Summary
The Id (inhibitor of differentiation or DNA binding) family of transcription regulators plays an important role in cell proliferation, differentiation and senescence. However, regulation of Id expression during these processes is poorly understood. Id proteins are known to undergo rapid turnover mediated by the ubiquitin-proteasome pathway. Anaphase-promoting complex has been shown to ubiquitinate Id2, but E3 ubiquitin ligase(s) that ubiquitinate other Id family members are not known. Here we report for the first time the identification of Smurf2 as the E3 ligase that ubiquitinates Id1 and Id3. Smurf2-mediated ubiquitination and consequent degradation of Id1 or Id3 plays an important role in the regulation of Id expression in senescent cells. Furthermore, we found that Id1 is the mediator through which Smurf2 regulates p16 expression, providing a mechanistic link between Smurf2 and p16 expression during senescence.
Keywords: Smurf2, Id1, ubiquitination, E3 ligase, p16, senescence
Introduction
The Id (inhibitor of differentiation or DNA binding) family of transcriptional regulators has a helix-loop-helix (HLH) domain, but lacks a basic DNA-binding domain. The four mammalian members (Id1, Id2, Id3 and Id4) associate with and antagonize the DNA binding ability of basic HLH (bHLH) transcription factors (Benezra et al. 1990; Perk et al. 2005). Acting as dominant negative transcriptional regulators, Id proteins are involved in diverse cellular processes including cell proliferation, senescence, differentiation, and angiogenesis (Iavarone et al. 1994; Lyden et al. 1999; Perk et al. 2005). Different cell types express unique combinations of the four Id family members. Id1 and Id3 show a widespread expression in many types of cells, and share a similar expression pattern during mouse embryonic development (Lyden et al. 1999), whereas the expression of Id2 and Id4 shows a more restricted pattern (Riechmann et al. 1994). Genetic studies of Id knockout mice reveal non-overlapping functions of the four Id genes in different cell types, with some functional redundancy between Id1 and Id3 (Lyden et al. 1999; Perk et al. 2005).
The expression of Id1 is decreased in many cell lineages during senescence (Hara et al. 1994; Nickoloff et al. 2000; Schwarze et al. 2002; Tang et al. 2002), quiescence (Christy et al. 1991; Barone et al. 1994; Hara et al. 1994; Nickoloff et al. 2000), or differentiation (Benezra et al. 1990; Sun et al. 1991; Kreider et al. 1992). Serum or growth factors induce Id1 expression in quiescent cells (Christy et al. 1991; Barone et al. 1994; Hara et al. 1994), and inhibition of Id1 blocks quiescent cells from re-entering into cell cycle (Barone et al. 1994; Hara et al. 1994). In contrast, serum stimulation does not induce Id1 expression in senescent cells (Hara et al. 1994), suggesting that the expression of Id1 is regulated differentially between quiescent and senescent cells. Senescence is activated by two major pathways, p53- p21CIP1/WAF1 (p21) and p16INK4a (p16)-pRb (Ide et al. 1983; Shay et al. 1991). SV40 T antigen, which inhibits p53 and pRb, can reinitiate DNA synthesis in senescent cells (Ide et al. 1983). A mutant SV40 T antigen that only inhibits p53 but not pRb is unable to stimulate DNA synthesis in senescent cells. However, this mutant SV40 T antigen in cooperation with Id1 can reinitiate DNA synthesis (Hara et al. 1996), suggesting that Id1 antagonizes the p16-pRb pathway. Consistent with this notion, Id1 is found to suppress p16 expression through its ability to sequester bHLH transcription factor E47 and prevent E47 from transactivating p16 (Alani et al. 2001; Zheng et al. 2004). Down-regulation of Id1 has been found to activate senescence and p16 expression (Alani et al. 2001; Zheng et al. 2004), whereas ectopic expression of Id1 delays senescence in human and mouse cells (Hara et al. 1996; Nickoloff et al. 2000; Tang et al. 2002; Cummings et al. 2008; Suh et al. 2008), suggesting that Id1 plays a critical role in replicative senescence. In addition, Id1 is implicated in regulating p16 expression during stress-induced senescence. Aberrant activation of Ras-Raf-MEK signaling induces senescence and p16 expression (Serrano et al. 1997). Phosphorylation of Ets family transcription factor Ets2 by Ras-Raf-MEK signaling leads to transactivation of p16, which is antagonized by Id1 through its association with Ets2 (Ohtani et al. 2001). DNA damage also induces senescence and p16 expression. In response to DNA damage, Id1 expression decreases in a p53-dependent manner. Importantly, overexpression of Id1 attenuates DNA damage-induced senescence (Qian & Chen 2008).
