Abstract
Iron acquisition in vivo by Actinobacillus pleuropneumoniae depends upon a functional TonB system. Tonpitak et al. (W. Tonpitak, S. Thiede, W. Oswald, N. Baltes, and G.-F. Gerlach, Infect. Immun. 68:1164-1170, 2000) have described one such system, associated with tbpBA encoding the transferrin receptor, and here we report a second, termed tonB2. This gene cluster (exbB2-exbD2-tonB2) is highly homologous to those in other Pasteurellaceae, unlike the earlier system described (now termed tonB1), suggesting that it is the indigenous system for this organism. Both tonB2 and tonB1 are upregulated upon iron restriction. TonB2, but not TonB1, was found to be essential for growth in vitro when the sole source of iron was hemin, porcine hemoglobin, or ferrichrome. In the case of iron provided as iron-loaded porcine transferrin, neither tonB mutant was viable. The tonB1 phenotype could be explained by a polar effect of the mutation on transcription of downstream tbp genes. We propose that TonB2 is crucial for the acquisition of iron provided in this form, interacting with accessory proteins of the TonB1 system that have been demonstrated to be necessary by Tonpitak et al. TonB2 appears to play a much more important role in A. pleuropneumoniae virulence than TonB1. In an acute porcine infection model, the tonB2 mutant was found to be highly attenuated, while the tonB1 mutant was not. We hypothesize that acquisition of the tonB1-tbp gene cluster confers a biological advantage through its capacity to utilize transferrin-iron but that TonB1 itself plays little or no part in this process.
All forms of life need iron. The element is a cofactor in a wide range of biological reactions in both eukaryotes and prokaryotes, but free iron is toxic, so it is sequestered in a variety of ways to ensure that it is readily available when needed. Mammals reduce the availability of iron to potential pathogens by the use of very-high-affinity iron-chelating molecules, such as lactoferrin, transferrin, and hemoglobin. Host-adapted pathogens have accordingly evolved means to use these iron-bearing molecules as an iron source, as well as in some cases to synthesize small-molecule chelators of their own (siderophores), which are secreted, trap iron, and are transported back into the cell. However, whether the bacteria use host chelators or their own siderophores, energy is required to transport the iron into the cell. This energy is generated by the proton motive force of the cytoplasmic membrane and is made available to proteins in the outer membrane by the action of the energy transducing protein TonB. This protein acts in concert with the products of associated genes exbB and exbD, together forming what is commonly referred to as the TonB system. The two exb genes encode integral cytoplasmic membrane proteins which anchor TonB in the periplasm, while tonB encodes the energy transducing protein, which spans the periplasmic space as a dimer (7) to interact with high-affinity outer membrane receptors. Upon receptor-ligand interaction, a series of conformational changes occur in the TonB-receptor complex, energy from the cytoplasmic membrane is transduced to the outer membrane receptors, and iron is transported into the cell. Reflecting the importance of iron to the survival of bacterial pathogens within the host environment, TonB has been shown to be essential for virulence in diverse organisms (24, 25, 32, 35).
Actinobacillus pleuropneumoniae causes porcine pleuropneumonia, a highly infectious disease of swine that results in large economic losses worldwide. A tonB system has been identified by Tonpitak et al. (34) immediately upstream of tbpB and tbpA, which encode the transferrin receptor. This cluster of genes has been shown to be required for virulence (2). In a mutagenesis study of A. pleuropneumoniae virulence reported elsewhere (29), two attenuated strains were identified, with mutations in what appeared to be different versions of a tonB gene: one was the gene identified by Tonpitak et al. (34) and the second was a novel gene. Further sequencing of the new locus revealed associated second copies of exbB and exbD. We have named the new system tonB2 and refer to the originally described gene cluster as the tonB1 system. In this paper, we describe the tonB2 system of A. pleuropneumoniae and explore the relative contribution of tonB1 and tonB2 to bacterial biology and virulence.
MATERIALS AND METHODS
Bacterial strains, plasmids, and primers.
The bacterial strains, plasmids, and primers used in this study are shown in Table 1.
TABLE 1.
Bacterial strains, plasmids, and primers
| Strain, plasmid, or primer | Description or sequencea | Source or reference |
|---|---|---|
| Strains | ||
| E. coli TOP10 | Invitrogen | |
| E. coli XL-1 GOLD | Stratagene | |
| E. coli S17-1 | 15 | |
| A. pleuropneumoniae 4074 | Serotype 1, Nalr | 13 |
| A. pleuropneumoniae 27A12 | A. pleuropneumoniae 4074 with mini-Tn10 transposon interrupting tonB1 | 29 |
| A. pleuropneumoniae 0F6 | A. pleuropneumoniae 4074 with mini-Tn10 transposon interrupting tonB2 | 29 |
| Plasmids | ||
| pBR322 | E. coli cloning vector (Ampr) | New England Biolabs |
| pBluescript KS | E. coli cloning vector (Ampr) | Stratagene |
| pJFF244-NX | E. coli-A. pleuropneumoniae shuttle vector | 9 |
| pF6 | pBR322 plus a 5-kb EcoRI-SphI fragment from A. pleuropneumoniae tonB2::Kan | This study |
| pB1 | pF6 with a 2-kb deletion of the BamHI fragment from the mini-Tn10 transposon | This study |
| pLURO1 | pBR322 plus a 2.5-kb PCR product from A. pleuropneumoniae 4074 (tonB2) | This study |
| pLURO2 | pBluescript KS plus a 2.2-kb PCR product from A. pleuropneumoniae 4074 (tonB1) | This study |
| pLURO3 | pJFF244-NX plus a 2.5-kb PCR product from A. pleuropneumoniae 4074 (tonB1) | This study |
| pCPCM5164 | Cosmid containing approximately 30 kb of A. pleuropneumoniae cm5 (serotype 1) DNA | 5 |
| Primers | ||
| tonB5EcoRI | CAGAAT TCCGGCAGCGACTAAACT TTC C | This study |
| tonB6BamHI | TAGGATCCGCACATGACG TAGAC | This study |
| tonB10 | CCGGAAGTGAAATCGGTGC | This study |
| tonB11 | CCACCATTTCCACTA TCG | This study |
| tonB16 | CGCCTTAATCGGTTT ATC | This study |
| tonB18 | AATTTCACCGGAACG GTC | This study |
| tonBGER1 | CTTGGTGCTGGTTAT GGC | This study |
| tonBGER4 | GAAAGTTACACTGCC TAC | This study |
| tonBGER16XbaI | GCTCTAGACGTCATCAACTTAGTCGTGCC | This study |
| tonBGER17SalI | CGCGTCGACCTATTTTCGTTAGCCCCG | This study |
| tbpB1 | GCTTGCTGTAGTAATCTGGA | This study |
| tbpB2 | GTTGGACCATAGAAGCCACC | This study |
Underlined sequences identify restriction sites.
