Summary
Aging is accompanied by alterations in epigenetic marks that control chromatin states, including histone acetylation and methylation. Enzymes that reversibly affect histone marks associated with active chromatin have recently been found to regulate aging in C. elegans. However, relatively little is known about the importance for aging of histone marks associated with repressed chromatin. Here we use a targeted RNAi screen in C. elegans to identify four histone demethylases that significantly regulate worm lifespan, UTX-1, RBR-2, LSD-1, and T26A5.5. Interestingly, UTX-1 belongs to a conserved family of histone demethylases specific for lysine 27 of histone H3 (H3K27me3), a mark associated with repressed chromatin. Both utx-1 knock-down and heterozygous mutation of utx-1 extend lifespan and increase the global levels of the H3K27me3 mark in worms. The H3K27me3 mark significantly drops in somatic cells during the normal aging process. UTX-1 regulates lifespan independently of the presence of the germline, but in a manner that depends on the insulin-FoxO signaling pathway. These findings identify the H3K27me3 histone demethylase UTX-1 as a novel regulator of worm lifespan in somatic cells.
Keywords: Histone demethylase, aging, lifespan, UTX, H3K27me3, epigenetic, Insulin pathway, FoxO transcription factor, germline, soma, C. elegans
Introduction
Aging is accompanied by a dramatic loss of epigenetic control over repressed regions of chromatin in various species (Wareham et al., 1987; Gaubatz & Cutler 1990; Kennedy et al., 1995; Smeal et al., 1996; Shen et al., 2008). Furthermore, cells isolated from old individuals (>80 years) or from patients with Hutchinson-Gilford Progeria Syndrome, a premature aging disorder, exhibit a reduction in repressed chromatin marks (Scaffidi & Misteli 2005; Scaffidi & Misteli 2006). Age-dependent loss of chromatin repression is correlated with alterations in gene expression patterns, which have been documented in species ranging from C. elegans to humans (Lee et al., 2000; Lund et al., 2002; Bennett-Baker et al., 2003; Lu et al., 2004). While there is an age-dependent loss of chromatin repression in a range of organisms, whether and how maintenance of repressed chromatin affects longevity is only starting to be elucidated.
The repressed or active state of chromatin is governed by an array of epigenetic modifications, including modification of the core histones through phosphorylation, ubiquitylation, acetylation, and methylation (Berger 2007). Histone methylation at specific residues is particularly important for repressed chromatin (Strahl & Allis 2000). For example, trimethylated lysine 27 on histone H3 (H3K27me3) is associated with facultative heterochromatin and transcriptionally silenced chromatin regions (Cao et al., 2002; Bernstein et al., 2006). On the other hand, trimethylated lysine 4 on histone H3 (H3K4me3) is associated with active chromatin and is present at transcriptional start sites (Bernstein et al., 2002; Santos-Rosa et al., 2002). Histone methylation is controlled by the counteracting action of two classes of enzymes: methyltransferases and demethylases (Shi & Whetstine 2007). The reversibility of histone methylation raises the possibility that this epigenetic mark is an important control point for different processes, including aging.
Regulators of epigenetic marks associated with active chromatin have been implicated in lifespan regulation. For example, SIR2 was found to be important for yeast replicative lifespan by regulating acetylated H4K16, a mark associated with active chromatin (Dang et al., 2009). The H3K4 demethylase LSD-1 has also been identified as regulator of worm lifespan (McColl et al., 2008). Furthermore, a targeted RNAi screen for methyltransferases that regulate longevity identified members of the H3K4me3 ASH-2 Trithorax complex (Greer et al., 2010). Indeed, attenuation of members of the ASH-2 H3K4me3 methyltransferase complex extends lifespan and decreases H3K4me3 levels in worms (Greer et al., 2010). Consistently, ectopic expression of RBR-2, an H3K4me3 demethylase that counteracts the effect of the ASH-2 methyltransferase complex, also extends worm lifespan (Greer et al., 2010). The ability of members of this H3K4me3 regulatory complex to regulate lifespan is dependent on the presence of an intact germline (Greer et al., 2010), but independent of the insulin-FoxO signaling pathway, a conserved pathway that has been well-characterized in the regulation of aging (Johnson 1990; Kenyon et al., 1993; Morris et al., 1996; Kimura et al., 1997; Lin et al., 1997; Ogg et al., 1997). Thus, marks associated with active chromatin are emerging as important regulators of the aging process. However, much less is known about epigenetic marks associated with repressive chromatin. While our previous targeted RNAi screen for longevity did not identify known methyltransferases of repressive marks (Greer et al., 2010), this screen was only focused on methyltransferases. Thus, the possibility remained that histone demethylases that reversibly erase repressive marks play an important role in lifespan.
In this study, we used a targeted RNAi screen focused on histone demethylases to identify four histone demethylases, UTX-1, RBR-2, LSD-1, and T26A5.5, that regulate C. elegans lifespan. We focused on UTX-1 because its mammalian counterpart, UTX, is known to function as a demethylase for the repressive mark H3K27me3 (Agger et al., 2007; Hong et al., 2007). We found that attenuation of utx-1 using RNAi or heterozygous mutants of utx-1 extends both mean and maximal worm lifespan and increases the global levels of H3K27me3. Interestingly, H3K27me3 levels strikingly decline during normal aging in somatic cells in worms. We also showed that the H3K27me3 demethylase UTX-1 controls lifespan independently of the germline, but genetically interacts with the insulin-FoxO pathway to regulate longevity. Our findings identify several histone demethylases as novel regulators of worm lifespan, including the H3K27me3 demethylase UTX-1. Our results further indicate that loss of repressed chromatin is associated with aging in somatic cells.
Results
A targeted RNAi screen identifies RBR-2, LSD-1, T26A5.5 and UTX-1 histone demethylases as regulators of lifespan in C. elegans
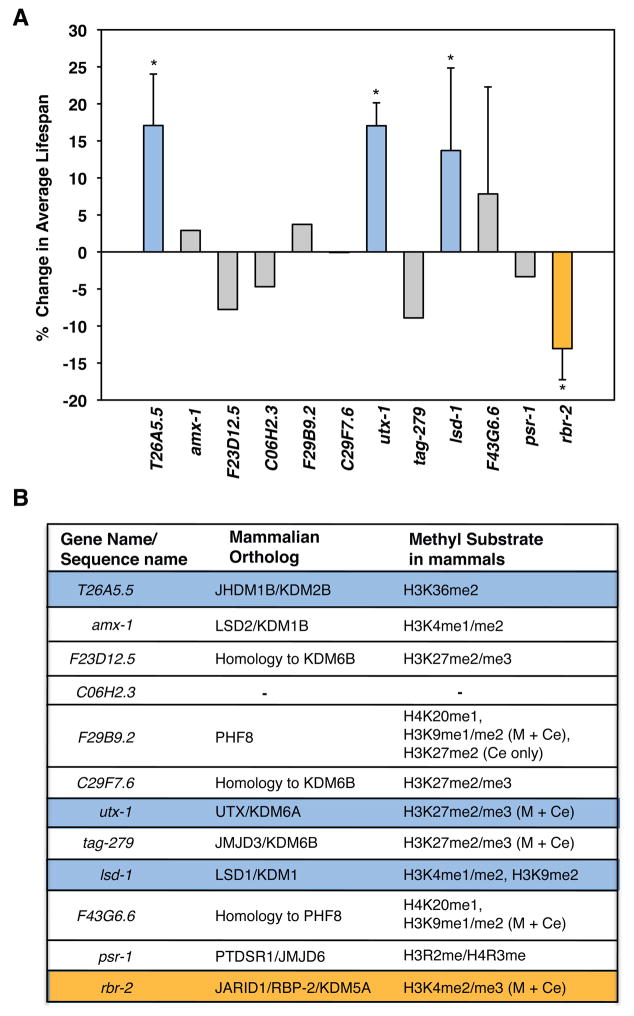
Histone methyltransferases have been implicated in the regulation of lifespan in both worms (Hamilton et al., 2005; Greer et al., 2010) and flies (Siebold et al., 2010). Because histone marks are dynamically controlled by both methyltransferases and demethylases, there is the potential that both classes of enzymes affect the aging process. At present, however, only two histone demethylases, RBR-2 and LSD-1, which are known to demethylate the activating marks H3K4me3 and H3K4me2, have been implicated in the regulation of lifespan (McColl et al., 2008; Greer et al., 2010). To identify additional demethylases that regulate lifespan, and to potentially uncover regulators of repressive marks important for the aging process, we performed a directed RNAi screen against histone demethylases in fertile worms, starting RNAi upon hatching at the first larval L1 stage. The histone demethylases were selected on the basis that they contain an amine oxidase or a Jumonji C-terminal (JmjC) domain, which are hallmarks of the two classes of enzymes responsible for catalysis of histone demethylation (Shi et al., 2004; Tsukada et al., 2006). Knockdown of rbr-2 significantly decreased lifespan whereas knock-down of lsd-1 extended lifespan, consistent with previous results (McColl et al., 2008; Greer et al., 2010). We also found that knock-down of two demethylases not previously known to regulate lifespan, T26A5.5 and utx-1, significantly increased lifespan (17.07% ±6.94 and 17.04% ±3.09 increases in lifespan, respectively) (Fig. 1A; Table S1 in Supporting Information). Thus, our targeted RNAi screen identified four histone demethylases that significantly altered lifespan of fertile worms, RBR-2, LSD-1, T26A5.5, and UTX-1, two of which (T26A5.5, and UTX-1) had not been previously identified to regulate the aging process.
