neuropharm. 2005;48:779–787. doi: 10.1016/j.neuropharm.2005.01.017. [DOI] [PubMed] [Google Scholar]
- Bate L, Gardiner M. Molecular genetics of human epilepsies. Expert Rev Mol Med. 1999;1999:1–22. doi: 10.1017/S1462399499001349. [DOI] [PubMed] [Google Scholar]
- Ben-Ari Y. Limbic seizure and brain damage produced by kainic acid: mechanisms and relevance to human temporal lobe epilepsy. Neuroscience. 1985;14:375–403. doi: 10.1016/0306-4522(85)90299-4. [DOI] [PubMed] [Google Scholar]
- Bergren SK, Chen S, Galecki A, Kearney JA. Genetic modifiers affecting severity of epilepsy caused by mutation of sodium channel Scn2a. Mamm Genome. 2005;16:683–690. doi: 10.1007/s00335-005-0049-4. [DOI] [PubMed] [Google Scholar]
- Bolivar VJ, Cook MN, Flaherty L. Mapping of quantitative trait loci with knockout/congenic strains. Genome Res. 2001;11:1549–1552. doi: 10.1101/gr.194001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boonplueang R, Akopian G, Stevenson FF, Kuhlenkamp JF, Lu SC, Walsh JP, Andersen JK. Increased susceptibility of glutathione peroxidase-1 transgenic mice to kainic acid-related seizure activity and hippocampal neuronal cell death. Exp Neurol. 2005;192:203–214. doi: 10.1016/j.expneurol.2004.12.017. [DOI] [PubMed] [Google Scholar]
- Buckmaster PS, Dudek FE. Neuron loss, granule cell axon organization, and functional changes in the dentate gyrus of epileptic kainate-treated rats. J Comp Neurol. 1997;385:385–404. [PubMed] [Google Scholar]
- Chandler KJ, Chandler RL, Broeckelmann EM, Hou Y, Southard-Smith EM, Mortlock DP. Relevance of BAC transgene copy number in mice: transgene copy number variation across multiple transgenic lines and correlations with transgene integrity and expression. Mamm Genome. 2007;18:693–708. doi: 10.1007/s00335-007-9056-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang BS, Lowenstein DH. Epilepsy. N Engl J Med. 2003;349:1257–1266. doi: 10.1056/NEJMra022308. [DOI] [PubMed] [Google Scholar]
- Cook MN, Bolivar VJ, McFadyen MP, Flaherty L. Behavioral differences among 129 substrains: implications for knockout and transgenic mice. Behav Neurosci. 2002;116:600–611. [PubMed] [Google Scholar]
- Crusio WE. Gene targeting studies: new methods, old problems. Trends Neurosci. 1996;19:186–187. doi: 10.1016/s0166-2236(96)20023-2. [DOI] [PubMed] [Google Scholar]
- Crusio WE. Flanking gene and genetic background problems in genetically manipulated mice. Biol Psychiatry. 2004;56:381–385. doi: 10.1016/j.biopsych.2003.12.026. [DOI] [PubMed] [Google Scholar]
- De Lanerolle NC, Eid T, von Campe G, Kovacs I, Spencer DD, Brines M. Glutamate receptor subunits GluR1 and GluR2/3 distribution shows reorganization in the human epileptogenic hippocampus. Eur J Neurosci. 1998;10:1687–1703. doi: 10.1046/j.1460-9568.1998.00171.x. [DOI] [PubMed] [Google Scholar]
- Deal KK, Cantrell VA, Chandler RL, Saunders TL, Mortlock DP, Southard-Smith EM. Distant regulatory elements in a Sox10-ßGEO BAC transgene are required for expression of Sox10 in the enteric nervous system and other neural crest-derived tissues. Dev Dynamics. 2006;235:1413–1432. doi: 10.1002/dvdy.20769. [DOI] [PubMed] [Google Scholar]
- Dikkes P, Hawkes C, Kar S, Lopez MF. Effect of kainic acid treatment on insulin-like growth factor-2 receptors in the IGF2-deficient adult mouse brain. Brain Res. 2007;1131:77–87. doi: 10.1016/j.brainres.2006.11.022. [DOI] [PubMed] [Google Scholar]