Despite the importance of Id1 in senescence regulation, the mechanism by which Id1 is regulated during senescence is not entirely clear. Id proteins are known to undergo rapid turnover, and ubiquitin-proteasome mediated degradation regulates the steady-state levels of Id proteins (Bounpheng et al. 1999; Trausch-Azar et al. 2004). However, the E3 ubiquitin ligase(s) that mediate ubiquitination of Id1 or Id3 have not been identified. Here we report the identification of Smurf2 as the E3 ligase that ubiquitinates Id1 and Id3. Smurf2-mediated ubiquitination of Id1/Id3 plays an important role in the decreased Id expression in senescent cells. Furthermore, ubiquitination and consequent degradation of Id1 by Smurf2 is responsible for Smurf2-mediated p16 regulation during senescence, providing a mechanistic link between Smurf2 and p16 during senescence.
Results
Smurf2 regulates steady-state protein level of Id1 and Id3
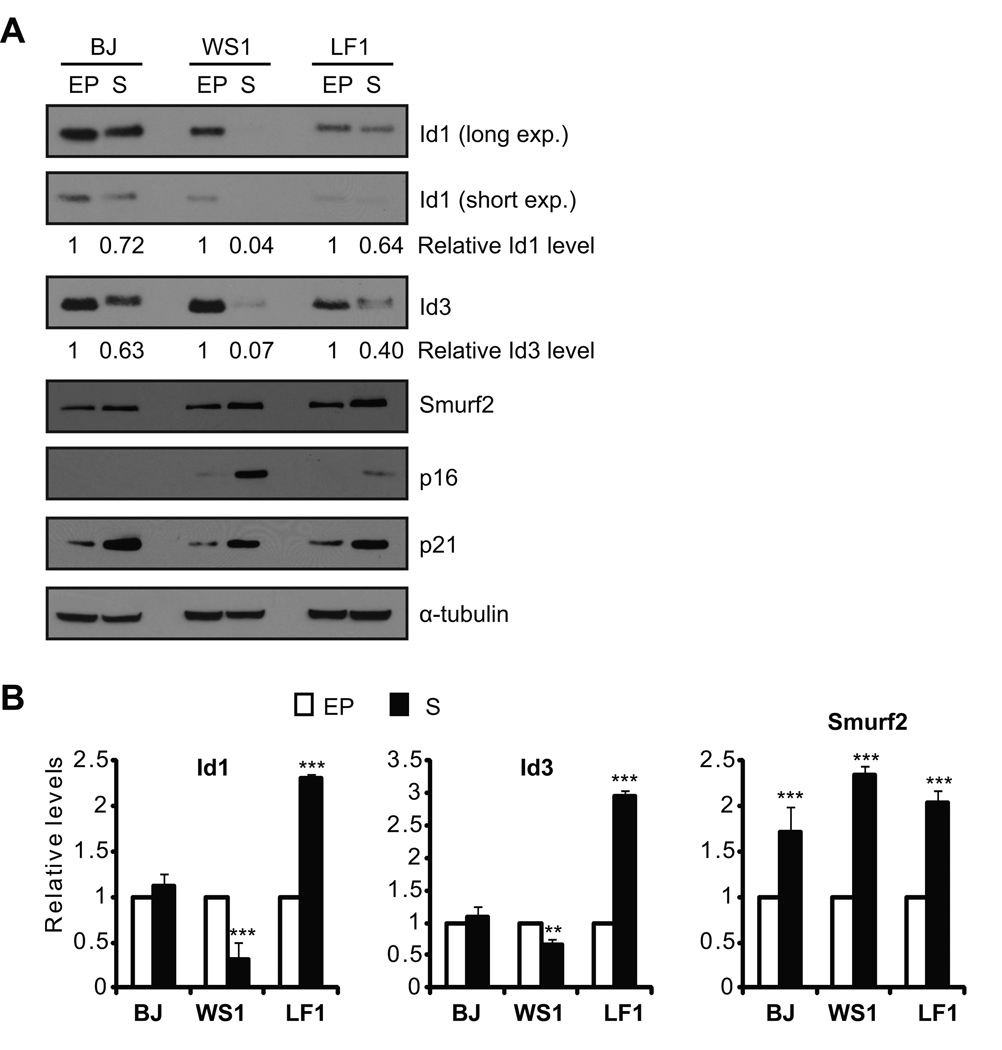
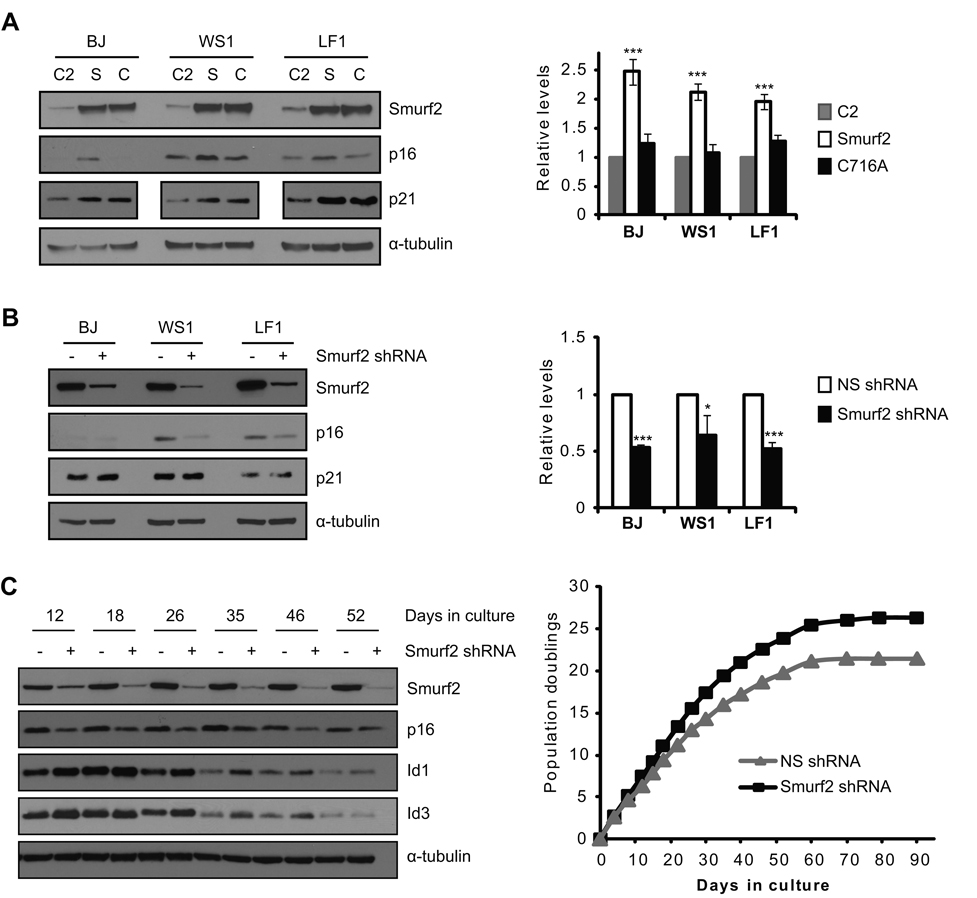
Human primary fibroblasts were passaged in culture until they entered replicative senescence, as indicated by elevated expression of p16 and p21 (Fig. 1A) and positive staining for senescence-associated β-galactosidase (Fig. S1). We found that steady-state protein levels of Id1 were decreased in senescent fibroblasts as compared to early passage proliferating cells (Fig. 1A), consistent with previous reports (Hara et al. 1994; Ohtani et al. 2001; Zheng et al. 2004). Similar results were obtained for the closely related Id3 (Fig. 1A). Surprisingly, transcripts of Id1 or Id3 in senescent cells showed a cell line-dependent change as compared to early passage cells. Only WS1 cells showed decreased Id1/Id3 transcripts during senescence, whereas Id1/Id3 transcripts in senescent BJ and LF1 cells did not change or even increased (Fig. 1B). These results suggest that regulation at protein level, possibly through protein degradation, plays an important role in the decreased expression of Id1/Id3 in senescent cells.
Fig. 1.
Decreased expression of Id1 and Id3 in senescent cells. (A) Western blot and (B) quantitative RT-PCR analyses of Id1, Id3 and Smurf2 in early passage (EP) and senescent (S) human fibroblasts. Id1 or Id3 proteins were quantitated using NIH ImageJ, and normalized with α-tubulin. Relative Id1 or Id3 protein levels in early passage cells were set to be 1. Relative transcript levels of Id1, Id3 or Smurf2 in early passage cells were set to be 1 after normalization with β-actin. Error bars were calculated from standard deviations of at least three independent experiments. Two-tailed and unpaired Student t–test was used in statistical analysis, and statistic significance is indicated as: * (P<0.05), ** (P<0.01), and *** (P<0.001).
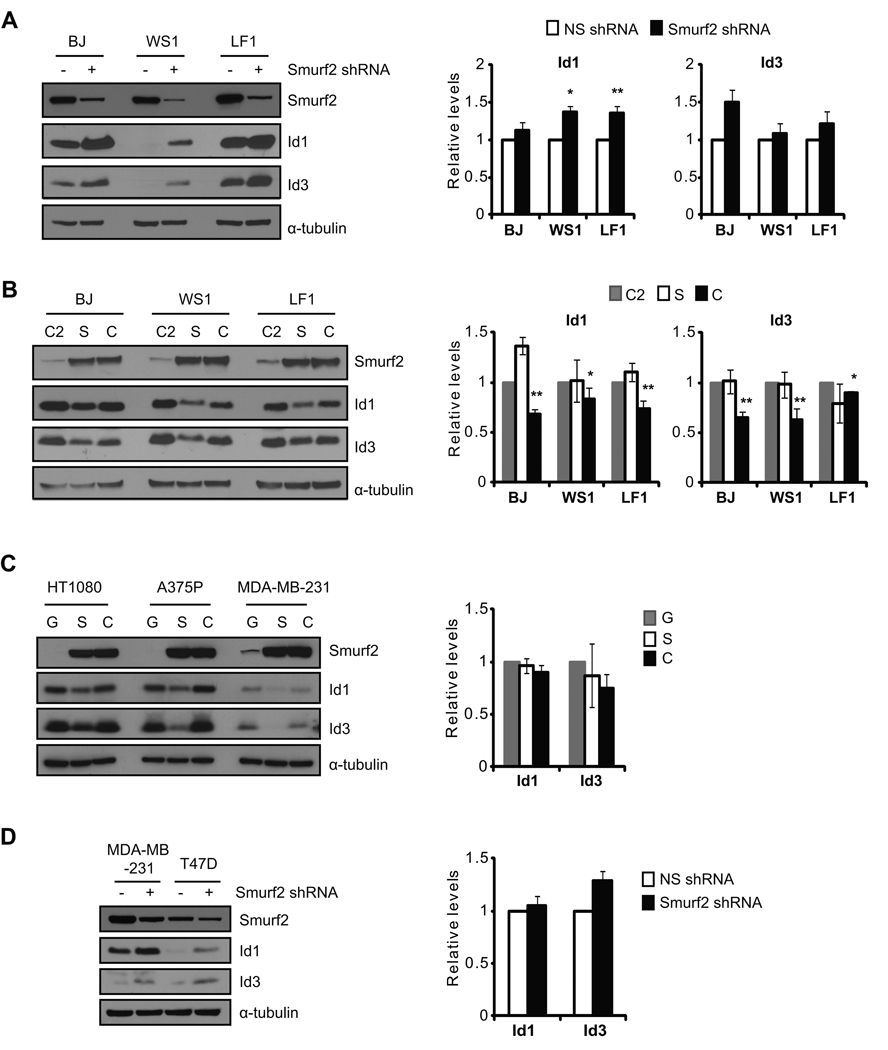
Previously we have found that the expression of Smurf2 increases in senescent cells (Zhang & Cohen 2004). The inverse correlation between Smurf2 and Id1/Id3 expression in senescent cells (Fig. 1) prompted us to ask whether Smurf2 is involved in the regulation of Id1/Id3 expression during senescence. Senescent human fibroblasts were infected with lentivirus expressing a short hairpin RNA (shRNA) to knockdown Smurf2 expression. We found that down-regulation of Smurf2 by shRNA led to elevated steady-state protein levels of Id1 and Id3 (Fig. 2A), suggesting that Smurf2 is required for the decreased Id1/Id3 expression in senescent cells. Quantitative RT-PCR results showed that Id1/Id3 transcripts were increased only slightly in cells with Smurf2 down-regulation (Fig. 2A).
Fig. 2.
Smurf2 regulates the steady-state protein levels of Id1 and Id3. The expression of Smurf2 was down-regulated by shRNA in (A) senescent fibroblasts or (D) tumor cells. Expression of Id1 and Id3 was analyzed in Western blot and quantitative RT-PCR. Relative transcript levels of Id1/Id3 in cells expressing a non-silencing shRNA (NS shRNA) were set to be 1 after normalized with β-actin. Smurf2 (S), C716A (C), C2 or GFP (G) was ectopically expressed in (B) early passage fibroblasts or (C) tumor cells. Expression of Id1 and Id3 was analyzed in Western blot and quantitative RT-PCR. Relative transcript levels of Id1/Id3 in cells expressing C2 or GFP were set to be 1 after normalization with β-actin. Error bars were calculated from standard deviations of at least three independent experiments. Two-tailed and unpaired Student t–test was used in statistical analysis, and statistic significance is indicated as: * (P<0.05), ** (P<0.01), and *** (P<0.001).