Media and growth conditions.
Escherichia coli strains were grown in Luria-Bertani broth, supplemented with antibiotics when appropriate (kanamycin, 100 μg/ml; ampicillin, 100 μg/ml; and chloramphenicol, 25 μg/ml), at 37°C. A. pleuropneumoniae strains were grown on brain heart infusion (BHI) plates supplemented with 10% Levinthal's base or in BHI broth supplemented with 0.01% β-NAD at 37°C. Selection of A. pleuropneumoniae was achieved by using chloramphenicol (2 μg/ml), kanamycin (50 μg/ml), or nalidixic acid (20 μg/ml) where appropriate. For iron restriction in broth cultures, 1,10-phenanthroline was added to a final concentration of 30 μM.
DNA manipulations and analysis.
Chromosomal DNA, plasmids, and RNA were extracted by use of the appropriate Qiagen kit. The QiaEasy DNase kit was also used to remove contaminating DNA during RNA extraction. DNA was digested and ligated with enzymes and reagents supplied by Roche Molecular Biochemicals, following the manufacturer's protocols. E. coli strains were transformed by standard methods. For Southern hybridization (27), a 250-bp probe was prepared based on the published partial sequence of tonB1 from A. pleuropneumoniae (GenBank accession number Y17916). Labeling was achieved through digoxigenin-11-dUTP incorporation during PCR with primers tonBGER1 and tonBGER4 (Table 1). Oligonucleotide primers for sequencing were manufactured by MWG-Biochem and sequencing was performed in an ABI 2000 sequencer.
Iron utilization bioassay.
A. pleuropneumoniae strains were grown to an optical density at 600 nm of 0.6, and 100 μl of culture was plated on BHI supplemented with 0.01% NAD and 200 μM ethylenediamine di(o-hydroxyphenylacetic) acid to give a lawn of 108 CFU. Paper disks loaded with phosphate-buffered saline (PBS) (10 μl), hemin (10 μl; 10 mg/ml), porcine hemoglobin (10 μl; 10 mg/ml), porcine transferrin (pTf) (75 μl; 40 mg/ml), or Fe(NO3)3 (10 μl; 500 μM) were placed onto the lawn. For testing of ferrichrome utilization, a 5-μl drop of ferrichrome solution (100 μM) was placed directly onto the bacterial lawn. Plates were incubated overnight at 37°C.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis and immunoblotting.
Whole-cell lysates of 5 × 107 CFU from wild-type and tonB1 mutant organisms cultured under iron-replete and iron-restricted conditions were boiled for 10 min in sample buffer containing β-mercaptoethanol and were separated in sodium dodecyl sulfate-10% polyacrylamide gels. Proteins were then transferred to a nitrocellulose membrane (Hybond-C Extra; Amersham Pharmacia) by electroblotting. Blots were blocked in 3% blocking solution (PBS containing 0.05% Tween 20 and 3% skim milk) for 1 h and washed three times (one time for 15 min and two times for 5 min each) in PBS containing 0.05% Tween 20 (PBS-T). The blots were then incubated with monoclonal antibody (MAb) 1.48 (4) at a concentration of 1 μg/ml in blocking solution for 1 h, followed by washing in PBS-T as described above. The secondary antibody, anti-mouse immunoglobulin G conjugated to horseradish peroxidase (DAKO), was diluted 1:500 in PBS-T, and the blots were incubated with it for 1 h and then washed with PBS-T (one time for 15 min and four times for 5 min each). Blots were developed by using the ECL-Plus system (as described by the manufacturer) and were exposed to ECL-Hyperfilm (Amersham Pharmacia).
PCR and RT-PCR.
PCR was carried out by using standard methods (27). Reverse transcription (RT)-PCR was achieved by using the OneStep RT-PCR kit (Qiagen) according to the manufacturer's instructions. Approximately 2 μg of RNA was used in each reaction for tonB2 assays, 300 pg was used for tonB1 assays, and 30 to 250 ng was used for tbpB assays. Negative control experiments to test for the presence of contaminating DNA were performed by incubating the RT-PCR mixture (prior to the addition of enzyme) with 4 μl of RNase A (100 mg/ml) for 15 min at 37°C or by using HotStart Taq (Qiagen) instead of the supplied enzyme mixture, thus eliminating the reverse transcriptase step.
Construction and manipulation of plasmids.
tonB1 was identified in a cosmid library of A. pleuropneumoniae serotype 1 by using a probe based on the published partial sequence (34), and tonB1 and associated genes were sequenced directly from the clone. Oligonucleotide primers tonBGER16XbaI and tonBGER17SalI were designed from this sequence and used to amplify a 2.2-kb fragment containing tonB1, exbB1, and part of exbD1 which was cloned into pBluescript KS (+) as pLURO1.
The flanking regions of tonB2 were cloned from A. pleuropneumoniae 0F6 (tonB2 mutant) into pF6, and pB1 was derived from this by removal of the inactivating kanamycin resistance cassette. For cloning of the tonB2 system (exbB2-exbD2-tonB2), oligonucleotide primers tonB5BamHI and tonB6EcoRI were designed and used in a PCR to produce a 2.5-kb fragment which was cloned into pBR322 as pLURO2.
