Abstract
Porphyromonas gingivalis is an important component of the complex plaque biofilm that is a direct precursor of periodontal disease. The major fimbriae are required for attachment to oral surfaces and are an important virulence factor. Fimbrillin (FimA) expression in P. gingivalis is inhibited by surface molecule of Streptococcus cristatus, an early colonizer of dental plaque. In this study, differential display PCR was used to identify P. gingivalis genes that are regulated in response to S. cristatus. Of several differentially expressed genes, pg2131 and pg2167 were upregulated by S. cristatus signaling molecules. A null mutant of pg2167 did not transcriptionally regulate fimA following exposure to S. cristatus. In fact, fimA transcription was enhanced in the pg2167 mutant, suggesting that pg2167 may act to repress fimA expression. In contrast, a mutation in pg2131 did not affect transcription of fimA in the presence of S. cristatus. However, production of fimbrillin was significantly diminished in the pg2131 mutant, implicating involvement in posttranscriptional regulation in fimbriation. These data suggest that P. gingivalis fimbriation is controlled by more than one regulation mechanism, involving both transcriptional and posttranscriptional processes.
Porphyromonas gingivalis is an important pathogen in severe and chronic manifestations of periodontal disease (18, 24). Colonization of the dental plaque biofilm by P. gingivalis is mediated by attachment to salivary molecules in the acquired pellicle (12) and to antecedent colonizing bacteria such as certain oral streptococci (10). P. gingivalis can adhere to Streptococcus gordonii in a fimbria (FimA)-dependent manner and subsequently accretes into biofilm microcolonies (9, 11). In contrast, P. gingivalis does not accumulate on substrata of Streptococcus mutans or S. cristatus. Indeed, a signaling event between S. cristatus and P. gingivalis results in downregulation of fimA transcription (21).
Expression of the fimA gene is modulated by a variety of environmental cues. FimA production is significantly decreased when P. gingivalis senses the elevated temperatures (39°C) which are characteristic of inflamed subgingival pockets (1, 23). Expression of fimA is also regulated at the transcriptional level in response to hemin concentrations (23). Despite identification of stimuli involved in fimA regulation, signal processing in P. gingivalis is not well understood. Our early studies indicate that fimbrillin itself, along with arginine (Rgp) and lysine (Kgp) proteases, is a necessary element in fimA transcription (20). A two-component regulatory system including fimS and fimR was also found to be involved in the fimbriation of P. gingivalis (7). Sequence analysis demonstrates that fimS is a homologue of histidine protein kinase genes and fimR is a homologue of the response regulator gene. Disruption of fimS or fimR results in significant reduction of fimbrillin production. The signals that initiate sensory information flow through this two-component system remain unknown. However, the evidence suggests that P. gingivalis possesses complex signal transduction systems, which allow the organism to process and respond to a variety of signals in order to maximize its chances of survival.
In the present study, we investigated the elements involved in the P. gingivalis signaling process in response to S. cristatus. Differential display reverse transcription-PCR identified several P. gingivalis genes that are differentially expressed in the course of interaction between P. gingivalis and S. cristatus. A mutation in one of these genes (pg2167) rendered P. gingivalis insensitive to the S. cristatus signaling molecule. Moreover, the mutant appears to overexpress FimA. In contrast, mutation of a second gene (pg2131) reduced FimA production significantly. However, reverse transcription (RT)-PCR showed that fimA expression was not affected at the transcriptional level in this mutant. These results suggest that the protein encoded by the differentially expressed gene pg2167 is an element acting in a repressor system for fimA expression and that PG2131 regulates fimA expression at the posttranscriptional level.