Figure 1. A targeted RNAi screen in fertile worms identifies four histone demethylases that significantly affect adult worm lifespan.
A) Percent change in average lifespan induced by RNAi knock-down of genes encoding specific demethylases compared to the empty vector control. Genes whose RNAi knock-down resulted in a 10% increase (solid blue bars) or decrease (solid orange bar) in lifespan were selected and their effect on lifespan was repeated at least once. Mean +/− SD of 2 independent experiments, *p<0.05. Mean lifespan and statistics for independent experiments are presented in Table S1. B) Orthologs of worm histone demethylases in mammals, and the histone marks they have been shown to regulate to date. (M) and (Ce) denote that the substrate specificity of the particular demethylase has been determined for mammals and C. elegans, respectively (Shi et al., 2004; Tsukada et al., 2006; Agger et al., 2007; Chang et al., 2007; Christensen et al., 2007; Hong et al., 2007; Klose et al., 2007; Lan et al., 2007; He et al., 2008; Karytinos et al., 2009; Kleine-Kohlbrecher et al., 2010). Blue or orange highlighted genes represent histone demethylases whose knock-down was found to increase or decrease lifespan in the RNAi screen, respectively.
RBR-2 is orthologous to the human H3K4me3 demethylase, RBP2 (JARID1A/KDM5A), and associates with the PRC2 complex to repress gene transcription (Pasini et al., 2008) (Fig. 1B). LSD-1 (T08D10.2) is orthologous to the lysine specific demethylase 1 (LSD1) protein in mammals which demethylates H3K4me2/me1 as well as H3K9me2 and also functions as a repressor of gene transcription (Shi et al., 2004) (Fig. 1B). T26A5.5 encodes an orthologue of the JmjC domain-containing protein JHDM1B/KDM2B which specifically demethylates H3K36me3 (Tsukada et al., 2006; He et al., 2008), a mark associated with active gene transcription (Morris et al., 2005; Rao et al., 2005) (Fig. 1B). UTX-1 is orthologous to the members of a mammalian phylogenetic subgroup that contains the histone demethylases JMJD3, UTX and UTY, with closest homology to UTX. Interestingly, mammalian UTX specifically demethylates the repressive mark H3K27me3 and is required for the transcription of multiple target genes (Agger et al., 2007; Hong et al., 2007; Wang et al., 2010) (Fig. 1B). The observation that several histone demethylases with different substrate specificities can control worm lifespan suggests interplay between epigenetic marks for the regulation of lifespan.
T26A5.5 and UTX-1 histone demethylases regulate adult lifespan in C. elegans
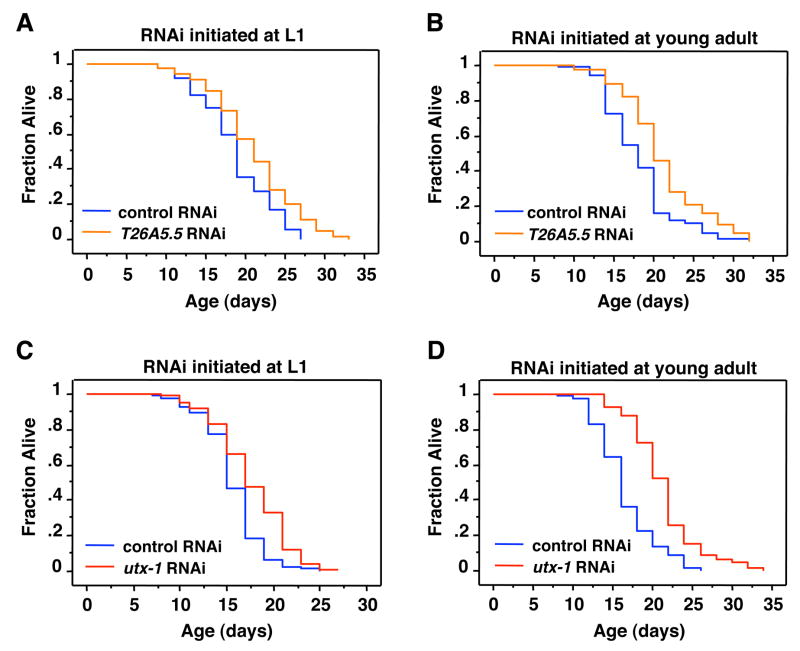
The RNAi screen was performed by adding RNAi at the larval stage L1, which could affect developmental processes. We next asked whether knock-down of the identified histone demethylases regulates lifespan by affecting adult aging, or as a consequence of perturbing development. Depletion of lsd-1 in adult worms increased lifespan (McColl et al., 2008) whereas depletion of rbr-2 in adult worms decreased lifespan (Greer et al., 2010), indicating that both LSD-1 and RBR-2 regulate lifespan by affecting adult aging. To test whether T26A5.5 and utx-1 also extended lifespan by slowing adult aging, we treated the worms with the corresponding RNAi at the young adult stage upon reaching sexual maturity. Knock-down of T26A5.5 initiated at the L1 (Fig. 2A; Table S1) and young adult stage (Fig. 2B; Table S2) both increased lifespan by ~12% and 15% respectively compared to empty vector control. Knock-down of utx-1 initiated at the L1 stage extended worm lifespan by ~13% (Fig. 2C; Fig. 1A; Tables S1 and S2). Interestingly, knock-down of utx-1 initiated at the young adult stage extended lifespan by ~29% (Fig. 2D; Table S2). These results indicate that T26A5.5 and utx-1 regulate lifespan in adult worms. The fact that we observed a greater difference in the degree of lifespan extension when utx-1 knock-down was initiated at adulthood compared to L1, suggests that utx-1 deficiency during development may partially impair the overall health of the worm.
Figure 2. Knock-down of two histone demethylases, T26A5.5. and UTX-1, extend lifespan when initiated either during development or in adulthood.
A–B) T26A5.5 RNAi extends lifespan at L1 (12%, p<0.0009) (A), and young adults (B) (16.6%, p<0.0005). C) utx-1 knock-down initiated at the L1 stage extends worm lifespan by 12.6% (p<0.0001) compared to control RNAi. D) utx-1 knock-down initiated after worms have reached adulthood extends lifespan by 29.29% (p<0.0001) compared to control RNAi. The efficacy of the RNAi-mediated knock-down of utx-1 was confirmed by quantitative RT-PCR (Supplementary Fig. 1). Mean lifespan and statistics for independent experiments are presented in Table S1 and Table S2.
The H3K27me3 demethylase UTX-1 regulates lifespan in worms
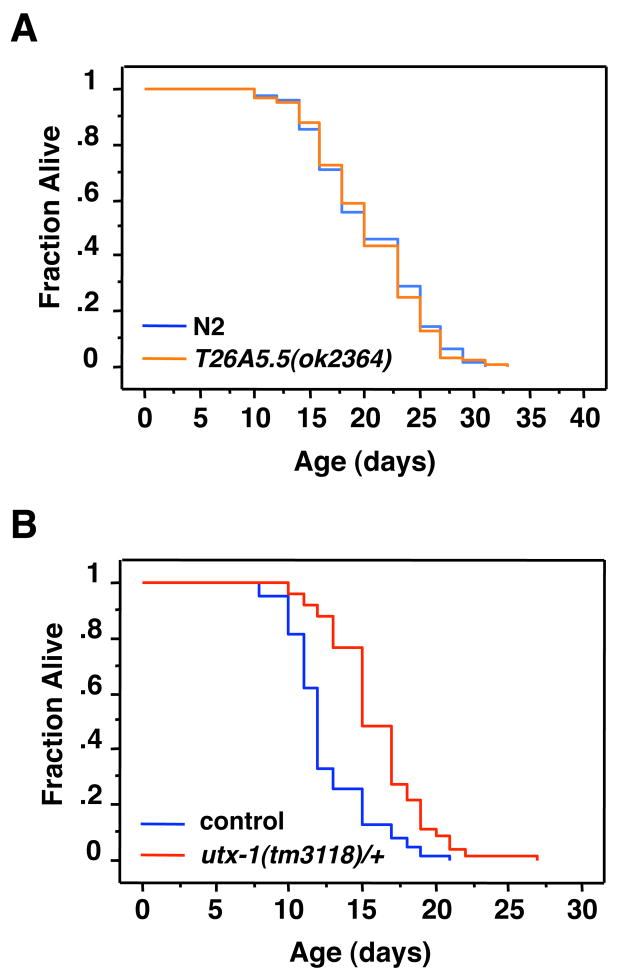
To further investigate whether T26A5.5 regulates aging, we assessed the lifespan ofT26A5.5(ok2364) mutant worms. Unlike the results of knocking-down T26A5.5 the lifespan of T26A5.5(ok2364) mutant worms was not significantly increased compared to wildtype worms (Fig. 3A; Table S2). The fact that the RNAi-mediated knock-down of T26A5.5 extends lifespan but T26A5.5(ok2364) mutant worms are not long-lived may be due to the allele of T26A5.5 used in this study, to the acute versus chronic removal of T26A5.5, or to the fact that T26A5.5(ok2364) may be a hypomorphic mutation that does not completely abrogate T26A5.5 activity.
Figure 3. Loss of one utx-1 allele is sufficient to extend worm lifespan.