For construction of a plasmid to complement the tonB1 mutant A. pleuropneumoniae 27A12, a 2-kb fragment containing tonB1 and exbB1 was amplified by PCR from wild-type A. pleuropneumoniae 4074 by using oligonucleotide primers tonBGER16XbaI and tonBGER17SalI and was cloned into the shuttle vector pJFF224-NX (9), resulting in plasmid pLURO3. pLURO3 was transformed into E. coli strain S17-1 and transferred into A. pleuropneumoniae 27A12 (tonB1 mutant) and A. pleuropneumoniae 0F6 (tonB2 mutant) by conjugation.
Determination of competitive indices and virulence studies.
For in vitro competitive growth experiments, mutant and wild-type strains taken from plates incubated overnight at 37°C were resuspended in 0.5 ml of PBS and used to establish 5-ml starter cultures in BHI-NAD broth. From these, a 10-ml mixed culture was set up, containing 107 CFU of each strain. Bacteria were enumerated at time zero and at 3 h by plating onto selective (for mutant organisms) and nonselective (for mutant plus wild-type organisms) media, and the competitive index (CI) was calculated by dividing the ratio of mutants to wild type in the output by the ratio of mutants to wild type in the input. A CI of 1 indicates no attenuation, while CIs of <0.2 were considered to reflect attenuation.
For an examination of the relative virulence of the mutants compared to the wild-type parent, tonB mutants and wild-type A. pleuropneumoniae were grown in Columbia broth with 5 μg of NAD/ml and 11 mM CaCl2. At an optical density at 600 nm of 0.3, bacteria were washed and diluted 1:50 in sterile HEPES saline (10 mM HEPES, 150 mM NaCl, 3 mM KCl, 1 mM CaCl2; pH 7.3), and suspensions were combined to a final concentration of approximately 3 × 106 CFU of each strain per ml. Two specific-pathogen-free, large White Cross piglets (7 weeks old) from a herd known to be free of A. pleuropneumoniae were anesthetized with 3 ml of Saffan (alfaxalone and alfadolone acetate; Schering-Plough Animal Health) administered intravenously into the jugular vein. Three milliliters (approximately 2 × 107 CFU in total) of bacterial suspension was inoculated directly into the trachea at the midpoint between the base of the larynx and the anterior point of the sternum. This dose represents ∼100 times the 50% infective dose for this model system (30). The animals were then allowed to recover and observed closely for signs of disease. Those which developed respiratory symptoms were humanely killed with pentobarbitone. Surviving animals were sacrificed 24 h after infection. At necropsy, lungs were evaluated for degree of pathology by macroscopic evidence of superficial fibrin deposition, palpable consolidation, hemorrhage, and necrosis. Experimental infection with virulent strains typically results in 25 to 75% lung involvement. For harvesting of bacteria from the lungs, the whole organ was homogenized in Hank's balanced salts solution. Serial dilutions were plated onto selective and nonselective media for enumeration. The CI was determined as described above.
Nucleotide sequence accession numbers.
DNA sequences containing the tonB1 gene and its associated promoter region and the tonB2 system and flanking genes have been submitted to GenBank (accession numbers AY428646 and AY428647).
RESULTS
Sequence analysis of the tonB2 system.
Three kilobases of DNA flanking the mutagenizing cassette in the newly identified tonB2 was cloned into plasmids pB1 and pLURO1 and sequenced. Seven open reading frames (ORFs) were identified, six of which encode products that closely match entries in GenBank (Fig. 1). Three of these genes were identified as exbB (encoding a deduced 150-amino-acid [aa] protein), exbD (encoding 129 aa), and tonB (encoding 285 aa), and their deduced products were 86, 85, and 62% identical to products of Haemophilus ducreyi exbB, exbD, and tonB (GenBank accession numbers O51808, O51809, and O51810, respectively). However, the deduced sequence of the product of tonB2 showed little similarity to that of TonB1 described by Tonpitak et al. (34). All of tonB1 was sequenced, using plasmids pLURO2 and pCPCM5164, and its encoded product was found to have only 18% identity to TonB2 (Fig. 2). BLAST analysis of TonB1 (246 aa) showed the protein to be most similar to TonB of Neisseria meningitidis (GenBank accession number NP 274733), with 31% identity.
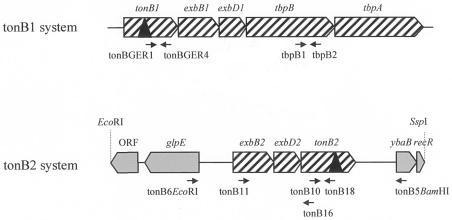
FIG. 1.
Genetic organization of two tonB systems in A. pleuropneumoniae, with tonB1 (as described by Tonpitak et al. [34]) shown at the top and tonB2 shown at the bottom. Hatched boxes represent genes in the same operon, while filled boxes represent other ORFs. Black triangles indicate the sites of insertional mutations in A. pleuropneumoniae strains 27A12 and 0F6. Arrows below the gene clusters represent the approximate binding positions of the indicated oligonucleotide primers.
FIG. 2.
Alignment of A. pleuropneumoniae TonB1 and TonB2 showing amino acid identities (lines) and biochemically conservative substitutions (dots). The identity for the proteins was 18%, and the similarity was 25%.
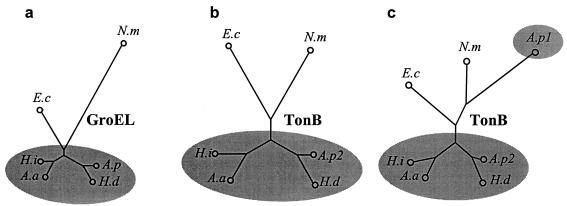
The multiple sequence comparison program UnrootedTree (http://cbrg.inf.ethz.ch/server/MultAlign.html) was then used to graphically display the relationship between the sequences of TonB from A. pleuropneumoniae, Actinobacillus actinomycetemcomitans, Haemophilus influenzae, H. ducreyi, E. coli, and N. meningitidis. A similar analysis can be carried out for any set of homologous proteins, and for comparison, one is shown for the chaperone GroEL. These phylogenetic trees are drawn on the principle that the length of the path joining loci representing protein sequences is in proportion to their degree of dissimilarity. Short and long distances thus signify, respectively, similar or divergent sequences. Figure 3 displays the relationships for these proteins. If TonB1 of A. pleuropneumoniae is excluded from the initial analysis, trees of very similar morphology arise (Fig. 3a and b), reflecting fundamental relationships between these organisms. Figure 3c includes the A. pleuropneumoniae TonB1 sequence, which is seen to be highly divergent, suggesting that it is this gene rather than tonB2 that has been acquired by lateral transfer.