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains and plasmids used are listed in Table 1. P. gingivalis ATCC 33277 was used as the parental strain for mutant construction. P. gingivalis strains were grown from frozen stocks in Trypticase soy broth (TSB) or on TSB blood agar plates supplemented with yeast extract (1 mg/ml), hemin (5 μg/ml), and menadione (1 μg/ml) at 37°C in an anaerobic chamber (85% N2, 10% H2, 5% CO2). Streptococcus strains were grown in Trypticase peptone broth supplemented with 0.5% glucose at 37°C under aerobic conditions. Escherichia coli DH5α was the host for plasmids. E. coli strains were grown in L broth at 37°C. Antibiotics were used when appropriate, at the following concentrations: gentamicin, 100 μg/ml; erythromycin, 20 μg/ml; ampicillin, 50 μg/ml; kanamycin, 50 μg/ml; and tetracycline, 20 μg/ml.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristicsa | Source or reference |
|---|---|---|
| S. gordonii G9B | Low-passage plaque isolate | Lab collection |
| S. cristatus CC5A | Low-passage plaque isolate | Lab collection |
| P. gingivalis | ||
| ATCC 33277 | Type strain | Lab collection |
| UPF | Derivative of P. gingivalis 33277 containing fimA::lacZ gene fusion in its chromosomal DNA, Emr | 22 |
| DPG3 | P. gingivalis mutant with fimA gene inactivated by insertion-duplication mutagenesis, Emr | 14 |
| 2131 | P. gingivalis mutant carrying an insertional mutation in pg2131 gene, Emr | This study |
| 2167 | P. gingivalis mutant carrying an insertional mutation in pg2167 gene, Emr | This study |
| E. coli | ||
| S17-1 | F pro recA1 derivative of E. coli 294 carrying a modified derivative of IncPα plasmid pRP4 (Tcs Kms) integrated in the chromosome, Tpr | 13 |
| DH5α | F− φ80dlacZΔ(lacZYA-argF)U169 endA1 supE44 recA1 relA1 | BRL |
| GH | Host strain for plasmid PCR-TRSAP | GenHunter |
| Plasmids | ||
| pVA3000 | Suicide vector for Bacteroides; Emr, 5.3 kb | 13 |
| pVA2131 | pVA300 with a 143-bp fragment of pg2131 gene at BamHI-XbaI sites | This study |
| pVA2167 | pVA300 with a 214-bp fragment of pg2167 gene at BamHI-XbaI sites | This study |
| PCRII-TOTO | Linearized plasmid with single 3 dT residues, Kmr Amr | Invitrogen |
| PCR-TRAP | Cloning PCR product by blunt-end ligation, cI, Tetr | GenHunter |
cI, phage lambda repressor gene; Kmr, Smr, Tetr, Emr, Tpr, Amr, resistance to kanamycin, streptomycin, tetracyline, erythromycin, trimethoprim, and ampicillin, respectively; Aps Tcs Kms, sensitive to amorcillin, tetracycline, and kanamycin.
Initial purification of S. cristatus surface proteins.
S. cristatus CC5A was cultured to late log phase, and cells were collected by centrifugation and resuspended in phosphate-buffered saline (PBS). A surface extract was prepared by sonication with a Sonic Dismembrator (Fisher Scientific; output control, 8; time, 30 s times three), and whole cells were removed by centrifugation (13,000 × g for 30 min) followed by filtration (0.2-μm pore size). The crude extract of CC5A was partially purified by ammonium sulfate fractionation; 2.5 ml of ammonium sulfate solution (90 to 95%) was slowly added to 10 ml of the crude extract. The mixture was incubated on ice for 5 min and centrifuged at 15,000 × g and 4°C for 5 min. The supernatant was transferred to a clean tube, and the precipitate pellet was resuspended in 1 ml of 20 mM Tris buffer, pH 8.0. This fraction was designated AS1 (≈15% saturation). Another 2.5 ml of ammonium sulfate was added to the supernatant, and the proteins precipitated out of solution were designated AS2 (≈30%). The procedure was repeated until the proteins were salted out in the presence of 70% ammonium sulfate.
All six AS fractions were dialyzed against PBS to remove the ammonium sulfate, and protein concentrations were determined by the Bio-Rad protein assay. AS fractions (25 μg) were mixed with 105 cells of P. gingivalis UPF, which contains a chromosomal fimA promoter-lacZ reporter construct, and spotted onto a TSB blood agar plate. The ability of AS fractions to inhibit fimA expression in P. gingivalis was determined with a β-galactosidase assay.
Differential display PCR.
