Abstract
Purpose
To gain a greater understanding of the potential of the Aurora kinase A inhibitor MLN8237 in the treatment of pediatric malignancies.
Methods
The activity of MLN8237 was evaluated against 28 neuroblastoma and Ewing sarcoma cell lines, and its in vivo efficacy was studied over a range of doses against 12 pediatric tumor xenograft models. Pharmacokinetic, pharmacodynamic, and genomic studies were undertaken.
Results
In vitro neuroblastoma cell lines were generally more sensitive to MLN8237 than Ewing sarcoma lines. MLN8237 demonstrated significant activity in vivo against solid tumor models at the maximum tolerated dose (MTD); however, only 2 of 6 neuroblastoma models had objective responses at 0.25MTD. In contrast, MLN8237 induced objective responses at its MTD and at 0.5MTD in three ALL models and in two out of three at 0.25MTD. Pharmacokinetic studies at 0.5MTD demonstrated a T max of 0.5 h, C max of 24.8 μM, AUC(0–24) of 60.3 μM h, and 12 h trough level of 1.2 μM. Mitotic indices increased 6–12 h after MLN8237 administration. AURKA copy number variation was frequent in xenografts, and expression was highly correlated with copy number.
Conclusions
Objective responses were more frequent in tumors with decreased AURKA copy number (5/8) compared to those with increased gene copy number (2/14). This report confirms the significant activity against both solid tumor and ALL xenografts at the MTD, with a steep dose response. These data support clinical development of MLN8237 in childhood cancer. Because of the steep dose–response relationship, such studies should target achieving trough levels of 1 μM or higher for sustained periods of treatment.
Electronic supplementary material
The online version of this article (doi:10.1007/s00280-011-1618-8) contains supplementary material, which is available to authorized users.
Keywords: Preclinical testing, Developmental therapeutics, MLN8237, Pediatric cancer
Introduction
One of the hallmarks of transformed/malignant cells is their limitless proliferation capacity and defective cell cycle checkpoints that, when functional, operate to detect errors in replication processes and direct cells into apoptosis [1, 2]. Thus, interfering with mitosis has proven to be a successful cancer treatment strategy [3]. Several components of the mitotic machinery have been identified as potential therapeutic targets, and antimitotic agents are already crucial in the chemotherapy of both adult and childhood malignancies. For instance, the microtubule-targeting Vinca alkaloids are a central component of curative regimens for many childhood solid tumors and leukemias. Other appealing targets include mitotic kinesins [3, 4], centromere components required for chromosome alignment and spindle complex formation [5], as well as Polo-like kinases and the Aurora kinases [6].
The Aurora serine/threonine protein kinases are a family of three kinases (Aurora A, Aurora B, and Aurora C) with different tissue and temporal expression profiles that play key roles in mitosis and meiosis, defects in which can lead to abnormal mitotic events and apoptosis induction [7]. The essential nature of Aurora kinase A is highlighted by the fact that genetically engineered null mice are embryonic lethal (dying at the blastocyst stage) [8]. Aurora kinase A activity is also required for centrosome duplication and separation, microtubule–kinetochore attachment, spindle checkpoint formation, cytokinesis [9, 10], the G2/M transition [11], and phosphorylation of Polo-like kinase 1 [12]. Further, Aurora kinase A has been implicated as an oncogenic driver in human cancers [13]. Aurora kinase A has been found to be overexpressed in cancer cells, and the AURKA gene locus is amplified in selected adult tumors [14]. However, limited information on the role of Aurora kinase A in pediatric cancers is available.
Aurora kinase inhibitors are the focus of several pharmaceutical development programs. Aurora kinase inhibitors with different specificities and activities as well as pharmacodynamic markers are currently being assessed, and some are already well advanced in clinical trials (reviewed in [15]). Most of these inhibitors show a broad range of activity, with AZD-1152 being an example of a selective Aurora kinase B inhibitor and MLN8054 (or its derived compound, MLN8237, used in the present study) an example of a selective Aurora kinase A inhibitor. The effects of Aurora kinase A inhibition are multiple, as corresponds to the varied nature of its substrates, and include abnormal spindle pole formation, proliferation reduction (with G2-M arrest), and polyploidy [16], followed by apoptosis induction. The latter could involve signaling mediated by p53, as Aurora kinase A has been shown to modify the phosphorylation status of p53 and histone H3 [17, 18] and to interact with the MYCN protein, limiting p53 ubiquitination and degradation by the proteasome in neuroblastoma cell lines [19, 20]. Although p53 is frequently non-functional in cancer cells, inhibition of Aurora kinase A by MLN8054 can lead to p73-dependent apoptosis in p53-deficient cells [21]. Aurora kinase A has also been reported to influence cell survival through the Akt pathway and by interfering with IkBα [22].
The primary focus of the Pediatric Preclinical Testing Program (PPTP) is to identify novel agents that have significant antitumor activity against models of childhood solid tumors and acute lymphoblastic leukemia (ALL) as one source of data to use in prioritizing clinical development of such agents in the pediatric setting. The PPTP has reported the single-agent evaluation of activity of the Aurora kinase A inhibitor MLN8237 against its panels of in vitro cell lines and in vivo xenograft models [23]. Both the neuroblastoma and ALL panels were particularly sensitive to the single-agent treatment. In fact, this Aurora kinase A inhibitor is the only drug out of more than 20 tested with preferential activity against the neuroblastoma panel. Despite these encouraging results, issues of how responsiveness relates to drug exposure in mice and humans, the dose range over which MLN8237 exerts significant antitumor activity, and the correlation of sensitivity to Aurora kinase A expression remain unanswered. Here, we report the in vitro activity of MLN8237 against an extended panel of neuroblastoma and Ewing sarcoma cell lines, and we report in vivo dose–response efficacy studies focusing on neuroblastoma and pediatric ALL xenografts, as well as assessment of pharmacokinetic, pharmacodynamic, and molecular parameters associated with these responses.
Materials and methods
In vitro testing
In vitro testing was performed using DIMSCAN, a semiautomatic fluorescence-based digital image microscopy system that quantifies viable (using fluorescein diacetate [FDA]) cells in tissue culture multiwell plates [24]. Cells were incubated in the presence of MLN8237 for 96 h at concentrations from 1 nM to 10 μM and analyzed as previously described [25]. Two measures of sensitivity were used; the absolute IC50, defined as the drug concentration inhibiting growth by 50% compared to controls, and the relative IC50 (previously termed EC50), defined as the drug concentration yielding 50% of the maximum inhibitory effect.
Cell lines for in vitro testing
The cell lines used in this study were obtained from the originator of the cell line or the Deutsche Sammlung von Mikroorganismen unde Zellkulturen (Braunschweig, Germany) or the American Type Culture Collection (ATCC, VA, U.S.A.) and were maintained in culture according to the corresponding initial report. All lines underwent DNA genotyping as described [26]. Short tandem repeat (STR) assay was used to verify each line against the Children’s Oncology Group STR database (this can be found in http://www.COGcell.org).
In vivo tumor growth inhibition studies
CB17SC scid −/− female mice (Taconic Farms, Germantown NY) were used to propagate subcutaneously implanted kidney/rhabdoid tumors, sarcomas (osteosarcoma, rhabdomyosarcoma), and neuroblastoma tumors as previously described [27–29]. Human leukemia cells were propagated by intravenous inoculation in female non-obese diabetic (NOD)/scid −/− mice as described previously [30]. Details of these tumor panels can be obtained at http://pptp.nchresearch.org/documents.html. Female mice were used irrespective of the patient gender from which the original tumor was derived. All mice were maintained under barrier conditions, and experiments were conducted using protocols and conditions approved by the institutional animal care and use committee of the appropriate consortium member. Ten mice (solid tumor models) and 8 mice (ALL models) were used in each control or treatment group. Tumor volumes (cm3) [solid tumor xenografts] or percentages of human CD45-positive cells out of the total leukocyte population in peripheral blood [hCD45, ALL xenografts] were determined as previously described [31]. An event was defined for the solid tumors as a quadrupling of tumor volume from the tumor volume at start of treatment, and for the ALL models when the proportion of hCD45 reached 25%. Event-free survival (EFS) was estimated for individual mice as the time required from treatment initiation to reach the defined event threshold.