A) T21A5.5(ok2364) mutant worms fail to extend lifespan compared to wild type (N2) worms (p=0.7993). B) utx-1(tm3118) heterozygous worms (stDp2(X;II)/+II; utx-1(tm3118)/uDf1 X) (indicated as utx-1(tm3118)/+) are long lived compared to the homozygous worms of the same balancer strain background (stDp2X;II)/+II; uDf1 X) (27–50%, p<0.0001) (control). Mean lifespan and statistics for independent experiments are presented in Table S2.
To confirm that UTX-1 regulated longevity, we assessed the lifespan of utx-1 mutants. Of the available strains carrying utx-1 mutations, neither utx-1(tm3118) nor utx-1(tm3136) are viable when homozygous (Kemphues et al., 1988), precluding the examination of lifespan. Thus, we determined the lifespan of the viable utx-1(tm3118) heterozygous worms. To maintain the stability of the heterozygous utx-1 mutation across several generations, we crossed the utx-1(tm3118)/+ strain with a heterozygous balancer strain TU899 [stDp(2X;II)/+ II; uDf1 X] carrying a mutation proximate to the genomic position of utx-1. The lifespan of utx-1 heterozygous worms [stDp2(X;II)/+ II; utx-1(tm3118) X] worms was compared to that of the balancer strain. While both of these strains had a higher incidence of matricide (also called bagging) than wild type (N2) worms, we observed that utx-1(3118) heterozygous worms have a significant increase in their mean lifespan compared to the control worms, ranging from 27–50% (Fig. 3B; Table S2). These results support our findings that utx-1 deficiency extends lifespan in worms. Thus, we focused on UTX-1 in the rest of the study.
UTX-1 regulates H3K27me3 levels in the germline and the soma of C. elegans
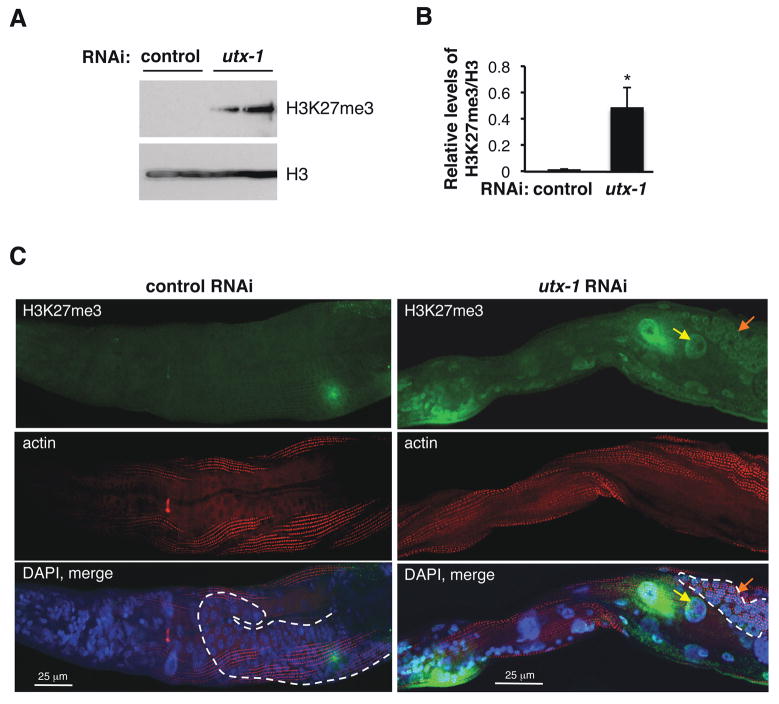
In mammals, UTX is known to specifically demethylate the H3K27me3 mark by acting antagonistically to the polycomb repressor complex 2 (PRC2) (Agger et al., 2007; Hong et al., 2007). A recent study in worms also revealed that utx-1 knock-down led to elevated H3K27me3 levels in whole worm lysates (Fisher et al., 2010). Western blot analysis using an antibody specifically directed to the H3K27me3 mark confirmed that utx-1 knock-down resulted in increased levels of H3K27me3 in whole worm lysates at the L3 stage (Fig. 4A, B). While the H3K27me3 signal from lysates of the RNAi control worms is not readily visible in Fig. 4A, it is important to note that wildtype worms are not entirely devoid of the H3K27me3 mark, as evidenced by longer exposure times (data not shown).
Figure 4. The demethylase UTX-1 regulates H3K27me3 levels in the germline and soma of worms.
A) Western blot on whole worm extracts of wild type (N2) worms at the L3 stage treated with empty vector control RNAi or utx-1 RNAi using an H3K27me3 antibody. An antibody to total histone H3 (H3) was used to control for equal loading. Each lane represents an independent cohort of ~200 worms. The blots presented are representative of independent experiments performed two times. B) Quantification of relative H3K27me3 levels compared to H3. Mean +/− SD of two independent experiments. *p<0.05 by paired t-tests. C) Whole worm immunofluorescence of worms treated with empty vector control RNAi or utx-1 RNAi and co-stained with an H3K27me3 antibody (green) and an actin antibody (red) as a control for antibody accessibilty. DAPI staining (blue) was used to visualize nuclei and is shown in the merged image from all three channels (bottom panel). The worms are oriented anterior (left) to posterior (right). Yellow arrow: intestinal cell; orange arrow: germline cell. These results are representative of four independent experiments. The dashed line indicates the germline. Scale bar, 25 μm.
To determine whether utx-1 knock-down led to an increase in H3K27me3 in all tissues or only in specific tissues, we performed whole worm immunofluorescence using the H3K27me3 antibody. utx-1 knock-down led to an increase in the H3K27me3 signal in all tissues, including both somatic cells and germline cells (Fig. 4C; Fig. S2A). We verified that antibody permeability was similar in control worms and utx-1 knocked-down worms by co-staining with an actin antibody (Fig. 4C) and that utx-1 knock-down did not significantly alter total histone H3 signal (S2B). These results show that UTX-1 is an H3K27me3 demethylase in worms, and that UTX-1 regulates H3K27 in both somatic and germline cells. These findings also indicate that an increase in the H3K27me3 mark, a mark associated with repressed chromatin, is correlated with extended lifespan in worms.
Longevity induced by utx-1 knock-down does not require an intact germline
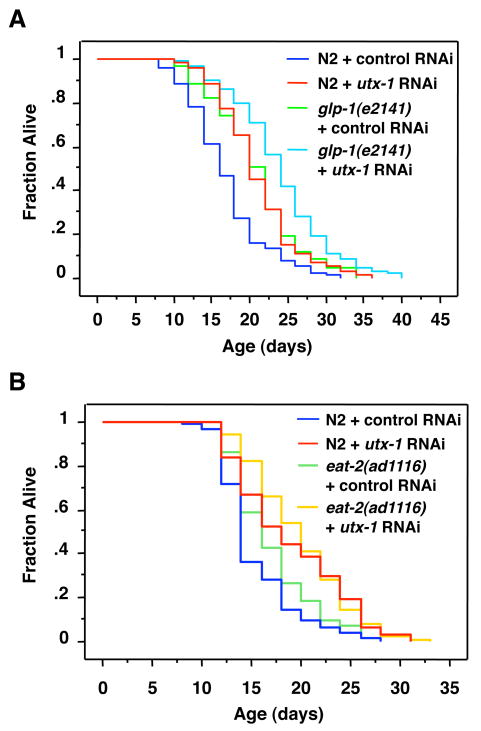
We have recently shown that an H3K4 trimethyltransferase complex regulates worm lifespan and does so in a manner that requires an intact germline (Greer et al., 2010). The H3K27me3 demethylase UTX was recently found to co-purify with the ASH-2 methyltransferase complex in mammalian cells (Issaeva et al., 2007), raising the possibility that the UTX-1 demethylase, like ASH-2, may regulate lifespan in a germline-dependent manner. To test whether the longevity induced by utx-1 knock-down also requires germline cells, we examined the effects of utx-1 knock-down in glp-1(e2141) mutant worms. When maintained at the restrictive temperature, the glp-1(e2141) worms fail to give rise to a functional germline (Priess et al., 1987) and exhibit an increased lifespan compared to wild type worms (Arantes-Oliveira et al., 2002). Surprisingly, we found that utx-1 knockdown further extends the lifespan of both long-lived glp-1(e2141) worms (15.9%, p<0.0006) and wild type (N2) worms (23.6%, p<0.0001) (Fig. 5A). Analysis by two-way ANOVA confirmed that there was no significant interaction between the glp-1(e2141) genotype and utx-1 RNAi for lifespan extension (p=0.574). Together, these results indicate that unlike the ASH-2 complex, UTX-1 regulates lifespan independently of the presence of an intact germline. These findings further imply that UTX-1 likely does not act together with the ASH-2 containing methyltransferase complex to regulate lifespan. These data also suggest that UTX-1 regulates lifespan by acting in somatic cells.
Figure 5. Longevity induced by utx-1 knock-down is independent of the germline glp-1 pathway and does not entirely require the eat-2 DR pathway.
A) glp-1(e2141) mutant worms were shifted from the permissive (15°C) to the restrictive temperature (25°C) at the L1 stage along with N2 worms. utx-1 knockdown initiated at the young adult stage further extends the lifespan of long-lived glp-1(e2141) mutants (15.9%, p=0.0006) and that of wild type (N2) worms (23.6%, p=0.0001) (p=0.574 by two-way ANOVA to test for a statistical interaction between the glp-1(e2141) genotype and utx-1 RNAi). B) utx-1 knockdown initiated at the young adult stage extends the lifespan of wild type (N2) worms (23.2%, p<0.0001) and further extends the lifespan of eat-2(ad1116) mutant worms by 16.7% (p<0.0001 by two-way ANOVA to test for a statistical interaction between the eat-2(ad1116) genotype and utx-1 RNAi). Mean lifespan and statistics for independent experiments are presented in Table S2.