To determine whether Smurf2 is sufficient to regulate Id expression, we ectopically expressed Smurf2 in early passage human fibroblasts, and examined the consequence of Smurf2 up-regulation on Id expression. Smurf2’s C2 domain and a ligase mutant C716A, in which the conserved cysteine at residue 716 is replaced by alanine to abolish its E3 ligase activity (Kavsak et al. 2000; Lin et al. 2000; Zhang et al. 2001), were similarly expressed as controls. As shown in Fig. 2B, ectopic expression of Smurf2 resulted in decreased protein levels of Id1 and Id3 as compared to C2 controls, indicating that Smurf2 is sufficient to down-regulate Id1/Id3 expression. Ectopic expression of the ligase mutant C716A at the similar level did not lead to a similar reduction in Id1/Id3 (Fig. 2B), indicating that down-regulation of Id1/Id3 by Smurf2 requires its E3 ligase activity. No significant change in Id1/Id3 transcripts was found in cells expressing Smurf2, while a small decrease in Id1/Id3 transcripts was found with C716A expression when compared to the C2 controls (Fig. 2B), suggesting that the decrease in steady-state protein levels of Id1/Id3 upon Smurf2 up-regulation is unlikely caused by transcriptional alteration.
We have shown previously that ectopic expression of Smurf2 or the ligase mutant C716A induces senescence in early passage cells (Zhang & Cohen 2004; Zhang et al. 2008). To investigate whether Smurf2-mediated regulation of Id proteins is unique during senescence or in fibroblasts, we expressed Smurf2 in human tumor cells (HT-1080, A375P, MDA-MB-231), in which Smurf2 does not induce senescence (Zhang et al. 2008). We found that Id1 and Id3 decreased in cells with ectopic expression of Smurf2, but remained largely unchanged when C716A was similarly expressed (Fig. 2C). Conversely, shRNA knockdown of Smurf2 in human breast cancer cells led to elevated Id1 and Id3 (Fig. 2D), similar to what we found in fibroblasts. Collectively, these results indicate that Smurf2 can regulate Id proteins in cells other than fibroblasts and regardless of whether senescence is activated. At the transcript level, no significant changes were found in Id1/Id3 when Smurf2 was altered in MDA-MB-231 cells (Fig. 2C, 2D), suggesting that Id1/Id3 is not regulated at transcript level, but rather at protein level by Smurf2.
Smurf2 has been implicated in regulating TGF-β signaling through ubiquitination of Smad1/2 and TGF-β receptors (Kavsak et al. 2000; Lin et al. 2000; Zhang et al. 2001). TGF-β and BMP signaling pathways have been implicated in regulating the transcription of Id1 (Hollnagel et al. 1999; Kang et al. 2003). To test whether Smurf2-mediated regulation of Id proteins depends on TGF-β or BMP signaling, we used MDA-MB-231 cells, in which both BMP and TGF-β signaling pathways are impaired by stably expressed shRNA targeting Smad4. We found that ectopic expression of Smurf2 led to decreased protein level of Id1 in these cells with Smad4 down-regulation (Fig. S2), suggesting that TGF-β or BMP signaling is not required for Smurf2-mediated regulation of Id1. This result is consistent with the notion that Smurf2 regulates Id1/Id3 at the protein level instead of the transcript level.
Ubiquitin-proteasome mediated degradation regulates the stability of Id proteins (Bounpheng et al. 1999; Trausch-Azar et al. 2004). The steady-state protein levels of Id1 and Id3 increased in MDA-MB-231 cells when treated with proteasome inhibitor MG132, but not with DMSO control (Fig. S3A). We next treated MDA-MB-231 cells expressing GFP, Smurf2 or C716A with MG132, and found that protein levels of Id1 and Id3 showed a time-dependent increase in all cell lines. Within 4 hours of MG132 treatment, the steady-state levels of Id1/Id3 in cells expressing Smurf2 were similar to those in cells expressing GFP or C716A, albeit starting with lower levels (Fig. S3B). Collectively, these results indicate that regulation of Id1/Id3 by Smurf2 at the protein level is likely through the ubiquitin-proteasome pathway, and further argue that Smurf2 is the E3 ligase responsible for ubiquitination and consequent degradation of Id1/Id3.
Smurf2 forms a complex with Id1 or Id3
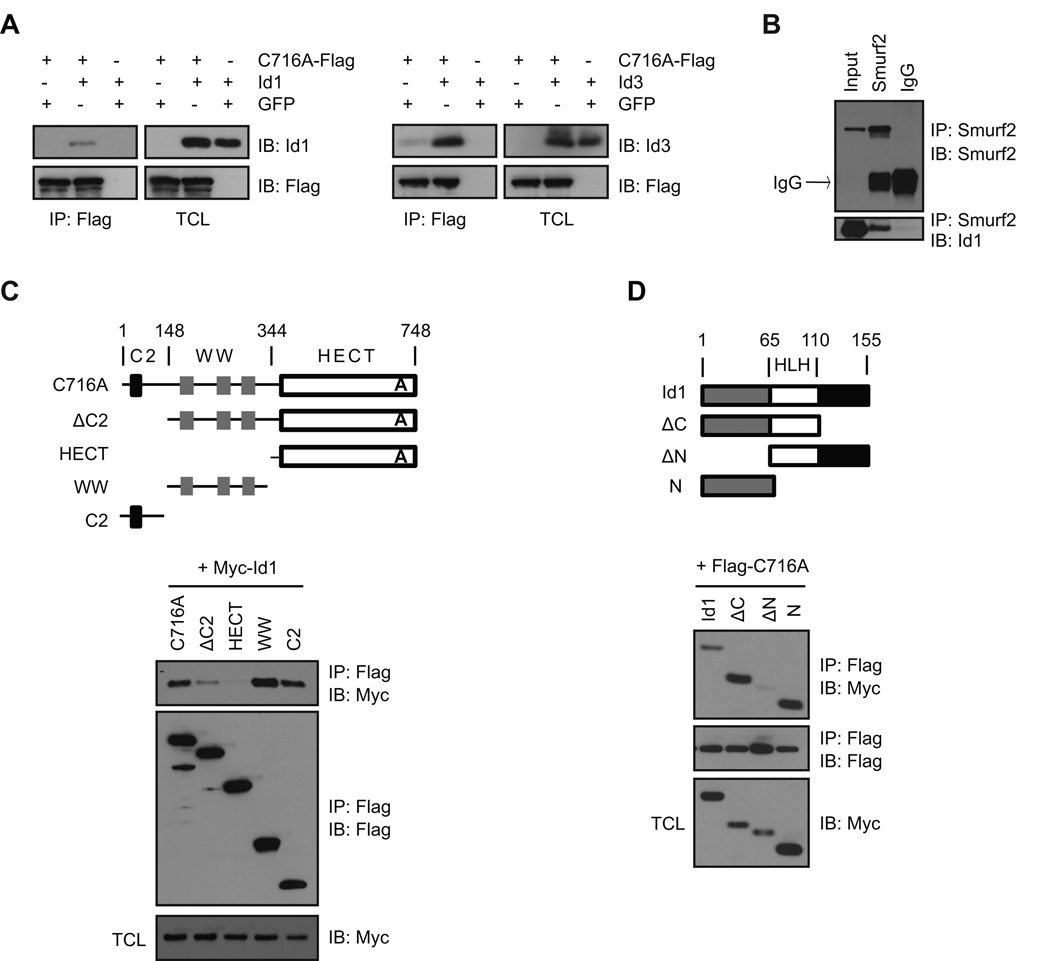
As the C2-WW-HECT class of E3 ligase interacts with protein substrates to catalyze ubiquitination, we wanted to know whether Smurf2 interacts with Id1 or Id3. The ability of Smurf2 to interact with Id proteins was first investigated in 293T cells transfected with Flag-tagged C716A together with Id1 or Id3. The ligase mutant C716A was used to limit potential degradation of Id proteins mediated by Smurf2 and facilitate the detection of Smurf2-Id interactions. As shown in Fig. 3A, Id1 or Id3 co-immunoprecipitated with Flag-tagged C716A, suggesting that Smurf2 forms a complex with Id1 or Id3. We next investigated the Smurf2-Id association at the endogenous levels of expression in MDA-MB-231 cells. Endogenous Id1 and Smurf2 were detected in the same immunocomplex (Fig. 3B), suggesting that Smurf2 interacts with Id1.
Fig. 3.