Determination of response
Responses were assessed using three activity measures as previously described [31]. For all the solid tumors on an individual basis, progressive disease (PD) was defined as <50% regression from initial volume during the study period and >25% increase in initial volume at the end of study period. Stable disease (SD) was defined as <50% regression from initial volume during the study period and ≤25% increase in initial volume at the end of the study. Partial response (PR) was defined as a tumor volume regression ≥50% for at least one time point but with measurable tumor (≥0.10 cm3). Complete response (CR) was defined as a disappearance of measurable tumor mass (<0.10 cm3) for at least one time point. A complete response was considered maintained (MCR) if the tumor volume was <0.10 cm3 at the end of the study period. Similarly, for the ALL xenografts, PD was defined as a continuous increase in hCD45 that reached event before the end of the 42-day monitoring period, SD as hCD45 that did not decrease to under 1% and did not reach the event threshold, PR was defined as a decrease in hCD45 to under 1% at only one time point, CR when the hCD45 stayed under 1% for two consecutive measures, and MCR when the hCD45 stayed under 1% for the last 3 weeks of the monitoring period. Tumor growth delay (TGD) values were calculated based on the numbers of days to event. For each individual mouse that had PD and had an event in the treatment groups, a TGD value was calculated by dividing the time to event for that mouse by the median time to event in the respective control group. Median times to event were estimated based on the Kaplan–Meier event-free survival distribution. For treatment groups only, if the tumor response was PD, then the score was further classified into PD1 or PD2 based on the TGD value: if the TGD value ≤1.5, that mouse was considered PD1 while if the TGD value >1.5, the mouse was considered PD2. Mice that had PD but did not have an event at the end of the study were coded as SD. A Median Group Response was obtained for each cohort based on the median of the scores of the individual mice. An in-depth description of the analysis methods is included in the Supplemental Response Definitions section. The three scores reflecting tumor regression (PR, CR and MCR) were considered Objective Responses (ORs).
Drug information and formulation
Millennium Pharmaceuticals, Inc., through the Cancer Therapy Evaluation Program (NCI), provided MLN8237 to the Pediatric Preclinical Testing Program. MLN8237 was dissolved in DMSO and diluted in culture media for in vitro tests or was suspended in 10% 2-hydroxypropyl-β-cyclodextrin and 1% sodium bicarbonate in water and administered to mice via oral gavage twice daily for 5 days repeated each week for a total of 6 weeks for the solid tumor xenografts, and for 3 weeks for the leukemia xenografts at doses of 20.8 mg/kg (the MTD), 10.4, 5.2, and 2.6 mg/kg.
Statistical methods
The exact log-rank test, as implemented using Proc StatXact for SAS®, was used to compare event-free survival (EFS) distributions between treatment and control groups. P values were two-sided and were not adjusted for multiple comparisons given the exploratory nature of the studies. The Mann–Whitney test was used to test for differences in the median IC50 values for the Ewing sarcoma and neuroblastoma cell lines.
Pharmacokinetic analysis
Blood was obtained by cardiac puncture 0, 0.5, 1, 2, 4, 6, 8, 12, and 24 h after dosing with MLN8237 (10 mg/kg) from 3 mice per time point. Plasma samples were prepared and analyzed for MLN8237 concentrations by an LC/MS/MS assay according to previously described methods for MLN8054 [16].
Pharmacodynamic analysis
Accumulation of mitotic cells was used as a pharmacodynamic measure of Aurora kinase A inhibition in NB-1771 tumor-bearing animals dosed with 20.8 mg/kg MLN8237. Tumors were collected from animals at 0, 2, 6, 8, 12, and 24 h following MLN8237 dosing from 3 mice per time point and were formalin-fixed and paraffin-embedded. Tumor sections (5 μm) were stained for two independent mitotic markers, MPM2 (Mitotic protein monoclonal #2) and histone H3 phosphorylated on serine 10 (pHistH3) using the Discovery® XT (Ventana Medical Systems, AZ, U.S.A.) automated slide stainer. Sections were deparaffinized with EZ prep™ solution, and antigen retrieval was completed with Cell Conditioning 1 solution, CC1 (Ventana Medical Systems). The sections were incubated for 60 min at room temperature with mouse MPM-2 antibody (1:100, Upstate Biotechnology, MA, U.S.A.) and rabbit anti-phospho-histone H3 polyclonal (1:25, Upstate Biotechnology). Biotin-conjugated anti-mouse antibody (Animal Research Kit, DAKO, CA, U.S.A.) was included to amplify the MPM2 signal. Conjugated fluorophores, including Alexa Fluor 488-conjugated streptavidin (1:100, Molecular Probes, OR, U.S.A.) or Rhodamine-Red-X-AffiniPure goat anti-rabbit IgG (1:25, Jackson ImmunoResearch, PA, U.S.A.), were incubated for 60 min at room temperature. Slides were washed in PBS and mounted with DAPI Vectashield Hard Set Mounting Medium (Vector Laboratories, CA, U.S.A.). Images were acquired using a Canon E300 microscope (Canon, U.S.A.) with an automated stage. Five images from each slide were captured using a 40× PlanFluor objective (Nikon Instruments, U.S.A.) and analyzed on the MetaMorph® image processing software (Universal Imaging Corp., PA, U.S.A.) that used a custom image processing application module. Mitotic indices were determined as the percentage of total cells that were positive for either pHisH3 or MPM2 staining.
Copy number analysis
Copy number analysis was performed using the Affymetrix Genome-Wide SNP Array 6.0. DNA (500 ng) from each sample was processed according to the manufacturer’s instructions. SNP 6.0 data were processed from CEL files to extract raw signal intensity values using dChip [32] PM-only model-based expression analysis. The signal data were then normalized using a reference-based normalization algorithm [33]. For each marker in each array, the log2 ratio of tumor versus the median signal obtained from 90 reference samples from St. Jude Children’s Research Hospital was calculated. Then, the segmentation algorithm [34] implemented in the DNAcopy package from Bioconductor [35] was applied to the above log2 ratio data to identify copy number alterations for each tumor sample. Copy number gains and losses were defined by genomic segments with log2 ratios >0.2 or <0.2, respectively.
Correlation analysis of gene expression and genomic copy number variation
Gene expression data were obtained previously using the Affymetrix HG-U133 Plus 2.0 array [36]. To estimate the variance in gene expression attributed to underlying copy number variation, a linear regression model was fitted to compare SNP data (segment log2 ratio) against expression data (log2 signal). For each probe set on the HG-U133 Plus 2.0 array, the correlation coefficient was calculated using each segment falling within a genomic window of ±5 kb up/downstream of the annotated gene. Because multiple segments and probe sets can arise within a given gene boundary, the segment and probe set with the highest correlation (R2) value were selected for subsequent analysis.
Results
MLN8237 is effective in vitro against both Ewing sarcoma and neuroblastoma cell lines
In order to evaluate the activity of MLN8237 against cell lines in vitro, an expanded panel of Ewing sarcoma (n = 11) and neuroblastoma (n = 17) cell lines was tested by DIMSCAN. The median relative IC50 for the Ewing sarcoma and neuroblastoma extended panels of cell lines was 32 nM, while the median absolute IC50 was 37 nM (Table 1). Corresponding ratios of the median relative and absolute IC50 values to the comparable values for each cell line tested are depicted in Table 1 and Supplemental Figure 1. The sensitivity of the Ewing sarcoma cell lines was generally less than the median for both measurements (ratio < 1), whereas neuroblastoma cell lines were generally more sensitive to MLN8237 (ratio > 1). Only one Ewing sarcoma cell line, CHLA-56, was completely resistant (IC50 > 10 μM) to MLN8237 exposure in vitro. The relative IC50 values were significantly lower for the neuroblastoma panel (23 nM) than for the Ewing sarcoma cell lines (58 nM, P = 0.0019), even after excluding the resistant line (CHLA-56) from this analysis (P = 0.0039). The cytotoxicity of MLN8237 (as assessed by minimum T/C values (Ymin) approaching 0) was variable, with a median Ymin value of 10.9%, and a range from 0.5 to 48% (Table 1). The median Ymin values did not differ between the Ewing cell lines (11.4%) and the neuroblastoma cell lines (10.4%).