P. gingivalis ATCC 33277 was cultivated on TSB blood agar plates with or without S. cristatus CC5A AS6 fraction (50 μg of protein) for 48 h. Cells were harvested and resuspended in Tris-EDTA buffer containing lysozyme (40 μg/100 μl). Total RNA was isolated with a Nucleospin nucleic acid purification kit (Clontech). To remove DNA, samples were treated with DNase I (GenHunter). Fluorescent differential display was performed at GenHunter (Nashville, Tenn.) with 24 arbitrary primers. Differentially displayed bands were excised from polyacrylamide gels and reamplified by PCR with the same set of primers. The PCR products were ligated into the pCR-TRAP cloning vector (GenHunter). Ligated plasmids were transformed into competent cells of Escherichia coli strain GH and plated on LB plates containing 20 μg of tetracycline per ml. The pCR-TRAP vector allows tetracycline-dependent positive selection of plasmids with DNA inserts. Only recombinant plasmids confer antibiotic resistance. The insertions were confirmed by PCR with the same set of primers. The inserts were then recloned into pCRII-Topo, and DNA was sequenced on both strands with an automated 377 DNA sequencer (Applied Biosystems/Perkin-Elmer, Foster City, Calif.). DNA sequence data were analyzed with the sequencing analysis 3.4.1 Alias (Applied Biosystems/Perkin-Elmer).
RT-PCR.
Reverse transcription-PCR was conducted with Superscript II RNase H reverse transcriptase (Invitrogen). For first-strand cDNA synthesis, 5 μg of total RNA was added to 20-μl reaction solution containing 40 units of RNase OUT recombinant RNase inhibitor and incubated at 42°C for 50 min. The reaction was stopped by heating at 70°C for 15 min. To remove RNA complementary to the cDNA, 1 μl of E. coli RNase H (Promega) was added; 10% of the first-stand reaction was used in standard PCRs. Specific primers (Table 2) were used in both first-strand cDNA synthesis and PCRs.
TABLE 2.
Synthetic oligonucleotide primers used for PCR and RT-PCR
| Primer | Sequence (5′ → 3′) | PCR product |
|---|---|---|
| pg2167for | TAGCCCACAATAAGGCTTTG | 224-bp fragment of pg2167 gene |
| pg2167rev | CGTGCATCATCATCGGCATA | |
| pg2131for | AGCTTACCAGGTAGGGAAAT | 143-bp fragment of pg2131 gene |
| pg2131rev | GAAGCATAGCCTACCATGGA | |
| pg1361for | CTTCATTCCGCACACTCGAT | 362-bp fragment of pg1361 gene |
| pg1361rev | CACCATACTCTGATCCATTC | |
| pg0707for | TTGGGCTTGAAAGTGGGTTA | 162-bp fragment of pg0707 gene |
| pg0707rev | ATGTAGCCTGTACGCGGCTG | |
| pg2167for2 | CTCTTCGGAATGTCTACG | 1.4-kb fragment of pg2167 gene |
| pg2167rev2 | TACAACTAACTCTTATCG | |
| RGP1 | TCAACACCGGTAGAGGAAAA | 900-bp fragment of rgp gene |
| RGP2 | AATGGTGCTGGCGATAATAG | |
| FE1 | CGGGATCCCGTGGTATTGAAGACCAGCAAT | 1,042-bp fragment of fimA gene |
| FE2 | GGAATTCCAAGTAGCATTCTGACCAACGAG |
Construction of P. gingivalis mutant strains.
Insertional mutation of differentially displayed genes was generated by standard DNA recombinant technology (17). Plasmid DNA was prepared with the Wizard Plus miniprep kit (Promega) according to the manufacturer's instruction. A fragment of pg2167 was amplified with PCR, and a 214 bp of PCR product was cloned into pCRII-Topo. The pg2167 fragment was then cloned into the BamHI and XbaI sites of suicide plasmid pVA3000 carrying the erythromycin resistance gene cassette ermAM-ermF (13) to create pVA2167. Plasmid pVA2167 was introduced into P. gingivalis by conjugation. The conjugative strain E. coli S17-1 was transformed with pVA2167 to generate the donor strain for mating with P. gingivalis. An overnight culture of E. coli donor was inoculated into 5 ml of L broth and grown aerobically for 2 to 3 h to reach an A600 of 0.3. An overnight culture of P. gingivalis ATCC 33277, the recipient, was grown anaerobically in TSB medium for 8 h. The donor and the recipient were mixed at a ratio of 1 to 5 and spotted onto HAWP filters (pore size, 0.45 μm; Millipore). The mating was initially done under aerobic conditions for 16 h and was followed by anaerobic growth for 8 h at 37°C. Transconjugants, designated P. gingivalis 2167, were selected on TSB blood plates containing erythromycin (20 μg/ml) and gentamicin (100 μg/ml). The same procedure was used to generate P. gingivalis 2131 with an insertional mutation in the pg2131 gene.