Smurf2 interacts with Id1/Id3. (A) 293T cells were transfected with constructs as indicated. Immunoprecipitation (IP) with anti-Flag antibody was followed by immunoblot (IB) with anti-Id antibody. (B) MDA-MB-231 cells were treated with MG-132 (20 µM) for 4 hrs before cell lysis. Immunoprecipitation with anti-Smurf2 antibody or IgG control was followed by Western blot. (C) and (D) Determination of domain(s) in Smurf2 and Id1 that mediates Smurf2-Id1 interaction. Deletion constructs of Flag-tagged Smurf2 and Myc-tagged Id1 were shown schematically in (C) and (D). 293T cells were transfected with constructs as indicated. Cell lysates were subjected to immunoprecipitation with anti-Flag antibody. Precipitated immunocomplex were analyzed in Western blot with anti-Myc antibody. TCL: total cell lysates.
To determine the domain(s) in Smurf2 that mediate its interaction with Id1, we made deletion constructs expressing different domains of Smurf2 tagged with Flag (Fig. 3C), and examined their ability to interact with Id1 in co-immunoprecipitation and Western analysis. As shown in Fig. 3C, deletion of C2 domain significantly decreased the interaction between Smurf2 and Id1, whereas deletion of C2 and WW domains (i.e., HECT) effectively abolished Smurf2-Id1 interaction. Furthermore, WW or C2 domain alone can interact with Id1, indicating that WW and C2 domains in Smurf2 mediate its interaction with Id1. Reciprocally, we determined the domain(s) in Id1 that interact with Smurf2 using deletion constructs expressing different regions of Id1 tagged with Myc (Fig. 3D). Because of the short half-life of Id1 protein (Trausch-Azar et al. 2004), MG132 was added 4 hours before cell lysis to stabilize Id1 and facilitate the detection of Smurf2-Id1 interaction. HLH domain or C-terminus of Id1 was not used, because their expression was very low even in the presence of MG132. As shown in Fig. 3D, N-terminus was sufficient to interact with Smurf2, whereas deletion of N-terminus of Id1 abolished the interaction between Id1 and Smurf2, indicating that N-terminus of Id1 is responsible for its interaction with Smurf2.
Smurf2 ubiquitinates Id1
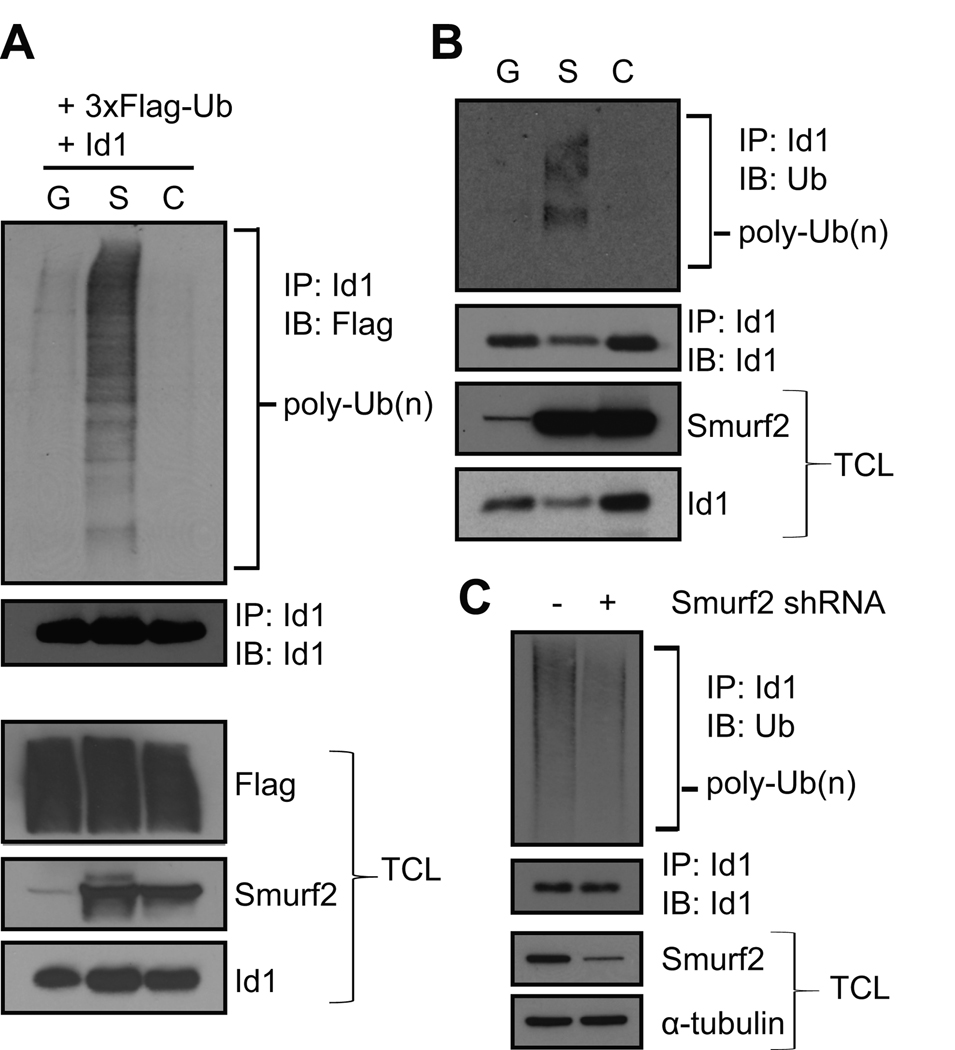
The ability of Smurf2 to regulate the steady-state protein level of Id1 suggests that Smurf2 is the E3 ubiquitin ligase mediating ubiquitination of Id1. To test this hypothesis, we transfected Id1, Flag-tagged ubiquitin and Smurf2 into 293T cells, and investigated whether Smurf2 induces ubiquitination of Id1. GFP and the ligase mutant C716A were used as controls. Ubiquitinated Id1 was detected using anti-Flag antibody in Western blot after Id1 was immunoprecipitated with anti-Id1 antibody. As shown in Fig. 4A, ubiquitination of Id1, as indicated by a smear of bands, was greatly increased in cells expressing Smurf2 as compared to GFP control. Catalytically inactive C716A lost the ability to ubiquitinate Id1, indicating that ubiquitination of Id1 requires the E3 ligase activity of Smurf2. We next investigated whether Smurf2 induces ubiquitination of endogenous Id1 in A375P cells, in which Smurf2, C716A or GFP was stably expressed. Anti-ubiquitin antibody was used to detect ubiquitinated Id1 in Western blot after Id1 was immunoprecipitated with anti-Id1 antibody. Fig. 4B shows that Smurf2, but not GFP or C716A, induced ubiquitination of endogenous Id1. Conversely, we used stably expressed shRNA to knockdown the expression of Smurf2 in senescent LF1 fibroblasts, and found that down-regulation of Smurf2 led to reduced ubiquitination of Id1 (Fig. 4C). Collectively, these results support the notion that Smurf2 ubiquitinates Id1.
Fig. 4.
Smurf2 induces ubiquitination of Id1. (A) 293T cells were transfected with constructs as indicated. MG-132 (20 µM) was added 2 hrs before cell lysis with RIPA buffer plus N-ethylmaleimide (NEM, 10 mM). Id1 was immunoprecipitated with anti-Id1 antibody, followed by immunoblotting of precipitated proteins with anti-Flag antibody. (B) A375P cells expressing Smurf2 (S), C716A (C) or GFP (G) were treated with MG-132 (20 µM) for 2 hrs before cell lysis with RIPA buffer plus NEM (10 mM). Endogenous Id1 was immunoprecipitated with anti-Id1 antibody, and ubiquitinated Id1 was detected in Western blot using anti-ubiquitin antibody. (C) The expression of Smurf2 in senescent LF1 fibroblasts was down-regulated by shRNA, and cells were treated with MG-132 (20 µM) for 4 hrs before cell lysis with RIPA buffer plus NEM (10 mM). Endogenous Id1 was immunoprecipitated with anti-Id1 antibody, and ubiquitinated Id1 was detected in Western blot using anti-ubiquitin antibody. TCL: total cell lysates.