Table 1.
Summary of in vitro sensitivity of Ewing sarcoma and neuroblastoma cell lines
| Cell line | Histology | Relative IC50 (nM) | Absolute IC50 (nM) | Median EC50 ratio | Median IC50 ratio | Ymin (%) |
|---|---|---|---|---|---|---|
| A-673 | Ewing sarcoma | 30 | 32 | 1.05 | 1.14 | 13.1 |
| TC-32 | Ewing sarcoma | 34 | 39 | 0.92 | 0.94 | 6.5 |
| TC-71 | Ewing sarcoma | 100 | 102 | 0.32 | 0.36 | 10.0 |
| SK-N-MC | Ewing sarcoma | 66 | 72 | 0.48 | 0.51 | 2.8 |
| CHLA-9 | Ewing sarcoma | 16 | 18 | 1.97 | 2.08 | 4.2 |
| CHLA-10 | Ewing sarcoma | 56 | 60 | 0.57 | 0.60 | 4.7 |
| CHLA-25 | Ewing sarcoma | 58 | 168 | 0.55 | 0.22 | 30.1 |
| CHLA-32 | Ewing sarcoma | 92 | 136 | 0.35 | 0.27 | 13.1 |
| CHLA-56 | Ewing sarcoma | 10,000 | 10,000 | 0.00 | 0.00 | 48.1 |
| CHLA-258 | Ewing sarcoma | 82 | 132 | 0.39 | 0.28 | 18.8 |
| COG-E-352 | Ewing sarcoma | 35 | 43 | 0.91 | 0.86 | 11.4 |
| CHLA-90 | Neuroblastoma | 48 | 61 | 0.67 | 0.60 | 16.3 |
| CHLA-119 | Neuroblastoma | 22 | 22 | 1.46 | 1.64 | 0.5 |
| CHLA-122 | Neuroblastoma | 17 | 19 | 1.82 | 1.96 | 0.6 |
| CHLA-136 | Neuroblastoma | 36 | 39 | 0.89 | 0.94 | 10.4 |
| CHLA-140 | Neuroblastoma | 14 | 26 | 2.23 | 1.39 | 29.4 |
| LA–N-6 | Neuroblastoma | 31 | 54 | 1.01 | 0.68 | 32.1 |
| NB-1643 | Neuroblastoma | 32 | 37 | 0.98 | 0.99 | 10.2 |
| NB-EBc1 | Neuroblastoma | 49 | 50 | 0.65 | 0.74 | 3.6 |
| SK-N-BE(1) | Neuroblastoma | 24 | 28 | 1.35 | 1.32 | 4.0 |
| SK-N-BE(2) | Neuroblastoma | 26 | 36 | 1.21 | 1.01 | 16.5 |
| SMS-KAN | Neuroblastoma | 32 | 34 | 0.99 | 1.08 | 13.5 |
| SMS-KANR | Neuroblastoma | 23 | 26 | 1.39 | 1.41 | 11.4 |
| SMS-KCN | Neuroblastoma | 17 | 19 | 1.86 | 1.97 | 10.4 |
| SMS-KCNR | Neuroblastoma | 9 | 10 | 3.42 | 3.65 | 6.6 |
| SMS-LHN | Neuroblastoma | 20 | 32 | 1.61 | 1.13 | 25.1 |
| SMS-MSN | Neuroblastoma | 17 | 22 | 1.92 | 1.66 | 16.1 |
| SMS-SAN | Neuroblastoma | 18 | 20 | 1.79 | 1.80 | 5.9 |
| Median | 32 | 37 | 1.00 | 1.00 | 10.9 | |
| Minimum | 9 | 10 | 0.00 | 0.00 | 0.5 | |
| Maximum | 10,000 | 10,000 | 3.42 | 3.65 | 48.1 |
MLN8237 induces significant cancer growth inhibition in vivo with limited toxicity at its MTD
We previously reported MLN8237 as highly effective against the PPTP’s neuroblastoma and ALL xenograft models [23]. With the aim of confirming these results, the efficacy of MLN8237 as a single agent at its MTD (20.8 mg/kg administered twice daily) was evaluated in 9 solid tumor (6 of them neuroblastoma) and 3 ALL xenograft models (Table 2). A complete summary of results is provided in Supplemental Table I, including total numbers of mice, number of mice that died (or were otherwise excluded), numbers of mice with events and average times to event, tumor growth delay, as well as numbers of responses and T/C values.
Table 2.
Anti-tumor Efficacy of MLN8237 in vivo
| Line description | Tumor type | Treatment groupa | Estimate of median time to event | P value (unadjusted) | EFS T/C | Median RTV/hCD45 at end of study | Tumor volume T/C | Median group response | T/C volume activity | EFS activity | Response activity |
|---|---|---|---|---|---|---|---|---|---|---|---|
| KT-10 | Wilms | B | >EP | <0.001 | >4.4 | 0.0 | 0.35 | MCR | Int | High | High |
| KT-10 | Wilms | C | 14.6 | 0.009 | 1.5 | >4 | 0.53 | PD1 | Low | Low | Low |
| KT-10 | Wilms | D | 16.0 | 0.110 | 1.7 | >4 | 0.71 | PD2 | Low | Low | Int |
| KT-10 | Wilms | E | 9.6 | 0.768 | 1.0 | >4 | 0.84 | PD1 | Low | Low | Low |
| Rh65 | ALV Rhabdomyosarcoma | B | >EP | <0.001 | >3.9 | 0.0 | 0.01 | MCR | High | High | High |
| Rh65 | ALV Rhabdomyosarcoma | C | >EP | <0.001 | >3.9 | 3.2 | 0.11 | PD2 | High | Int | Int |
| Rh65 | ALV Rhabdomyosarcoma | D | 39.3 | <0.001 | 3.7 | >4 | 0.19 | PD2 | Int | Int | Int |
| Rh65 | ALV Rhabdomyosarcoma | E | 22.9 | <0.001 | 2.1 | >4 | 0.31 | PD2 | Int | Int | Int |
| NB-SD | Neuroblastoma | B | >EP | <0.001 | >4.4 | 0.9 | 0.47 | SD | Low | High | Int |
| NB-SD | Neuroblastoma | C | 37.5 | <0.001 | 3.9 | >4 | 0.50 | PD2 | Low | Int | Int |
| NB-SD | Neuroblastoma | D | 20.9 | <0.001 | 2.2 | >4 | 0.60 | PD2 | Low | Int | Int |
| NB-SD | Neuroblastoma | E | 15.2 | <0.001 | 1.6 | >4 | 0.59 | PD1 | Low | Low | Low |
| NB-1771 | Neuroblastoma | B | >EP | <0.001 | >2.1 | 0.0 | 0.02 | MCR | High | High | High |
| NB-1771 | Neuroblastoma | C | >EP | <0.001 | >2.1 | 0.0 | 0.08 | MCR | High | High | High |
| NB-1771 | Neuroblastoma | D | >EP | <0.001 | >2.1 | 2.2 | 0.38 | PD2 | Int | Int | Int |
| NB-1771 | Neuroblastoma | E | 38.2 | <0.001 | 1.9 | >4 | 0.38 | PD2 | Int | Low | Int |
| NB-EBc1 | Neuroblastoma | B | >EP | <0.001 | >7.2 | 0.3 | 0.15 | MCR | Int | High | High |
| NB-EBc1 | Neuroblastoma | C | 17.9 | <0.001 | 3.1 | >4 | 0.22 | PD2 | Int | Int | Int |
| NB-EBc1 | Neuroblastoma | D | 11.3 | <0.001 | 2.0 | >4 | 0.44 | PD2 | Int | Low | Int |