FIG. 3.
Schematic trees displaying the phylogenetic relatedness of protein sequences. (a) GroEL; (b) TonB (without A. pleuropneumoniae TonB1); (c) TonB (with A. pleuropneumoniae TonB1). For a full explanation, see the text. Sequences were obtained from GenBank or this work for the organisms. Abbreviations: A.a, A. actinomycetemcomitans; A.p, A. pleuropneumoniae; E.c, E. coli; H.i, H. influenzae; H.d, H. ducreyi; and N.m, N. meningitidis; A.p1, TonB1; A.p2, TonB2. Line lengths representing phylogenetic relatedness are to scale within but not between trees. The shaded areas enclose sequences from Pasteurellaceae.
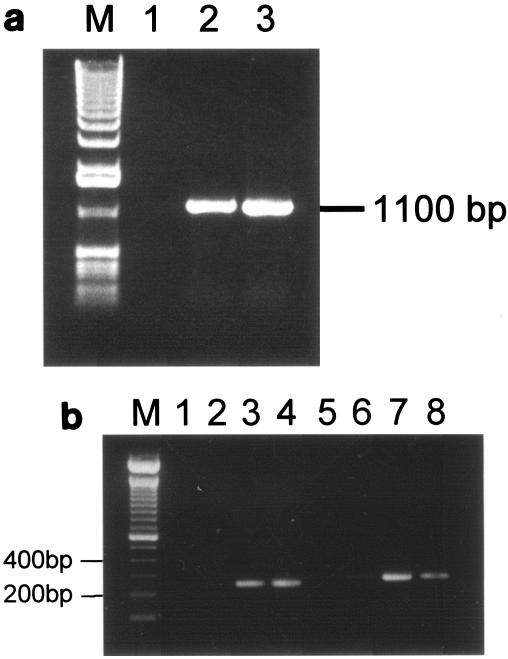
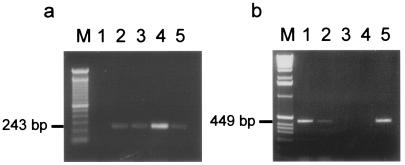
The genes of the tonB1 system, namely tonB1, exbB1, and exbD1, are transcriptionally linked to the transferrin binding proteins encoded by tbpB and tbpA (34). In contrast, an analysis of the sequence surrounding the tonB2 system failed to identify any genes that are putatively involved in iron metabolism. Immediately upstream of exbB2, carried on the opposite strand, lies an ORF that is 62% identical to glpE, the product of which is part of the sn-glycerol-3-phosphate (glp) regulon. Downstream of tonB2, carried on the same strand, is an ORF encoding a deduced product that is 92% identical to that of H. influenzae ybaB, a putative protein of unknown function encoded by a conserved gene found adjacent to recR in various genomes (e.g., E. coli, Clostridium perfringens, Mycobacterium tuberculosis, Vibrio cholerae, and Pasteurella multocida). We have found recR immediately downstream of ybaB in A. pleuropneumoniae also. The exbB2, exbD2, and tonB2 genes are positioned closely together, with just 44 bases separating exbB2 from exbD2 and 9 bases separating exbD2 from tonB2, suggesting an operon formation. To determine if the three genes are indeed cotranscribed, we performed RT-PCR analysis using primers tonB11 and tonB16, specific to the exbB2 and tonB2 genes, respectively. The resulting product of 1,100 bp confirmed that all three genes are present on the same mRNA transcript (Fig. 4a).
FIG. 4.
RT-PCR analysis. (a) Products of RT-PCR of wild-type A. pleuropneumoniae grown under iron-restricted conditions, with primers tonB11 (exbB2) and tonB16 (tonB2) and templates as follows. Lane 1, total RNA treated with RNase A (negative control); lane 2, total RNA; lane 3, chromosomal DNA (positive control). M, size marker. (b) RT-PCR analysis of tonB expression in wild-type A. pleuropneumoniae, with primers tonBGER1 and tonBGER4 for tonB1 (lanes 1 to 4) and primers tonB10 and tonB18 for tonB2 (lanes 5 to 8), to generate bands between 200 and 400 bp as indicated. Templates were as follows. Lanes 1 and 5, RNA treated with RNase A, extracted from bacteria grown under iron-restricted conditions (negative control); lanes 2 and 6, RNA from bacteria grown under iron-replete conditions; lanes 3 and 7, RNA from bacteria grown under iron-restricted conditions; lanes 4 and 8, whole cellular DNA (positive control).
Presence of tonB1 and tonB2 in 14 serotypes of A. pleuropneumoniae.
Tonpitak et al. (34) showed previously that the tonB1 system is present in 12 serotypes of A. pleuropneumoniae. We have used PCR to confirm this (data not shown), extending the analysis to strains of serotypes 1 to 14 (12, 13, 16, 17, 18, 19, 20, 21, 26), and to show that the tonB2 system is also present in all 14 serotypes. The primers used were tonBGER1 and tonBGER4 (specific for tonB1) and tonB10 and tonB18 (specific for tonB2) (Fig. 1).
Both tonB1 and tonB2 are upregulated during iron restriction.
RT-PCR analysis of the tonB1 system by Tonpitak et al. (34) revealed it to be upregulated under iron-restricted conditions. To investigate the regulation of the tonB2 system, we performed RT-PCR on RNA from iron-replete and iron-restricted cultures of A. pleuropneumoniae 4074. Expression of tonB2 was only seen for the iron-restricted cultures (Fig. 4b). The same was observed for tonB1, but whereas 2 μg of total RNA was required to obtain a RT-PCR product from tonB2, only 0.015% of this amount (300 pg) was required to obtain a product with tonB1.