- Duncan GE, Inada K, Koller BH, Moy SS. Increased sensitivity to kainic acid in a genetic model of reduced NMDA receptor function. Brain Res. 2010;1307:166–176. doi: 10.1016/j.brainres.2009.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisener-Dorman AF, Lawrence DA, Bolivar VJ. Cautionary insights on knockout mouse studies: the gene or not the gene? Brain Behav Immun. 2009;23:318–324. doi: 10.1016/j.bbi.2008.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmslie F, Gardiner M. Genetics of the epilepsies. Curr Opin Neurol. 1995;8:126–129. doi: 10.1097/00019052-199504000-00007. [DOI] [PubMed] [Google Scholar]
- Engel T, Murphy BM, Hatazaki S, Jimenez-Mateos EM, Concannon CC, Woods I, Prehn JHM, Henshall DC. Reduced hippocampal damage and epileptic seizures after status epilepticus in mice lacking proapoptotic Puma. FASEB J. 2010;24:853–861. doi: 10.1096/fj.09-145870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferraro TN, Golden GT, Smith GG, Berrettini WH. Differential susceptibility to seizures induced by systemic kainic acid treatment in mature DBA/2J and C57BL/6J mice. Epilepsia. 1995;36:301–307. doi: 10.1111/j.1528-1157.1995.tb00999.x. [DOI] [PubMed] [Google Scholar]
- Ferraro TN, Golden GT, Smith GG, Schork NJ, St Jean P, Ballas C, Choi H, Berrettini WH. Mapping murine loci for seizure response to kainic acid. Mamm Genome. 1997;8:200–208. doi: 10.1007/s003359900389. [DOI] [PubMed] [Google Scholar]
- Ferraro TN, Golden GT, Snyder R, Laibinis M, Smith GG, Buono RJ, Berrettini WH. Genetic influences on electrical seizure threshold. Brain Res. 1998;813:207–210. doi: 10.1016/s0006-8993(98)01013-0. [DOI] [PubMed] [Google Scholar]
- Ferraro TN, Golden GT, Smith GG, Longman RL, Snyder RL, DeMuth D, Szpilzak I, Mulholland N, Eng E, Lohoff FW, Buono RJ, Berrettini WH. Quantitative genetic study of maximal electroshock seizure threshold in mice: evidence for a major seizure susceptibility locus on distal chromosome 1. Genomics. 2001;75:35–42. doi: 10.1006/geno.2001.6577. [DOI] [PubMed] [Google Scholar]
- Ferraro TN, Golden GT, Smith GG, Martin JF, Lohoff FW, Gieringer TA, Zamboni D, Schwebel CL, Press DM, Kratzer SO, Zhao H, Berrettini WH, Buono RJ. Fine mapping of a seizure susceptibility locus on mouse Chromosome 1: nomination of Kcnj10 as a causative gene. Mamm Genome. 2004;15:239–251. doi: 10.1007/s00335-003-2270-3. [DOI] [PubMed] [Google Scholar]
- Ferraro TN, Smith GG, Schwebel CL, Lohoff FW, Furlong P, Berrettini WH, Buono RJ. Quantitative trait locus for seizure susceptibility on mouse chromosome 5 confirmed with reciprocal congenic strains. Physiol Genomics. 2007a;31:458–462. doi: 10.1152/physiolgenomics.00123.2007. [DOI] [PubMed] [Google Scholar]
- Ferraro TN, Golden GT, Dahl JP, Smith GG, Schwebel CL, MacDonald R, Lohoff FW, Berrettini WH, Buono RJ. Analysis of a quantitative trait locus for seizure susceptibility in mice using bacterial artificial chromosome-mediated gene transfer. Epilepsia. 2007b;48:1667–1677. doi: 10.1111/j.1528-1167.2007.01126.x. [DOI] [PubMed] [Google Scholar]
- Ferraro TN, Smith GG, Schwebel CL, Doyle GA, Ruiz SE, Oleynick JV, Lohoff FW, Berrettini WH, Buono RJ. Confirmation of multiple seizure susceptibility QTLs on chromosome 15 in C57BL/6J and DBA/2J inbred mice. Physiol Genomics. 2010;42A:1–7. doi: 10.1152/physiolgenomics.00096.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferraro TN, Smith GG, Ballard D, Zhao H, Schwebel CL, Gupta A, Rappaport EF, Ruiz SE, Lohoff FW, Doyle GA, Berrettini WH, Buono RJ. Quantitative trait loci for electrical seizure threshold mapped in C57BLKS/J and C57BL/10SnJ mice. Genes Brain Behav. 2011;10:309–315. doi: 10.1111/j.1601-183X.2010.00668.x. [DOI] [PubMed] [Google Scholar]