| NB-EBc1 | Neuroblastoma | E | 9.6 | 0.010 | 1.7 | >4 | 0.57 | PD2 | Low | Low | Int |
| NB-1643 | Neuroblastoma | B | >EP | <0.001 | >2.4 | 0.0 | 0.14 | MCR | High | High | High |
| NB-1643 | Neuroblastoma | C | >EP | <0.001 | >2.4 | 0.0 | 0.30 | MCR | Int | High | High |
| NB-1643 | Neuroblastoma | D | >EP | <0.001 | >2.4 | 0.4 | 0.38 | PR | Int | High | High |
| NB-1643 | Neuroblastoma | E | 20.3 | 0.203 | 1.2 | >4 | 1.01 | PD1 | Low | Low | Low |
| NB-1382 | Neuroblastoma | B | >EP | <0.001 | >1.7 | 2.1 | 0.31 | PD2 | Int | NE | Int |
| SK-N-AS | Neuroblastoma | B | 7.6 | 0.077 | 1.4 | >4 | 0.71 | PD1 | Low | Low | Low |
| OS-1 | Osteosarcoma | B | >EP | <0.001 | >1.5 | 0.8 | 0.45 | SD | Int | NE | Int |
| OS-1 | Osteosarcoma | C | >EP | <0.001 | >1.5 | 2.0 | 0.56 | PD2 | Low | NE | Int |
| OS-1 | Osteosarcoma | D | >EP | 0.022 | >1.5 | 3.4 | 0.70 | PD2 | Low | NE | Int |
| OS-1 | Osteosarcoma | E | >EP | 0.026 | >1.5 | 3.5 | 0.75 | PD2 | Low | NE | Int |
| ALL-2 | ALL B-precursor | B | >EP | <0.001 | >3.8 | 0.3 | – | MCR | High | High | |
| ALL-2 | ALL B-precursor | C | >EP | <0.001 | >3.8 | 3.1 | – | CR | Int | High | |
| ALL-2 | ALL B-precursor | D | >EP | <0.001 | >3.8 | >25 | – | CR | Int | High | |
| ALL-2 | ALL B-precursor | E | 25.2 | <0.001 | 2.3 | >25 | – | PD2 | Int | Int | |
| ALL-8 | ALL T-cell | B | >EP | 0.002 | >3.3 | 0.2 | – | MCR | High | High | |
| ALL-8 | ALL T-cell | C | >EP | <0.001 | >3.3 | 6.2 | – | CR | Int | High | |
| ALL-8 | ALL T-cell | D | 31.6 | 0.001 | 2.5 | >25 | – | PR | Int | High | |
| ALL-8 | ALL T-cell | E | 23.0 | 0.052 | 1.8 | >25 | – | PD2 | Low | Int | |
| ALL-19 | ALL B-precursor | B | 41.8 | 0.026 | 16.8 | >25 | – | CR | Int | High | |
| ALL-19 | ALL B-precursor | C | 37.2 | 0.012 | 14.9 | >25 | – | CR | Int | High | |
| ALL-19 | ALL B-precursor | D | 22.9 | 0.041 | 9.2 | >25 | – | PD2 | Int | Int | |
| ALL-19 | ALL B-precursor | E | 2.6 | 0.742 | 1.0 | >25 | – | PD1 | Low | Low |
aTreatment groups correspond to the following doses: A controls, B 20.8 mg kg, C 10.4 mg/kg, D 5.2 mg/kg, and E 2.6 mg/kg, >EP greater than evaluation period
Toxicity was limited in the solid tumor study. Six of 180 mice died during the study (2.8%), 1 of 90 in the control arms (1.1%), and 5 of 90 in the MLN8237 treatment arms (5.6%). Toxicity was greater in the leukemia models (5 of 24 mice in the treated arm versus 1 of 25 controls), but none of the groups met criteria setup for exclusion from analysis (>25% mortality).
Antitumor effects were evaluated using the PPTP activity measures for time to event (EFS T/C), tumor growth delay (tumor volume T/C), and Median Group Response and are summarized in Table 2. MLN8237 induced significant differences in EFS distributions compared to controls in all solid tumor models except SK-N-AS, and in all three ALL models. Eight out of 11 evaluable lines met the criteria for high activity with EFS T/C values greater than 2 and with final tumor volumes less than the initial treatment volumes. The drug treatment at its MTD resulted in 5 objective responses (partial or complete tumor regressions) out of 9 models in the solid tumor panel and in objective responses for all 3 of the ALL xenografts tested.
MLN8237 efficacy against solid tumors shows a steep dose–response in vivo
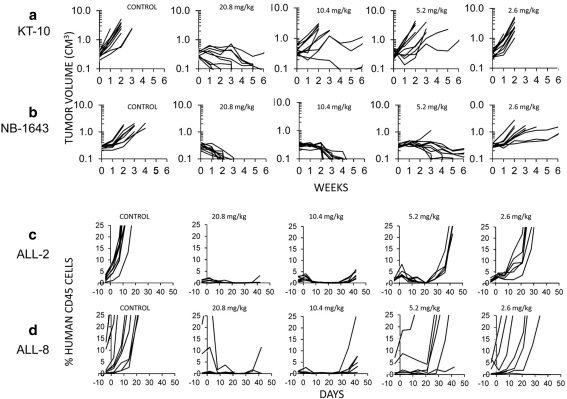
To investigate the efficacy of MLN8237 over a range of doses, we evaluated the efficacy of the drug in vivo at the MTD and 0.5, 0.25, and 0.125 of the MTD dose in six solid tumors and 3 ALL models that demonstrated stable disease or regression at the highest dose level. The in vivo testing results for the objective response measure of activity are presented in Supplemental Fig. 2 in a ‘heat-map’ format as well as a ‘COMPARE’-like format, based on the scoring criteria described in the “Material and methods” and the Supplemental Response Definitions section. The latter analysis demonstrates relative tumor sensitivities around the midpoint score of 5 (SD). At the 0.5MTD dose (10.4 mg/kg), only two of six solid tumor models demonstrated objective responses (NB-1771, NB-1643), indicating a steep dose–response relationship for MLN8237 (Table 2). Dose–response relationships for KT-10, for which antitumor activity was observed only at the highest dose, and for NB-1643, for which MLN8237 exhibited broad-range activity, are shown in Fig. 1 (panels a and b). By contrast, for the ALL panel, MLN8237 induced CR in each of three ALL models at 0.5MTD, and even at 0.25MTD, two out of three xenografts were classified as objective responses (one CR, one PR; Fig. 1, panels c and d), suggesting that the leukemia xenografts are more sensitive to MLN8237 than the solid tumor models.
Fig. 1.