Role of TonB2 and TonB1 in the uptake of iron from hemin, porcine transferrin, and porcine hemoglobin.
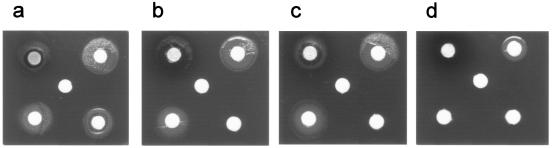
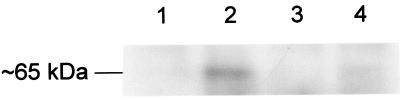
Initial experiments to define the role of TonB2 in iron acquisition were conducted in iron-restricted broth. While the wild-type strain grew under iron-restricted conditions, albeit slowly, the tonB2 strain did not grow at all. However, the addition of iron (III) nitrate to both cultures restored growth to the levels seen under iron-replete conditions (data not presented). To explore this further, we plated A. pleuropneumoniae strains 4074 (wild type), 27A12 (tonB1 mutant), and 0F6 (tonB2 mutant) onto iron-restricted medium, with iron-loaded pTf (Fe-pTf), porcine hemoglobin, or hemin provided as the sole iron source. The tonB2 mutant failed to grow under any of these conditions. The tonB1 mutant failed to grow when supplied with Fe-pTf but grew normally when supplied with either hemoglobin or hemin (Fig. 5). Plasmid pLURO3 (providing tonB1 and exbB1) was unable to complement either tonB mutation in the plate assays (Fig. 5 and data not presented), despite RT-PCR evidence of transcription of the plasmid tonB1 gene at a high level (Fig. 6a). While this indicates that TonB1 cannot substitute functionally for TonB2, alternative explanations may be offered for the tonB1 phenotype: either the gene does not encode an active TonB, the mutation in chromosomal tonB1 has a polar effect, or both. By using quantitative RT-PCR, tonB1 DNA immediately downstream of the Kanr (aph3′) cassette was shown to be transcribed at the same level as in the wild type (Fig. 6a). However, when primers tbpB1 and tbpB2 (Fig. 1) were used in quantitative RT-PCRs with RNAs extracted from cultures of wild-type and tonB1 mutant strains grown under iron-restricted conditions, the expression of tbpB was shown to be greatly decreased in the tonB1 mutant (Fig. 6b). This result was confirmed by further experiments with serially diluted template (data not shown) and was substantiated by Western blotting. MAb 1.48 was used to detect TbpB in wild-type and tonB1 mutant strains. A substantial band of reactivity was only seen for the wild-type strain (Fig. 7). The tonB1 mutation clearly does significantly attenuate the transcription of downstream genes.
FIG. 5.
Zones of bacterial growth on iron-restricted solid medium around disks impregnated with different iron sources (clockwise from top left) as follows: hemin, Fe(NO3)3, Fe-pTf, porcine hemoglobin, and PBS (center). Panels show growth of wild-type A. pleuropneumoniae (a), the tonB1 mutant strain (b), the tonB1 mutant strain transformed with pLURO3 (c), and the tonB2 mutant strain (d).
FIG. 6.
RT-PCR analysis of tonB1 and tbpB expression. (a) RT-PCR analysis of tonB1 expression using primers tonBGER1 and tonBGER4. Lanes contain products from RNA templates under different growth conditions as follows: 1, wild type, iron replete; 2, wild type, iron restricted; 3, tonB1 mutant, iron restricted; 4, tonB1::pLURO3, iron restricted; 5, tonB2 mutant, iron restricted. M, size marker. (b) RT-PCR analysis of tbpB expression in A. pleuropneumoniae grown under conditions of iron restriction, using primers tbpB1 and tbpB2. Lanes contain products from RNA or DNA templates as follows: 1, wild-type RNA; 2, tonB1 mutant RNA; 3, wild-type RNA treated with RNase A (negative control); 4, tonB1 mutant RNA treated with RNase A (negative control); 5, wild-type DNA (positive control).
FIG. 7.
Immunoblotting analysis of wild-type and tonB1 mutants probed with MAb 1.48. Lanes contain separated whole-cell lysates of strains under different growth conditions as follows: 1, wild type, iron replete; 2, wild type, iron restricted; 3, tonB1 mutant, iron replete; 4, tonB1 mutant, iron restricted. The position of TbpB is indicated.
TonB2 is the energy coupling mechanism for ferrichrome uptake.
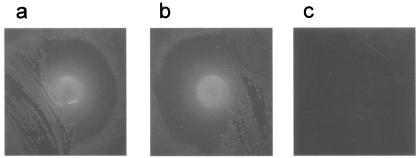
Baltes and colleagues have recently investigated ferrichrome-mediated iron uptake by A. pleuropneumoniae via the siderophore receptor FhuA (3). They found that a nonpolar mutation in the exbB1 gene did not affect this phenotype, from which we can infer that the tonB1 system is not involved. To test if the tonB2 system was involved, we applied ferrichrome to bacterial lawns grown under iron-restricted conditions as described above. Both the wild-type and tonB1 mutant strains were able to utilize ferrichrome, but the tonB2 mutant was not (Fig. 8), indicating the importance of the tonB2 system in ferrichrome uptake. All three strains were able to utilize Fe(NO3)3 equally well (results not shown).
FIG. 8.
Bacterial growth visualized as colonies on the dark background of ferrichrome-stained agar, demonstrating ferrichrome utilization by A. pleuropneumoniae strains. (a) Wild type; (b) tonB1 mutant; (c) tonB2 mutant.
TonB2 is required for virulence.