- Frankel WN, Taylor L, Beyer B, Tempel BL, White HS. Electroconvulsive thresholds of inbred mouse strains. Genomics. 2001;74:306–312. doi: 10.1006/geno.2001.6564. [DOI] [PubMed] [Google Scholar]
- Gambardella A, Labate A, Giallonardo A, Aguglia U. Familial mesial temporal lobe epilepsies: clinical and genetic features. Epilepsia. 2009;50(Suppl 5):55–57. doi: 10.1111/j.1528-1167.2009.02123.x. [DOI] [PubMed] [Google Scholar]
- Gant JC, Thibault O, Blalock EM, Yang J, Bachstetter A, Kotick J, Schauwecker PE, Hauser KF, Smith GM, Mervis R, Li Y, Barnes GN. Decreased number of interneurons and increased seizures in neuropilin 2 deficient mice: Implications for autism and epilepsy. Epilepsia. 2009;50:629–645. doi: 10.1111/j.1528-1167.2008.01725.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardiner M, Lehesjoki AE. Genetics of the epilepsies. Curr Opin Neurol. 2000;13:157–164. doi: 10.1097/00019052-200004000-00008. [DOI] [PubMed] [Google Scholar]
- Gerlai R. Gene-targeting studies of mammalian behavior: is it the mutation or the background genotype? Trends Neurosci. 1996;19:177–181. doi: 10.1016/s0166-2236(96)20020-7. [DOI] [PubMed] [Google Scholar]
- Gerlai R. Gene targeting: technical confounds and potential solutions in behavioral brain research. Behav Brain Res. 2001;125:13–21. doi: 10.1016/s0166-4328(01)00282-0. [DOI] [PubMed] [Google Scholar]
- Gorter JA, van Vliet EA, Aronica E, Lopes da Silva FH. Progression of spontaneous seizures after status epilepticus is associated with mossy fibre sprouting and extensive bilateral loss of hilar parvalbumin and somatostatin-immunoreactive neurons. Eur J Neurosci. 2001;13:657–669. doi: 10.1046/j.1460-9568.2001.01428.x. [DOI] [PubMed] [Google Scholar]
- Gurnett CA, Hedera P. New ideas in epilepsy genetics. Arch Neurol. 2007;64:324–328. doi: 10.1001/archneur.64.3.324. [DOI] [PubMed] [Google Scholar]
- Hellier JL, Patrylo PR, Buckmaster PS, Dudek FE. Recurrent spontaneous motor seizures after repeated low-dose systemic treatment with kainate: assessment of a rat model of temporal lobe epilepsy. Epilepsy Res. 1998;31:73–84. doi: 10.1016/s0920-1211(98)00017-5. [DOI] [PubMed] [Google Scholar]
- Hitt BD, Jaramillo TC, Chetkovich DM, Vassar R. BACE-/- mice exhibit seizure activity that does not correlate with sodium channel level or axonal localization. Mol Neurodegen. 2010;5:31–35. doi: 10.1186/1750-1326-5-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houser CR, Miyashiro JE, Swartz BE, Walsh GO, Rich JR, Delgado-Escueta AV. Altered patterns of dynorphin immunoreactivity suggest mossy fiber reorganization in human hippocampal epilepsy. J Neurosci. 1990;10:267–282. doi: 10.1523/JNEUROSCI.10-01-00267.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jourquin J, Tremblay E, Bernard A, Charton G, Chaillan FA, Marchetti E, Roman FS, Soloway PD, Dive V, Yioitakis A, Khrestchatisky M, Rivera S. Tissue inhibitor of metalloproteinases-1 (TIMP-1) modulates neuronal death, axonal plasticity, and learning and memory. Eur J Neurosci. 2005;22:2569–2578. doi: 10.1111/j.1460-9568.2005.04426.x. [DOI] [PubMed] [Google Scholar]