Longevity induced by utx-1 knock-down does not entirely require the DR pathway induced by eat-2
Environmental interventions such as dietary restriction (DR) extend the mean and maximal lifespan in worms, flies, rodents [for review, (Mair & Dillin 2008)], and even primates (Colman et al., 2009). There are several ways to elicit DR in worms (Klass 1977; Hosono et al., 1989; Houthoofd et al., 2002; Kaeberlein et al., 2006; Lee et al., 2006; Bishop & Guarente 2007; Greer et al., 2007; Honjoh et al., 2009). eat-2(ad1116) mutant worms provide a genetic way to mimic DR as they exhibit a significantly reduced rate of pharyngeal pumping and have an extended lifespan (Avery 1993; Lakowski & Hekimi 1996). We tested whether utx-1 functions in the same genetic pathway as eat-2 mutants to regulate lifespan. As shown in Fig. 5B, utx-1 knock-down further extended the lifespan of both long-lived eat-2(ad1116) mutant worms (16.7% (p<0.0001) and wild type (N2) worms (23.2%, p<0.0001), suggesting that utx-1 and eat-2 do not function in the same genetic pathway to regulate lifespan. However, analysis by two-way ANOVA revealed a significant interaction between the eat-2(ad1116) genotype and utx-1 RNAi (p<0.0001). Although interpretation of these results is limited by the absence of independent replication and by variation in longevity in strains with different genetic background (Gems & Riddle 2000), these results suggest that utx-1 regulates lifespan in a manner that does not require the DR pathway induced by the eat-2 mutation, but that utx-1 may partially overlap with this DR pathway.
Longevity induced by utx-1 deficiency requires an intact insulin-FoxO signaling pathway
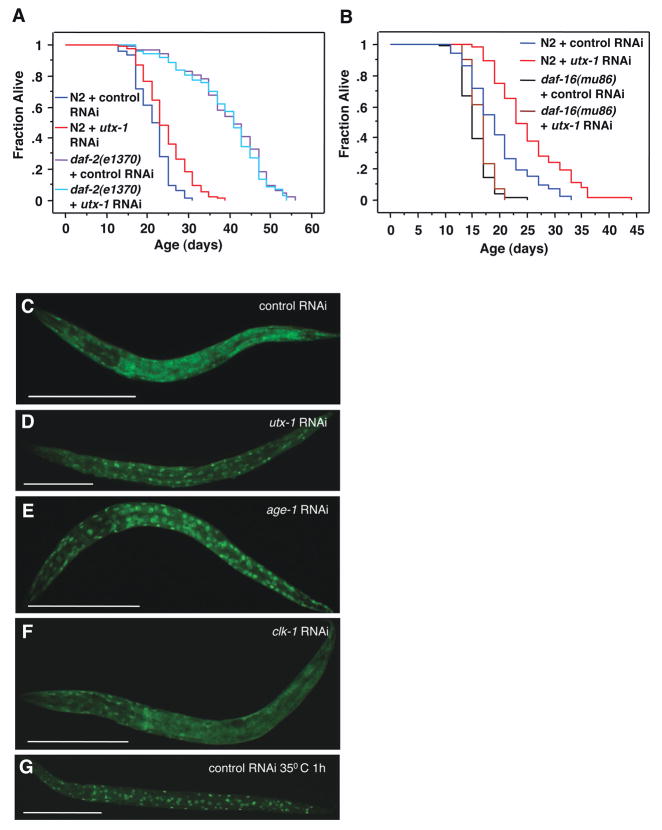
Mutations that decrease the activity of the insulin receptor DAF-2 more than double the lifespan of the worm (Kenyon et al., 1993; Kimura et al., 1997). The long lifespan of daf-2 is further extended by deficiencies in the germline pathway (Hsin & Kenyon 1999), similar to that of utx-1 deficient worms. Thus, we asked if longevity induced by utx-1 knock-down is dependent on the insulin/IGF-1 signaling pathway. utx-1 knock-down extends the lifespan of wild type (N2) worms (14%, p<0.0001), but does not further extend the long lifespan of daf-2(e1370) mutant worms (−1.4%, p=0.5367) (Fig. 6A). Analysis by two-way ANOVA revealed a statistically significant interaction between the daf-2(e1370) genotype and utx-1 RNAi (p=0.039). Although epistasis experiments for longevity can be difficult to interpret when the mutant allele, such daf-2(e1370), is not null (Gems & Partridge 2008), these results suggest that the longevity induced by utx-1 knock-down requires intact insulin/IGF-1 signaling.
Figure 6. Longevity induced by utx-1 deficiency requires the insulin-FoxO pathway.
A) utx-1 knock-down did not further extend the lifespan of the long lived daf-2(e1370) mutant worms (-1.42%, p=0.5267) (p=0.039 by two-way ANOVA to test for an interaction between the daf-2(e1370) genotype and utx-1 RNAi). B) utx-1 knock-down did not significantly increase the lifespan of the short lived daf-16(mu86) mutant worms (2.4%, p=0.6034) (p<0.0001 by two-way ANOVA to test for a statistical interaction between the daf-16(mu86) genotype and utx-1 RNAi). Mean lifespan and statistics for independent experiments are presented in Supplementary Table. C–F) Representative pictures of FoxO/DAF-16 localization in DAF-16::GFP transgenic worms. The percentage of worms with nuclear FoxO/DAF-16 is indicated in parenthesis, with n = total number of individuals examined. The worms are oriented anterior (left) to posterior (right). C) control RNAi (20%, n=20); D) utx-1 RNAi (80%, n=20) E) age-1 RNAi (100%, n=20); F) clk-1 RNAi (20%, n=20); and G) control RNAi, 35°C heat shock for 1h (100%, n=50). Scale bar, 100 μm.
We further tested the requirement of the insulin pathway in longevity regulation by UTX-1. Extended longevity of the daf-2 mutants is in large part mediated by DAF-16, the worm FoxO transcription factor (Lin et al., 1997; Ogg et al., 1997). We next examined the effect of utx-1 knock-down in daf-16(mu86) mutant worms, which carry a null mutation for the FoxO gene. While utx-1 knockdown significantly extended the lifespan of wild type (N2) worms (28.2%, p<0.0001), utx-1 knock-down no longer extended the lifespan of the daf-16(mu86) mutant worms (2.4%, p=0.6034). Analysis by two-way ANOVA revealed a significant interaction between the daf-16(mu86) genotype and utx-1 RNAi (p<0.0001) (Fig. 6B). These results indicate that the longevity induced by utx-1 RNAi requires the pro-longevity transcription factor FoxO/DAF-16.
We next asked whether knock-down of utx-1 affects the nuclear translocation of FoxO/DAF-16, using a strain of worms carrying an integrated DAF-16::GFP transgene (Henderson & Johnson 2001). In standard culture conditions, DAF-16::GFP was predominately cytoplasmic (Fig. 6C). Interestingly, utx-1 knockdown increased DAF-16::GFP nuclear localization (Fig. 6D), similar to the knockdown of age-1, an insulin signaling pathway effector (phosphatidylinositol-3 kinase/PI3K) (Rahman et al., 2010) (Fig. 6E), or to 1h heat shock at 35°C (Henderson & Johnson 2001) (Fig. 6G). The effect of utx-1 knock-down on FoxO/DAF-16 nuclear localization was relatively specific, in that knock-down of another gene involved in longevity, clk-1, did not result in increased nuclear localization of DAF-16::GFP, as shown previously (Henderson & Johnson 2001) (Fig. 6F). Together with our genetic interaction results, these findings suggest that UTX-1 regulates lifespan at least in part by modulating FoxO/DAF-16 nuclear localization. UTX-1 may normally prevent the FoxO/DAF-16 transcription factor from accessing its target genes.