Smurf2 regulates p16 expression in senescent cells
Since Id1 has been shown to repress p16 expression (Alani et al. 2001; Ohtani et al. 2001; Zheng et al. 2004), our finding of Smurf2 ubiquitinating Id1 led us to hypothesize that Smurf2 regulates p16 expression during senescence. To test this hypothesis, we ectopically expressed Smurf2 or C716A in early passage human fibroblasts, and examined the consequence of Smurf2 up-regulation on p16 expression. Smurf2’s C2 domain, which does not induce senescence (Zhang & Cohen 2004), was expressed as a control. As shown in Fig. 5A, ectopic expression of Smurf2 led to increased levels of p16 protein and transcript as compared to C2 control, indicating that Smurf2 is sufficient to induce p16 expression. Furthermore, ectopic expression of C716A at a similar level as Smurf2 did not lead to increased p16 expression (Fig. 5A), indicating that up-regulation of p16 by Smurf2 requires its E3 ubiquitin ligase activity. Similar results were obtained with ectopic expression of Smurf2 in telomerase-immortalized human fibroblasts (Fig. S4). We have shown previously that ectopic expression of Smurf2 or C716A induces p21 expression and senescence in early passage cells (Zhang & Cohen 2004; Zhang et al. 2008). This ligase-dependent regulation of p16 by Smurf2 contrasts with the previously identified ligase-independent regulation of p21 by Smurf2 (Zhang & Cohen 2004) (Fig. 5A). Together, they provide a plausible explanation as how Smurf2 recruits both the p53-p21 and p16-pRb pathways to induce senescence (Zhang & Cohen 2004).
Fig. 5.
Smurf2 regulates the expression of p16. (A) Smurf2 (S), C716A (C) or C2 control was expressed in early passage human fibroblasts. Expression of p16 was analyzed in Western blot and quantitative RT-PCR. Relative transcript levels of p16 in cells expressing C2 were set to be 1 after normalization with β-actin. (B) The expression of Smurf2 was knockdown by shRNA in senescent human fibroblasts. Expression of p16 was analyzed in Western blot and quantitative RT-PCR. Relative transcript levels of p16 in cells expressing a non-silencing shRNA (NS shRNA) were set to be 1 after normalization with β-actin. Error bars were calculated from standard deviations of at least three independent experiments. Two-tailed and unpaired Student t–test was used in statistical analysis, and statistic significance is indicated as: * (P<0.05), ** (P<0.01), and *** (P<0.001). (C) The expression of Smurf2 was knocked down by stably expressed shRNA in mid-passage LF1 fibroblasts. The expression of Smurf2, p16, Id1 and Id3 was determined in Western blot, and replicative lifespan was determined by serial passage of cells expressing Smurf2 shRNA or non-silencing shRNA control.
To investigate whether Smurf2 is required for elevated p16 expression during senescence, lentivirus expressing shRNA targeting Smurf2 was used to infect senescent human fibroblasts. We found that down-regulation of Smurf2 led to decreased p16 expression (Fig. 5B), suggesting that Smurf2 is necessary for p16 elevation in senescent cells. In contrast, p21 expression was largely unchanged upon Smurf2 down-regulation (Fig. 5B), whereas p21 expression was elevated by ectopic expression of Smurf2 or C716A (Fig. 5A) (Zhang & Cohen 2004; Zhang et al. 2008), suggesting that Smurf2 is sufficient but not required to regulate p21 expression during senescence. To investigate the consequence of Smurf2 down-regulation on senescence activation, we stably expressed shRNA to knockdown Smurf2 expression in mid-passage LF1 fibroblasts and determined their replicative lifespan in culture. Down-regulation of Smurf2 resulted in elevated Id1/Id3 and decreased p16 expression in each cell passage as compared to non-silencing shRNA controls (Fig. 5C). More importantly, down-regulation of Smurf2 resulted in delayed entry of these cells into senescence, as indicated by increased population doublings (Fig. 5C) and delayed appearance of positive staining for senescence-associated β-galactosidase (Fig. S5), suggesting that Smurf2 is critical in senescence regulation.
Id1 is responsible for Smurf2-mediated regulation of p16 expression
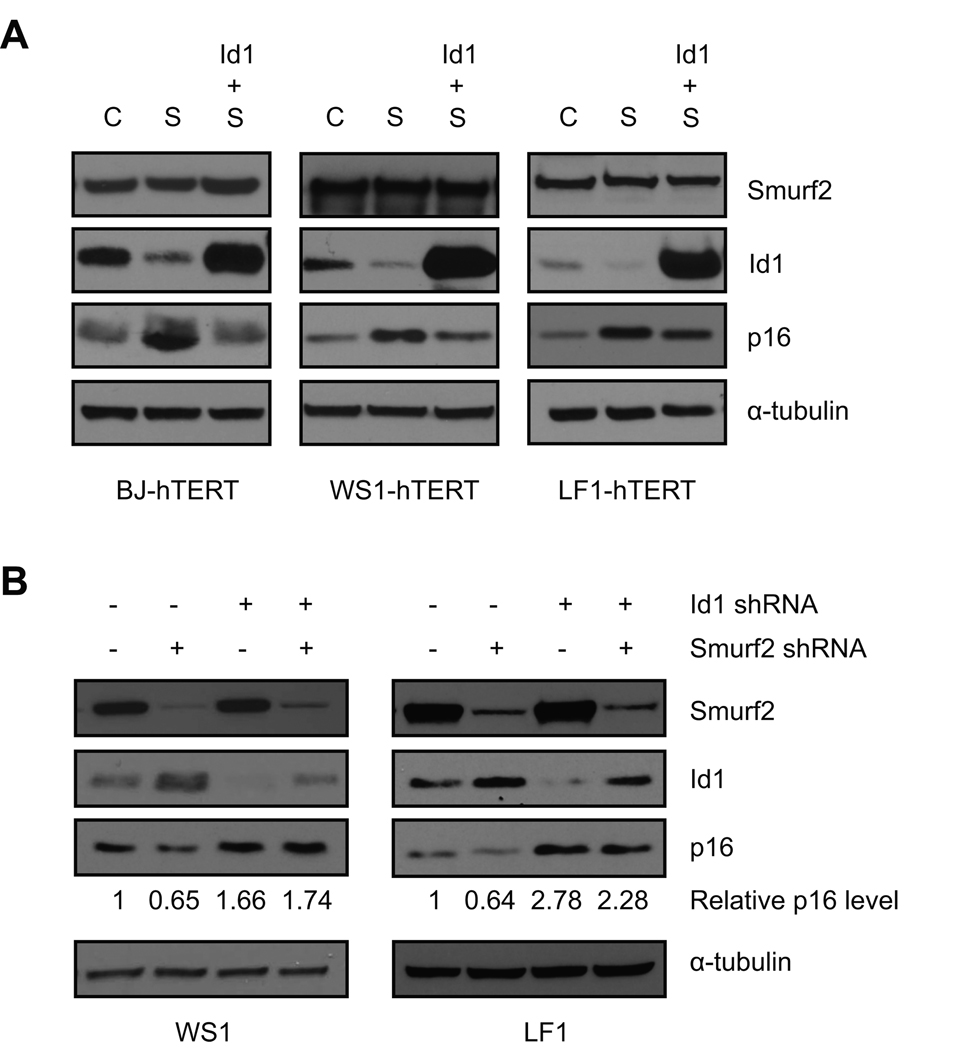
To test whether Id1 is the mediator through which Smurf2 regulates p16 expression, we first investigated whether Id1 can antagonize the ability of Smurf2 to induce p16 expression. As compared to ligase mutant C716A, ectopic expression of Smurf2 led to decreased Id1 and elevated p16 expression (Fig. 6A, lanes 2 vs. 1). When Smurf2 was expressed together with Id1, which compensated for the reduction in endogenous Id1 proteins caused by Smurf2 up-regulation, we found that induction of p16 expression mediated by Smurf2 was largely abrogated (Fig. 6A, lanes 3 vs. 2), suggesting that Id1 is sufficient to antagonize the ability of Smurf2 to induce p16 expression. Next we asked whether Id1 is required for Smurf2-mediated regulation of p16. The expression of Id1 was down-regulated by stably expressed shRNA in late passage human fibroblasts. Consistent with a previous report (Zheng et al. 2004), shRNA knockdown of Id1 expression led to elevated p16 (Fig. 6B, lanes 1 vs. 3). In control cells expressing a non-silencing shRNA, knockdown of Smurf2 expression by shRNA led to decreased expression of p16 (Fig. 6B, lanes 1 vs. 2). In contrast, in cells with Id1 down-regulation, Smurf2-mediated regulation of p16 expression was largely abrogated (Fig. 6B, lanes 3 vs. 4), suggesting that Id1 is necessary for the inhibition of p16 expression as a result of Smurf2 down-regulation. Collectively, these results indicate that Id1 is the mediator through which Smurf2 regulates p16 expression.
Fig. 6.
Id1 mediates the regulation of p16 expression by Smurf2. (A) Smurf2 (S), C716A (C) and Id1 were expressed in TERT-immortalized human fibroblasts as indicated. Expression of p16 was analyzed in Western blot. (B) Expression of Id1 was knockdown by stably expressed shRNA in late passage human fibroblasts. In cells with Id1 down-regulation or control cells expressing non-silencing shRNA (−), shRNA targeting Smurf2 was expressed using lentiviral infection. Expression of p16 was analyzed in Western blot. p16 proteins were quantitated using NIH ImageJ, and normalized with α-tubulin. Relative p16 protein levels in non-silencing controls were set to be 1.
