Summary
LDL receptor-related proteins 5 and 6 (LRP5/6) are co-receptors for Wnt growth factors, and also bind Dkk proteins, secreted inhibitors of Wnt signaling. The LRP5/6 ectodomain contains four β-propeller/EGF-like domain repeats. The first two repeats (LRP6(1-2)) bind to several Wnt variants, whereas LRP6(3-4) binds other Wnts. We present the crystal structure of the Dkk1 C-terminal domain bound to LRP6(3-4), and show that the Dkk1 N-terminal domain binds to LRP6(1-2), demonstrating that a single Dkk1 molecule can bind to both portions of the LRP6 ectodomain and thereby inhibit different Wnts. Small-angle x-ray scattering analysis of LRP6(1-4) bound to a non-inhibitory antibody fragment or to full-length Dkk1 shows that in both cases the ectodomain adopts a curved conformation that places the first three repeats at a similar height relative to the membrane. Thus, Wnts bound to either portion of the LRP6 ectodomain likely bear a similar spatial relationship to Frizzled co-receptors.
Wnt growth factors have essential roles in specifying cell fate during embryogenesis and the renewal of tissues in the adult (Clevers, 2006; Logan and Nusse, 2004; Reya and Clevers, 2005). In the Wnt/β-catenin pathway, Wnts bind to two co-receptors: 7-transmembrane helix Frizzled (Fzd) proteins, and a single-pass transmembrane receptor, LDL receptor-related protein 5 or 6 (LRP5/6) (Clevers, 2006; Logan and Nusse, 2004; MacDonald et al., 2009). Wnt binding to Fzd and LRP5/6 leads to phosphorylation of the LRP5/6 cytoplasmic tail, which inhibits β-catenin destruction; the stabilized β-catenin acts as a transcriptional coactivator of Wnt target genes. Inappropriate activation of this pathway is associated with a number of cancers and other diseases (Clevers, 2006; Logan and Nusse, 2004; MacDonald et al., 2009).
The importance of LRP5/6 in Wnt signaling is highlighted by natural and experimentally derived mutations. Mutants of the Drosophila Lrp5/6 ortholog Arrow are phenotypically similar to wingless (dWnt-1) mutants (Wehrli et al., 2000). In mice, deletion of both LRP5 and LRP6 causes embryonic lethality due to failure of gastrulation (Kelly et al., 2004). Deletion of LRP6 results in perinatal lethality with midbrain and hindbrain defects, posterior truncation, and abnormal limb development, whereas deletion of LRP5 leads to osteoporosis and other metabolic defects (Kato et al., 2002; Pinson et al., 2000). Missense mutations in LRP5 associated with autosomal recessive osteoporosis-pseudoglioma syndrome (OPPG) compromise Wnt signaling (Gong et al., 2001). Missense mutations in the LRP5 ectodomain are also associated with autosomal dominant and recessive familial exudative vitreoretinopathy (FEVR), although the biochemical consequences of these changes has not been reported (Jiao et al., 2004; Qin et al., 2005; Toomes et al., 2004).
The LRP5/6 ectodomain comprises four repeating units of a six-bladed β-propeller connected to an EGF-like domain, followed by three LDLR-type A repeats (Figure 1A). A study using purified proteins demonstrated that Wnt9b binds to an LRP6 construct comprising the first two propeller/EGF repeats, designated here LRP6(1-2), whereas Wnt3a binds to LRP6(3-4) (Bourhis et al., 2010). Deletion mutagenesis and antibody blocking experiments have implicated LRP6(1-2) in binding to Wnts 1, 2, 2b, 6, 8a, 9a, 9b and 10b, whereas LRP6(3-4) is required for Wnt3a binding (Ai et al., 2005; Gong et al., 2010; Itasaki et al., 2003; Mao et al., 2001a; Zhang et al., 2004). Antibodies to different regions of LRP6 can inhibit Wnt signaling, presumably by competing with Wnts directly or inhibiting formation of ternary receptor complexes, whereas others enhance signaling, possibly by receptor clustering (Binnerts et al., 2009; Gong et al., 2010; Yasui et al., 2010).
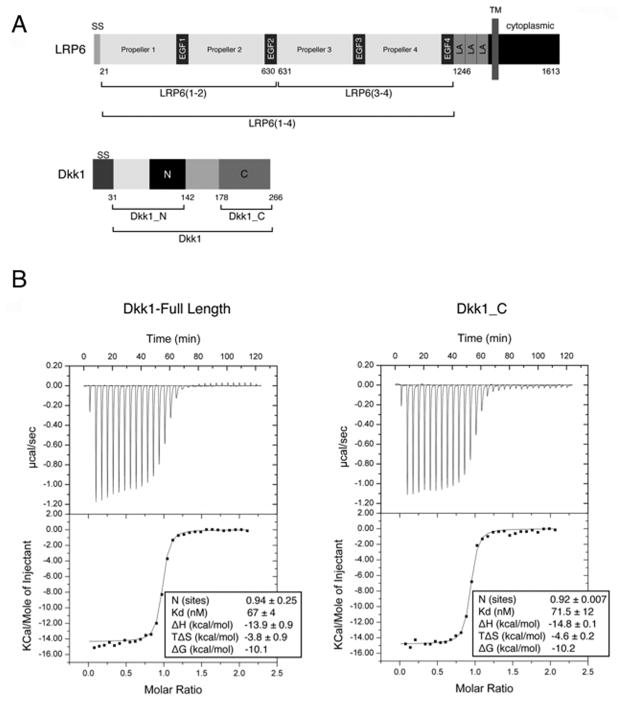
Figure 1. Dkk1_C mediates binding to LRP6(3-4).
(A) Primary structures of human LRP6 and Dkk1. The conserved cysteine-rich N- and C-terminal domains of Dkk1 are denoted “N” and “C”. SS, signal sequence; LA, LDLR type A repeat, TM, transmembrane segment. Boundaries of constructs used in this study are indicated below each protein.
(B) ITC binding of LRP6(3-4) to either full length Dkk1 (left) or Dkk1_C (right). See also Table S1.
Dickkopf (Dkk) proteins are secreted modulators of Wnt signaling that bind to LRP5/6 with high affinity (Bourhis et al., 2010; Niehrs, 2006). Deletion of Dkk1 results in embryonic lethality including loss of anterior head structures and fused vertebrae (Mukhopadhyay et al., 2001), and Dkk2 null mice show osteopenia and blindness (Li et al., 2005a; Mukhopadhyay et al., 2006). High bone mass (HBM) disease arises from missense mutations in LRP5 repeat 1 that reduce or ablate the ability of inhibitors, including Dkks, to down-regulate Wnt signaling (Ai et al., 2005; Balemans et al., 2007). Dkks also bind to the cell-surface receptor Kremen, which appears to control internalization of LRP5/6 under some circumstances (Mao and Niehrs, 2003; Mao et al., 2002; Semenov et al., 2008; Wang et al., 2008).
Each of the four vertebrate Dkk family members consists of two conserved cysteine-rich domains, designated here Dkk_N and Dkk_C, connected by a linker of ~50 residues in Dkks 1, 2, and 4 (Figure 1A). Dkk1_C and Dkk2_C alone antagonize Wnt signaling (Brott and Sokol, 2002; Li et al., 2002; Mao and Niehrs, 2003), consistent with the absence of Dkk_N in Dkks of lower organisms such as Hydra (Guder et al., 2006). Dkk1 binds to both LRP6(1-2) and LRP6(3-4) (Bafico et al., 2001; Binnerts et al., 2009; Bourhis et al., 2010; Li et al., 2005b; Liu et al., 2009; Mao et al., 2001a; Zhang et al., 2004), but the regions of Dkk1 required for these interactions are unknown.
Here we describe the crystal structure of human LRP6(3-4) bound to human Dkk1_C, and a low-resolution picture of the full LRP6(1-4) region derived from small-angle x-ray scattering (SAXS). We show that Dkk1 acts as a bipartite inhibitor of Wnt binding, with Dkk1_N binding to LRP6(1-2) while Dkk1_C binds to LRP6(3-4). The LRP6(1-4) region adopts a twisted, curved conformation that likely places its multiple Wnt binding surfaces at comparable heights from the membrane. The low resolution envelopes of LRP6(1-4) bound to a Fab fragment of a monoclonal antibody or to Dkk1 indicate that the receptor adopts similar conformations in both cases.
Results
The Dkk1 C-terminal domain specifies binding to LRP6(3-4)
The three LDLR-A motifs at the C-terminus of the LRP6 ectodomain do not affect Wnt signaling nor its inhibition by Dkk1 (Mao et al., 2001a), so our studies employed only the β-propeller/EGF repeats. LRP6(3-4), The second two repeats, LRP6(3-4) (Figure 1A), are required for Dkk1 inhibition of Wnt signaling. Isothermal titration calorimetry (ITC) (Figure 1B) revealed that Dkk1 forms a 1:1 complex with LRP6(3-4), consistent with quantitative N-terminal sequencing (Table S1A), and binds with a Kd of 67 nM (Figure 1B), in good agreement with biolayer interferometry measurements (Bourhis et al., 2010). Only the conserved C-terminal cysteine-rich region of Dkk1, starting at residue 178 and designated Dkk1_C, was protected when the LRP6(3-4)–Dkk1 complex was digested with trypsin (data not shown). ITC revealed that Dkk1_C binds to LRP6(3-4) with the same thermodynamics as full-length Dkk1 (Figure 1B), demonstrating that that Dkk1_C harbors the full set of residues that bind to LRP6(3-4).