Global somatic H3K27me3 levels decrease with age in germline-deficient worms
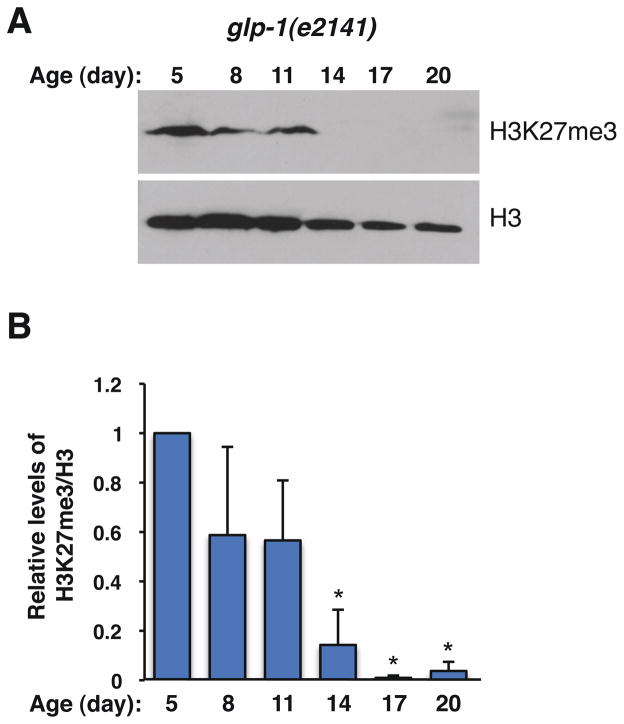
Because deficiency in the H3K27me3 demethylase UTX-1 leads to both an extended lifespan and an increase in global H3K27me3 levels, we asked if the levels of H3K27me3 changed during the normal aging process in the worm. We initially assessed the H3K27me3 mark in wild type worms maintained on solid media. We observed a slight decrease in the H3K27me3 mark after day 12, which corresponds to middle age in these worms (Fig. S3A, B). However, the interpretation of these experiments was limited by the fact that growth on solid media made it difficult to obtain enough worms at later stages of life (e.g. day 20, when more than 80% of worms are already dead), thereby resulting in variability in H3K27me3 levels. To obtain sufficient amounts of synchronized worms, especially for later ages, we grew worms in liquid cultures. Given that UTX-1 regulates lifespan in a germline-independent manner, we sought to monitor H3K27me3 levels in somatic tissues by assessing this mark in germline-deficient worms (glp-1(e2141) mutant worms at the restrictive temperature) (Fig. 7). Western blot experiments revealed that H3K27me3 levels were not significantly affected between youth and middle age in these worms (11 days in liquid culture) (Fig. 7A, B). Interestingly, H3K27me3 levels drastically dropped to almost undetectable levels in glp-1(e2141) worms older than day 11 (day 14, 17 and 20) (Fig. 7A, B). The late-stage loss of the H3K27me3 mark in glp-1(e2141) worms is consistent with our observation that increased H3K27me3 is associated with longevity. These results also suggest that high somatic levels of H3K27me3 are a biomarker of youthfulness. Collectively, these data support the possibility that utx-1 deficiency extends lifespan by maintaining high levels of H3K27me3, perhaps allowing a better control of chromatin repression.
Figure 7. Somatic H3K27me3 levels decrease with age in germline-deficient worms.
A) glp-1(e2141) worms were switched to the restrictive temperature (25°C) at L1 and were aged in liquid cultures. Whole lysates from worms of the indicated ages were blotted with antibodies to H3K27me3 and total histone H3 (H3) as a control for loading. Each lane represents an independent cohort of ~1000 worms. The blots presented are representative of 3 independent experiments. B) Quantification of H3K27me3 levels relative to histone H3 levels in glp-1(e2141) worms at different ages. Mean +/− SEM of 3 independent experiments. *p<0.05 by paired t-tests.
Discussion
Our targeted screen for histone demethylases regulating lifespan in C. elegans confirms a role for the histone demethylases RBR-2 and LSD-1 in the control of longevity, and identifies potential novel regulators of lifespan (T26A5.5 and UTX-1). In particular, our study reveals that the H3K27me3 demethylase, UTX-1, regulates lifespan in an insulin pathway dependent manner. Because histone demethylases and their functions are highly conserved in more complex animals, including mammals, their effect on lifespan in C. elegans may likely be extended to other species.
The reason UTX-1 was not identified earlier in previous large-scale RNAi screens (Lee et al., 2003; Hansen et al., 2005; Chen et al., 2007; Curran & Ruvkun 2007) is unclear. It is possible that the increase in lifespan upon utx-1 knockdown was not large enough to be reproducibly detected in a large screen. One clear difference between this screen and its predecessors is that our screen was performed using fertile worms in the absence of the DNA synthesis inhibitor, 5-fluorodeoxyuridine (FUdR). While longevity induced by utx-1 knock-down is independent of the worm’s fertility and should not be affected by FUdR, this drug can increase the lifespan of wild type worms (Aitlhadj & Sturzenbaum, 2010). Thus, FUdR treatment may have masked the effects on lifespan of utx-1 RNAi in previous screens.
The H3K27me3 mark is associated with regions of facultative heterochromatin. By demethylating the H3K27me3 mark, UTX may relieve chromatin repression (Agger et al., 2007). As shown in this study, knock-down of utx-1 resulted in a corresponding increase in H3K27me3 levels. Coupled with our observation that utx-1 knock-down extends lifespan, these results suggest that the rate of aging may be subject to the regulation of the H3K27me3 mark. Indeed, a loss of epigenetic control over transcriptional silencing has been observed during aging (Wareham et al., 1987; Gaubatz & Cutler 1990; Kennedy et al., 1995; Smeal et al., 1996; Shen et al., 2008) and may be explained, at least in part, by the drop in H3K27me3 levels we observed in the late stages of the worm’s life. Thus, the RNAi-mediated reduction of UTX-1 may promote longevity through the continued maintenance of the repressive H3K27me3 mark, preventing spurious and/or detrimental gene transcription late in life. Our data do not exclude the possibility that utx-1 deficiency at the beginning of adult life sets a different level of H3K27me3, which may have consequences on longevity later in life. Changes in H3K27 methylation status may also be an indirect consequence of UTX-1 depletion in worms. For example, sir-2.1 depletion has been found to indirectly increase H3K27 methylation (Wirth et al., 2009).
The specific genes that may be derepressed by loss of H3K27me3 during aging are not known yet. UTX is thought to control the expression of HOX genes in mammalian cells (Agger et al., 2007; Lan et al., 2007), and a recent genome-wide study identified 2000 genes that are occupied by UTX in mammalian cells, including the retinoblastoma (Rb) gene (Wang et al., 2010). In fact, it is likely that histone demethylases, such as UTX-1, regulate the expression of many genes, making it difficult to identify precisely which ones are important for longevity. In worms, the requirement of the insulin-FoxO pathway for longevity induced by UTX-1 deficiency raises the intriguing possibility that UTX-1 directly influences the expression of regulators of the insulin-FoxO pathway. This is consistent with the observation that utx-1 knock-down triggers FoxO nuclear translocation. Collectively, our results suggest that utx-1 is genetically upstream of FoxO/daf-16, perhaps directly regulating genes that affect the activity of the insulin signaling pathway. However, our study does not exclude the possibility that UTX-1 regulation and H3K27me3 levels are also affected by insulin-FoxO signaling.
In Drosophila, the heterozygous mutation of E(Z), a member of the PRC2 and H3K27 trimethyltransferase complex, has been recently found to extend longevity (Siebold et al., 2010). One explanation for the fact that attenuation of an H3K27me3 methyltransferase (E(Z)) in flies or of an H3K27me3 demethylase (UTX-1) in worms both extend lifespan is that UTX-1 and E(Z) may not function in the same tissue or cell in the organism to regulate lifespan. Moreover, UTX-1 may not work in opposition of every single E(Z) target gene. It is also possible that optimal levels of H3K27me3 are required for proper lifespan extension and that either excess or dearth of H3K27me3 are detrimental for optimal fitness and lifespan. While there is a striking degree of conservation in the histone methylation pathway across species, it is also possible that there exist species-specific differences in how epigenetic marks regulate lifespan.
H3K27 demethylation was recently found to be accompanied with H3K4 trimethylation in mammalian cells (Issaeva et al., 2007) and in C. elegans (Fisher et al., 2010). The H3K4me3 methyltransferase trithorax complex, which contains the subunits ASH-2, WDR5, and the H3K4 specific trimethyltransferase MLL2, co-purifies with UTX in mammalian cells (Issaeva et al., 2007). In C. elegans, homologous proteins responsible for H3K4 trimethylation, ASH-2, WDR5/TAG-125, and the methyltransferase SET-2 was recently found to regulate lifespan in a germline dependent manner (Greer et al., 2010) As we have shown in this study however, knock-down of utx-1 did not require the presence of the germline to extend lifespan. Furthermore, utx-1 knock-down extends lifespan in a manner that depends on the insulin-FoxO pathway, while the longevity induced by set-2 knock-down was only partially dependent upon daf-16/FoxO to regulate lifespan (Greer et al., 2010). Collectively, our results suggest that the UTX-1 demethylase and the SET-2 trimethyltransferase complex impact lifespan by acting in distinct tissues, somatic versus germline, respectively. It is possible, however, that there is coordinated regulation of H3K4 trimethylation and H3K27 demethylation at genes that regulate lifespan. Indeed, evidence from a recent study in C. elegans suggests that UTX-1 and the SET-16 methyltransferase function together in an MLL-like complex (Fisher et al., 2010). Although SET-16 does not appear to regulate lifespan under the conditions tested (Greer et al., 2010), UTX-1 and SET-16 may still cooperate to regulate lifespan under specific circumstances. UTX-1 could also associate with other methyltransferase complexes to regulate aging within the soma. In fact, two other methyltransferases (SET-9 and SET-15) regulate lifespan in a germline-independent manner (Greer et al., 2010), raising the possibility that UTX-1 regulates lifespan together with one or both of these methyltransferases. Our results suggest that different chromatin modifying complexes, involving both methyltransferases and demethylases, regulate lifespan in the germline and in the soma. Understanding the interplay between these reversible epigenetic modifications in different tissues will give insights into mechanisms that slow – or possibly reverse – the aging process in multicellular organisms.