Western blot analysis.
P. gingivalis surface proteins were collected by sonication and centrifugation as described previously (21). Protein concentrations of the samples were determined with the Bio-Rad protein assay. Samples were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (12% gel) along with prestained molecular size standards (Bio-Rad) and transferred to nitrocellulose membranes (Gibco-BRL) with a Mini Transblot electrophoretic transfer cell (Bio-Rad Laboratories) at 100 V for 1 h. The membrane was treated with 30 ml of blocking solution (3% bovine serum albumin in PBS containing 0.1% Tween 20, pH 7.4) for 1 h and incubated for 1 h with antifimbrillin antibodies (20) diluted 1:1,000 in PBS containing 0.1% Tween 20, pH 7.4. The membrane was then rinsed twice and washed three times for 15 min each with 0.1% Tween 20 in PBS. The membrane was incubated with horseradish peroxidase-conjugated anti-rabbit secondary antibodies (Amersham Biosciences) for 1 h and rinsed and washed as described above. Antigen-antibody reactivity was visualized by enhanced chemiluminescence (Amersham Pharmacia Biotech).
Enzyme-linked immunosorbent assay.
Microtiter plates (Costar) were coated with surface proteins extracted from (i) P. gingivalis ATCC 33277, (ii) P. gingivalis 2167, (iii) P. gingivalis 2131, and (iv) P. gingivalis DPG3. Plates were sealed, incubated overnight at 4°C, and then washed five times with PBS containing 0.1% Tween 20, pH 7.4. Plates were blocked with 3% bovine serum albumin in PBS-Tween for 1 h at 37°C. Antifimbrillin antibodies (1:500) were then incubated at room temperature for 1 h, and the plates were washed and incubated with horseradish peroxidase-conjugated antibody to rabbit IgG (Amersham Biosciences) in incubation buffer for 1 h at room temperature. After the wells were washed five times with PBS, peroxidase substrate was added to each well. The optical density of the reaction mixture was determined at 405 nm with a microplate reader (Molecular Design). All samples were run in duplicate.
β-Galactosidase assays.
Expression of the lacZ gene under control of the fimA promoter was measured by the standard spectrophotometric β-galactosidase assay with o-nitrophenyl-β-d-galactopyranoside (ONPG) as the substrate, as described by Miller (15). P. gingivalis UPF was cultured anaerobically with Trypticase blood plates under a variety of conditions. Bacteria were recovered from late-log-phase cultures and tested at an A600 of 0.4 to 0.6.
RESULTS
Enrichment of S. cristatus protein that inhibits fimA expression in P. gingivalis.
Initial fractionation of the complex protein extract of S. cristatus CC5A was done by ammonium sulfate precipitation. The inhibitory activity for P. gingivalis fimA expression was determined by a β-galactosidase assay. When added to P. gingivalis growth media, the proteins precipitated by 30, 37, and 45% saturated (NH4)2SO4 did not alter the level of fimA transcription. The fraction precipitated with 52% saturated (NH4)2SO4 inhibited fimA expression by twofold. The highest inhibitory activity was found in AS6, which was precipitated with (NH4)2SO4 at 70% saturation. In the presence of AS6, the fimA transcriptional level was decreased 10-fold. To examine the role of AS6 in the growth of P. gingivalis, 25, 50, and 100 μg of AS6 were mixed with 105 cells of P. gingivalis UPF. After 48 h the growth rate of the bacteria was determined by optical density at A600. The results showed that the addition of AS6 fraction up to 100 μg did not affect the growth rate of P. gingivalis. The 25-μg AS6 fraction of CC5A surface proteins was then used for identification of differentially expressed genes in P. gingivalis.