Discussion
To our knowledge, Smurf2 is the first E3 ubiquitin ligase identified to ubiquitinate Id1 and Id3. We found that Smurf2-mediated ubiquitination and consequent degradation of Id1/Id3 played an important role in the decreased Id expression in senescent cells. As Smurf2 is up-regulated in senescent cells but not in quiescent cells (Zhang & Cohen 2004), regulation of Id1 by Smurf2 likely contributes to the differential regulation of Id1 expression between quiescent and senescent cells (Christy et al. 1991; Barone et al. 1994; Hara et al. 1994). Previously we have found that the expression of Smurf2 increases in response to telomere shortening during senescence, and it recruits both the p53-p21 and p16-pRb pathways to activate senescence. While we have found that Smurf2 regulates p21 expression independent of its ligase activity (Zhang & Cohen 2004), whether and how Smurf2 activation leads to the p16-pRb pathway was not known. Our current finding that ubiquitination and degradation of Id1 by Smurf2 is responsible for Smurf2-mediated regulation of p16 expression provides a mechanistic link between Smurf2 and p16 in senescence regulation. Together, the ligase-independent regulation of p21 and ligase-dependent regulation of p16 by Smurf2 provide a plausible explanation as how Smurf2 recruits both the p53-p21 and p16-pRb pathways to induce senescence.
The expression of p16 is increased in senescent cells and with aging in many tissues of human and rodent (Collins & Sedivy 2003; Kim & Sharpless 2006). Elevated expression of p16 is sufficient to induce senescence (McConnell et al. 1998; Vogt et al. 1998), while inactivation of p16 delays senescence in some human cells (Bond et al. 2004; Brookes et al. 2004), suggesting that p16 plays a critical role in senescence regulation. Furthermore, age-dependent increase of p16 expression is associated with a decline in stem cell self-renewal in brain, pancreas, and hematopoietic system. Stem cells derived from old mice lacking p16 have enhanced regenerative capacity (Janzen et al. 2006; Krishnamurthy et al. 2006; Molofsky et al. 2006), whereas the opposite is true in mice with increased p16 expression (Krishnamurthy et al. 2006). Despite the importance of p16 in senescence and aging, it is not entirely clear how p16 expression is regulated during senescence or aging (Collins & Sedivy 2003; Gil & Peters 2006; Kim & Sharpless 2006). Polycomb complex proteins have been shown to inhibit INK4a/Arf locus, which encodes p16 and Arf. This regulation may be responsible for coordinated expression of p16 and Arf during senescence and aging (Gil & Peters 2006; Kim & Sharpless 2006). However, the expression of p16 is also regulated independently of Arf expression. Id1 has been found to specifically suppress p16 but not Arf expression through its ability to sequester E47 or Ets2 and prevent them from transactivating p16 (Alani et al. 2001; Ohtani et al. 2001; Zheng et al. 2004). Decreased expression of Id1, rather than E47 or Ets2 whose expression remains unchanged or decreased in senescent cells (Hara et al. 1994; Ohtani et al. 2001; Zheng et al. 2004), is likely responsible for p16 elevation during senescence. Only a weak correlation was found between transcripts of Id1 and p16 during aging in mouse or rat tissues (Krishnamurthy et al. 2004), but our finding of the discrepancy between Id1 transcript and protein in senescent cells highlights the importance to understand the regulation of Id1 at both the transcript and protein levels. More studies are needed to determine whether Id1 protein levels correlate with p16 expression during aging, and whether Id1 plays a role in regulating p16 expression in aging. It will be of great interest to investigate whether the expression of Smurf2 increases with age, and whether Smurf2-Id1 regulates p16 expression during aging in vivo.
Id proteins undergo rapid turnover, and ubiquitin-proteasome mediated degradation regulates the stability of Id proteins (Bounpheng et al. 1999; Fajerman et al. 2004; Trausch-Azar et al. 2004). In addition to being ubiquitinated at internal lysine residues, Id1 and Id2 also undergo N-terminal ubiquitination via α-NH2 group of the N-terminal residue. It is thought that the N-terminus serves as not only an anchor for ubiquitination, but also a binding site for an E3 ubiquitin ligase (Fajerman et al. 2004; Trausch-Azar et al. 2004). Anaphase-promoting complex (APC/C) has been shown to ubiquitinate Id2 (Lasorella et al. 2006). However, it is unlikely that APC/C ubiquitinates Id2 through N-terminal ubiquitination, because APC/C-mediated ubiquitination of Id2 does not require the N-terminus of Id2, and Id2 mutant that is defective in N-terminal ubiquitination can be efficiently ubiquitinated by APC/C. Instead, ubiquitination of Id2 by APC/C requires a destruction box (D-box) in the C-terminus of Id2. Mutations in this D-box abolish Id2’s ability to interact with APC/C and its ubiquitination by APC/C. Although this D-box is present in Id1 and Id4, it is absent in Id3. Consistently, APC/C does not regulate the steady-state level of Id3 (Lasorella et al. 2006). Our finding of Smurf2 interacting with the N-terminus of Id1 (Fig. 3) suggests a possible role for Smurf2 in N-terminal ubiquitination of Id1. To our knowledge, Smurf2 is the first E3 ubiquitin ligase identified to ubiquitinate Id1. Whether Smurf2 mediates N-terminal ubiquitination of Id proteins needs further investigation. As Id1 has been implicated in many cellular processes (Perk et al. 2005), it is of great interest to understand whether Smurf2 plays an expanded role in these processes beyond its previously identified functions in regulating senescence and TGF-β signaling.
Experimental Procedures
Cell lines and cell culture
Human fibroblasts (BJ and WS1), fibrosarcoma (HT-1080), melanoma (A375P), and breast cancer (MDA-MB-231 and T47D) cell lines were purchased from American Type Culture Collection (Manassas, VA). Telomerase-immortalized BJ-hTERT and WS1-hTERT were described previously (Zhang & Cohen 2004). Human fibroblasts LF1 and LF1-hTERT were kindly provided by Dr. John Sedivy (Brown University). MDA-MB-231 cells expressing a Smad4 shRNA were provided by Dr. Yibin Kang (Princeton University). Cells were cultured in Dulbecco’s modified Eagle medium (Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (Hyclone, Logan, UT) in a humidified chamber containing 5% CO2 at 37°C.
Plasmid constructs and lentivirus production
Lentiviral constructs expressing Smurf2, C716A, C2 or GFP were described previously (Zhang et al. 2008). Deletion constructs of Flag-tagged Smurf2 were generated by inserting PCR-amplified fragments into pLenti-CMV-Hyg (provided by Dr. Paul Kowalski, University of Toronto). Id1 or Id3 (a gift from Dr. Yibin Kang, Princeton University) is inserted into pLenti-CMV-BSD (from Dr. Paul Kowalski). Deletion constructs of Myc-tagged Id1 were generated by inserting PCR fragments into pCMV-Myc (Clontech, Mountain View, CA). All constructs were verified by sequencing. Lentiviral shRNA constructs targeting Smurf2 (V2LHS_10399), Id1 (V3LHS_397680) and a non-silencing shRNA (RHS4346) were purchased from Open Biosystems (Huntsville, AL).
Lentiviral packaging and infection were carried out as described previously (Zhang et al. 2008). Briefly, lentiviral vectors were co-transfected into 293T cells with a plasmid (pMD2.VSV-G) encoding vesicular stomatitis virus glycoprotein (VSV-G) and a plasmid (pCMVdR8.74) encoding packaging proteins. VSV-G pseudotyped virus were collected 48 hr after transfection and used to infected target cells in the presence of 4 µg/ml polybrene (Sigma, St. Louis, MO). Two days later, infected cells were selected with 0.8 µg/mL puromycin (Sigma), 100 µg/mL hygromycin B (Calbiochem, San Diego, CA), or 10 µg/mL blasticidin (Invitrogen).
Replicative life span determination
Cells were plated at 1 × 105 to 3 × 105 cells per 10-cm dish and subcultured before they reached high cell density. Population doubling (PD) per passage was calculated as log2 (number of cells at time of subculture/number of cells plated). Cumulative PD was plotted against total time in culture to assess replicative life span. Cells at selected passages were stained for senescence-associated β-galactosidase activity as previously described (Zhang & Cohen 2004).
Quantitative RT–PCR
Total RNA was isolated using RNeasy Mini kit (Qiagen, Valencia, CA), and reverse-transcribed using Superscript II (Invitrogen) according to manufacturer’s instruction. Real-time PCR was carried out using SYBR Green PCR kit (Bio-Rad, Hercules, CA). The following primers were used: β–actin (5’-TTGCCGACAGGATGCAGAAGGA-3’ and 5’-AGGTGGACAGCGAGGCCAGGAT-3’); Id1 (5’-GGCTGTTACTCACGCCTCAAG-3’ and 5’-CCAACTGAAGGTCCCTGATGTAG-3’); Id3 (5’-AGTCCCGAGAGGCACTCAG-3’ and 5’-GCTCCTTTTGTCGTTGGAGATG-3’); Smurf2 (5’-TGCTGCAGGCCCGAGACTCTTTACC-3’ and 5’-CTCGTCGACGTCATTCCACAGCAAA-3’); p16 (5’-GCCCAACGCACCGAATAGT-3’ and 5’-CGATGGCCCAGCTCCTCAG-3’).