Experimental Procedures
Worm strains and RNA interference
Wild type (N2) and daf-16(mu86) strains were provided by Dr. Man-Wah Tan. The daf-2(e1370) and eat-2(ad1116) were provided by Dr. Cynthia Kenyon. The utx-1(tm3118) strain was provided by Dr. Shohei Mitani. TU899 stDp2II(X;II)+ II; uDf1 X balancer strain, T26A5.5(ok2364), and the glp-1(e2141ts) strains were provided by Dr. Theresa Stiernagle and the Caenorhabditis Genome Center. The Is DAF-16::GFP (TJ356) (Henderson & Johnson 2001) was provided by Dr. Stuart Kim. All mutant strains were backcrossed three times to our lab’s N2 strain, except eat-2(ad1116) which was backcrossed three times to the Kenyon lab’s N2 strain), Is DAF-16::GFP (TJ356) which was backrossed five times to the Kim lab’s N2 strain, and TU899 stDp2II(X;II)+ II; uDf1 X, which was not backcrossed. HT115 (DE3) bacteria transformed with vectors expressing RNAi to the genes of interest were obtained from the Ahringer library (a gift from Dr. M.-W. Tan) or the Open Biosystems library (a gift from Dr. K. Shen). RNAi constructs were validated by sequencing. RNAi bacteria were grown at 37°C and seeded onto standard nematode growth medium (NGM) plates containing Ampicillin (100 mg·ml−1) and IPTG (0.4 mM). Adult worms were placed on standard NGM plates and removed after 4–6 h or bleached to obtain synchronized populations of worms. L1 worms obtained from these synchronized populations were placed on NGM plates containing Ampicillin (100 mg·ml−1) and IPTG (0.4 mM) seeded with the respective bacteria. Worms placed on RNAi at different time points were treated with empty vector control at the L1 stage and shifted to the respective RNAi containing bacteria at the appropriate time.
Lifespan assays
Worm lifespan assays were performed at 20°C as described previously (Greer et al., 2007), unless noted otherwise. Worms were transferred to new plates every other day and were scored as dead or alive. Worms were scored dead if they did not respond to repeated prods with a platinum pick. Worms were scored as censored if they died due to bagging, vulval rupture, or if they crawled off the plate. Data from the censored worms were included up to the day of censorship. For each lifespan assay, 90 worms per condition total were divided evenly among three plates (30 worms per plate) with exception for the initial RNAi screen, which was performed with 30–60 worms per condition. RNAi treatments that produced a greater than 10% relative change in lifespan were further validated using 90 worms per condition. This 10% cutoff was chosen arbitrarily, and RNAi treatments that resulted in less than 10% change in lifespan in the initial screen might turn out to significantly regulate lifespan, when using larger populations of worms. The results and statistical analyses are presented in the Supplementary Tables.
Quantitative RT-PCR
Two hundred worms were picked to NGM plates with OP50 overnight two days in a row. Worms were then picked to bacteria free NGM plates and washed three times with M9 buffer (KH2PO4, 22mM; K2HPO4, 34 mM; NaCl, 86 mM; MgSO4, 1mM). Worm pellets were resuspended in Trizol (Invitrogen), followed by six freeze-thaw cycles in liquid nitrogen. One μg of total RNA was reverse transcribed with oligo dT primers using Superscript II reverse transcriptase (Invitrogen), according to the manufacturer’s protocol. Real time PCR was performed on a Bio-rad iCycler using iQ SYBR green (Bio-rad) with the following primers: pan-actin F: TCGGTATGGGACAGAAGGAC, pan-actin R: CATCCCAGTTGGTGACGATA, utx-1 F: TTCGATGTACTTCGGGTTAGG, utx-1 R: TCTTGTGAATGCCTCGATTG. The experiments were conducted in duplicate and the results were expressed as 2(-(utx-1 number of cycles – pan-actin number of cycles)).
Western blot analysis
Worms were synchronously grown to appropriate stages and washed off of plates with M9 buffer (KH2PO4, 22 mM; K2HPO4, 34 mM; NaCl, 86 mM; MgSO4, 1 mM). Worms were washed several times in M9 buffer to remove any remaining bacteria and then snap frozen in liquid nitrogen. Laemmli sample buffer (SDS, 2.36%; glycerol, 9.43%; β-mercaptoethanol, 5%; Tris pH 6.8, 0.0945 M; bromophenol blue, 0.001%) was added to samples and they were repeatedly snap frozen in liquid nitrogen and thawed at room temperature to break cuticle walls. Worm extracts were sonicated 3 times for 30 seconds at ~15W (VirSonic 600) and boiled at 100°C for 2 minutes before being resolved on SDS-PAGE (14%) and transferred to nitrocellulose membranes. The membranes were incubated with primary antibodies to H3K27me3 (Upstate 07449, 1:2500), or histone H3 (Abcam ab1791, 1:1000). The primary antibodies were visualized using horseradish peroxidase-conjugated anti-rabbit secondary antibody (Calbiochem, 1:7500) and ECL Plus (Amersham Biosciences).
Age-matched cohorts of N2 worms were grown on NGM plates and transferred to new plates accordingly to exclude contaminating progeny. Living worms from age-matched cohorts were washed in M9 buffer and snap frozen in liquid nitrogen. Whole lysates were then extracted from worms at each stage (4, 6, 8, 10, 12, 14, and 16 days old), as described above. Cohorts of glp-1(e2141) mutant worms were grown in liquid culture at the restrictive temperature (25°C) to generate large quantities of worms and to eliminate confounding methylation signals which may arise from the germline or progeny. Worms were grown in S Medium supplemented with E. coli OP50 as a food source and shaken vigorously to ensure oxygenation (Lewis & Fleming 1995). Age-matched cohorts were harvested from liquid culture, centrifuged in 60% sucrose solution to remove bacteria and dead worms, washed in M9 buffer, and snap frozen. Whole lysates were then extracted from worms at each stage (5, 8, 11, 14, 17, and 20 days old), as described above. The blots were scanned and the bands were quantified using ImageJ 1.42q software. Background-subtracted optical density (OD) values for H3K27me3 were normalized to the background-subtracted OD values for total histone H3 protein.
Whole worm immunofluorescence
Worms were washed several times to remove bacteria and resuspended in fixing solution (160 mM KCl, 100 mM Tris HCl pH 7.4, 40 mM NaCl, 20 mM Na2EGTA, 1 mM EDTA, 10 mM spermidine HCl, 30 mM Pipes pH 7.4, 1% Triton X-100, 50% methanol, 2% formaldehyde) and subjected to two rounds of snap freezing in liquid N2. The worms were fixed at 4°C for 30 min and washed briefly in T buffer (100 mM Tris HCl pH 7.4, 1 mM EDTA, 1% Triton X-100) before a 1 h incubation in T buffer supplemented with 1% β-mercaptoethanol at 37°C. The worms were washed with borate buffer (25 mM H3BO3, 12.5 mM NaOH, pH 9.5) and then incubated in borate buffer containing 10 mM DTT for 15 min. Worms were blocked in PBST (PBS, pH 7.4, 0.5% Triton X-100, 1 mM EDTA) containing 1% BSA for 30 min and incubated overnight first with the actin antibody (Chemicon MAB1501R, 1:100), followed by goat anti-mouse Alexa Fluor 488 antibody (Invitrogen, A21042, 1:100), and with the H3K27me3 antibody (Upstate 07449, 1:100) followed with goat anti-rabbit Alexa Fluor 594 antibody (Invitrogen, A11012, 1:100). DAPI (2 μg/ml) was added to visualize nuclei. For histone H3 staining, worms were incubated with the histone H3 antibody (Abcam ab1791, 1:1000) followed with goat anti-rabbit Alexa Fluor 594 antibody as described above. The worms were mounted on a microscope slide and individual optical planes were visualized using a Leica SP2 confocal system. Image acquisition parameters were identical across conditions, so that the fluorescence signal could be compared. DAPI and Alexa Fluor 488 signals were sequentially imaged to eliminate the signal from overlapping emission.
Visualization of DAF-16 localization
The DAF-16::GFP transgenic worms were birthed on plates seeded with RNAi bacteria and grown to adulthood. Worms were then transferred to 1 ml of fixing solution and incubated for 5 min. Fixed worms were then washed with M9 buffer, mounted on a microscope slide, and imaged using a Zeiss Axioskop 2 plus fluorescence microscope with a 20X objective. Twenty to fifty animals per condition were scored as having predominantely nuclear versus cytoplasmic DAF-16::GFP.
Statistical analysis
Statistical analyses of lifespan were performed on Kaplan-Meier survival curves in StatView 5.0.01 by Logrank (Mantel-Cox) tests. For statistical comparison of independent replicates, the Fisher’s combined probability test was performed. To compare the interaction between genotype and RNAi treatment, two-way ANOVA tests were performed in Prism 5 using the mean and standard error values obtained from the Kaplan-Meier survival curves. The values from the Kaplan-Meier curves, Fisher’s combined probability tests, and two-way ANOVA tests are included in the Supplementary Tables.
Supplementary Material
Acknowledgments
We are grateful to A. Fire, S. Kim, S. Mitani, A. Villeneuve, M.W. Tan, and S. Strome for gifts of strains, reagents, antibodies, and for advice. We thank T. Stiernagle and the Caenorhabditis Genetic Center for their assistance with strain nomenclature as well as for providing multiple strains. We thank J. Maniar, T. Kawli, E. Van Nostrand, and A. Sanchez-Blanco for their valuable advice and discussion. We thank all the members of the Brunet laboratory for their critical reading of the manuscript. This work was supported by NIH R01-AG31198 to A.B.; T.J.M. was supported by NIH T32-HG000044 and by NIH F32-AG037254; E.L.G. was supported by NIH T32-CA009302, an NSF graduate fellowship, and by NIH ARRA-AG31198.