Identification of P. gingivalis genes regulated by S. cristatus surface proteins.
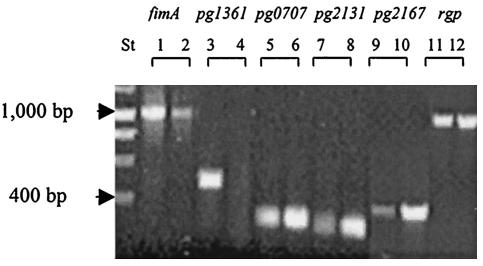
To screen for P. gingivalis genes involved in intergeneric communication, the organism was cultured with and without the CC5A AS6 fraction. Differential display analysis was used to identify specific changes in gene expression in response to S. cristatus CC5A surface proteins. Several genes were differentially expressed in P. gingivalis (Fig. 1). Among them, four genes were cloned and subjected to further investigation. Three genes were significantly induced and one was repressed by CC5A proteins. RT-PCR was performed to verify the differential expression of these genes. As shown in Fig. 2, these experiments confirmed that the genes were induced or suppressed in response CC5A proteins. In agreement with our previous report (21), expression of rgp, a gene encoding an arginine-specific cysteine proteinase, was not affected by CC5A AS6. As a control for specificity of the response to CC5A, P. gingivalis was also cultured with a surface extract from Streptococcus gordonii G9B. RT-PCR did not detect differential expression of these genes in response to S. gordonii G9B (data not shown).
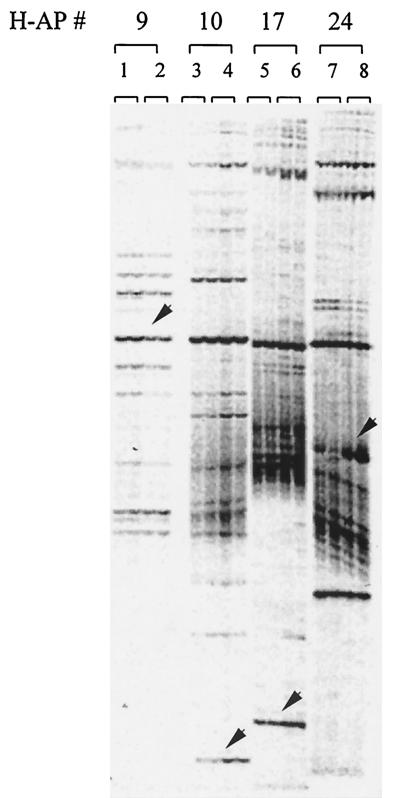
FIG. 1.
Differential gene expression in P. gingivalis. Total RNA was isolated from P. gingivalis 33277 grown on TSB plates with (lanes 2, 4, 6, and 8) or without S. cristatus fraction AS6 (lanes 1, 3, 5, and 7) and subjected to differential display analysis. Arbitrary primer (H-AP) combinations were used in reverse transcription reactions. The fluorescent differential display gel shows the band pattern obtained from duplicate reactions. The arrows indicate the bands corresponding to cDNA fragments that were differentially expressed in the presence of S. cristatus CC5A proteins and that were cloned and sequenced.
FIG. 2.
RT-PCR expression analysis of differentially expressed genes. Total RNAs were extracted from P. gingivalis grown without S. cristatus CC5A proteins (lanes 1, 3, 5, 7, and 9) and from P. gingivalis grown with CC5A proteins (lanes 2, 4, 6, 8, and 10). The differentially expressed genes and fimA were amplified under the same conditions. DNA markers (lane St) are indicated by arrows.
To identify the differentially expressed genes, the cDNA fragments were cloned and sequenced. Searches of the TIGR Comprehensive Microbial Resource with the nucleotide sequence revealed that the genes had significant sequence identity to PG1361 (identities 390 of 414, 94%), PG0707 (identities 144 of 169, 85%), PG2131 (identities 136 of 144, 94%), and PG2167 (identities 219 of 228, 96%). PG1361 is a putative dipeptidyl aminopeptidase IV and was shown to be negatively regulated in response to CC5A proteins. Of the positively regulated genes, PG0707 is a hypothetical protein; PG2131 is a 60-kDa protein; and PG2167 is an immunoreactive 53-kDa antigen. Of these, PG2131 and PG2167 were selected for further investigation.