Western blot analysis
Total cell lysates were collected using RIPA buffer (50 mM Tris–HCl pH 7.5, 150 mM NaCl, 1% Triton X-100, 0.1% SDS, 0.5% deoxycholic acid, 0.02% sodium azide) with freshly added complete protease inhibitors (Roche, Indianapolis, IN). Protein lysates (20 µg) were separated by SDS–PAGE Criterion X-gel (Bio-Rad) and transferred to nitrocellulose membranes (GE Osmonics, Minnetonka, MN). Immunoblots were analyzed by Western blotting and visualized using a Western lightening chemiluminescence detection kit (PerkinElmer, Waltham, MA). Primary antibodies used in this study were Smurf2, p16 (Epitomics, Burlingame, CA), Id1, Id3 (BioCheck, Foster City, CA), Flag (M2), α-tubulin (DM1A, Sigma), C-Myc (9E10, Invitrogen), and ubiquitin (P4D1, Santa Cruz Biotechnology, Santa Cruz, CA).
Co-immunoprecipitation analysis
Cells were lysed in NP40 lysis buffer (20 mM Tris–HCl, 150 mM NaCl, 2 nM EDTA, 1% Nonidet P-40) with freshly added complete protease inhibitors (Roche). Lysates were pre-cleared with protein A-agarose (Invitrogen) and incubated with anti-Smurf2 antibody or matched rabbit IgG at 4°C overnight. Protein A-agarose was then added to incubate for three more hours at 4°C. In co-immunoprecipitation assay with Flag-tagged proteins, lysates were incubated with anti-Flag M2 affinity gel (Sigma) overnight at 4°C. Immunoprecipitates were washed four times with NP40 lysis buffer, and analyzed in Western blot with specific antibodies.
Ubiquitination assay
293T cells were transfected with constructs that express Id1, 3xFlag-ubiquitin (kindly provided by Dr. Quan Lu, Harvard School of Public Health) and Smurf2. GFP and C716A were used as controls. To detect ubiquitination of endogenous Id1, A375P cells stably expressing GFP, Smurf2 or C716A, or senescent LF1 fibroblasts expressing Smurf2 shRNA or non-silencing shRNA were used. Cells were treated with MG132 (20 µM, Sigma) for 2–4 hrs before cell lysis. Cell were collected in RIPA buffer plus N-ethylmaleimide (NEM, 10 mM, Fisher Scientific, Pittsburgh, PA), and cell lysates were incubated with anti-Id1 antibody overnight at 4°C followed by incubation with protein A-agarose (Invitrogen). Immunoprecipitates were washed with RIPA buffer three times, and analyzed in Western blot with anti-Flag or anti-ubiquitin antibody to detect ubiquitin conjugation.
Statistical analysis
Data are presented as mean ± SD. Two-tailed and unpaired Student t-test is used for all comparisons, with P<0.05 considered as statistically significant.
Supplementary Material
Acknowledgements
We thank Drs. Yibin Kang, Paul Kowalski, Quan Lu and John Sedivy for kindly providing reagents. This work was supported by grants from Our Danny Cancer Fund to Y.K., the National Cancer Institute (R01CA131210), American Cancer Society (IRG 93-033) and The Ellison Medical Foundation (AG-NS-0347-06) to H.Z.
Footnotes
Author contributions
Y.K. and H.Z. designed the experiments, Y.K. and H.C. performed the experiments, Y.K, H.C. and H.Z analyzed the data, and Y.K. and H.Z. wrote the manuscript.
Supporting Information
Additional supporting information may be found in the online version of this article:
Fig. S1 Senescence-associated β-galactosidase staining in senescent and early passage fibroblasts.
Fig. S2 Regulation of Id1 by Smurf2 does not require TGF-β or BMP signaling.
Fig. S3 Id proteins are stabilized in the presence of proteasome inhibitor MG132.
Fig. S4 Smurf2 regulates the expression of p16 in TERT-immortalized cells.
Fig. S5 Down-regulation of Smurf2 leads to delayed senescence entry.
Contributor Information
Yahui Kong, Email: yahui.kong@umassmed.edu.
Hang Cui, Email: hang.cui@umassmed.edu.
References
- Alani RM, Young AZ, Shifflett CB. Id1 regulation of cellular senescence through transcriptional repression of p16/Ink4a. Proc Natl Acad Sci U S A. 2001;98:7812–7816. doi: 10.1073/pnas.141235398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barone MV, Pepperkok R, Peverali FA, Philipson L. Id proteins control growth induction in mammalian cells. Proc Natl Acad Sci U S A. 1994;91:4985–4988. doi: 10.1073/pnas.91.11.4985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benezra R, Davis RL, Lockshon D, Turner DL, Weintraub H. The protein Id: a negative regulator of helix-loop-helix DNA binding proteins. Cell. 1990;61:49–59. doi: 10.1016/0092-8674(90)90214-y. [DOI] [PubMed] [Google Scholar]
- Bond J, Jones C, Haughton M, DeMicco C, Kipling D, Wynford-Thomas D. Direct evidence from siRNA-directed "knock down" that p16(INK4a) is required for human fibroblast senescence and for limiting ras-induced epithelial cell proliferation. Exp Cell Res. 2004;292:151–156. doi: 10.1016/j.yexcr.2003.09.021. [DOI] [PubMed] [Google Scholar]
- Bounpheng MA, Dimas JJ, Dodds SG, Christy BA. Degradation of Id proteins by the ubiquitin-proteasome pathway. Faseb J. 1999;13:2257–2264. [PubMed] [Google Scholar]
- Brookes S, Rowe J, Gutierrez Del Arroyo A, Bond J, Peters G. Contribution of p16(INK4a) to replicative senescence of human fibroblasts. Exp Cell Res. 2004;298:549–559. doi: 10.1016/j.yexcr.2004.04.035. [DOI] [PubMed] [Google Scholar]
- Christy BA, Sanders LK, Lau LF, Copeland NG, Jenkins NA, Nathans D. An Id-related helix-loop-helix protein encoded by a growth factor-inducible gene. Proc Natl Acad Sci U S A. 1991;88:1815–1819. doi: 10.1073/pnas.88.5.1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins CJ, Sedivy JM. Involvement of the INK4a/Arf gene locus in senescence. Aging Cell. 2003;2:145–150. doi: 10.1046/j.1474-9728.2003.00048.x. [DOI] [PubMed] [Google Scholar]
- Cummings SD, Ryu B, Samuels MA, Yu X, Meeker AK, Healey MA, Alani RM. Id1 delays senescence of primary human melanocytes. Mol Carcinog. 2008;47:653–659. doi: 10.1002/mc.20422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fajerman I, Schwartz AL, Ciechanover A. Degradation of the Id2 developmental regulator: targeting via N-terminal ubiquitination. Biochem Biophys Res Commun. 2004;314:505–512. doi: 10.1016/j.bbrc.2003.12.116. [DOI] [PubMed] [Google Scholar]
- Gil J, Peters G. Regulation of the INK4b-ARF-INK4a tumour suppressor locus: all for one or one for all. Nat Rev Mol Cell Biol. 2006;7:667–677. doi: 10.1038/nrm1987. [DOI] [PubMed] [Google Scholar]
- Hara E, Uzman JA, Dimri GP, Nehlin JO, Testori A, Campisi J. The helix-loop-helix protein Id-1 and a retinoblastoma protein binding mutant of SV40 T antigen synergize to reactivate DNA synthesis in senescent human fibroblasts. Dev Genet. 1996;18:161–172. doi: 10.1002/(SICI)1520-6408(1996)18:2<161::AID-DVG9>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- Hara E, Yamaguchi T, Nojima H, Ide T, Campisi J, Okayama H, Oda K. Id-related genes encoding helix-loop-helix proteins are required for G1 progression and are repressed in senescent human fibroblasts. J Biol Chem. 1994;269:2139–2145. [PubMed] [Google Scholar]
- Hollnagel A, Oehlmann V, Heymer J, Ruther U, Nordheim A. Id genes are direct targets of bone morphogenetic protein induction in embryonic stem cells. J Biol Chem. 1999;274:19838–19845. doi: 10.1074/jbc.274.28.19838. [DOI] [PubMed] [Google Scholar]
- Iavarone A, Garg P, Lasorella A, Hsu J, Israel MA. The helix-loop-helix protein Id-2 enhances cell proliferation and binds to the retinoblastoma protein. Genes Dev. 1994;8:1270–1284. doi: 10.1101/gad.8.11.1270. [DOI] [PubMed] [Google Scholar]