References
- Agger K, Cloos PA, Christensen J, Pasini D, Rose S, Rappsilber J, Issaeva I, Canaani E, Salcini AE, Helin K. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature. 2007;449:731–734. doi: 10.1038/nature06145. [DOI] [PubMed] [Google Scholar]
- Aitlhadj L, Sturzenbaum SR. The use of FUdR can cause prolonged longevity in mutant nematodes. Mech Ageing Dev. 2010;131:364–365. doi: 10.1016/j.mad.2010.03.002. [DOI] [PubMed] [Google Scholar]
- Arantes-Oliveira N, Apfeld J, Dillin A, Kenyon C. Regulation of life-span by germ-line stem cells in Caenorhabditis elegans. Science. 2002;295:502–505. doi: 10.1126/science.1065768. [DOI] [PubMed] [Google Scholar]
- Avery L. The genetics of feeding in Caenorhabditis elegans. Genetics. 1993;133:897–917. doi: 10.1093/genetics/133.4.897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett-Baker PE, Wilkowski J, Burke DT. Age-associated activation of epigenetically repressed genes in the mouse. Genetics. 2003;165:2055–2062. doi: 10.1093/genetics/165.4.2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007;447:407–412. doi: 10.1038/nature05915. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Humphrey EL, Erlich RL, Schneider R, Bouman P, Liu JS, Kouzarides T, Schreiber SL. Methylation of histone H3 Lys 4 in coding regions of active genes. Proc Natl Acad Sci U S A. 2002;99:8695–8700. doi: 10.1073/pnas.082249499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, Jaenisch R, Wagschal A, Feil R, Schreiber SL, Lander ES. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Bishop NA, Guarente L. Two neurons mediate diet-restriction-induced longevity in C. elegans. Nature. 2007;447:545–549. doi: 10.1038/nature05904. [DOI] [PubMed] [Google Scholar]
- Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- Chang B, Chen Y, Zhao Y, Bruick RK. JMJD6 is a histone arginine demethylase. Science. 2007;318:444–447. doi: 10.1126/science.1145801. [DOI] [PubMed] [Google Scholar]
- Chen D, Pan KZ, Palter JE, Kapahi P. Longevity determined by developmental arrest genes in Caenorhabditis elegans. Aging Cell. 2007;6:525–533. doi: 10.1111/j.1474-9726.2007.00305.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen J, Agger K, Cloos PA, Pasini D, Rose S, Sennels L, Rappsilber J, Hansen KH, Salcini AE, Helin K. RBP2 belongs to a family of demethylases, specific for tri-and dimethylated lysine 4 on histone 3. Cell. 2007;128:1063–1076. doi: 10.1016/j.cell.2007.02.003. [DOI] [PubMed] [Google Scholar]
- Colman RJ, Anderson RM, Johnson SC, Kastman EK, Kosmatka KJ, Beasley TM, Allison DB, Cruzen C, Simmons HA, Kemnitz JW, Weindruch R. Caloric restriction delays disease onset and mortality in rhesus monkeys. Science. 2009;325:201–204. doi: 10.1126/science.1173635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curran SP, Ruvkun G. Lifespan regulation by evolutionarily conserved genes essential for viability. PLoS Genet. 2007;3:e56. doi: 10.1371/journal.pgen.0030056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang W, Steffen KK, Perry R, Dorsey JA, Johnson FB, Shilatifard A, Kaeberlein M, Kennedy BK, Berger SL. Histone H4 lysine 16 acetylation regulates cellular lifespan. Nature. 2009;459:802–807. doi: 10.1038/nature08085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher K, Southall SM, Wilson JR, Poulin GB. Methylation and demethylation activities of a C. elegans MLL-like complex attenuate RAS signalling. Dev Biol. 2010;341:142–153. doi: 10.1016/j.ydbio.2010.02.023. [DOI] [PubMed] [Google Scholar]
- Gaubatz JW, Cutler RG. Mouse satellite DNA is transcribed in senescent cardiac muscle. J Biol Chem. 1990;265:17753–17758. [PubMed] [Google Scholar]
- Gems D, Partridge L. Stress-response hormesis and aging: “that which does not kill us makes us stronger”. Cell Metab. 2008;7:200–203. doi: 10.1016/j.cmet.2008.01.001. [DOI] [PubMed] [Google Scholar]
- Gems D, Riddle DL. Defining wild-type life span in Caenorhabditis elegans. J Gerontol A Biol Sci Med Sci. 2000;55:B215–219. doi: 10.1093/gerona/55.5.b215. [DOI] [PubMed] [Google Scholar]
- Greer EL, Dowlatshahi D, Banko MR, Villen J, Hoang K, Blanchard D, Gygi SP, Brunet A. An AMPK-FOXO pathway mediates longevity induced by a novel method of dietary restriction in C. elegans. Curr Biol. 2007;17:1646–1656. doi: 10.1016/j.cub.2007.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer EL, Maures TJ, Hauswirth AG, Green EM, Leeman DS, Maro GS, Banko MR, Gozani O, Brunet A. Members of the Histone H3 Lysine 4 Trimethylation Complex Regulate Lifespan in a Germline-dependent Manner in C. elegans. Nature. 2010 doi: 10.1038/nature09195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton B, Dong Y, Shindo M, Liu W, Odell I, Ruvkun G, Lee SS. A systematic RNAi screen for longevity genes in C. elegans. Genes Dev. 2005;19:1544–1555. doi: 10.1101/gad.1308205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen M, Hsu AL, Dillin A, Kenyon C. New genes tied to endocrine, metabolic, and dietary regulation of lifespan from a Caenorhabditis elegans genomic RNAi screen. PLoS Genet. 2005;1:119–128. doi: 10.1371/journal.pgen.0010017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J, Kallin EM, Tsukada Y, Zhang Y. The H3K36 demethylase Jhdm1b/Kdm2b regulates cell proliferation and senescence through p15(Ink4b) Nat Struct Mol Biol. 2008;15:1169–1175. doi: 10.1038/nsmb.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson ST, Johnson TE. daf-16 integrates developmental and environmental inputs to mediate aging in the nematode Caenorhabditis elegans. Curr Biol. 2001;11:1975–1980. doi: 10.1016/s0960-9822(01)00594-2. [DOI] [PubMed] [Google Scholar]
- Hong S, Cho YW, Yu LR, Yu H, Veenstra TD, Ge K. Identification of JmjC domain-containing UTX and JMJD3 as histone H3 lysine 27 demethylases. Proc Natl Acad Sci U S A. 2007;104:18439–18444. doi: 10.1073/pnas.0707292104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honjoh S, Yamamoto T, Uno M, Nishida E. Signalling through RHEB-1 mediates intermittent fasting-induced longevity in C. elegans. Nature. 2009;457:726–730. doi: 10.1038/nature07583. [DOI] [PubMed] [Google Scholar]
- Hosono R, Nishimoto S, Kuno S. Alterations of life span in the nematode Caenorhabditis elegans under monoxenic culture conditions. Exp Gerontol. 1989;24:251–264. doi: 10.1016/0531-5565(89)90016-8. [DOI] [PubMed] [Google Scholar]
- Houthoofd K, Braeckman BP, Lenaerts I, Brys K, De Vreese A, Van Eygen S, Vanfleteren JR. Axenic growth up-regulates mass-specific metabolic rate, stress resistance, and extends life span in Caenorhabditis elegans. Exp Gerontol. 2002;37:1371–1378. doi: 10.1016/s0531-5565(02)00173-0. [DOI] [PubMed] [Google Scholar]
- Hsin H, Kenyon C. Signals from the reproductive system regulate the lifespan of C. elegans. Nature. 1999;399:362–366. doi: 10.1038/20694. [DOI] [PubMed] [Google Scholar]
- Issaeva I, Zonis Y, Rozovskaia T, Orlovsky K, Croce CM, Nakamura T, Mazo A, Eisenbach L, Canaani E. Knockdown of ALR (MLL2) reveals ALR target genes and leads to alterations in cell adhesion and growth. Mol Cell Biol. 2007;27:1889–1903. doi: 10.1128/MCB.01506-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson TE. Increased life-span of age-1 mutants in Caenorhabditis elegans and lower Gompertz rate of aging. Science. 1990;249:908–912. doi: 10.1126/science.2392681. [DOI] [PubMed] [Google Scholar]
- Kaeberlein TL, Smith ED, Tsuchiya M, Welton KL, Thomas JH, Fields S, Kennedy BK, Kaeberlein M. Lifespan extension in Caenorhabditis elegans by complete removal of food. Aging Cell. 2006;5:487–494. doi: 10.1111/j.1474-9726.2006.00238.x. [DOI] [PubMed] [Google Scholar]
- Karytinos A, Forneris F, Profumo A, Ciossani G, Battaglioli E, Binda C, Mattevi A. A novel mammalian flavin-dependent histone demethylase. J Biol Chem. 2009;284:17775–17782. doi: 10.1074/jbc.M109.003087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemphues KJ, Kusch M, Wolf N. Maternal-effect lethal mutations on linkage group II of Caenorhabditis elegans. Genetics. 1988;120:977–986. doi: 10.1093/genetics/120.4.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy BK, Austriaco NR, Jr, Zhang J, Guarente L. Mutation in the silencing gene SIR4 can delay aging in S. cerevisiae. Cell. 1995;80:485–496. doi: 10.1016/0092-8674(95)90499-9. [DOI] [PubMed] [Google Scholar]