For investigation of the role of TonB2 in the virulence of A. pleuropneumoniae, competitive growth experiments were performed in vitro and in vivo. In vitro, the tonB2 mutant was not attenuated (CI = 2.703), but it was significantly attenuated in vivo in the porcine intratracheal infection model (CI = 0.022). In contrast, the tonB1 mutant was not found to be attenuated in vitro (CI = 0.902) or in vivo (CI = 1.54) at the same infecting dose. For confirmation of the necessity of TonB2 for virulence, the mutant was also used in pure culture to challenge two animals. The same dose (107 organisms) and route of infection (intratracheal) were used as those for the CI experiments. Neither pig exhibited any symptoms of illness, and upon necropsy, the lungs contained no signs of disease. Bacteria could only be recovered from the lungs in very low numbers (50 CFU/ml, or approximately 10,000 per total lung). In this acute infection model, bacterial virulence depends on the presence of TonB2.
DISCUSSION
A. pleuropneumoniae possesses two tonB systems, that originally described by Tonpitak et al. (34), now renamed the system, tonB1, and a tonB2 system consisting of the exbB2, exbD2, and tonB2 genes. The gene order at the tonB2 locus, exbB-exbD-tonB, matches that found in the tonB loci of Pasteurellaceae such as H. ducreyi (8), Mannheimia haemolytica (10), and H. influenzae (GenBank accession number NC 000907), and the inferred sequence of TonB2 more closely matches the protein sequences from these species than any others. In contrast, the gene order of the previously described tonB1 system, tonB-exbB-exbD, resembles that in various non-Pasteurellaceae (e.g., N. meningitidis [31] and Pseudomonas [36]), and the inferred protein sequence is significantly less similar to those of the Pasteurella and Haemophilus proteins. These observations suggest that tonB2 is the system indigenous to A. pleuropneumoniae, while the tonB1 system may have been acquired through lateral transfer from an unidentified donor (although its presence in all serotypes argues strongly against recent acquisition). This is reflected in the phylogenetic relatedness analysis.
While most organisms possess a single tonB system, multiple systems (each consisting of tonB, with or without accessory exb genes) are being increasingly recognized as more and more genomes are sequenced in their entirety (1, 22, 23, 28, 33, 36). In most cases, the benefit conferred by having more than one tonB system has not been established. In some organisms, the chromosomal location of one or more tonB genes close to genes encoding specific iron uptake systems hints at a dedication of the individual TonBs to different functions. For example, in Campylobacter jejuni, tonB1 and tonB3 are each located on the chromosome adjacent to genes encoding putative siderophore receptors (11, 23). Recent studies have focused on the two tonB systems of V. cholerae (14, 28). These appear to have particular affinities for different iron sources, but there is apparent redundancy, in that mutation of genes of the tonB1 or tonB2 locus leaves a wild-type capacity to utilize certain iron sources, indicating that tonB2, exbB2, and exbD2 can functionally replace tonB1, exbB1, and exbD1 (and vice versa) (22). In other organisms, while one TonB appears to have a straightforward function in iron acquisition, the role of the other has frustrated definition. In Pseudomonas aeruginosa strain PAO1, while TonB1 is needed for ferric siderophore and heme uptake, TonB2 appears not to be essential for iron acquisition, at least in vitro (36). However, deletion of tonB2 from a tonB1 mutant yields a Pseudomonas strain incapable, unlike its parent, of growth in an iron-restricted minimal medium, indicating that in this bacterial species TonB2 must, to some extent, be able to substitute for TonB1.
A different form of cross-substitution of TonB proteins appears to occur in A. pleuropneumoniae. The tonB2 system of A. pleuropneumoniae, not associated on the chromosome with any identifiable iron uptake genes, is absolutely necessary for the assimilation of iron associated with hemin, hemoglobin, and ferrichrome. The tonB1 system appears to be unnecessary for utilization of these iron sources, and tonB1 supplied on plasmid pLURO3 failed to complement the tonB2 mutation. TonB2 is also essential for the uptake of iron from Fe-pTf, but our observations on the effect of mutation in the tonB1 system add intriguing complexity. Our tonB1 mutant (with an intact tonB2 system) failed to grow in vitro when Fe-pTf was the sole iron source. At first sight, the simple explanation would appear to be that the mutation has a polar effect on the transcription of downstream genes. The level of tbpB transcription is substantially reduced in the tonB1 mutant, and very little TbpB could be detected by immunoblotting. However, the situation is clearly more complex. Tonpitak et al. have shown that an in-frame deletion of exbB1 abolishes the ability to utilize Fe-pTf, yet the expression of tbpB remains at a wild-type level (34). ExbB1 of the tonB1 system is clearly essential for this TonB-coupled iron uptake pathway. Taken together, these data indicate that elements of both tonB systems are necessary for Fe-pTf utilization. The simplest conceptual model would couple TonB2 with the Exb proteins encoded at the tonB1 locus; another possibility might be that TonB1 and TonB2 interact as a heterodimer to bridge the periplasm between the ExbBD complex at the inner membrane and the TbpAB complex in the outer membrane.
Considering this in the context of the phylogenetic analysis, we speculate that the acquisition of a once-functional tonB1 system in association with tbpBA conferred the significant biological advantage of the capacity to assimilate iron from Fe-pTf and that an inactivating mutation(s) in tonB1 may have arrived subsequently, driving the engagement of TonB2 in this pathway. Interestingly, in wild-type A. pleuropneumoniae, the level of transcription of tonB1 (and hence tbpBA, expressed from the same mRNA [34]) was found to be substantially higher than that of tonB2. It may be that an upregulating promoter mutation(s) occurred in response to the putative mutation in tonB1 that was not sufficient to compensate and leave TonB1 engaged in the Fe-pTf pathway, but had the advantageous side effect of increasing tbp transcription.