MLN8237 in vivo activity against individual solid tumor xenografts (KT-10; NB-1643 xenograft panel a and b respectively) or ALL xenografts (ALL-2; ALL-8, panel c and d respectively). Results show growth of individual tumors in control, or mice treated with 2.6, 5.2, 10.4, or 20.8 mg/kg twice daily, 5-days per week for 6 weeks for solid tumors or 3 weeks for ALL
Pharmacokinetic and pharmacodynamic markers
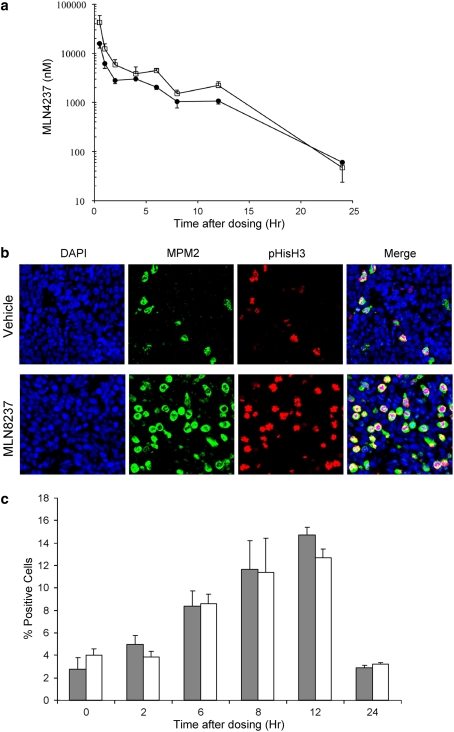
Pharmacokinetic parameters for MLN8237 in mice were assessed to evaluate whether the drug levels associated with the high level of anticancer activity observed for the xenograft models are attainable in the clinical setting. The systemic exposure of MLN8237 was evaluated by dosing non-tumored scid mice with a single dose of 10.4 or 20.8 mg/kg MLN8237 and collecting blood at various time points to determine MLN8237 plasma concentrations. At the 20.8 mg/kg dose, MLN8237 was rapidly absorbed with a T max of 0.5 h and a corresponding C max of 42.5 μM. The AUC0–24 h was 78.4 μM h, and the 12 h trough level was 1.8 μM. For the 10 mg/kg dose, the C max was 15.8 μM, and the AUC0–24 h was 39 μM h (Fig. 2a).
Fig. 2.
Pharmacokinetic and pharmacodynamic activity of MLN8237. a MLN8237 (10.4 mg/kg (filled circle) or 20.8 mg/kg (open square)) was dosed orally in non-tumored scid mice, and blood was isolated at various times thereafter. MLN8237 concentrations were determined in plasma from 3 different animals per time point; means ± standard error of the means are shown; b Representative immunofluorescence images of tumor sections from NB-1771 xenografts stained with antibodies against MPM2 and pHistoH3 12 h after in vivo administration of vehicle control (upper panel) or MLN8237 (20.8 mg/kg, lower panel); c The percentage of cells positive for the mitotic markers MPM2 (dark bars) or pHistH3 (white bars) were determined from 3 different animals at multiple time points; means ± standard deviation are shown. Mice bearing the human neuroblastoma tumor NB-1771 were dosed once orally with MLN8237 at 20.8 mg/kg
Pharmacodynamic markers of MLN8237 on target effects were investigated in mice bearing the NB-1771 tumor xenograft by assaying for a transient accumulation of mitotic cells that occurs subsequent to Aurora kinase A inhibition. The mitotic index was estimated in tumors collected from mice that received a single 20.8 mg/kg dose of MLN8237 by determining the percentage of cells positive for two distinct mitotic markers, MPM2 and pHistH3. Representative photomicrographs of NB-1771 tumor sections stained for MPM2 and H3 pHistH3 are shown in Fig. 2b. The mitotic indices as evaluated through these two markers increased (approximately twofold) within 6 h following MLN8237 dosing, peaked at 12 h (three to fivefold increase), and returned to baseline levels 24 h after dosing (Fig. 2c). There was concordance between both pharmacodynamic markers, with very similar profiles of mitotic indices obtained with each marker.
Molecular markers
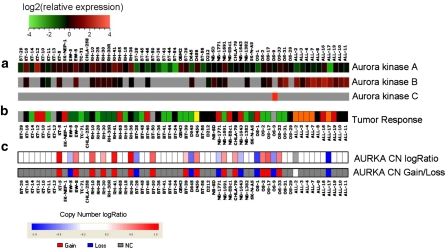
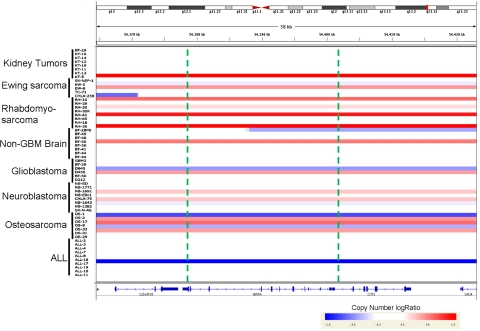
Aurora kinases are overexpressed in Ewing sarcoma as a consequence of the EWS-FLI1 gene fusion [37] while the gene expression of Aurora kinase A in neuroblastoma is not augmented [36]. mRNA expression levels of the Aurora kinases were previously assessed using the Affymetrix platform [36] and are shown in Fig. 3 (panel a) for the xenografts tested in vivo (panel b) by the PPTP against MLN8237 at its MTD as a single agent [23]. The ALL and neuroblastoma xenograft panels showed relatively low levels of expression of Aurora kinase A among all of the xenograft tested. From the 60 samples tested for in vivo sensitivity, 22 (37%) showed significant copy number variation at the Aurora kinase A locus (Figs. 3 panel c, 4). In many instances, copy number alteration at the Aurora kinase A locus (AURKA) was attributed to large genomic regions, even entire chromosomal arms, undergoing amplification or deletion on chromosome 20 (Supplemental Fig. 3). Frequently, the gene dosage of Aurora kinase A showed clear correlation with variation in expression across the PPTP lines (Fig. 3). For example, copy loss in the BT-28, D645, OS-1, and ALL-17 was associated with substantially lower expression in those lines. The correlation of gene expression variation with AURKA copy number status was very strong (Pearson R = 0.573) for the PPTP models. Indeed, this high positive correlation placed the Aurora kinase A locus among the top 1.6% of all genes tested, indicating that its gene expression is strongly influenced by gene dosage. Copy number loss was noted in 8 models, and their response to therapy ranged from PD1 (insensitive, EW-5, BT-28) to CR or MCR (sensitive, ALL-17, NB-1771, NB-1643, OS-1). Conversely, copy gain was observed in approximately one half of the rhabdomyosarcoma lines, suggesting that at least some of the relatively high expression across the entire rhabdomyosarcoma group may have arisen due to copy gain at the Aurora kinase A locus. With the exception of Rh65 (MCR), which does not exhibit increased AURKA copy number, the rhabdomyosarcomas were poorly sensitive to MLN8237. Of the 14 tumors exhibiting copy number gain, there were only 2 that had objective responses to MLN8237 at the MTD.
Fig. 3.
Gene expression, copy number analysis of the Aurora kinase genes, and drug sensitivity of the PPTP in vivo models. a Relative gene expression of Aurora kinases A, B, and C as determined by Affymetrix gene expression arrays; b Tumor sensitivity to MLN8237 administered at the MTD (data from ref [23]) presented as a categorical heat map. The colored heat map depicts group response scores: MCR (red), CR (orange), PR (yellow), SD (gray), PD2 (light green), PD1 (dark green), Not evaluated (black) (see Supplemental Fig. 2, and Median Group Response scoring in the Supplemental Response Definitions section); c Copy number assessment of Aurora kinase A (AURKA) from the Affymetrix SNP 6.0 array. The upper panel shows a continuous heat map representation of copy number log2 ratio, while the lower panel shows a categorical representation of copy gain (red), copy loss (blue), copy diploid (gray), or no data (white) (color figure online)
Fig. 4.
Copy number analysis using the Affymetrix SNP 6.0 array. Copy number representation of the in vivo tested panel according to log2 ratio of segments identified showing copy number status across the Aurora kinase A locus. The location of the Aurora kinase A locus on chromosome 20 is indicated by a red bar across the top panel, and green vertical lines indicate the boundaries of the AURKA locus (color figure online)
Discussion
The main goal of the PPTP is to prioritize drugs being developed predominantly for adult cancer treatment for expedited clinical trials in children with relapsed/refractory cancers. MLN8237, which has 200-fold specificity for Aurora kinase A inhibition versus Aurora kinase B [16], showed high-level activity at its MTD in its initial PPTP evaluation; therefore, it was critical to validate and extend these previous results. This was done by evaluating MLN8237 against an extensive number of Ewing sarcoma and neuroblastoma cancer lines in vitro, and by assessing its activity in vivo against neuroblastoma and ALL xenografts across a range of doses with pharmacokinetic and pharmacodynamic correlation.