The crystal structure of the LRP6(3-4)–Dkk1_C complex was determined at 2.8 Å resolution (Table 1). Surprisingly, the asymmetric unit of the crystal contains two copies of LRP6(3-4) and a single copy of Dkk1_C. The crystallized material was purified by mixing LRP6(3-4) with an excess of Dkk_C at concentrations >100 x Kd to insure that all of the LRP6(3-4) would be bound to Dkk1_C. The mixture was applied to a size exclusion chromatography column, and the two proteins co-eluted in a volume corresponding to ~80 kDa, the expected size of a 1:1 complex. However, quantitative N-terminal Edman sequencing of the fractions containing the two proteins revealed a molar ratio of 2 LRP6(3-4):1 Dkk_C (Table S1B). These results suggested that the purified (and crystallized) material is an equimolar mixture of a 1:1 LRP6(3-4)–Dkk1_C complex and unbound LRP6(3-4) that was unresolved on the size exclusion column; a 2 LRP6(3-4):1 Dkk_C complex would be expected to migrate at ~150 kDa. Native PAGE confirmed that the material used for crystallization was an equimolar mixture of complex and unbound LRP6(3-4) (data not shown). As the complex was prepared under conditions that should have insured 100% complex formation, the presence of bound and unbound LRP6(3-4) cannot be readily explained.
Table 1.
Crystallographic data.
| Native | HgCl2 | |
|---|---|---|
| Data collection* | ||
| Space group | P212121 | P212121 |
| Unit cell lengths a,b,c (Å) | 96.0, 108.0, 173.1 | 96.2, 107.6, 172.4 |
| Wavelength (Å) | 1.0039 | 1.0039 |
| Resolution (Å) | 45.8 –2.80 (2.90-2.80) | 45.8 - 3.08 (3.19-3.08) |
| Unique reflections | 45,089 | 32,773 |
| Multiplicity | 3.4 (3.4) | 3.6 (3.6) |
| Completeness, % | 99.7 (99.9) | 97.0 (94.7) |
| <I/ σ (I)> | 23.8 (2.6) | 12.2 (1.6) |
| Rmergea (%) | 6.1 (51.9) | 10.4 (70.6) |
| Model refinement | ||
| Resolution, Å | 45.8-2.80 | |
| No. reflections work/test set | 42,788/2,235 | |
| Number of residues | ||
| protein | 1,298 | |
| carbohydrate | 16 | |
| water | 56 | |
| Rcrystb (%) | 19.4 | |
| Rfreeb (%) | 25.1 | |
| Rmsd bonds (Å) | 0.003 | |
| Rmsd angles (°) | 0.69 | |
| Ramachandran plotc | ||
| % in favored regions | 93.2 | |
| % in additional allowed regions | 6.7 | |
| % outliers | 0.1 | |
| Average temperature factor (Å2) | ||
| Lrp6(3-4) chain A | 66.1 | |
| Lrp6(3-4) chain B | 56.8 | |
| Dkk1_C | 61.0 | |
| Solvent | 42.8 | |
Data were measured at 100 K at beamline 11-1 at the Stanford Synchrotron Radiation Laboratory (SSRL), and were integrated and scaled with HKL2000 (Otwinowski and Minor, 1997). Values in parentheses are for the highest resolution shell. Rmsd, root-mean square deviation.
Rmerge=ΣhΣI|(II(h)−<I(h)>| / ΣhΣI(h), where II(h) is the Ith measurement of reflection h, and <I(h)> is the weighted mean of all measurements of h.
R = Σh|Fobs(h)| − |Fcalc(h)| | / Σh|Fobs(h)|. Rcryst and Rfree were calculated using the working and test reflection sets, respectively.
As defined in Molprobity (Chen et al., 2010).
All residues of LRP6(3-4) are visible in both copies in the crystal, except for the loop comprising residues 1006–1012. In both copies, at least one sugar is visible at each of the five predicted N-linked glycosylation sites (Figure S1). The model of Dkk1_C comprises residues 182–264, except that the loop spanning residues 250–258 is disordered.
Structure of LRP6(3-4) and Dkk1_C
Each repeat of LRP6(3-4) consists of a six-bladed β-propeller attached by a 7–8 residue linker to an EGF-like domain that packs tightly aginst the propeller (Figure 2A). Each blade consists of four anti-parallel β-strands, with the first strand, A, occupying the position closest to the pseudo six-fold symmetry axis of the propeller (Figure 2A). The B strands of blades 2–6 harbor the Tyr-Trp-Thr-Asp (YWTD) repeat motif that characterizes these structures (Figure S1). As in other YWTD β-propeller structures, the amino acid sequence of the barrel begins with strand B of blade 1, and the last β-strand in the sequence is the A strand of blade 1, thereby closing the barrel (Jeon et al., 2001) (Figure S1).
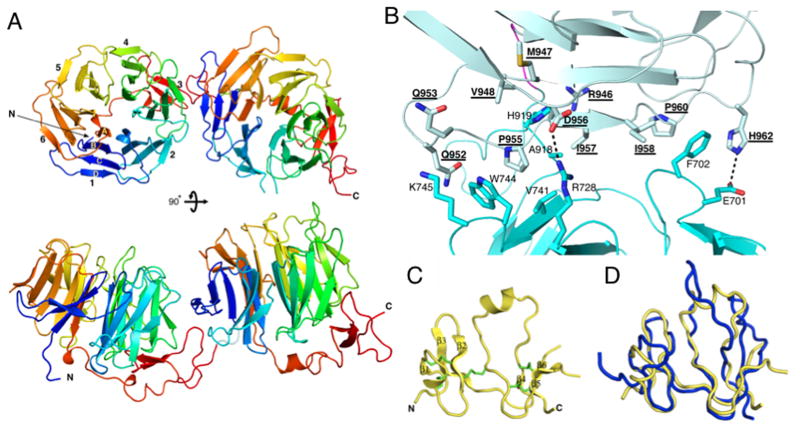
Figure 2. Structures of LRP6(3-4) and Dkk1_C.
(A) The LRP6(3-4) structure, viewed down the pseudo-six fold symmetry axis of propeller 3 (top) or towards the side (bottom). Blades are numbered in repeat 3, and individual strands are labeled in blade 1.
(B) Closeup of the repeat 3-4 interface. Interacting side chains are shown in stick representation. Polar interactions are shown with dashed lines. Repeat 4 residue labels are underlined. The complete list of interactions is given in Table S2. See also Figure S1.
(C) Ribbon representation of Dkk1_C. Disulfide bridges are show in green. The loop between β-strands 5 and 6 is disordered. See also Figure S2
(D) Superposition of human Dkk1_C structure (gold) with the NMR solution structure of mouse Dkk2_C (blue) (PDB entry 2JTK).
The structures of the two LRP6 β-propeller/EGF repeats are very similar to one another and to the single repeats found in the LDL receptor and the basement membrane protein nidogen (Jeon et al., 2001; Takagi et al., 2003). The two copies of repeat 3 or those of repeat 4 superimpose closely (root-mean-square deviation (rmsd) = 1.2 Å). The rmsd of repeats 3 and 4 to the LDLR repeat are 1.7 Å (283 residues, 34.3% identity) and 2.1 Å (278 residues, 25.9% identity), respectively. The largest variation amongst these β-propeller/EGF repeat structures occurs in the connection between the propeller and the EGF-like domain. Nonetheless, the EGF-like domain retains a very similar disposition with respect to the β-propeller in all cases.
The LRP6(3-4) structure provides a view of tandem β-propeller/EGF repeat modules. Just four resides connect the end of the repeat 3 EGF-like domain and the first β-strand of the repeat 4 propeller, and the two repeats interact extensively with one another. The inter-repeat interface buries 1819 Å2 of surface area with a mixture of polar and non-polar interactions (Figure 2B; Table S2). The two crystallographically independent copies show limited flexibility: if only repeats 3 are superimposed, then repeats 4 differ by a rotation of about 7°.
The Dkk1_C structure can be described as two sub-domains, each consisting of three anti-parallel β-strands, that are connected by a long loop which contains a short α-helix (Figure 2C). The fold is stabilized by five disulfide bridges. The molecule is rather flat, with approximate dimensions 30 x 30 x 15 Å. The overall structure is similar to that of the mouse Dkk2_C determined by NMR (Chen et al., 2008) (rmsd = 2.4 Å for 63 residues; 61.9% identity) (Figure 2D), but there are two significant differences. First, a portion of the long connection between strands 4 and 5 is relatively flexible in the unbound, solution NMR structure, but in the crystal structure this region interacts with LRP6. This region is almost identical in sequence between the two proteins, implying that the difference in structure is due to binding to LRP6. Conversely, the loop between β strands 5 and 6 in the NMR structure would clash with the LRP6 propeller, whereas this region is disordered in the present structure.