Finally, we examined the effect of tonB mutations on bacterial virulence. The tonB2 mutant was highly attenuated in the capacity to cause acute porcine pleuropneumonia following intratracheal challenge with 107 CFU, consistent with the findings of others in studies of tonB from other pathogens (24, 25, 32, 35). In contrast, at the same intratracheal infecting dose, the tonB1 mutant appeared to be as virulent as the wild type. At a lower intratracheal dose of 105 CFU, however, the same tonB1 mutant has been found to be attenuated (29). Other data have informed us about the contribution of the tonB systems to the virulence of A. pleuropneumoniae. Baltes et al. infected pigs by the aerosol route with a strain containing a nonpolar mutation in exbB1, exposing animals to ∼102 CFU/liter of aerosol for 45 min, and found the strain to be highly attenuated in the capacity to persist in the respiratory tract and to cause chronic infection (2). Such contrasting observations with high and low infecting doses (analogous to observations made with urease mutants of A. pleuropneumoniae [6]) may be reconciled in several ways. At a high intratracheal infecting dose, with which relatively limited bacterial replication occurs before the rapidly fatal outcome, the capacity to use Fe-pTf may simply no longer be a determinant of virulence. In such circumstances, the capacity to assimilate iron from other sources, dependent on intact tonB2, is apparently necessary and sufficient for wild-type virulence. Alternatively, the limited capacity to utilize Fe-pTf that likely remains in the polar mutant may be sufficient in these circumstances. At a low intratracheal infecting dose, on the other hand, the polar tonB1 mutant, which has significantly reduced tbp transcription, is attenuated. This may reflect a greater need for the Fe-pTf pathway in circumstances in which more bacterial replication in vivo is required. When the lowest dose (and most natural) aerosol infection model was employed by Baltes et al., an intact exbB1 appeared essential to establish a persistent infection (2). It may be inferred that under these circumstances, in which prolonged bacterial replication must occur under the tightly iron-restricted conditions prevalent in the respiratory tract, the Fe-pTf pathway is essential. Alternatively, there may be another pathway(s) altogether that has not been identified for which ExbB1 (and perhaps TonB1) is essential for virulence at a low, though not a high, intratracheal dose.
Acknowledgments
We thank Peter Heegaard for his kind gift of MAb 1.48.
This work was supported by grants to P.R.L., A.N.R., and J.S.K. from the Wellcome Trust and the United Kingdom Biotechnology and Biological Sciences Research Council.
Editor: V. J. DiRita
REFERENCES
- 1.Alm, R. A., L. S. Ling, D. T. Moir, B. L. King, E. D. Brown, P. C. Doig, D. R. Smith, B. Noonan, B. C. Guild, B. L. deJonge, G. Carmel, P. J. Tummino, A. Caruso, M. Uria-Nickelsen, D. M. Mills, C. Ives, R. Gibson, D. Merberg, S. D. Mills, Q. Jiang, D. E. Taylor, G. F. Vovis, and T. J. Trust. 1999. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature 397:176-180. [DOI] [PubMed] [Google Scholar]
- 2.Baltes, N., W. Tonpitak, G. F. Gerlach, I. Hennig-Pauka, A. Hoffmann-Moujahid, M. Ganter, and H. J. Rothkotter. 2001. Actinobacillus pleuropneumoniae iron transport and urease activity: effects on bacterial virulence and host immune response. Infect. Immun. 69:472-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baltes, N., W. Tonpitak, I. Hennig-Pauka, A. D. Gruber, and G. F. Gerlach. 2003. Actinobacillus pleuropneumoniae serotype 7 siderophore receptor FhuA is not required for virulence. FEMS Microbiol. Lett. 220:41-48. [DOI] [PubMed] [Google Scholar]
- 4.Bog, Y. S., L. O. Andresen, L. Bastholm, F. Elling, O. Angen, and P. M. Heegaard. 2001. The transferrin receptor of Actinobacillus pleuropneumoniae: quantitation of expression and structural characterization using a peptide-specific monoclonal antibody. Vet. Microbiol. 81:51-64. [DOI] [PubMed] [Google Scholar]
- 5.Bossé, J. T., and J. I. MacInnes. 1997. Genetic and biochemical analyses of Actinobacillus pleuropneumoniae urease. Infect. Immun. 65:4389-4394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bossé, J. T., and J. I. MacInnes. 2000. Urease activity may contribute to the ability of Actinobacillus pleuropneumoniae to establish infection. Can J. Vet. Res. 64:145-150. [PMC free article] [PubMed] [Google Scholar]
- 7.Chang, C., A. Mooser, A. Pluckthun, and A. Wlodawer. 2001. Crystal structure of the dimeric C-terminal domain of TonB reveals a novel fold. J. Biol. Chem. 276:27535-27540. [DOI] [PubMed] [Google Scholar]
- 8.Elkins, C., P. A. Totten, B. Olsen, and C. E. Thomas. 1998. Role of the Haemophilus ducreyi Ton system in internalization of heme from hemoglobin. Infect. Immun. 66:151-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frey, J. 1992. Construction of a broad host range shuttle vector for gene cloning and expression in Actinobacillus pleuropneumoniae and other Pasteurellaceae. Res. Microbiol. 143:263-269. [DOI] [PubMed] [Google Scholar]
- 10.Graham, M. R., and R. Y. Lo. 2002. A putative iron-regulated TonB-dependent receptor of Mannheimia (Pasteurella) haemolytica A1: possible mechanism for phase variation. Vet. Microbiol. 84:53-67. [DOI] [PubMed] [Google Scholar]
- 11.Guerry, P., J. Perez-Casal, R. Yao, A. McVeigh, and T. J. Trust. 1997. A genetic locus involved in iron utilization unique to some Campylobacter strains. J. Bacteriol. 179:3997-4002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kamp, E. M., J. K. Popma, and L. A. Van Leengoed. 1987. Serotyping of Haemophilus pleuropneumoniae in the Netherlands: with emphasis on heterogeneity within serotype 1 and (proposed) serotype 9. Vet. Microbiol. 13:249-257. [DOI] [PubMed] [Google Scholar]