Aurora kinase inhibitors have to date shown only modest clinical activity against solid tumors in adults, although more pronounced activity has been reported in leukemia patients [14]. There are limited data available to support Aurora kinase A as a relevant molecular target in pediatric cancers besides the report by Shang et al. [38] and the PPTP’s previous report of MLN8237 Stage 1 testing [23]. In this latter publication, high levels of activity were obtained against several solid tumor models and against ALL xenografts of both T and B lineage. The most intriguing set of results was that MLN8237 performed more impressively than other investigational drugs, and even established drugs, against the neuroblastoma panel as a single agent at its MTD.
The Aurora kinases play critical roles in cell division, and alteration of their expression and function has been associated with oncogenesis. Knockdown of Aurora kinase A using RNA interference results in mitotic spindle defects, mitotic delay, and apoptosis in human cells [39], while overexpression leads to transformation of normal cells [40]. Also, Aurora kinase A is amplified or overexpressed in some adult cancers [41–43], which supports its potential exploitation as a cancer therapeutic target [14]. Similarly, the overexpression of Aurora kinase A has been postulated as predictive of susceptibility to inhibition of the specific kinase activity. Thus, Ewing sarcomas, with genetic alterations that enhance Aurora kinase A expression [37], should have higher sensitivity than the lower expressing neuroblastoma or ALL panels. The results presented in this study confirm our previous results of high-level activity for MLN8237 against neuroblastoma and ALL xenografts, which express markedly lower Aurora kinase A levels compared to other PPTP xenografts [23], thereby calling into question the premise that overexpression of Aurora kinase A is associated with more effective cell kill upon kinase inhibition. Although the Ewing sarcoma xenografts had slightly increased expression of AURKA compared to the median for all xenografts, our study did not confirm enhances in sensitivity to MLN8237 in vitro or in vivo. Indeed, the gene copy number analysis for AURKA appears to support an inverse relationship between Aurora kinase A expression and sensitivity. Increased copy number was present in half of the rhabdomyosarcomas and in 14 of the solid tumors. Loss of copy number was detected in 7 solid tumors and ALL-17. Further, the correlation between gene expression variation and copy number variation was strong, placing this locus in the top 1.6% of all genes tested. Although there is no absolute relationship between copy number variation and tumor sensitivity, of the 14 solid tumors with increased copy number, there were only two that showed sensitivity to MLN8237 (1 PR, 1 SD). In contrast, of the eight models demonstrating decreased copy number, there were five sensitive models (3 MCR, 1 CR, and 1 SD).
The in vitro activity of MLN8237 against the Ewing sarcoma and neuroblastoma extended panels (n = 11 and n = 17, respectively) is consistent with the PPTP’s Stage 1 results for MLN8237, which showed median relative and absolute IC50 values against all of the cell lines in the PPTP in vitro panel of 49 and 61 nM, respectively [23]. The larger number of Ewing and neuroblastoma cell lines described in this report compared to those studied in Stage 1 testing allowed detection of significantly lower IC50 values for the neuroblastoma cell lines compared to the Ewing sarcoma cell lines. Further, one Ewing sarcoma cell line (CHLA-56) was resistant to MLN8237 (IC50 > 10 μM). The identification of this highly resistant cell line places it as a valuable tool for identifying resistance mechanisms and warrants further investigation. Recently, a functional Aurora kinase A mutation (T217D) that renders the kinase impervious to MLN8054 and MLN8237 inhibition has been reported [44] and points to a mechanism of resistance independent from levels of expression.
The efficacy of MLN8237 treatment in vivo at its MTD was confirmed against the xenograft panel included in this report. Out of 10 xenografts also evaluated in the previous report, only one (OS-1) was scored more than one response category apart from its previous score [23]. We have confirmed the high level of activity of MLN8237 against xenograft models of neuroblastoma and ALL, when administered as a single agent at its MTD. This further demonstrates the potential relevance of Aurora kinase A inhibition for neuroblastoma cancer treatment. However, the efficacy of MLN8237 (as indicated by the Median Group Response) was reduced or lost for most of the solid tumor models with dose reduction (Fig. 1). Thus, at 0.5MTD, only two xenografts exhibited an objective response, and at 0.25MTD, only one xenograft was classified as PR. In contrast, the dose–response relationship for the ALL xenografts was not as steep, with all three models exhibiting objective responses at 0.5MTD and only one not reaching an objective response upon further reduction to 0.25MTD.
Data for the pharmacokinetics of MLN8237 in patients have recently been presented [45]. In patients receiving 50 mg BID, the C max and AUC0–24 h were 1.3 and 40 μM h, respectively. At the recommended phase 2 dose of 50 mg BID for 7 days, average trough concentrations exceeded 1 μM, the efficacious concentration estimated in previous preclinical work. In mice receiving MLN8237 at 10 mg/kg, the C max and AUC0–24 h were 16 and 39μM h, respectively, with the 12 h level being 1.2 μM. Thus, results presented here suggest that drug exposures achievable in patients may induce responses in only the most sensitive of tumors and that dose intensity and scheduling may be critical as a minority of the solid tumor models exhibited objective responses at this level of drug exposure.
When comparing the plasma exposure of MLN8237 to the pharmacodynamic response, the peak of pharmacodynamic activity (~8–12 h) was delayed relative to the peak plasma exposure (0.5 h). This is consistent with previous observations using the Aurora kinase A inhibitor MLN8054 in a colon tumor xenograft [16] and is likely due to the time it takes for a sufficient number of cells to transit the cell cycle and accumulate in mitosis subsequent to Aurora kinase A inhibition as well as to the time during which MLN8237 drug levels are above a threshold level required for Aurora kinase A inhibition. The comparable mitotic indices estimated using MPM2 and pHistH3 as mitotic markers are consistent with specific inhibition of Aurora kinase A by MLN8237 in vivo, as histone H3 is phosphorylated by Aurora kinase B [46].
A likely critical step in the development of MLN8237 for use in the treatment of pediatric cancers is the development of effective drug combinations. The limited activity observed at reduced doses of MLN8237 as a single agent against most solid tumor xenografts may be overcome if synergistic interactions with other drugs can be identified. Combinations of MLN8237 with established drugs against in vivo models of pediatric solid tumors and ALL are under evaluation by the PPTP.
The cumulative evidence of anti-tumor activity observed in preclinical testing together with the results presented here provides strong rationale for expeditious evaluation of MLN8237 in the childhood cancer setting. A pediatric phase 1/2 trial was opened in the Children’s Oncology Group Phase 1 Consortium during 2008. As results from that clinical trial emerge, it will be crucial to correlate the observed anti-tumor activities with pharmacokinetic measurements to assess whether drug levels are in the range associated with substantial preclinical activity.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
This work was supported by NO1-CM-42216, NO1-CM91001-03, CA21765, and CA108786 from the National Cancer Institute and used MLN8237 supplied by Millennium Pharmaceuticals, Inc. Sherry Ansher, Catherine A. Billups, Joshua Courtright, Mila Dolotin, Edward Favours, Henry S. Friedman, Debbie Payne-Turner, Charles Stopford, Chandra Tucker, Joe Zeidner, Ellen Zhang, and Jian Zhang contributed to this work in addition to the authors. Children’s Cancer Institute Australia for Medical Research is affiliated with the University of New South Wales and Sydney Children’s Hospital.