The LRP6(3-4)–Dkk1 interaction
A single Dkk1_C molecule is sandwiched between the two independent copies of LRP6(3-4) in the asymmetric unit of the crystal (Figure 3A). In both copies of LRP6, the same face of the repeat 3 propeller interacts with Dkk1_C, and in both cases the Dkk1_C molecule is not centered on the propeller but offset towards blades 3, 4 and 5. This face of the propeller presents a concave, amphitheater-like structure that serves as the binding surface for ligands, as seen in the nidogen–laminin complex (Takagi et al., 2003), and the intramolecular interaction with LDLR-A repeats in the low pH LDL receptor (Rudenko et al., 2002). In the LRP6 repeat 3 propeller, this surface is electrostatically negative (Figure 3B), consistent with the positively charged nature of both Dkk_C (calculated pI = 9.2) and most Wnts (average pI = 8.9 for the 19 human Wnts). In contrast, the equivalent face of the repeat 4 barrel is more positively charged (Figure 3B), and is exposed in this structure.
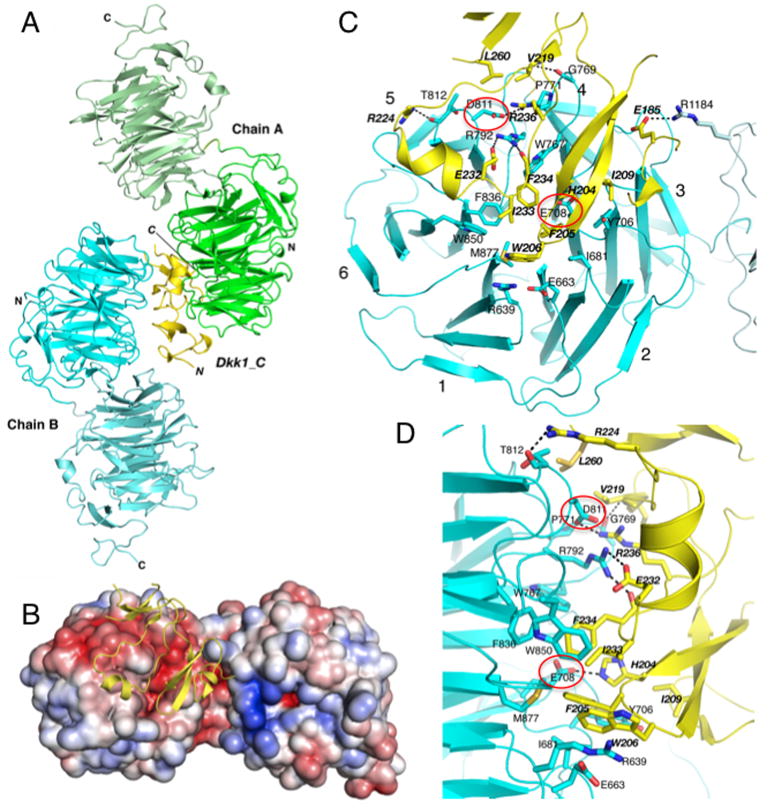
Figure 3. Dkk1_C interacts with both copies of LRP6(3-4) in the asymmetric unit.
(A) Overall structure of the asymmetric unit. LRP6(3-4) copy A is shown in green and copy B in cyan, with repeats 3 and 4 respectively shown in darker and ligher shades. The single copy of Dkk1_C is shown in gold.
(B) Electrostatic surface of LRP6(3-4) copy B bound to Dkk1_C. Red represents regions of negative charge, blue positive, contoured from −5 to +5 kBT/e.
(C,D) Closeup view of the LRP6(3-4) copy B interface with Dkk1_C, looking down the pseudo-six fold axis of LRP6 propeller 3 (C) or viewed from the side (D). Interacting residues are shown in stick representation. Polar interactions are shown with dashed lines. Dkk1 residue labels are italicized. The complete list of interactions is given in Table S3. Glu708 and Asp811 are highlighted in red ovals. See also Figures S1, S2 and S5. The copy A interface is shown in Figure S3 and contacts listed in Table S4.
The interface between Dkk1_C and copy B of LRP6(3-4) buries 2033 Å2 of surface area. The core of the interface features a cluster of aromatic and non-polar side chain interactions near the center of the LRP6 repeat 3 β-propeller (Figure 3C,D; Table S3). In addition, Dkk1 His204 forms a hydrogen bond with LRP6 Glu708, Glu232 forms a salt bridge with Arg792, and several side chains also form hydrogen bonds with backbone amide nitrogen and carbonyl oxygen atoms. A second cluster of interactions occurs towards the rim of the LRP6 amphitheater, at blades 4 and 5: Dkk1 Val219 and Leu260 pack against Pro771 and Thr812, Arg236 forms a salt bridge with Asp811, and Arg224 forms a hydrogen bond with Asn813 and the backbone at 812. Finally, there is a single salt bridge made by Dkk1 Glu185 and LRP6 Arg1184, the only contact between Dkk1 and LRP6 repeat 4.
In contrast to the extensive non-polar packing interactions between Dkk1 and LRP6 chain B, the interface with copy A involves mostly polar Dkk1 residues and is somewhat smaller (1830 Å2). These Dkk1 residues interact with many of the same LRP6 side chains that form the interface with the aromatic cluster on the other face of Dkk1_C (Figure S3; Table S4). Most notable is the interaction of Lys226 methylene groups with the same aromatic and aliphatic residues that mediate contacts between copy B and the Dkk1 aromatic cluster; the terminal amine is neutralized by an electrostatically negative region in the center of the amphitheater. Only one non-polar side chain of Dkk1, Leu231, contacts LRP6 in this interface, packing against the aromatic ring of Tyr875. In addition to these interactions, LRP6 side chains Arg751, Arg792, and Trp850 form hydrogen bonds with the main chain of Dkk1.
To determine the biologically relevant interface, we mutated several Dkk1 residues that contact LRP6(3-4) in the crystal and tested their ability to inhibit Wnt3a signaling (Figure 4A). Mutations in the interface with copy B, including E232K, W206A, F234K, abolished or significantly reduced inhibition, consistent with the behavior of charge reversal mutations H204E, K211E, R236E and H261E in this interface described previously for mouse Dkk1 (human Dkk1 numbering shown) (Chen et al., 2008). In contrast, mutation of residues in the interface with copy A had only small (K226E) or no (H229E) effects on the ability of Dkk1 to inhibit signaling, as did two other published mutations of mouse Dkk1 in this interface, K226A and R191E (human numbering) (Wang et al., 2008). This polar face of Dkk1_C has been shown to interact with Kremen proteins (Wang et al., 2008), which may explain the small effect of K226E on Wnt reporter activity, as mutation of this residue to either Ala or Glu was shown to have no effect on LRP6 binding (Wang et al., 2008).
Figure 4. Effect of Dkk1 and LRP6 mutations on Wnt3a activity.
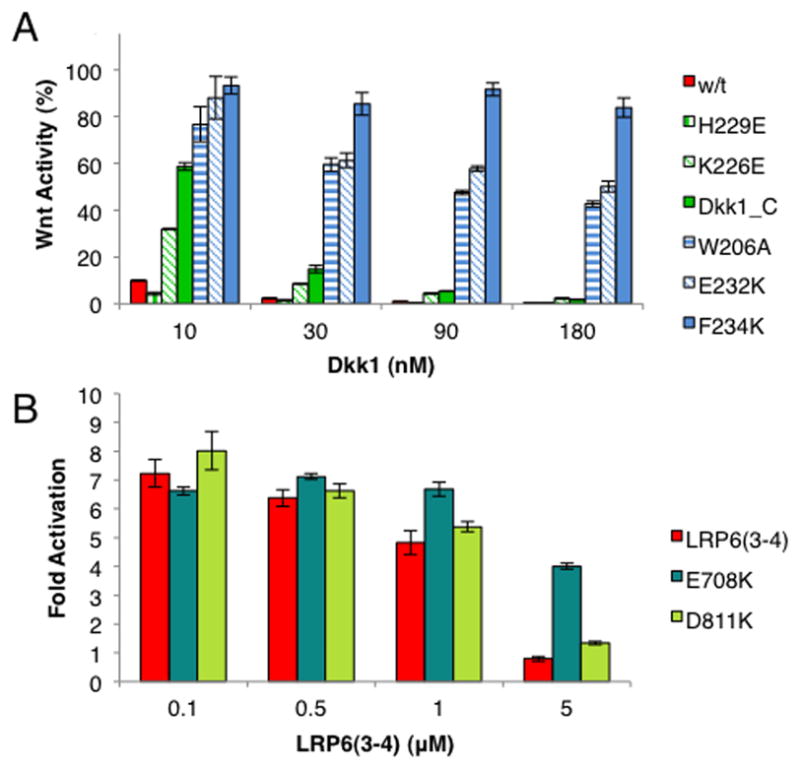
LSL cells were treated with Wnt3a conditioned media plus Dkk1_C, wild-type Dkk1 or a mutant. The activity from treatment with Wnt3a CM is taken to be 100%. Error bars denote standard deviation.
(A) Dkk1 mutants.
(B) LRP6(3-4) mutants.