- Kenyon C, Chang J, Gensch E, Rudner A, Tabtiang R. A C. elegans mutant that lives twice as long as wild type. Nature. 1993;366:461–464. doi: 10.1038/366461a0. [DOI] [PubMed] [Google Scholar]
- Kimura KD, Tissenbaum HA, Liu Y, Ruvkun G. daf-2, an insulin receptor-like gene that regulates longevity and diapause in Caenorhabditis elegans. Science. 1997;277:942–946. doi: 10.1126/science.277.5328.942. [DOI] [PubMed] [Google Scholar]
- Klass MR. Aging in the nematode Caenorhabditis elegans: major biological and environmental factors influencing life span. Mech Ageing Dev. 1977;6:413–429. doi: 10.1016/0047-6374(77)90043-4. [DOI] [PubMed] [Google Scholar]
- Kleine-Kohlbrecher D, Christensen J, Vandamme J, Abarrategui I, Bak M, Tommerup N, Shi X, Gozani O, Rappsilber J, Salcini AE, Helin K. A functional link between the histone demethylase PHF8 and the transcription factor ZNF711 in X-linked mental retardation. Mol Cell. 2010;38:165–178. doi: 10.1016/j.molcel.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose RJ, Yan Q, Tothova Z, Yamane K, Erdjument-Bromage H, Tempst P, Gilliland DG, Zhang Y, Kaelin WG., Jr The retinoblastoma binding protein RBP2 is an H3K4 demethylase. Cell. 2007;128:889–900. doi: 10.1016/j.cell.2007.02.013. [DOI] [PubMed] [Google Scholar]
- Lakowski B, Hekimi S. Determination of life-span in Caenorhabditis elegans by four clock genes. Science. 1996;272:1010–1013. doi: 10.1126/science.272.5264.1010. [DOI] [PubMed] [Google Scholar]
- Lan F, Bayliss PE, Rinn JL, Whetstine JR, Wang JK, Chen S, Iwase S, Alpatov R, Issaeva I, Canaani E, Roberts TM, Chang HY, Shi Y. A histone H3 lysine 27 demethylase regulates animal posterior development. Nature. 2007;449:689–694. doi: 10.1038/nature06192. [DOI] [PubMed] [Google Scholar]
- Lee CK, Weindruch R, Prolla TA. Gene-expression profile of the ageing brain in mice. Nat Genet. 2000;25:294–297. doi: 10.1038/77046. [DOI] [PubMed] [Google Scholar]
- Lee GD, Wilson MA, Zhu M, Wolkow CA, de Cabo R, Ingram DK, Zou S. Dietary deprivation extends lifespan in Caenorhabditis elegans. Aging Cell. 2006;5:515–524. doi: 10.1111/j.1474-9726.2006.00241.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SS, Lee RY, Fraser AG, Kamath RS, Ahringer J, Ruvkun G. A systematic RNAi screen identifies a critical role for mitochondria in C. elegans longevity. Nat Genet. 2003;33:40–48. doi: 10.1038/ng1056. [DOI] [PubMed] [Google Scholar]
- Lewis JA, Fleming JT. Basic culture methods. Methods Cell Biol. 1995;48:3–29. [PubMed] [Google Scholar]
- Lin K, Dorman JB, Rodan A, Kenyon C. daf-16: An HNF-3/forkhead family member that can function to double the life-span of Caenorhabditis elegans. Science. 1997;278:1319–1322. doi: 10.1126/science.278.5341.1319. [DOI] [PubMed] [Google Scholar]
- Lu T, Pan Y, Kao SY, Li C, Kohane I, Chan J, Yankner BA. Gene regulation and DNA damage in the ageing human brain. Nature. 2004;429:883–891. doi: 10.1038/nature02661. [DOI] [PubMed] [Google Scholar]
- Lund J, Tedesco P, Duke K, Wang J, Kim SK, Johnson TE. Transcriptional profile of aging in C. elegans. Curr Biol. 2002;12:1566–1573. doi: 10.1016/s0960-9822(02)01146-6. [DOI] [PubMed] [Google Scholar]
- Mair W, Dillin A. Aging and survival: the genetics of life span extension by dietary restriction. Annu Rev Biochem. 2008;77:727–754. doi: 10.1146/annurev.biochem.77.061206.171059. [DOI] [PubMed] [Google Scholar]
- McColl G, Killilea DW, Hubbard AE, Vantipalli MC, Melov S, Lithgow GJ. Pharmacogenetic analysis of lithium-induced delayed aging in Caenorhabditis elegans. J Biol Chem. 2008;283:350–357. doi: 10.1074/jbc.M705028200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris JZ, Tissenbaum HA, Ruvkun G. A phosphatidylinositol-3-OH kinase family member regulating longevity and diapause in Caenorhabditis elegans. Nature. 1996;382:536–539. doi: 10.1038/382536a0. [DOI] [PubMed] [Google Scholar]
- Morris SA, Shibata Y, Noma K, Tsukamoto Y, Warren E, Temple B, Grewal SI, Strahl BD. Histone H3 K36 methylation is associated with transcription elongation in Schizosaccharomyces pombe. Eukaryot Cell. 2005;4:1446–1454. doi: 10.1128/EC.4.8.1446-1454.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogg S, Paradis S, Gottlieb S, Patterson GI, Lee L, Tissenbaum HA, Ruvkun G. The Fork head transcription factor DAF-16 transduces insulin-like metabolic and longevity signals in C. elegans. Nature. 1997;389:994–999. doi: 10.1038/40194. [DOI] [PubMed] [Google Scholar]
- Pasini D, Hansen KH, Christensen J, Agger K, Cloos PA, Helin K. Coordinated regulation of transcriptional repression by the RBP2 H3K4 demethylase and Polycomb-Repressive Complex 2. Genes Dev. 2008;22:1345–1355. doi: 10.1101/gad.470008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priess JR, Schnabel H, Schnabel R. The glp-1 locus and cellular interactions in early C. elegans embryos. Cell. 1987;51:601–611. doi: 10.1016/0092-8674(87)90129-2. [DOI] [PubMed] [Google Scholar]
- Rao B, Shibata Y, Strahl BD, Lieb JD. Dimethylation of histone H3 at lysine 36 demarcates regulatory and nonregulatory chromatin genome-wide. Mol Cell Biol. 2005;25:9447–9459. doi: 10.1128/MCB.25.21.9447-9459.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos-Rosa H, Schneider R, Bannister AJ, Sherriff J, Bernstein BE, Emre NC, Schreiber SL, Mellor J, Kouzarides T. Active genes are tri-methylated at K4 of histone H3. Nature. 2002;419:407–411. doi: 10.1038/nature01080. [DOI] [PubMed] [Google Scholar]
- Scaffidi P, Misteli T. Reversal of the cellular phenotype in the premature aging disease Hutchinson-Gilford progeria syndrome. Nat Med. 2005;11:440–445. doi: 10.1038/nm1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scaffidi P, Misteli T. Lamin A-dependent nuclear defects in human aging. Science. 2006;312:1059–1063. doi: 10.1126/science.1127168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen S, Liu A, Li J, Wolubah C, Casaccia-Bonnefil P. Epigenetic memory loss in aging oligodendrocytes in the corpus callosum. Neurobiol Aging. 2008;29:452–463. doi: 10.1016/j.neurobiolaging.2006.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- Shi Y, Whetstine JR. Dynamic regulation of histone lysine methylation by demethylases. Mol Cell. 2007;25:1–14. doi: 10.1016/j.molcel.2006.12.010. [DOI] [PubMed] [Google Scholar]
- Siebold AP, Banerjee R, Tie F, Kiss DL, Moskowitz J, Harte PJ. Polycomb Repressive Complex 2 and Trithorax modulate Drosophila longevity and stress resistance. Proc Natl Acad Sci U S A. 2010;107:169–174. doi: 10.1073/pnas.0907739107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smeal T, Claus J, Kennedy B, Cole F, Guarente L. Loss of transcriptional silencing causes sterility in old mother cells of S. cerevisiae. Cell. 1996;84:633–642. doi: 10.1016/s0092-8674(00)81038-7. [DOI] [PubMed] [Google Scholar]
- Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- Tsukada Y, Fang J, Erdjument-Bromage H, Warren ME, Borchers CH, Tempst P, Zhang Y. Histone demethylation by a family of JmjC domain-containing proteins. Nature. 2006;439:811–816. doi: 10.1038/nature04433. [DOI] [PubMed] [Google Scholar]
- Wang JK, Tsai MC, Poulin G, Adler AS, Chen S, Liu H, Shi Y, Chang HY. The histone demethylase UTX enables RB-dependent cell fate control. Genes Dev. 2010;24:327–332. doi: 10.1101/gad.1882610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wareham KA, Lyon MF, Glenister PH, Williams ED. Age related reactivation of an X-linked gene. Nature. 1987;327:725–727. doi: 10.1038/327725a0. [DOI] [PubMed] [Google Scholar]
- Wirth M, Paap F, Fischle W, Wenzel D, Agafonov DE, Samatov TR, Wisniewski JR, Jedrusik-Bode M. HIS-24 linker histone and SIR-2.1 deacetylase induce H3K27me3 in the Caenorhabditis elegans germ line. Mol Cell Biol. 2009;29:3700–3709. doi: 10.1128/MCB.00018-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.