Conflict of interest
JW, MM, and JE are employees of Millennium Pharmaceuticals, Inc. The authors consider that there are no actual or perceived conflicts of interest.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- 1.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100(1):57–70. doi: 10.1016/S0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 2.Weaver B, Cleveland D. Decoding the links between mitosis, cancer, and chemotherapy: the mitotic checkpoint, adaptation, and cell death. Cancer Cell. 2005;8(1):7–12. doi: 10.1016/j.ccr.2005.06.011. [DOI] [PubMed] [Google Scholar]
- 3.Jackson JR, Patrick DR, Dar MM, Huang PS. Targeted anti-mitotic therapies: can we improve on tubulin agents? Nat Rev Cancer. 2007;7(2):107–117. doi: 10.1038/nrc2049. [DOI] [PubMed] [Google Scholar]
- 4.Vijapurkar U, Wang W, Herbst R. Potentiation of kinesin spindle protein inhibitor-induced cell death by modulation of mitochondrial and death receptor apoptotic pathways. Cancer Res. 2007;67(1):237–245. doi: 10.1158/0008-5472.CAN-06-2406. [DOI] [PubMed] [Google Scholar]
- 5.Henderson MC, Shaw YJ, Wang H, Han H, Hurley LH, Flynn G, Dorr RT, Von Hoff DD. UA62784, a novel inhibitor of centromere protein E kinesin-like protein. Mol Cancer Ther. 2009;8(1):36–44. doi: 10.1158/1535-7163.MCT-08-0789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Garuti L, Roberti M, Bottegoni G. Small molecule aurora kinases inhibitors. Curr Med Chem. 2009;16(16):1949–1963. doi: 10.2174/092986709788682227. [DOI] [PubMed] [Google Scholar]
- 7.Warner SL, Gray PJ, Von Hoff DD. Tubulin-associated drug targets: aurora kinases, Polo-like kinases, and others. Semin Oncol. 2006;33(4):436–448. doi: 10.1053/j.seminoncol.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 8.Cowley DO, Rivera-Perez JA, Schliekelman M, He YJ, Oliver TG, Lu L, O’Quinn R, Salmon ED, Magnuson T, Van Dyke T. Aurora-A kinase is essential for bipolar spindle formation and early development. Mol Cell Biol. 2009;29(4):1059–1071. doi: 10.1128/MCB.01062-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wysong DR, Chakravarty A, Hoar K, Ecsedy JA. The inhibition of Aurora A abrogates the mitotic delay induced by microtubule perturbing agents. Cell Cycle. 2009;8(6):876–888. doi: 10.4161/cc.8.6.7897. [DOI] [PubMed] [Google Scholar]
- 10.Vader G, Lens SM. The Aurora kinase family in cell division and cancer. Biochim Biophys Acta. 2008;1786(1):60–72. doi: 10.1016/j.bbcan.2008.07.003. [DOI] [PubMed] [Google Scholar]
- 11.Macurek L, Lindqvist A, Lim D, Lampson MA, Klompmaker R, Freire R, Clouin C, Taylor SS, Yaffe MB, Medema RH. Polo-like kinase-1 is activated by aurora A to promote checkpoint recovery. Nature. 2008;455(7209):119–123. doi: 10.1038/nature07185. [DOI] [PubMed] [Google Scholar]
- 12.Seki A, Coppinger JA, Jang CY, Yates JR, Fang G. Bora and the kinase Aurora a cooperatively activate the kinase Plk1 and control mitotic entry. Science. 2008;320(5883):1655–1658. doi: 10.1126/science.1157425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou H, Kuang J, Zhong L, Kuo W, Gray J, Sahin A, Brinkley B, Sen S. Tumour amplified kinase STK15/BTAK induces centrosome amplification, aneuploidy and transformation. Nat Genet. 1998;20(2):189–193. doi: 10.1038/2496. [DOI] [PubMed] [Google Scholar]
- 14.Gautschi O, Heighway J, Mack PC, Purnell PR, Lara PN, Jr, Gandara DR. Aurora kinases as anticancer drug targets. Clin Cancer Res. 2008;14(6):1639–1648. doi: 10.1158/1078-0432.CCR-07-2179. [DOI] [PubMed] [Google Scholar]
- 15.Carpinelli P, Moll J. Aurora kinase inhibitors: identification and preclinical validation of their biomarkers. Exp Opin Therap Targets. 2008;12(1):69–80. doi: 10.1517/14728222.12.1.69. [DOI] [PubMed] [Google Scholar]
- 16.Manfredi MG, Ecsedy JA, Meetze KA, Balani SK, Burenkova O, Chen W, Galvin KM, Hoar KM, Huck JJ, LeRoy PJ, Ray ET, Sells TB, Stringer B, Stroud SG, Vos TJ, Weatherhead GS, Wysong DR, Zhang M, Bolen JB, Claiborne CF. Antitumor activity of MLN8054, an orally active small-molecule inhibitor of Aurora A kinase. PNAS. 2007;104(10):4106–4111. doi: 10.1073/pnas.0608798104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu Q, Kaneko S, Yang L, Feldman RI, Nicosia SV, Chen J, Cheng JQ. Aurora-A abrogation of p53 DNA binding and transactivation activity by phosphorylation of serine 215. J Biol Chem. 2004;279(50):52175–52182. doi: 10.1074/jbc.M406802200. [DOI] [PubMed] [Google Scholar]
- 18.Crosio C, Fimia G, Loury R, Kimura M, Okano Y, Zhou H, Sen S, Allis C, Sassone-Corsi P. Mitotic phosphorylation of histone H3: spatio-temporal regulation by mammalian aurora kinases. Mol Cell Biol. 2002;22(3):874–885. doi: 10.1128/MCB.22.3.874-885.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maris JM. Unholy matrimony: aurora A and N-Myc as malignant partners in neuroblastoma. Cancer Cell. 2009;15(1):5–6. doi: 10.1016/j.ccr.2008.12.008. [DOI] [PubMed] [Google Scholar]
- 20.Otto T, Horn S, Brockmann M, Eilers U, Schuttrumpf L, Popov N, Kenney AM, Schulte JH, Beijersbergen R, Christiansen H, Berwanger B, Eilers M. Stabilization of N-Myc is a critical function of aurora A in human neuroblastoma. Cancer Cell. 2009;15(1):67–78. doi: 10.1016/j.ccr.2008.12.005. [DOI] [PubMed] [Google Scholar]
- 21.Dar AA, Belkhiri A, Ecsedy J, Zaika A, El-Rifai W. Aurora kinase A inhibition leads to p73-dependent apoptosis in p53-deficient cancer cells. Cancer Res. 2008;68(21):8998–9004. doi: 10.1158/0008-5472.CAN-08-2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yao J-e, Yan M, Guan Z, Pan C-b, Xia L-p, Li C-x, Wang L-h, Long Z-j, Zhao Y, Li M-w, Zheng F-m, Xu J, Lin D-j, Liu Q. Aurora-A down-regulates IkappaBalpha via Akt activation and interacts with insulin-like growth factor-1 induced phosphatidylinositol 3-kinase pathway for cancer cell survival. Mol Cancer. 2009;8(1):95. doi: 10.1186/1476-4598-8-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maris JM, Morton CL, Gorlick R, Kolb EA, Lock R, Carol H, Keir ST, Reynolds CP, Kang MH, Wu J, Smith MA, Houghton PJ. Initial testing of the aurora kinase a inhibitor MLN8237 by the pediatric preclinical testing program (PPTP) Pediatr Blood Cancer. 2010;55(1):26–34. doi: 10.1002/pbc.22430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Frgala T, Kalous O, Proffitt RT, Reynolds CP. A fluorescence microplate cytotoxicity assay with a 4-log dynamic range that identifies synergistic drug combinations. Mol Cancer Ther. 2007;6(3):886–897. doi: 10.1158/1535-7163.MCT-04-0331. [DOI] [PubMed] [Google Scholar]
- 25.Houghton PJ, Morton CL, Kolb EA, Lock R, Carol H, Reynolds CP, Keshelava N, Maris JM, Keir ST, Wu J, Smith MA. Initial testing (stage 1) of the proteasome inhibitor bortezomib by the pediatric preclinical testing program. Pediatr Blood Cancer. 2008;50(1):37–45. doi: 10.1002/pbc.21214. [DOI] [PubMed] [Google Scholar]