The mutational data and the 1:1 stoichiometry observed for the LRP6(3-4)–Dkk1_C interaction indicate that the interface between Dkk1 and LRP6 chain B in the crystal represents the high-affinity and likely biologically relevant interaction between these two proteins. The behavior of the K226E mutant may suggest a role for the polar interface with chain A at the cell surface, although SAXS measurements at high concentrations do not support a 2 LRP6: 1 Dkk1 stoichiometry (see below).
Conservation of the LRP5/6 interface with Dkk
Dkk1_C residues that directly contact LRP6 are conserved in Dkk2 and Dkk4, as expected from the ability of these Dkk isoforms to inhibit Wnt signaling, as well as the Hydra homolog (Figure S2). In contrast most of these positions differ in Dkk3, consistent with earlier studies indicating that Dkk3 is a divergent family member that does not inhibit Wnt signalling (Niehrs, 2006).
The amino acid sequences of human LRP5 and LRP6 are 71% identical, and almost all LRP6 residues that interact with Dkk1_C are identical in LRP5 (Figure S1). The four propeller/EGF repeats of human LRP5/6 are very similar to one another, with few relative insertions and deletions: within LRP6, repeat 3 shares 47% sequence identity with repeats 1 and 2, and while its identity to repeat 4 is only 33%, the structures of repeats 3 and 4 are very similar. Consistent with its lower sequence homology, the concave barrel surface of repeat 4 is poorly conserved, both in electrostatic character (Figure 3B) and in the residues that form the interface with Dkk1. For example, the hydrophobic cluster in the amphitheater of propeller 3 (Figure 3C) is present in repeats 1 and 2, but not 4.
A key difference amongst repeats occurs at the position equivalent to Asp811 in repeat 3, which forms a salt bridge with Arg236 of Dkk1: a lysine is present at this position in repeats 1 and 2. Since R236E in Dkk1 ablates binding (Chen et al., 2008), the equivalent charge reversal caused by the presence of lysine on the LRP6 surface would be expected to prevent Dkk1_C binding. Interestingly, an arginine is present in this position in repeat 3 of the Drosophila LRP5/6 homolog Arrow, which correlates with the absence of Dkk proteins in this organism. Also, several substitutions in repeats 1 and 2 of LRP5 or 6 would appear to diminish binding to Dkk1_C. Ile681, which forms a non-polar contact with Dkk1 Phe205, is Val in repeat 1 but Asp in repeat 2. Tyr706 is replaced by Ser, His or Asp in repeats 1 and 2, and Arg792 is replaced by Trp in repeats 1 and 2.
Wnt3a and Dkk1 bind to an overlapping surface of LRP6
The simplest model for inhibition of Wnt binding by Dkks is direct competition for a common binding site on LRP5/6. We introduced charge reversal mutations E708K and D811K into LRP6(3-4) that, based on the effects of Dkk1 H204E and R236E mutations (Chen et al., 2008), would disrupt the polar interactions with Dkk1, and compared the ability of purified wild-type, E708K and D811K LRP6(3-4) to inhibit signaling by competing with cell surface LRP6 for Wnt3a. Glu708 lies at the heart of the LRP6 repeat 3 amphitheater, and Asp811 is near the rim of blade 5 (Figure 3C,D). Inhibition by wild-type LRP6(3-4) required a concentration of 5 μM, similar to previous reports using this assay (Bourhis et al., 2010; Liu et al., 2009). The D811K mutant behaved similarly to wild-type, but E708K showed only weak inhibition at the highest concentration tested (Figure 4B). These data suggest that the footprint of Wnt3a partially overlaps that of Dkk1_C such that they directly compete for LRP6. Dkk1_C is a weaker inhibitor of Wnt3a signaling than full-length Dkk1 (Figure 4A), which may be due to its weaker affinity for LRP6. However, Wnt3a binds to LRP6(1-4) 20 times more strongly than to LRP6(3-4) (Bourhis et al., 2010), suggesting that there are additional interactions of Wnt3a involving the LRP6(1-2) region that are affected only by full-length Dkk1 and not Dkk1_C, or that LRP6(1-2) influences the conformation of LRP6(3-4).
Dkk1 is a bipartite inhibitor of LRP–Wnt interactions
Purified Wnt9b and Wnt3a bind to independent sites on LRP6: Wnt9b binds to LRP6(1-2), whereas Wnt3a binds to LRP6(3-4), and both can bind simultaneously to LRP6(1-4) (Bourhis et al., 2010). Full-length Dkk1 inhibits both Wnts from binding to their cognate LRP6 fragment (Bourhis et al., 2010). Dkk1 binds to LRP6(1-4) with a Kd of 3 nM, whereas binding to LRP6(1-2) or LRP6(3-4) is 21 or 7 times weaker, respectively (Bourhis et al., 2010). One interpretation of these data is that the same portion of Dkk1 binds to homologous surfaces on either half of the LRP6 ectodomain, such that two copies of Dkk1 bind to one LRP6(1-4). However, the two proteins appear to form a 1:1 complex (see below, and (Bafico et al., 2001; Mao et al., 2001a)), and sequence and mutational data (see above) indicate that Dkk1_C is unlikely to bind to repeats 1 or 2. These observations suggest that the higher affinity of full-length Dkk1 binding is due to the N-terminal portion of Dkk1.
We prepared a Dkk1_N construct spanning residues 31–142 (Figure 1A). Dkk1_N, but not Dkk1_C, associated with LRP6(1-2) on a size exclusion chromatography column (Figure 5A). ITC and native gel shift assays (Figures 5B,S4) showed that Dkk1_N binds to LRP6(1-2) with an apparent Kd of 66 nM, with a 1:1 stoichiometry (Figures 5B,S4). Conversely, we observe no binding of Dkk1_N to LRP6(3-4) by gel filtration or native gel shift assays (data not shown). These data indicate that full-length Dkk1 binds to LRP6 in a bipartite manner: the N-terminal region interacts with the repeat 1-2 region, whereas the C-terminal region binds to repeat 3.
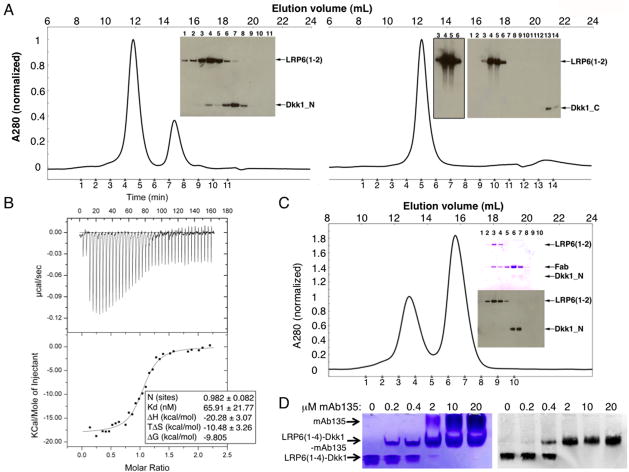
Figure 5. Dkk1_N and Dkk1_C bind to distinct regions of the LRP6 ectodomain.
(A) LRP6(1-2) at 2.5 μM was mixed with 12.5 μM Dkk1_N (left) or 12.5 μM Dkk1_C (right) and run on a Superdex 200 column. An anti-His6 western blot was used to visualize the proteins in the indicated fractions. Dkk1_C transfers poorly for western blotting, as seen by the difference in band intensities versus Dkk1_N even though the same amount of protein is present in both experiments. The overexposed western blot of lanes 3-6 shown in the Dkk1_C experiment demonstrates that no Dkk1_C co-eluted with LRP6(1-2).
(B) ITC measurement of Dkk1_N binding to LRP6(1-2). See also Figure S4 for alternative binding assay.
(C) LRP6(1-2) at 2.5 μM was mixed with 2.5 μM Dkk1_N and 12.5 μM Fab135 and run on a Superdex 200 column. Fractions were analyzed by SDS-PAGE. The upper gel was stained with Coomassie blue to visualize LRP6(1-2) and Fab135; the lower gel was analyzed by western blot with anti-His6 to visualize LRP6(1-2) and Dkk1.
(D) mAb135 and Dkk1 can both bind to LRP6(1-4). LRP6(1-4)–Dkk1 complex at 2 μM was incubated with the indicated concentration of mAb135, run on native PAGE, and analyzed by Coomassie blue staining (left) and western blot with anti-Dkk1 (right). The antibody shifts the LRP6(1-4)–Dkk1 complex upward to the middle band, which still contains Dkk1 as shown by the western blot.