- Kaminski RM, Shippenberg TS, Witkin JM, Rocha BA. Genetic deletion of the norepinephrine transporter decreases vulnerability to seizures. Neuroscience Lett. 2005;382:51–55. doi: 10.1016/j.neulet.2005.02.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kibar Z, Gauthier S, Seung-Hwan L, Vidal S, Gros P. Rescue of the neural tube defect of loop-tail mice by a BAC clone containing the Ltap gene. Genomics. 2003;82:397–400. doi: 10.1016/s0888-7543(03)00113-7. [DOI] [PubMed] [Google Scholar]
- Kong S, Lorenzana A, Deng Q, McNeill TH, Schauwecker PE. Variation in Galr1 expression determines susceptibility to excitotoxin-induced cell death in mice. Genes Brain Behav. 2008;7:587–598. doi: 10.1111/j.1601-183X.2008.00395.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosobud AE, Crabbe JC. Genetic correlations among inbred strain sensitivities to convulsions induced by 9 convulsant drugs. Brain Res. 1990;526:8–16. doi: 10.1016/0006-8993(90)90243-5. [DOI] [PubMed] [Google Scholar]
- Lander ES, Schork NJ. Genetic dissection of complex traits. Science. 1994;265:2037–2048. doi: 10.1126/science.8091226. [DOI] [PubMed] [Google Scholar]
- Leite JP, Garcia-Cairasco N, Cavalheiro EA. New insights from the use of pilocarpine and kainate models. Epilepsy Res. 2002;50:93–103. doi: 10.1016/s0920-1211(02)00072-4. [DOI] [PubMed] [Google Scholar]
- Linder CC. The influence of genetic background on spontaneous and genetically engineered mouse models of complex diseases. Lab Anim. 2001;30:34–39. [PubMed] [Google Scholar]
- Linder CC. Genetic variables that influence phenotyope. ILAR J. 2006;47:132–140. doi: 10.1093/ilar.47.2.132. [DOI] [PubMed] [Google Scholar]
- Lopez PHH, Ahmad AS, Mehta NR, Toner M, Rowland EA, Zhang J, Doré S, Schnaar RL. Myelin-associated glycoprotein protects neurons from excitotoxicity. J Neurochem. 2011;116:900–908. doi: 10.1111/j.1471-4159.2010.07069.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzana A, Chancer Z, Schauwecker PE. A quantitative trait locus on chromosome 18 is a critical determinant of excitotoxic cell death susceptibility. Eur J Neurosci. 2007;25:1998–2008. doi: 10.1111/j.1460-9568.2007.05443.x. [DOI] [PubMed] [Google Scholar]
- Lu M-O, Zhang X-M, Mix E, Quezada HC, Jin T, Zhu J, Adem A. TNF-α receptor 1 deficiency enhances kainic acid-induced hippocampus injury in mice. J Neurosci Res. 2008;86:1608–1614. doi: 10.1002/jnr.21600. [DOI] [PubMed] [Google Scholar]
- McKhann GM, 2nd, Wenzel HJ, Robbins CA, Sosunov AA, Schwartzkroin PA. Mouse strain differences in kainic acid sensitivity, seizure behavior, mortality and hippocampal pathology. Neuroscience. 2003;122:551–561. doi: 10.1016/s0306-4522(03)00562-1. [DOI] [PubMed] [Google Scholar]
- McLin JP, Steward O. Comparison of seizure phenotype and neurodegeneration induced by systemic kainic acid in inbred, outbred, and hybrid mouse strains. Eur J Neurosci. 2006;24:2191–2202. doi: 10.1111/j.1460-9568.2006.05111.x. [DOI] [PubMed] [Google Scholar]
- McNamara JO, Morrisett R, Nadler JV. Recent advances in understanding mechanisms of the kindling model. Adv Neurol. 1992;57:555–560. [PubMed] [Google Scholar]
- Mathern GW, Babb TL, Pretorius JK, Melendez M, Lévesque MF. The pathophysiologic relationships between lesion pathology, intracranial ictal EEG onsets, and hippocampal neuron losses in temporal lobe epilepsy. Epilepsy Res. 1995;21:133–147. doi: 10.1016/0920-1211(95)00014-2. [DOI] [PubMed] [Google Scholar]