- Ide T, Tsuji Y, Ishibashi S, Mitsui Y. Reinitiation of host DNA synthesis in senescent human diploid cells by infection with Simian virus 40. Exp Cell Res. 1983;143:343–349. doi: 10.1016/0014-4827(83)90060-5. [DOI] [PubMed] [Google Scholar]
- Janzen V, Forkert R, Fleming HE, Saito Y, Waring MT, Dombkowski DM, Cheng T, DePinho RA, Sharpless NE, Scadden DT. Stem-cell ageing modified by the cyclin-dependent kinase inhibitor p16INK4a. Nature. 2006;443:421–426. doi: 10.1038/nature05159. [DOI] [PubMed] [Google Scholar]
- Kang Y, Chen CR, Massague J. A self-enabling TGFbeta response coupled to stress signaling: Smad engages stress response factor ATF3 for Id1 repression in epithelial cells. Mol Cell. 2003;11:915–926. doi: 10.1016/s1097-2765(03)00109-6. [DOI] [PubMed] [Google Scholar]
- Kavsak P, Rasmussen RK, Causing CG, Bonni S, Zhu H, Thomsen GH, Wrana JL. Smad7 binds to Smurf2 to form an E3 ubiquitin ligase that targets the TGF beta receptor for degradation. Mol Cell. 2000;6:1365–1375. doi: 10.1016/s1097-2765(00)00134-9. [DOI] [PubMed] [Google Scholar]
- Kim WY, Sharpless NE. The regulation of INK4/ARF in cancer and aging. Cell. 2006;127:265–275. doi: 10.1016/j.cell.2006.10.003. [DOI] [PubMed] [Google Scholar]
- Kreider BL, Benezra R, Rovera G, Kadesch T. Inhibition of myeloid differentiation by the helix-loop-helix protein Id. Science. 1992;255:1700–1702. doi: 10.1126/science.1372755. [DOI] [PubMed] [Google Scholar]
- Krishnamurthy J, Ramsey MR, Ligon KL, Torrice C, Koh A, Bonner-Weir S, Sharpless NE. p16INK4a induces an age-dependent decline in islet regenerative potential. Nature. 2006;443:453–457. doi: 10.1038/nature05092. [DOI] [PubMed] [Google Scholar]
- Krishnamurthy J, Torrice C, Ramsey MR, Kovalev GI, Al-Regaiey K, Su L, Sharpless NE. Ink4a/Arf expression is a biomarker of aging. J Clin Invest. 2004;114:1299–1307. doi: 10.1172/JCI22475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasorella A, Stegmuller J, Guardavaccaro D, Liu G, Carro MS, Rothschild G, de la Torre-Ubieta L, Pagano M, Bonni A, Iavarone A. Degradation of Id2 by the anaphase-promoting complex couples cell cycle exit and axonal growth. Nature. 2006;442:471–474. doi: 10.1038/nature04895. [DOI] [PubMed] [Google Scholar]
- Lin X, Liang M, Feng XH. Smurf2 is a ubiquitin E3 ligase mediating proteasome-dependent degradation of Smad2 in transforming growth factor-beta signaling. J Biol Chem. 2000;275:36818–36822. doi: 10.1074/jbc.C000580200. [DOI] [PubMed] [Google Scholar]
- Lyden D, Young AZ, Zagzag D, Yan W, Gerald W, O'Reilly R, Bader BL, Hynes RO, Zhuang Y, Manova K, Benezra R. Id1 and Id3 are required for neurogenesis, angiogenesis and vascularization of tumour xenografts. Nature. 1999;401:670–677. doi: 10.1038/44334. [DOI] [PubMed] [Google Scholar]
- McConnell BB, Starborg M, Brookes S, Peters G. Inhibitors of cyclin-dependent kinases induce features of replicative senescence in early passage human diploid fibroblasts. Curr Biol. 1998;8:351–354. doi: 10.1016/s0960-9822(98)70137-x. [DOI] [PubMed] [Google Scholar]
- Molofsky AV, Slutsky SG, Joseph NM, He S, Pardal R, Krishnamurthy J, Sharpless NE, Morrison SJ. Increasing p16INK4a expression decreases forebrain progenitors and neurogenesis during ageing. Nature. 2006;443:448–452. doi: 10.1038/nature05091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickoloff BJ, Chaturvedi V, Bacon P, Qin JZ, Denning MF, Diaz MO. Id-1 delays senescence but does not immortalize keratinocytes. J Biol Chem. 2000;275:27501–27504. doi: 10.1074/jbc.C000311200. [DOI] [PubMed] [Google Scholar]
- Ohtani N, Zebedee Z, Huot TJ, Stinson JA, Sugimoto M, Ohashi Y, Sharrocks AD, Peters G, Hara E. Opposing effects of Ets and Id proteins on p16INK4a expression during cellular senescence. Nature. 2001;409:1067–1070. doi: 10.1038/35059131. [DOI] [PubMed] [Google Scholar]
- Perk J, Iavarone A, Benezra R. Id family of helix-loop-helix proteins in cancer. Nat Rev Cancer. 2005;5:603–614. doi: 10.1038/nrc1673. [DOI] [PubMed] [Google Scholar]
- Qian Y, Chen X. ID1, inhibitor of differentiation/DNA binding, is an effector of the p53-dependent DNA damage response pathway. J Biol Chem. 2008;283:22410–22416. doi: 10.1074/jbc.M800643200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riechmann V, van Cruchten I, Sablitzky F. The expression pattern of Id4, a novel dominant negative helix-loop-helix protein, is distinct from Id1, Id2 and Id3. Nucleic Acids Res. 1994;22:749–755. doi: 10.1093/nar/22.5.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarze SR, DePrimo SE, Grabert LM, Fu VX, Brooks JD, Jarrard DF. Novel pathways associated with bypassing cellular senescence in human prostate epithelial cells. J Biol Chem. 2002;277:14877–14883. doi: 10.1074/jbc.M200373200. [DOI] [PubMed] [Google Scholar]
- Serrano M, Lin AW, McCurrach ME, Beach D, Lowe SW. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell. 1997;88:593–602. doi: 10.1016/s0092-8674(00)81902-9. [DOI] [PubMed] [Google Scholar]
- Shay JW, Pereira-Smith OM, Wright WE. A role for both RB and p53 in the regulation of human cellular senescence. Exp Cell Res. 1991;196:33–39. doi: 10.1016/0014-4827(91)90453-2. [DOI] [PubMed] [Google Scholar]
- Suh HC, Leeanansaksiri W, Ji M, Klarmann KD, Renn K, Gooya J, Smith D, McNiece I, Lugthart S, Valk PJ, Delwel R, Keller JR. Id1 immortalizes hematopoietic progenitors in vitro and promotes a myeloproliferative disease in vivo. Oncogene. 2008;27:5612–5623. doi: 10.1038/onc.2008.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun XH, Copeland NG, Jenkins NA, Baltimore D. Id proteins Id1 and Id2 selectively inhibit DNA binding by one class of helix-loop-helix proteins. Mol Cell Biol. 1991;11:5603–5611. doi: 10.1128/mcb.11.11.5603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J, Gordon GM, Nickoloff BJ, Foreman KE. The helix-loop-helix protein id-1 delays onset of replicative senescence in human endothelial cells. Lab Invest. 2002;82:1073–1079. doi: 10.1097/01.lab.0000022223.65962.3a. [DOI] [PubMed] [Google Scholar]
- Trausch-Azar JS, Lingbeck J, Ciechanover A, Schwartz AL. Ubiquitin-Proteasome-mediated degradation of Id1 is modulated by MyoD. J Biol Chem. 2004;279:32614–32619. doi: 10.1074/jbc.M403794200. [DOI] [PubMed] [Google Scholar]
- Vogt M, Haggblom C, Yeargin J, Christiansen-Weber T, Haas M. Independent induction of senescence by p16INK4a and p21CIP1 in spontaneously immortalized human fibroblasts. Cell Growth Differ. 1998;9:139–146. [PubMed] [Google Scholar]
- Zhang H, Cohen SN. Smurf2 up-regulation activates telomere-dependent senescence. Genes Dev. 2004;18:3028–3040. doi: 10.1101/gad.1253004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Teng Y, Kong Y, Kowalski PE, Cohen SN. Suppression of human tumor cell proliferation by Smurf2-induced senescence. J Cell Physiol. 2008;215:613–620. doi: 10.1002/jcp.21337. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Chang C, Gehling DJ, Hemmati-Brivanlou A, Derynck R. Regulation of Smad degradation and activity by Smurf2, an E3 ubiquitin ligase. Proc Natl Acad Sci U S A. 2001;98:974–979. doi: 10.1073/pnas.98.3.974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng W, Wang H, Xue L, Zhang Z, Tong T. Regulation of cellular senescence and p16(INK4a) expression by Id1 and E47 proteins in human diploid fibroblast. J Biol Chem. 2004;279:31524–31532. doi: 10.1074/jbc.M400365200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.