- 13.Kilian, M., J. Nicolet, and E. L. Biberstein. 1978. Biochemical and serological characterization of Haemophilus pleuropneumoniae (Matthews and Pattison, 1961) Shope 1964 and proposal of a neotype strain. Int. J. Syst. Bacteriol. 28:20-26. [Google Scholar]
- 14.Mey, A. R., and S. M. Payne. 2003. Analysis of residues determining specificity of Vibrio cholerae TonB1 for its receptors. J. Bacteriol. 185:1195-1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miller, V. L., and J. J. Mekalanos. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants of Vibrio cholerae requires toxR. J. Bacteriol. 170:2575-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nielsen, R. 1986. Serological characterization of Actinobacillus pleuropneumoniae strains and proposal of a new serotype: serotype 12. Acta Vet. Scand. 27:453-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nielsen, R. 1985. Serological characterization of Haemophilus pleuropneumoniae (Actinobacillus pleuropneumoniae) strains and proposal of a new serotype: serotype 9. Acta Vet. Scand. 26:501-512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nielsen, R. 1985. Serological characterization of Haemophilus pleuropneumoniae (Actinobacillus pleuropneumoniae) strains and proposal of a new serotype: serotype 10. Acta Vet. Scand. 26:581-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nielsen, R. 1986. Serology of Haemophilus (Actinobacillus) pleuropneumoniae serotype 5 strains: establishment of subtypes a and b. Acta Vet. Scand. 27:49-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nielsen, R., L. O. Andresen, T. Plambeck, J. P. Nielsen, L. T. Krarup, and S. E. Jorsal. 1997. Serological characterization of Actinobacillus pleuropneumoniae biotype 2 strains isolated from pigs in two Danish herds. Vet. Microbiol. 54:35-46. [DOI] [PubMed] [Google Scholar]
- 21.Nielsen, R., and P. J. O'Connor. 1984. Serological characterization of 8 Haemophilus pleuropneumoniae strains and proposal of a new serotype: serotype 8. Acta Vet. Scand. 25:96-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Occhino, D. A., E. E. Wyckoff, D. P. Henderson, T. J. Wrona, and S. M. Payne. 1998. Vibrio cholerae iron transport: haem transport genes are linked to one of two sets of tonB, exbB, and exbD genes. Mol. Microbiol. 29:1493-1507. [DOI] [PubMed] [Google Scholar]
- 23.Parkhill, J., B. W. Wren, K. Mungall, J. M. Ketley, C. Churcher, D. Basham, T. Chillingworth, R. M. Davies, T. Feltwell, S. Holroyd, K. Jagels, A. V. Karlyshev, S. Moule, M. J. Pallen, C. W. Penn, M. A. Quail, M. A. Rajandream, K. M. Rutherford, A. H. van Vliet, S. Whitehead, and B. G. Barrell. 2000. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature 403:665-668. [DOI] [PubMed] [Google Scholar]
- 24.Pradel, E., N. Guiso, F. D. Menozzi, and C. Locht. 2000. Bordetella pertussis TonB, a Bvg-independent virulence determinant. Infect. Immun. 68:1919-1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reeves, S. A., A. G. Torres, and S. M. Payne. 2000. TonB is required for intracellular growth and virulence of Shigella dysenteriae. Infect. Immun. 68:6329-6336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rosendal, S., and D. A. Boyd. 1982. Haemophilus pleuropneumoniae serotyping. J. Clin. Microbiol. 16:840-843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 28.Seliger, S. S., A. R. Mey, A. M. Valle, and S. M. Payne. 2001. The two TonB systems of Vibrio cholerae: redundant and specific functions. Mol. Microbiol. 39:801-812. [DOI] [PubMed] [Google Scholar]
- 29.Sheehan, B. J., J. T. Bosse, A. J. Beddek, A. N. Rycroft, J. S. Kroll, and P. R. Langford. 2003. Identification of Actinobacillus pleuropneumoniae genes important for survival during infection in its natural host. Infect. Immun. 71:3960-3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sheehan, B. J., P. R. Langford, A. N. Rycroft, and J. S. Kroll. 2000. [Cu, Zn]-superoxide dismutase mutants of the swine pathogen Actinobacillus pleuropneumoniae are unattenuated in infections of the natural host. Infect. Immun. 68:4778-4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stojiljkovic, I., and D. Perkins-Balding. 2002. Processing of heme and heme-containing proteins by bacteria. DNA Cell Biol. 21:281-295. [DOI] [PubMed] [Google Scholar]
- 32.Takase, H., H. Nitanai, K. Hoshino, and T. Otani. 2000. Requirement of the Pseudomonas aeruginosa tonB gene for high-affinity iron acquisition and infection. Infect. Immun. 68:4498-4504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tomb, J. F., O. White, A. R. Kerlavage, R. A. Clayton, G. G. Sutton, R. D. Fleischmann, K. A. Ketchum, H. P. Klenk, S. Gill, B. A. Dougherty, K. Nelson, J. Quackenbush, L. Zhou, E. F. Kirkness, S. Peterson, B. Loftus, D. Richardson, R. Dodson, H. G. Khalak, A. Glodek, K. McKenney, L. M. Fitzgerald, N. Lee, M. D. Adams, E. K. Hickey, D. E. Berg, J. D. Gocayne, T. R. Utterback, J. D. Peterson, J. M. Kelley, M. D. Cotton, J. M. Weidman, C. Fujii, C. Bowman, L. Watthey, E. Wallin, W. S. Hayes, M. Borodovsky, P. D. Karp, H. O. Smith, C. M. Fraser, and J. C. Venter. 1997. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 388:539-547. [DOI] [PubMed] [Google Scholar]
- 34.Tonpitak, W., S. Thiede, W. Oswald, N. Baltes, and G. F. Gerlach. 2000. Actinobacillus pleuropneumoniae iron transport: a set of exbBD genes is transcriptionally linked to the tbpB gene and required for utilization of transferrin-bound iron. Infect. Immun. 68:1164-1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Torres, A. G., P. Redford, R. A. Welch, and S. M. Payne. 2001. TonB-dependent systems of uropathogenic Escherichia coli: aerobactin and heme transport and TonB are required for virulence in the mouse. Infect. Immun. 69:6179-6185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao, Q., and K. Poole. 2000. A second tonB gene in Pseudomonas aeruginosa is linked to the exbB and exbD genes. FEMS Microbiol. Lett. 184:127-132. [DOI] [PubMed] [Google Scholar]