- Matsuo N, Takao K, Nakanishi K, Yamasaki N, Tanda K, Miyakawa T. Behavioral profiles of three C57BL/6 substrains. Front Behav Neurosci. 2010;4:29. doi: 10.3389/fnbeh.2010.00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Means GD, Boyd Y, Willis CR, Derry JMJ. Transgenic rescue of the tattered phenotype by using a BAC encoding Ebp. Mamm Genome. 2001;12:323–325. doi: 10.1007/s003350010262. [DOI] [PubMed] [Google Scholar]
- Meldrum BS. Excitatory amino acids in epilepsy and potential novel therapies. Epilepsy Res. 1992;12:189–196. doi: 10.1016/0920-1211(92)90040-z. [DOI] [PubMed] [Google Scholar]
- Mohajeri MH, Madani R, Saini K, Lipp HP, Nitsch RM, Wolfer DP. The impact of genetic background on neurodegeneration and behavior in seizured mice. Genes Brain Behav. 2004;3:228–239. doi: 10.1111/j.1601-1848.2004.00073.x. [DOI] [PubMed] [Google Scholar]
- Montkowski A, Poettig M, Mederer A, Holslover F. Behavioural performance in three substrains of mouse strain 129. Brain Res. 1997;762:12–18. doi: 10.1016/s0006-8993(97)00370-3. [DOI] [PubMed] [Google Scholar]
- Moore KJ, Nagle DL. Complex trait analysis in the mouse: The strengths, limitations and the promise yet to come. Annu Rev Genet. 2000;34:653–686. doi: 10.1146/annurev.genet.34.1.653. [DOI] [PubMed] [Google Scholar]
- Morrison RS, Wenzel HJ, Kinoshita Y, Robbins CA, Donehower LA, Schwartzkroin PA. Loss of the p53 tumor suppressor gene protects neurons from kainate-induced cell death. J Neurosci. 1996;16:1337–1345. doi: 10.1523/JNEUROSCI.16-04-01337.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller CJ, Gröticke I, Hoffmann K, Schughart K, Löscher W. Differences in sensitivity to the convulsant pilocarpine in substrains and sublines of C57BL/6 mice. Genes Brain Behav. 2009;8:481–492. doi: 10.1111/j.1601-183X.2009.00490.x. [DOI] [PubMed] [Google Scholar]
- Ottman R. Genetic epidemiology of epilepsy. Epidemiol Rev. 1997;19:120–128. doi: 10.1093/oxfordjournals.epirev.a017934. [DOI] [PubMed] [Google Scholar]
- Qian J, Xu K, Yoo J, Chen TT, Andrews G, Noebels JL. Knockout of Zn transporters Zip-1 and Zip-3 attenuates seizure-induced CA1 neurodegeneration. J Neurosci. 2011;31:97–104. doi: 10.1523/JNEUROSCI.5162-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangel A, Burgaya F, Gavín R, Soriano E, Aguzzi A, del Río J. Enhanced susceptibility of Prnp-deficient mice to kainate-induced seizures, neuronal apoptosis, and death: role of AMPA/kainate receptors. J Neurosci Res. 2007;85:2741–2755. doi: 10.1002/jnr.21215. [DOI] [PubMed] [Google Scholar]
- Rees MI. The genetics of epilepsy – the past, the present and future. Seizure. 2010;19:680–683. doi: 10.1016/j.seizure.2010.10.029. [DOI] [PubMed] [Google Scholar]
- Sanford LP, Kallapur S, Ormsby I, Doetschman T. Methods in Molecular Biology, Vol. 158: Gene Knockout Protocols. Humana Press; New Jersey: Influence of genetic background on knockout mouse phenotypes. [DOI] [PubMed] [Google Scholar]
- Schauwecker PE, Steward O. Genetic determinants of susceptibility to excitotoxic cell death: implications for gene targeting approaches. Proc Natl Acad Sci USA. 1997;94:4103–4108. doi: 10.1073/pnas.94.8.4103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauwecker PE. Seizure-induced neuronal death is associated with induction of c-Jun N-terminal kinase and is dependent on genetic background. Brain Res. 2000;884:116–128. doi: 10.1016/s0006-8993(00)02888-2. [DOI] [PubMed] [Google Scholar]