- 26.Kang M, Smith MA, Morton CL, Keshelava N, Houghton PJ, Reynolds CP (2011) National cancer institute pediatric preclinical testing program: model description for in vitro cytotoxicity testing. Pediatr Blood Cancer 56:239–249 [DOI] [PMC free article] [PubMed]
- 27.Friedman HS, Colvin OM, Skapek SX, Ludeman SM, Elion GB, Schold SC, Jr, Jacobsen PF, Muhlbaier LH, Bigner DD. Experimental chemotherapy of human medulloblastoma cell lines and transplantable xenografts with bifunctional alkylating agents. Cancer Res. 1988;48(15):4189–4195. [PubMed] [Google Scholar]
- 28.Graham C, Tucker C, Creech J, Favours E, Billups CA, Liu T, Fouladi M, Freeman BB, 3rd, Stewart CF, Houghton PJ. Evaluation of the antitumor efficacy, pharmacokinetics, and pharmacodynamics of the histone deacetylase inhibitor depsipeptide in childhood cancer models in vivo. Clin Cancer Res. 2006;12(1):223–234. doi: 10.1158/1078-0432.CCR-05-1225. [DOI] [PubMed] [Google Scholar]
- 29.Peterson JK, Tucker C, Favours E, Cheshire PJ, Creech J, Billups CA, Smykla R, Lee FY, Houghton PJ (2005) In vivo evaluation of ixabepilone (BMS247550), a novel epothilone B derivative, against pediatric cancer models. Clin Cancer Res 11(19 Pt 1):6950–6958 [DOI] [PubMed]
- 30.Liem NL, Papa RA, Milross CG, Schmid MA, Tajbakhsh M, Choi S, Ramirez CD, Rice AM, Haber M, Norris MD, MacKenzie KL, Lock RB. Characterization of childhood acute lymphoblastic leukemia xenograft models for the preclinical evaluation of new therapies. Blood. 2004;103(10):3905–3914. doi: 10.1182/blood-2003-08-2911. [DOI] [PubMed] [Google Scholar]
- 31.Houghton PJ, Morton CL, Tucker C, Payne D, Favours E, Cole C, Gorlick R, Kolb EA, Zhang W, Lock R, Carol H, Tajbakhsh M, Reynolds CP, Maris JM, Courtright J, Keir ST, Friedman HS, Stopford C, Zeidner J, Wu J, Liu T, Billups CA, Khan J, Ansher S, Zhang J, Smith MA. The pediatric preclinical testing program: description of models and early testing results. Pediatr Blood Cancer. 2007;49(7):928–940. doi: 10.1002/pbc.21078. [DOI] [PubMed] [Google Scholar]
- 32.Zhao X, Li C, Paez JG, Chin K, Janne PA, Chen TH, Girard L, Minna J, Christiani D, Leo C, Gray JW, Sellers WR, Meyerson M. An integrated view of copy number and allelic alterations in the cancer genome using single nucleotide polymorphism arrays. Cancer Res. 2004;64(9):3060–3071. doi: 10.1158/0008-5472.CAN-03-3308. [DOI] [PubMed] [Google Scholar]
- 33.Pounds S, Cheng C, Mullighan C, Raimondi SC, Shurtleff S, Downing JR. Reference alignment of SNP microarray signals for copy number analysis of tumors. Bioinformatics. 2009;25(3):315–321. doi: 10.1093/bioinformatics/btn624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Olshen AB, Venkatraman ES, Lucito R, Wigler M. Circular binary segmentation for the analysis of array-based DNA copy number data. Biostatistics. 2004;5(4):557–572. doi: 10.1093/biostatistics/kxh008. [DOI] [PubMed] [Google Scholar]
- 35.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn T, Huber W, Iacus S, Irizarry R, Leisch F, Li C, Maechler M, Rossini AJ, Sawitzki G, Smith C, Smyth G, Tierney L, Yang JY, Zhang J (2004) Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5(10):R80. doi:10.1186/gb-2004-5-10-r80 [DOI] [PMC free article] [PubMed]
- 36.Neale G, Su X, Morton CL, Phelps D, Gorlick R, Lock RB, Reynolds CP, Maris JM, Friedman HS, Dome J, Khoury J, Triche TJ, Seeger RC, Gilbertson R, Khan J, Smith MA, Houghton PJ. Molecular characterization of the pediatric preclinical testing panel. Clin Cancer Res. 2008;14(14):4572–4583. doi: 10.1158/1078-0432.CCR-07-5090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wakahara K, Ohno T, Kimura M, Masuda T, Nozawa S, Dohjima T, Yamamoto T, Nagano A, Kawai G, Matsuhashi A, Saitoh M, Takigami I, Okano Y, Shimizu K. EWS-Fli1 Up-regulates expression of the aurora A and Aurora B Kinases. Mol Cancer Res. 2008;6(12):1937–1945. doi: 10.1158/1541-7786.MCR-08-0054. [DOI] [PubMed] [Google Scholar]
- 38.Shang X, Burlingame SM, Okcu MF, Ge N, Russell HV, Egler RA, David RD, Vasudevan SA, Yang J, Nuchtern JG. Aurora A is a negative prognostic factor and a new therapeutic target in human neuroblastoma. Mol Cancer Ther. 2009;8(8):2461–2469. doi: 10.1158/1535-7163.MCT-08-0857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hirota T, Kunitoku N, Sasayama T, Marumoto T, Zhang D, Nitta M, Hatakeyama K, Saya H. Aurora-A and an interacting activator, the LIM protein Ajuba, are required for mitotic commitment in human cells. Cell. 2003;114(5):585–598. doi: 10.1016/S0092-8674(03)00642-1. [DOI] [PubMed] [Google Scholar]
- 40.Katayama H, Brinkley WR, Sen S. The Aurora kinases: role in cell transformation and tumorigenesis. Cancer Metastasis Rev. 2003;22(4):451–464. doi: 10.1023/A:1023789416385. [DOI] [PubMed] [Google Scholar]
- 41.Sen S, Zhou H, Zhang RD, Yoon DS, Vakar-Lopez F, Ito S, Jiang F, Johnston D, Grossman HB, Ruifrok AC, Katz RL, Brinkley W, Czerniak B. Amplification/overexpression of a mitotic kinase gene in human bladder cancer. J Natl Cancer Inst. 2002;94(17):1320–1329. doi: 10.1093/jnci/94.17.1320. [DOI] [PubMed] [Google Scholar]
- 42.Bischoff JR, Anderson L, Zhu Y, Mossie K, Ng L, Souza B, Schryver B, Flanagan P, Clairvoyant F, Ginther C, Chan CS, Novotny M, Slamon DJ, Plowman GD. A homologue of Drosophila aurora kinase is oncogenic and amplified in human colorectal cancers. EMBO J. 1998;17(11):3052–3065. doi: 10.1093/emboj/17.11.3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Anand S, Penrhyn-Lowe S, Venkitaraman AR. AURORA-A amplification overrides the mitotic spindle assembly checkpoint, inducing resistance to Taxol. Cancer Cell. 2003;3(1):51–62. doi: 10.1016/S1535-6108(02)00235-0. [DOI] [PubMed] [Google Scholar]
- 44.Sloane DA, Trikic MZ, Chu MLH, Lamers MBAC, Mason CS, Mueller I, Savory WJ, Williams DH, Eyers PA. Drug-resistant aurora A mutants for cellular target validation of the small molecule kinase inhibitors MLN8054 and MLN8237. ACS Chem Biol. 2010;5(6):563–576. doi: 10.1021/cb100053q. [DOI] [PubMed] [Google Scholar]
- 45.Dees EC, Infante JR, Burris HA, Astsaturov IA, Stinchcombe T, Liu H, Galvin K, Venkatakrishnan K, Fingert HJ, Cohen RB (2010) Phase I study of the investigational drug MLN8237, an Aurora A kinase (AAK) inhibitor, in patients (pts) with solid tumors. J Clin Oncol 28(15 suppl) (May 20 Supplement), Abstr #3010
- 46.Giet R, Glover DM. Drosophila aurora B kinase is required for histone H3 phosphorylation and condensin recruitment during chromosome condensation and to organize the central spindle during cytokinesis. J Cell Biol. 2001;152(4):669–682. doi: 10.1083/jcb.152.4.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.