LRP5 point mutations associated with HBM disease occur in positions on the repeat 1 propeller equivalent to residues in repeat 3 that either directly interact or position other amino acids to interact with Dkk_C (Figures S1, S5) (Ai et al., 2005; Balemans et al., 2007; Chen et al., 2008; Takagi et al., 2003; Zhang et al., 2004). The conservation of both sequence and length of the repeat 1 propellers of LRP5 and LRP6 (Figure S1), and the observation that the G158V mutation in LRP6, homologous to the LRP5 HBM mutation G171V, has equivalent effects on Dkk1 and Sclerostin interactions (Ai et al., 2005; Ellies et al., 2006), allowed us to test whether this surface is involved in Dkk1_N binding. The anti-LRP6 monoclonal antibody mAb135 enhances Wnt3a signaling, and binds to an epitope that includes LRP6 Ser243 (Binnerts et al., 2009). Ser243 is located on the rim of repeat 1 amphitheater adjacent to a conserved Trp that in repeat 3 mediates interactions with Dkk1_C (Figure S1). The Fab fragment of this antibody (Fab135) displaced Dkk1_N from LRP6(1-2) (Figure 5C), consistent with the binding of Dkk1_N to the repeat 1 amphitheater. Moreover, as expected from the high-affinity Dkk1_C–LRP6(3-4) interaction, mAb135 bound to, but did not compete full-length Dkk1 from, LRP6(1-4) (Figure 5D). Since Dkk1 binds to LRP6(3-4) more weakly than to LRP6(1-4), the ability of mAb135 to inhibit internalization (Binnerts et al., 2009) may be due to weakening the overall affinity of Dkk1 for LRP6 such that formation of ternary complexes with Kremen is less likely.
The observation that mAb135 does not displace Dkk1 from LRP6(1-4) implies that the two proteins bind with a 1:1 stoichiometry: if two Dkk1 molecules bound simultaneously to LRP6(1-4), we would expect a loss of the total Dkk1 band intensity with increasing antibody concentration in Figure 5D. We examined the stoichiometry of the complex directly with small-angle x-ray scattering (SAXS) (Figures 6A, S6A). The scattering intensity at zero scattering angle I(0) allows accurate determination of molecular mass (Orthaber et al., 2000). Including the N-linked carbohydrate, the calculated masses of LRP6(1-4) and Dkk1 are approximately 152 and 27.5 kDa. I(0) analysis of the LRP6(1-4)–Dkk1 data reveals a molecular mass of 185 ± 5 kDa (Fig. S6A). Thus, the SAXS data reveal a 1:1 stoichiometry for the LRP6(1-4)–Dkk1 complex at the highest concentrations experimentally accessible.
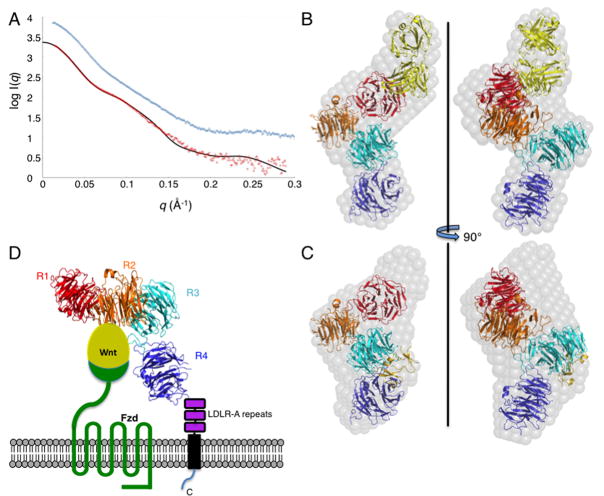
Figure 6. SAXS reconstructions of LRP6(1-4) bound to Fab135 or Dkk1.
(A) SAXS data for the LRP6(1-4)–Fab135 (red) and LRP6(1-4)–Dkk1 (blue) complexes, offset on the y axis for clarity. The solid line shows the scattering curve calculated from the LRP6(1-4)–Fab135 model shown in (B), χ2=3.1. See also Figure S6.
(B) Ab initio reconstruction (grey spheres) of the LRP6(1-4)–Fab135 complex, with a model of the LRP6(1-4)–Fab135 superimposed. LRP6 repeats 1, 2, 3 and 4 are shown in red, orange, teal, and blue, respectively, and the Fab is shown in yellow.
(C) Ab initio reconstruction of the LRP6(1-4)–Dkk1 complex. The model of the LRP6(1-4) region shown in (B) is superimposed on the envelope. The position of Dkk1_C after superposition of the LRP6(3-4)–Dkk1_C crystal structure on the SAXS model is shown in gold.
(D) Alternative view of LRP6(1-4) SAXS-derived model, oriented with repeats 1-3 roughly coplanar and the C-terminus of repeat 4 near the bottom, illustrating that the Wnt-binding regions of the receptor likely have similar heights with respect to the membrane and Fzd. Wnt, Fzd, and the three LDLR-A repeats, the transmembrane anchor, and the cytoplasmic domain of LRP5/6 are shown schematically.
Structure of the LRP6(1-4) region
We used SAXS to determine the overall conformation of the LRP6 repeat 1-4 region. The uncomplexed protein was not sufficiently well behaved for SAXS measurements, nor was its complex with Dkk1_C. However, both LRP6(1-4)–Fab135 and LRP6(1-4)–full length Dkk1 complexes gave excellent SAXS data (Figures 6A, S6). Ab initio reconstructions of envelopes for both complexes (Figure 6B,C) suggested a bent and twisted conformation for LRP6(1-4).
The LRP6(1-4)–Fab complex structure was modeled using the LRP6 repeat structures determined here and a Fab from the Protein Data Bank. No attempt was made to model the ~12 kDa of N-linked carbohydrate on LRP6(1-4). One end of the envelope could easily accommodate the crystal structure of LRP6(3-4), and the other end was readily fit with the Fab model. A homology model of LRP6(1-2) was made from the crystal structure of LRP6(3-4) with MODELLER (Sali and Blundell, 1993). LRP6 repeat 1 was positioned such that Ser243 was located near the antigen combining site of the Fab. Significant changes in the relative positions of repeats 1 and 2, and 2 and 3, were needed to fit the envelope. Some residues that mediate contacts between repeats 3 and 4 are not conserved in the 1-2 and 2-3 interfaces: for example, Phe702, Trp744, and His919 are replaced by much smaller residues, including Gly in the case of His919 (Figures 2B, S1 and Table S2). Morever, a proline present in the repeat 2–3 and 3–4 connections is replaced by glycine in the 1–2 linker. These differences could allow for different relative positions of the repeats than that observed for 3 and 4, and may also indicate that there is some flexibility between these repeats. Because SAXS models are underdetermined, several LRP6(1-4) models were consistent with the data, but all had the same curved, twisted shape (Figure 6B).
The structure of the first 181 residues of Dkk1 is not known, so we could not model the LRP6(1-4)–Dkk1 SAXS data in detail. Nonetheless, the LRP6(1-4) model made for the Fab135 complex fits into the ab initio envelope of the Dkk1 complex (Figure 6C). Superposition of the Dkk1_C–repeat 3 complex coordinates onto repeat 3 of the SAXS model shows Dkk1_C is accomodated in the envelope. Another significant unmodeled volume lies near the amphitheater surface of the repeat 1 propeller. In the SAXS-derived models, the repeat 1 amphitheater is roughly 80 Å away from the N-terminus of Dkk1_C. The linker between the two cysteine-rich N and C-terminal domains of Dkk1, 2 and 4 is approximately 50 residues long (Fig. S2), which could easily span this distance. Also, the Kremen-binding surface of Dkk1_C (Wang et al., 2008) faces outward and would be able to interact with Kremen on the cell surface.
Discussion
Our results, combined with the studies of (Bourhis et al., 2010), indicate that the two conserved regions of Dkk1 can inhibit all Wnts from binding to LRP5/6. LRP5/6 repeat 3, which binds to Dkk1_C, is needed for Dkk1 binding and Wnt1 antagonism (Mao et al., 2001a; Zhang et al., 2004), yet Wnt1 and many other Wnt variants appear to bind the LRP5/6(1-2) region (Gong et al., 2010). Moreover, Dkk1_N by itself does not appear to inhibit signaling (Brott and Sokol, 2002; Mao and Niehrs, 2003). Since Dkk1_N binds to the LRP6(1-2) region, the interaction of Dkk1_C with repeat 3 likely provides a high-affinity anchor that makes Dkk1_N an effective inhibitor of Wnt binding at physiological concentrations, consistent with the 21x stronger binding of Dkk1 to LRP6(1-4) compared to LRP6(1-2) (Bourhis et al., 2010). Signaling by Wnts 1 and 8, which appear to bind LRP6(1-2) (Gong et al., 2010), can be inhibited by Dkk1_C although not as strongly as by full-length Dkk1 (Brott and Sokol, 2002; Mao and Niehrs, 2003). We suggest that interaction with Kremen receptors and receptor internalization, or inhibition of receptor dimerization, mediate these effects (Binnerts et al., 2009; Gong et al., 2010).
The Dkk_N region is not as strongly conserved as Dkk_C (e.g., 40% identity between human Dkk1_N and Dkk2_N, versus 67% for Dkk1_C and Dkk2_C), and studies with deletion and chimeric constructs suggest that the differences between Dkk1 and 2 reside in Dkk_N (Brott and Sokol, 2002). The distinct biological effects of Dkk1, 2, and 4 (Brott and Sokol, 2002; Krupnik et al., 1999) may thus arise from differences in their interactions with the LRP5/6(1-2) region.