- Schauwecker PE. Complications associated with genetic background effects in models of experimental epilepsy. Prog Brain Res. 2002;135:139–148. doi: 10.1016/s0079-6123(02)35014-3. [DOI] [PubMed] [Google Scholar]
- Schauwecker PE. Genetic basis of kainate-induced excitotoxicity in mice: phenotypic modulation of seizure-induced cell death. Epilepsy Res. 2003;55:201–210. doi: 10.1016/s0920-1211(03)00115-3. [DOI] [PubMed] [Google Scholar]
- Schauwecker PE, Williams RW, Santos JB. Genetic control of sensitivity to hippocampal cell death induced by kainic acid: a quantitative trait loci analysis. J Comp Neurol. 2004;477:96–107. doi: 10.1002/cne.20245. [DOI] [PubMed] [Google Scholar]
- Schauwecker PE. Congenic strains provide evidence that a mapped locus on chromosome 15 influences excitotoxic cell death. Genes Brain Behav. 2011;10:100–110. doi: 10.1111/j.1601-183X.2010.00644.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheffer IE, Berkovic SF. The genetics of human epilepsy. Trends Pharmacol Sci. 2003;24:428–433. doi: 10.1016/S0165-6147(03)00194-9. [DOI] [PubMed] [Google Scholar]
- Schunk E, Aigner C, Stefanova N, Wenning G, Herzog H, Schwarzer C. Kappa opioid receptor activation blocks progressive neurodegeneration after kainic acid injection. Hippocampus. 2011;21:1010–1020. doi: 10.1002/hipo.20813. [DOI] [PubMed] [Google Scholar]
- Serratosa JM. Idiopathic epilepsies with a complex mode of inheritance. Epilepsia. 1999;40(Suppl 3):12–16. doi: 10.1111/j.1528-1157.1999.tb00893.x. [DOI] [PubMed] [Google Scholar]
- Shannon HE, Yang L. Seizure susceptibility of neuropeptide-Y null mutant mice in amygdala kindling and chemical-induced seizure models. Epilepsy Res. 2004;61:49–62. doi: 10.1016/j.eplepsyres.2004.06.002. [DOI] [PubMed] [Google Scholar]
- Shuttleworth CW, Connor JA. Strain-dependent differences in calcium signaling predict excitotoxicity in murine hippocampal neurons. J Neurosci. 2001;21:4225–4236. doi: 10.1523/JNEUROSCI.21-12-04225.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver LM. Mouse Genetics: Concepts and Applications. Oxford University Press; New York: 1995. [Google Scholar]
- Simpson EM, Linder CC, Sargent EE, Davisson MT, Mobraaten LE, Sharp JJ. Genetic variation among 129 substrains and its importance for targeted mutagenesis in mice. Nat Genet. 1997;16:19–27. doi: 10.1038/ng0597-19. [DOI] [PubMed] [Google Scholar]
- Steinlein OK. Gene defects in idiopathic epilepsy. Rev Neurol. 1999;155:450–453. [PubMed] [Google Scholar]
- Steinlein OK. Gene polymorphisms and their role in epilepsy treatment and prognosis. Naunyn Schmiedebergs Arch Pharmacol. 2010;382:109–118. doi: 10.1007/s00210-010-0531-8. [DOI] [PubMed] [Google Scholar]
- Todorova MT, Mantis JG, Le M, Kim CY, Seyfried TN. Genetic and environmental interactions determine seizure susceptibility in epileptic EL mice. Genes Brain Behav. 2006;5:518–527. doi: 10.1111/j.1601-183X.2006.00204.x. [DOI] [PubMed] [Google Scholar]
- Tripathi PP, Sgadò P, Scali M, Viaggi C, Casarosa S, Simon HH, Vaglini F, Corsini GU, Bozzi Y. Increased susceptibility to kainic acid-induced seizures in Engrailed-2 knockout mice. Neuroscience. 2009;159:842–849. doi: 10.1016/j.neuroscience.2009.01.007. [DOI] [PubMed] [Google Scholar]
- Tsang S, Sun Z, Luke B, Stewart C, Lum N, Gregory M, Wu X, Subleski M, Jenkins NA, Copeland NG, Munroe DJ. A comprehensive SNP-based genetic analysis of inbred mouse strains. Mamm Genome. 2005;16:476–480. doi: 10.1007/s00335-005-0001-7. [DOI] [PubMed] [Google Scholar]
- Turski WA, Cavalheiro EA, Schwarz M, Czuczwar SJ, Kleinrok Z, Turski L. Limbic seizures produced by pilocarpine in rats: behavioural, electroencephalographic and neuropathological study. Behav Brain Res. 1983;9:315–335. doi: 10.1016/0166-4328(83)90136-5. [DOI] [PubMed] [Google Scholar]
- Weiergräber M, Henry M, Radhakrishnan K, Hescheler J, Schneider T. Hippocampal seizure resistance and reduced neuronal excitotoxicity in mice lacking the CaV2.3 E/R-type voltage-gated calcium channel. J Neurophysiol. 2007;97:3660–3669. doi: 10.1152/jn.01193.2006. [DOI] [PubMed] [Google Scholar]
- Weil MM, Brown BW, Serachitopol DM. Genotype selection to rapidly breed congenic strains. Genetics. 1997;146:1061–1069. doi: 10.1093/genetics/146.3.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams PA, Dou P, Dudek FE. Epilepsy and synaptic reorganization in a perinatal rat model of hypoxia-ischemia. Epilepsia. 2004;45:1210–1218. doi: 10.1111/j.0013-9580.2004.60403.x. [DOI] [PubMed] [Google Scholar]
- Winawer MR, Makarenko N, McCloskey DP, Hintz TM, Nair N, Palmer AA, Scharfman HE. Acute and chronic responses to the convulsant pilocarpine in DBA/2J and A/J mice. Neuroscience. 2007;149:465–475. doi: 10.1016/j.neuroscience.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfer DP, Lipp HP. Dissecting the behaviour of transgenic mice: is it the mutation, the genetic background, or the environment? Exp Psych. 2000;85:627–634. [PubMed] [Google Scholar]
- Wolfer DP, Crusio WE, Lipp HP. Knockout mice: simple solutions to the problems of genetic background and flanking genes. Trends Neurosci. 2002;25:336–340. doi: 10.1016/s0166-2236(02)02192-6. [DOI] [PubMed] [Google Scholar]
- Wu G, Lu Z-H, Wang J, Wang Y, Xie X, Meyenhofer MF, Ledeen RW. Enhanced susceptibility to kainate-induced seizures, neuronal apoptosis, and death in mice lacking gangliotetraose gangliosides: protection with LIGA 20, a membrane-permeant analog of GM1. J Neurosci. 2005;25:11014–11022. doi: 10.1523/JNEUROSCI.3635-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi H, Noguchi Y, Kitamura K, Sato M. Deficiency of Vlgr1 resulted in deafness and susceptibility to audiogenic seizures while the degree of hearing impairment was not correlated with seizure severity in C57BL/6- and 129-backcrossed lines of Vlgr1 knockout mice. Neurosci Lett. 2009;461:190–195. doi: 10.1016/j.neulet.2009.06.012. [DOI] [PubMed] [Google Scholar]
- Yang Y, Mahaffey CL, Bérubé N, Maddatu TP, Cox GA, Frankel WN. Complex seizure disorder caused by Brunol4 deficiency in mice. PLoS Genet. 2007;3:e124. doi: 10.1371/journal.pgen.0030124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu ZL, Jiang JM, Wu DH, Xie HJ, Jiang JJ, Zhou L, Peng L, Bao GS. Febrile seizures are associated with mutation of seizure-related (SEZ) 6, a brain-specific gene. J Neurosci Res. 2007;85:166–172. doi: 10.1002/jnr.21103. [DOI] [PubMed] [Google Scholar]
- Zurita E, Chagoyen M, Cantero M, Alonso R, González-Neira A, López-Jiménez A, López-Moreno JA, Landel CP, Benitez J, Pazos F, Montoliu L. Genetic polymorphisms among C57BL/6 mouse inbred strains. Transgenic Res. 2010;20:481–489. doi: 10.1007/s11248-010-9403-8. [DOI] [PubMed] [Google Scholar]