Dkks likely inhibit Wnt signaling by directly competing with Wnts for binding sites on LRP5/6. However, HBM mutations in LRP5 diminish Dkk1, but not Wnt, binding to LRP5 (Ai et al., 2005; Balemans et al., 2007), indicating that the Wnt binding site on LRP5/6(1-2) is distinct from this portion of the repeat 1 surface (Figure S5) required for Dkk1_N binding. The other portion of the repeat 1 amphitheater might contribute to the Wnt interface, as well as the repeat 2 propeller. LRP5 missense mutations R494Q, R570W and V667M that cause OPPG and show diminished Wnt signaling (Gong et al., 2001) lie dispersed on the side of the repeat 2 βpropeller barrel. A naturally occurring mutation, R611C in LRP6 repeat 2 impairs Wnt3a-mediated signaling (Mani et al., 2007). This position is equivalent to Val913 and Arg1227 of repeats 3 and 4, which mediate interactions between the β-propeller and EGF-like domains within a repeat and likely stabilize their relative positions. We speculate that these mutations may directly prevent Wnt interactions with repeat 2, or they might alter the receptor conformation so as to make the Wnt binding site inaccessible or prevent receptor clustering.
Deletion of the entire extracellular region of LRP6 results in a constitutively active receptor, indicating that unliganded LRP5/6 is autoinhibited (Liu et al., 2003; Mao et al., 2001a; Mao et al., 2001b). Thus, Dkks may stabilize an autoinhibited conformation, whereas a Wnt might stabilize a distinct structure. The conformation of LRP6 bound to mAb135 should be “active” since the antibody enhances Wnt3a signaling (Binnerts et al., 2009). The LRP6(1-4) model derived from the Fab135 SAXS data appears compatible with the LRP6(1-4)–Dkk1 envelope, so potential conformational differences would appear to be small. Note, however, that the uncertainties of these low-resolution (~40 Å) reconstructions do not allow us to conclude that the receptor conformation the same in both cases.
Wnt signaling appears to depend upon the ability of a particular Wnt to bind simultaneously to LRP5/6 and a Fzd (He et al., 2004; MacDonald et al., 2009). The orientation and distance of the LRP5/6(1-4) region with respect to the membrane cannot be assessed without knowledge of the intervening LDLR-A repeats, but the overall conformation seen in the SAXS model of the Fab complex suggests that repeats 1, 2 and 3 could lie at roughly the same distance from the membrane (Figure 6D). Thus, a Wnt bound to either portion of the LRP5/6 ectodomain could access the Fzd CRD. The overall conformation of the LRP6 ectodomain also suggests that it may be able to engage two Wnt/Fzd complexes simultaneously (Bourhis et al., 2010), but further biophysical and functional studies will be needed to assess this possibility.
Experimental Procedures
Protein constructs, expression and purification
All constructs were cloned into the pACGP67 baculovirus transfer vector (BD Biosciences). A C-terminal His10 tag was added to the C-terminus of each LRP6 construct, and a C-terminal His6 tag was present on the end of the Dkk1 constructs. Sf9 cells were infected with LRP6 or Dkk1 viruses and grown for 72 hours. The cells were pelleted, and the supernatant was adjusted to 1 mM NiSO4, 5 mM CaCl2, 50 mM Tris pH 8.0, filtered, loaded onto a Ni2+-NTA agarose column (Qiagen), and eluted with 50 mM Tris pH 8.0, 300 mM NaCl, 250 mM imidazole.
The LRP6 constructs were further purified on MonoQ (GE Healthcare) in 50 mM Tris pH 8.5, 50 mM NaCl and eluted with a linear NaCl gradient. The eluates were subsequently purified by gel filtration on Superdex 200 (GE Healthcare) in 50 mM Tris pH 8.0, 200 mM NaCl. The buffers for LRP6(1-4) and LRP6(1-2) also included 5% (v/v) glycerol. Dkk1 constructs were purified by gel filtration in the same buffer (full-length on Superdex 200, Dkk1_N and Dkk1_C on Superdex 75). After gel filtration, Dkk1_C was further purified on Hi-Trap HP-SP (GE Healthcare) in 50 mM Tris pH 7.5, 50 mM NaCl and eluted with a linear NaCl gradient. Dkk1 and LRP6(3-4) point mutants were created by site-directed mutagenesis (Quikchange; Stratagene) and purified as their wild-type counterparts.
mAb135 was generated from the hybridoma previously described (Binnerts et al., 2009), and purified by Protein A affinity chromatography. The Fab fragment was prepared by cleaving mAb135 with immobilized papain (Pierce) and separated on a Protein A column.
Isothermal titration calorimetry
ITC data were measured at 25°C in 25 mM Tris, pH 8.0 and 200 mM NaCl, using a Microcal VP-ITC calorimeter. Full-length Dkk1 or Dkk1_C at 200–260 μM were injected into a solution of 25–30 μM LRP6(3-4). Dkk1_N at 30 μM was injected into a solution of 2.8 μM LRP6(1-2). Data were analyzed with the Origin 7 software (Originlab). Measurements of each complex were performed twice and the average values ± standard deviations are shown in the figures.
Crystallization, diffraction data and structure determination
Small crystals were grown by hanging drop vapor diffusion, using 10 mg/ml LRP6(3-4)–Dkk1_C complex in 50 mM Tris pH 8.0 and 200 mM NaCl and a reservior solution containing 10–15% PEG3350, 100 mM Tris pH 8.5 and 100 mM LiSO4 and used for repeated rounds of streak seeding with the same reservior. Crystals were cryoprotected in a solution of mother liquor containing 15–20% glycerol. Diffraction data are summarized in Table 1.
Initial phases were obtained by molecular replacement with PHASER (McCoy et al., 2007), using four copies of the LDLR β-propeller/EGF structure (PDB 1IJQ) (Jeon et al., 2001) as a search model. Density-modified SIRAS phases calculated from a weak two-site HgCl2 derivative using CNS (Brünger et al., 1998) at 4 Å resolution confirmed the location of the four repeats. Significantly, the two Hg sites were located next to the single free cysteine residue in LRP6(3-4), Cys1032, present in each of the two LRP6(3-4) copies. A phase combined map using these SIRAS phases with the molecular replacement phases was examined along with the MR-phased map. The model was built with Coot (Emsley and Cowtan, 2004) and refined with Phenix (Adams et al., 2002) and BUSTER (Blanc et al., 2004). After several rounds of refining the two LRP6(3-4) molecules, interpretable electron density for the single Dkk1_C became evident. Non-crystallographic symmetry restraints were imposed in initial rounds of refinement but dropped in the later stages. Buried surface area calculations were calculated with CNS, and coordinate superpositions were computed in Coot.
Wnt signaling assays
L-cells stably transfected with TOPFLASH and LacZ constructs (LSL cells) were seeded in 96 well plates in DMEM containing 10% FBS. The cells were subsequently treated with 10x diluted Wnt-3a conditioned media and the appropriate Dkk1 and/or LRP6(3-4) construct. Luciferase reporter activity was measured in a Veritas Luminometer (Turner Biosystems). Assays were carried out in triplicate, and relative luciferase units were normalized to LacZ.
Gel filtration and native PAGE binding assays
Purified proteins or protein complexes were incubated with potential binding partners for 1 h at room temperature or overnight at 4°C. For analytical gel filtration, the mixture was applied to a Superdex 200 HR10/30 column (GE Healthcare) in the buffers noted above, and fractions analyzed by SDS-PAGE. For native PAGE, samples were mixed with 5X native gel loading buffer (250 mM Tris-HCl, pH 6.8, 0.5% w/v bromophenol blue, 50% glycerol), loaded into a 4–15% Tris-glycine gel (Bio-Rad) in Tris-glycine running buffer, and run at 100 V at 4°C for 2 h. Gels were analyzed by Coomassie blue staining, or by western blotting with anti-His6 HRP conjugated antibodies (Qiagen) or anti-Dkk1 polyclonal antibodies (R&D systems).
SAXS Data Acquisition and analysis
SAXS data were measured on SSRL beamline 4-2 in the range 0.00965 Å−1 ≤ q ≤ 0.542 A−1 (q = 4πsin(θ)/λ) from solutions of LRP6(1-4)–Fab135 and LRP6(1-4)–Dkk1 complexes at four concentrations between 0.375 and 3 mg/ml, and 0.625 and 5 mg/ml, respectively. Samples were loaded into a 1.5 mm quartz capillary flow cell maintained at 20°C, and 15 x 1 s exposures were measured for each concentration. The raw scattering data were normalized to the incident beam intensity and buffer scattering subtracted. Individual scattering curves were visually inspected prior to averaging to insure that radiation damage was minimal. Scattering curves from different concentrations were scaled and merged with PRIMUS (Konarev et al., 2003) to produce a low-noise curve (Figure 6A).
Ab initio shape determinations were computed with DAMMIF (Franke and Svergun, 2009) using data in the range 0.012 Å−1 ≤ q ≤ 0.15 Å−1. Twelve independent models for each complex were aligned and averaged using SUPCOMB and DAMAVER to minimize the normalized spatial discrepancy (NSD) between runs (Kozin and Svergun, 2001; Volkov and Svergun, 2003). The mean±variation NSD values were 0.83±0.057 and 0.75±0.047 for the Fab135 and Dkk1 complexes, respectively. The averaged models were filtered to remove low occupancy and loosely connected atoms based on the experimentally determined excluded volume of the particle with DAMFILT. PDB entry 1N8Z was used for the Fab model. Scattering amplitudes were calculated from the models with CRYSOL (Svergun et al., 1995).
The molecular mass of the LRP6(1-4)–Dkk1 complex was obtained from I(0) using water as the calibration standard and assuming a protein partial specific volume of 0.7586 cm3/g (Orthaber et al., 2000) (Fig. S6). The calculated protein mass is 167 kDa (140.2 LRP6(1-4) + 26.5 Dkk1). The precise composition of N-linked carbohydrates on the LRP6(1-4) and Dkk1 produced in Sf9 cells is unknown, but the major species seen by mass spectrometry of Sf9 expressed LRP6(3-4), which has 5 N-linked sites, is approximately 6.5 kDa larger than the calculated protein mass (data not shown). Thus, a reasonable estimate for the total mass of carbohydrate in the complex is 13 kDa (10 sites on LRP6(1-4), one on Dkk1; assuming on average 6.5 sugar residues per site (Harrison and Jarvis, 2006)). The mass obtained from I(0) is proportional to (scattering contrast x partial specific volume)−2. The scattering contrast and partial specific volume of carbohydrates are approximately 1.8x and 0.8x that of proteins, respectively (Durschlag, 1989; Koch et al., 2003), so the molecular mass contribution from the carbohydrate will be overestimated by ~6 kDa using protein scattering contrast and partial specific volume values.
Supplementary Material
Highlights.
Crystal structure of a two-repeat fragment of LRP6 bound to the Wnt inhibitor Dkk1
Discrete regions of Dkk1 bind specifically to the two halves of the LRP6 ectodomain
The LRP6 ectodomain adopts a highly curved and twisted conformation
Acknowledgments
We thank Dick Winant for the quantitative Edman sequencing analysis, Kimberly Mulligan and Roel Nusse for providing the LSL cell reporter system, and Tsutomu Matsui and Thomas Weiss for SAXS beamline support and discussions. This work was supported by a Canadian Institutes of Health Research postdoctoral fellowship (V.E.A.), and U.S. National Institutes of Health grants R21AG33596 and R01AG39420 (W.I.W.). Beamlines at SSRL are supported by the U.S. Department of Energy and the National Institutes of Health.
Footnotes
Databases
Coordinates and structure factors for the LRP6(3-4)–Dkk1_C complex have been deposited in the Protein Data Bank, code 3S2K.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- Ai M, Holmen SL, Van Hul W, Williams BO, Warman ML. Reduced affinity to and inhibition by DKK1 form a common mechanism by which high bone mass-associated missense mutations in LRP5 affect canonical Wnt signaling. Mol Cell Biol. 2005;25:4946–4955. doi: 10.1128/MCB.25.12.4946-4955.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bafico A, Liu G, Yaniv A, Gazit A, Aaronson SA. Novel mechanism of Wnt signalling inhibition mediated by Dickkopf-1 interaction with LRP6/Arrow. Nat Cell Biol. 2001;3:683–686. doi: 10.1038/35083081. [DOI] [PubMed] [Google Scholar]
- Balemans W, Devogelaer JP, Cleiren E, Piters E, Caussin E, Van Hul W. Novel LRP5 missense mutation in a patient with a high bone mass phenotype results in decreased DKK1-mediated inhibition of Wnt signaling. J Bone Miner Res. 2007;22:708–716. doi: 10.1359/jbmr.070211. [DOI] [PubMed] [Google Scholar]
- Binnerts ME, Tomasevic N, Bright JM, Leung J, Ahn VE, Kim KA, Zhan X, Liu S, Yonkovich S, Williams J, et al. The first propeller domain of LRP6 regulates sensitivity to DKK1. Mol Biol Cell. 2009;20:3552–3560. doi: 10.1091/mbc.E08-12-1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc E, Roversi P, Vonrhein C, Flensburg C, Lea SM, Bricogne G. Refinement of severely incomplete structures with maximum likelihood in BUSTER-TNT. Acta Crystallogr D. 2004;60:2210–2221. doi: 10.1107/S0907444904016427. [DOI] [PubMed] [Google Scholar]
- Bourhis E, Tam C, Franke Y, Bazan JF, Ernst J, Hwang J, Costa M, Cochran AG, Hannoush RN. Reconstitution of a frizzled8.Wnt3a.LRP6 signaling complex reveals multiple Wnt and Dkk1 binding sites on LRP6. J Biol Chem. 2010;285:9172–9179. doi: 10.1074/jbc.M109.092130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brott BK, Sokol SY. Regulation of Wnt/LRP signaling by distinct domains of Dickkopf proteins. Mol Cell Biol. 2002;22:6100–6110. doi: 10.1128/MCB.22.17.6100-6110.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brünger AT, Adams PD, Clore GM, Gros P, Grosse-Kunstleve RW, Jiang J-S, Kuszewski J, Nilges M, Pannu NS, Read RJ, et al. Crystallography and NMR System (CNS): A new software system for macromolecular structure determination. Acta Cryst. 1998;D54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Chen L, Wang K, Shao Y, Huang J, Li X, Shan J, Wu D, Zhang JJ. Structural insight into the mechanisms of Wnt signaling antagonism by Dkk. J Biol Chem. 2008;283:23364–23370. doi: 10.1074/jbc.M802375200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clevers H. Wnt/β-catenin signaling in development and disease. Cell. 2006;127:469–480. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- Durschlag H. Determination of the partial specific volume of conjugated proteins. Colloid Polym Sci. 1989;267:1139–1150. [Google Scholar]
- Ellies DL, Viviano B, McCarthy J, Rey JP, Itasaki N, Saunders S, Krumlauf R. Bone density ligand, Sclerostin, directly interacts with LRP5 but not LRP5(G171V) to modulate Wnt activity. J Bone Miner Res. 2006;21:1738–1749. doi: 10.1359/jbmr.060810. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Franke D, Svergun DI. DAMMIF, a program for rapid ab-initio shape determination in small-angle scattering. J Appl Crystallogr. 2009;42:342–346. doi: 10.1107/S0021889809000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Y, Bourhis E, Chiu C, Stawicki S, DeAlmeida VI, Liu BY, Phamluong K, Cao TC, Carano RA, Ernst JA, et al. Wnt isoform-specific interactions with coreceptor specify inhibition or potentiation of signaling by LRP6 antibodies. PLoS One. 2010;5:e12682. doi: 10.1371/journal.pone.0012682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Y, Slee RB, Fukai N, Rawadi G, Roman-Roman S, Reginato AM, Wang H, Cundy T, Glorieux FH, Lev D, et al. LDL receptor-related protein 5 (LRP5) affects bone accrual and eye development. Cell. 2001;107:513–523. doi: 10.1016/s0092-8674(01)00571-2. [DOI] [PubMed] [Google Scholar]
- Guder C, Pinho S, Nacak TG, Schmidt HA, Hobmayer B, Niehrs C, Holstein TW. An ancient Wnt-Dickkopf antagonism in Hydra. Development. 2006;133:901–911. doi: 10.1242/dev.02265. [DOI] [PubMed] [Google Scholar]
- Harrison RL, Jarvis DL. Protein N-glycosylation in the baculovirus-insect cell expression system and engineering of insect cells to produce "mammalianized" recombinant glycoproteins. Adv Virus Res. 2006;68:159–191. doi: 10.1016/S0065-3527(06)68005-6. [DOI] [PubMed] [Google Scholar]
- He X, Semenov M, Tamai K, Zeng X. LDL receptor-related proteins 5 and 6 in Wnt/beta-catenin signaling: arrows point the way. Development. 2004;131:1663–1677. doi: 10.1242/dev.01117. [DOI] [PubMed] [Google Scholar]
- Itasaki N, Jones CM, Mercurio S, Rowe A, Domingos PM, Smith JC, Krumlauf R. Wise, a context-dependent activator and inhibitor of Wnt signalling. Development. 2003;130:4295–4305. doi: 10.1242/dev.00674. [DOI] [PubMed] [Google Scholar]
- Jeon H, Meng W, Takagi J, Eck MJ, Springer TA, Blacklow SC. Implications for familial hypercholesterolemia from the structure of the LDL receptor YWTD-EGF domain pair. Nat Struct Biol. 2001;8:499–504. doi: 10.1038/88556. [DOI] [PubMed] [Google Scholar]
- Jiao X, Ventruto V, Trese MT, Shastry BS, Hejtmancik JF. Autosomal recessive familial exudative vitreoretinopathy is associated with mutations in LRP5. Am J Hum Genet. 2004;75:878–884. doi: 10.1086/425080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M, Patel MS, Levasseur R, Lobov I, Chang BH, Glass DA, 2nd, Hartmann C, Li L, Hwang TH, Brayton CF, et al. Cbfa1-independent decrease in osteoblast proliferation, osteopenia, and persistent embryonic eye vascularization in mice deficient in Lrp5, a Wnt coreceptor. J Cell Biol. 2002;157:303–314. doi: 10.1083/jcb.200201089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly OG, Pinson KI, Skarnes WC. The Wnt co-receptors Lrp5 and Lrp6 are essential for gastrulation in mice. Development. 2004;131:2803–2815. doi: 10.1242/dev.01137. [DOI] [PubMed] [Google Scholar]
- Koch MHJ, Vachette P, Svergun DI. Small-angle scattering: a view on the properties, structures and structural changes of biological macromolecules in solution. Q Rev Biophys. 2003;36:147–222. doi: 10.1017/s0033583503003871. [DOI] [PubMed] [Google Scholar]
- Konarev PV, Volkov VV, Sokolova AV, Koch MHJ, Svergun DI. PRIMUS: a windows PC-based system for small-angel scattering data analysis. J Appl Crystallogr. 2003;36:1277–1282. [Google Scholar]
- Kozin MB, Svergun DI. Automated matching of high- and low-resolution structural models. J Appl Cryst. 2001;34:33–41. [Google Scholar]
- Krupnik VE, Sharp JD, Jiang C, Robison K, Chickering TW, Amaravadi L, Brown DE, Guyot D, Mays G, Leiby K, et al. Functional and structural diversity of the human Dickkopf gene family. Gene. 1999;238:301–313. doi: 10.1016/s0378-1119(99)00365-0. [DOI] [PubMed] [Google Scholar]
- Li L, Mao J, Sun L, Liu W, Wu D. Second cysteine-rich domain of Dickkopf-2 activates canonical Wnt signaling pathway via LRP-6 independently of dishevelled. J Biol Chem. 2002;277:5977–5981. doi: 10.1074/jbc.M111131200. [DOI] [PubMed] [Google Scholar]
- Li X, Liu P, Liu W, Maye P, Zhang J, Zhang Y, Hurley M, Guo C, Boskey A, Sun L, et al. Dkk2 has a role in terminal osteoblast differentiation and mineralized matrix formation. Nat Genet. 2005a;37:945–952. doi: 10.1038/ng1614. [DOI] [PubMed] [Google Scholar]
- Li X, Zhang Y, Kang H, Liu W, Liu P, Zhang J, Harris SE, Wu D. Sclerostin binds to LRP5/6 and antagonizes canonical Wnt signaling. J Biol Chem. 2005b;280:19883–19887. doi: 10.1074/jbc.M413274200. [DOI] [PubMed] [Google Scholar]
- Liu CC, Pearson C, Bu G. Cooperative folding and ligand-binding properties of LRP6 beta-propeller domains. J Biol Chem. 2009;284:15299–15307. doi: 10.1074/jbc.M807285200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G, Bafico A, Harris VK, Aaronson SA. A novel mechanism for Wnt activation of canonical signaling through the LRP6 receptor. Mol Biol Cell. 2003;23:5825–5835. doi: 10.1128/MCB.23.16.5825-5835.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan CY, Nusse R. The Wnt signaling pathway in development and disease. Annu Rev Cell Dev Biol. 2004;20:781–810. doi: 10.1146/annurev.cellbio.20.010403.113126. [DOI] [PubMed] [Google Scholar]
- MacDonald BT, Tamai K, He X. Wnt/beta-catenin signaling: components, mechanisms, and diseases. Dev Cell. 2009;17:9–26. doi: 10.1016/j.devcel.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mani A, Radhakrishnan J, Wang H, Mani MA, Nelson-Williams C, Carew KS, Mane S, Najmabadi H, Wu D, Lifton RP. LRP6 mutation in a family with early coronary disease and metabolic risk factors. Science. 2007;315:1278–1282. doi: 10.1126/science.1136370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao B, Niehrs C. Kremen2 modulates Dickkopf2 activity during Wnt/LRP6 signaling. Gene. 2003;302:179–183. doi: 10.1016/s0378-1119(02)01106-x. [DOI] [PubMed] [Google Scholar]
- Mao B, Wu W, Davidson G, Marhold J, Li M, Mechler BM, Delius H, Hoppe D, Stannek P, Walter C, et al. Kremen proteins are Dickkopf receptors that regulate Wnt/beta-catenin signalling. Nature. 2002;417:664–667. doi: 10.1038/nature756. [DOI] [PubMed] [Google Scholar]
- Mao B, Wu W, Li Y, Hoppe D, Stannek P, Glinka A, Niehrs C. LDL-receptor-related protein 6 is a receptor for Dickkopf proteins. Nature. 2001a;411:321–325. doi: 10.1038/35077108. [DOI] [PubMed] [Google Scholar]
- Mao J, Wang J, Liu B, Pan W, Farr GH, 3rd, Flynn C, Yuan H, Takada S, Kimelman D, Li L, et al. Low-density lipoprotein receptor-related protein-5 binds to Axin and regulates the canonical Wnt signaling pathway. Mol Cell. 2001b;7:801–809. doi: 10.1016/s1097-2765(01)00224-6. [DOI] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay M, Gorivodsky M, Shtrom S, Grinberg A, Niehrs C, Morasso MI, Westphal H. Dkk2 plays an essential role in the corneal fate of the ocular surface epithelium. Development. 2006;133:2149–2154. doi: 10.1242/dev.02381. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay M, Shtrom S, Rodriguez-Esteban C, Chen L, Tsukui T, Gomer L, Dorward DW, Glinka A, Grinberg A, Huang SP, et al. Dickkopf1 is required for embryonic head induction and limb morphogenesis in the mouse. Dev Cell. 2001;1:423–434. doi: 10.1016/s1534-5807(01)00041-7. [DOI] [PubMed] [Google Scholar]
- Niehrs C. Function and biological roles of the Dickkopf family of Wnt modulators. Oncogene. 2006;25:7469–7481. doi: 10.1038/sj.onc.1210054. [DOI] [PubMed] [Google Scholar]
- Orthaber D, Bergmann A, Glatter O. SAXS experiments on absolute scale with Kratky systems using water as a secondary standard. J Appl Crystallogr. 2000;33:218–225. [Google Scholar]
- Pinson KI, Brennan J, Monkley S, Avery BJ, Skarnes WC. An LDL-receptor-related protein mediates Wnt signalling in mice. Nature. 2000;407:535–538. doi: 10.1038/35035124. [DOI] [PubMed] [Google Scholar]
- Qin M, Hayashi H, Oshima K, Thaira T, Hayashi K, Kondo H. Complexity of the genotype-phenotype correlation in familial exudative vitreoretinopathy with mutations in the LRP5 and/or FZD4 genes. Human Mutation. 2005;26:104–112. doi: 10.1002/humu.20191. [DOI] [PubMed] [Google Scholar]
- Reya T, Clevers H. Wnt signalling in stem cells and cancer. Nature. 2005;434:843–850. doi: 10.1038/nature03319. [DOI] [PubMed] [Google Scholar]
- Rudenko G, Henry L, Henderson K, Ichtchenko K, Brown MS, Goldstein JL, Deisenhofer J. Structure of the LDL receptor extracellular domain at endosomal pH. Science. 2002;298:2353–2358. doi: 10.1126/science.1078124. [DOI] [PubMed] [Google Scholar]
- Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- Semenov MV, Zhang X, He X. DKK1 antagonizes Wnt signaling without promotion of LRP6 internalization and degradation. J Biol Chem. 2008;283:21427–21432. doi: 10.1074/jbc.M800014200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svergun D, Barberato C, Koch MHJ. CRYSOL: a program to evaluate x-ray solution scattering of biological macromolecules from atomic coordinates. J Appl Cryst. 1995;28:768–773. [Google Scholar]
- Takagi J, Yang Y, Liu JH, Wang JH, Springer TA. Complex between nidogen and laminin fragments reveals a paradigmatic beta-propeller interface. Nature. 2003;424:969–974. doi: 10.1038/nature01873. [DOI] [PubMed] [Google Scholar]
- Toomes C, Bottomley HM, Jackson RM, Towns KV, Scott S, Mackey DA, Craig JE, Jiang L, Yang Z, Trembath R, et al. Mutations in LRP5 or FZD4 underlie the common familial exudative vitreoretinopathy locus on chromosome 11q. Am J Hum Genet. 2004;74:721–730. doi: 10.1086/383202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkov VV, Svergun DI. Uniqueness of ab initio shape determination in small-angle scattering. J Appl Crystallogr. 2003;36:860–864. doi: 10.1107/S0021889809000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Zhang Y, Li X, Chen L, Wang H, Wu J, Zheng J, Wu D. Characterization of the Kremen-binding site on Dkk1 and elucidation of the role of Kremen in Dkk-mediated Wnt antagonism. J Biol Chem. 2008;283:23371–23375. doi: 10.1074/jbc.M802376200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehrli M, Dougan ST, Caldwell K, O'Keefe L, Schwartz S, Vaizel-Ohayon D, Schejter E, Tomlinson A, DiNardo S. arrow encodes an LDL-receptor-related protein essential for Wingless signalling. Nature. 2000;407:527–530. doi: 10.1038/35035110. [DOI] [PubMed] [Google Scholar]
- Yasui N, Mihara E, Nampo M, Tamura-Kawakami K, Unno H, Matsumoto K, Takagi J. Detection of endogenous LRP6 expressed on human cells by monoclonal antibodies specific for the native conformation. J Immunol Methods. 2010;352:153–160. doi: 10.1016/j.jim.2009.11.016. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Wang Y, Li X, Zhang J, Mao J, Li Z, Zheng J, Li L, Harris SE, Wu D. The LRP5 High-Bone-Mass G171V mutation disrupts LRP5 interaction with Mesd. Mol Cell Biol. 2004;24:4677–4684. doi: 10.1128/MCB.24.11.4677-4684.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.