Role of pg2167 in fimA expression.
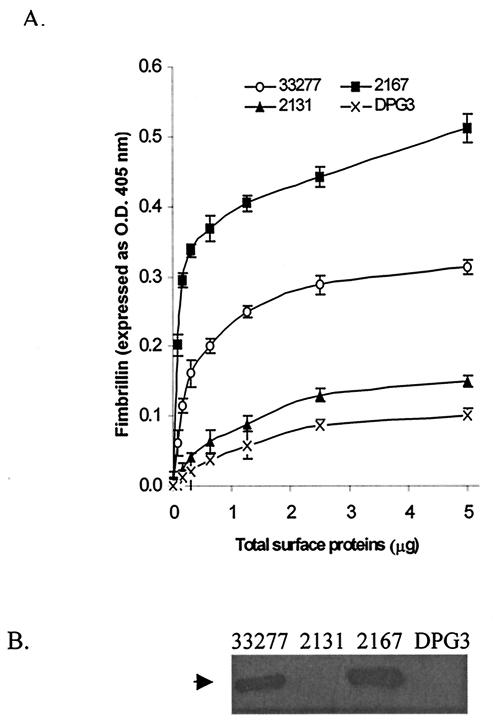
An insertional mutation of pg2167 was constructed with plasmid pVA2167 carrying a 224-bp fragment of the pg2167 gene and the erythromycin resistance gene cassette ermAM-ermF. The insertional mutation was confirmed by PCR with primers pg2167for2 and pg2167rev2. A 1.4-kb PCR product was detected when wild-type 33277 chromosomal DNA was used as the template, but no PCR product was detected when P. gingivalis 2167 DNA was used as the template under the same PCR conditions. RT-PCR was performed to determine alterations of fimA expression due to the mutation in the pg2167 gene. The level of fimA transcription in mutant strain 2167 did not change significantly in response to S. cristatus surface proteins (AS6 fraction), in contrast to the regulation in the parent strain (Fig. 3). However, when grown at different temperatures, both wild-type 33277 and the mutant 2167 showed enhanced transcriptional levels of the fimA gene at 34°C and reduced fimA transcription at 39°C (Fig. 3), indicating that the temperature-dependent control of fimA transcription remains in tact in this mutant. In addition, the RT-PCR results also indicated that P. gingivalis 2167 appeared to make more fimA message than the wild-type strain when grown under the same conditions. Furthermore, ELISA and Western blot analyses of total protein from the wild-type 33277 and the 2167 mutant with fimbrillin-specific polyclonal antibody demonstrated that fimbrillin production in P. gingivalis 2167 was at least twofold more than in P. gingivalis 33277 (Fig. 4). These results suggest that pg2167 may play a role in a repressor system for fimA expression.
FIG. 3.
RT-PCR analysis of gene expression in P. gingivalis 2167 and parental strain 33277. Total RNAs were extracted from P. gingivalis 33277 and 2167 grown with or without the AS6 fraction of CC5A. Primers FE1 and FE2 were used in RT-PCR to create a 1,024-bp fimA fragment. Lane 1, 33277 grown at 34°C; lane 2, 33277 grown at 39°C; lane 3, 2167 grown at 34°C; lane 4, 2167 grown at 39°C; lane 5, 33277 grown with S. cristatus CC5A surface protein AS6; lane 6, 33277 grown without CC5A proteins; lane 7, 2167 grown with CC5A proteins; lane 8, 2167 grown without CC5A proteins. DNA markers (lane St) are indicated by arrows.
FIG. 4.
Fimbrillin production in P. gingivalis. (A) Fimbrillin measured by ELISA. P. gingivalis cells (109) were suspended in PBS and sonicated three times for 20 s each. Protein extract was collected by centrifugation. Fimbrillin levels in wild-type 33277 and mutants 2131, 2167, and DPG3 were determined by ELISA with antiserum against fimbrillin. (B) Western blot analysis of fimbrillin production. Protein extracts (1 μg) from P. gingivalis 33277, 2131, 2167, and DPG3 (used for ELISA), resolved by SDS-10% PAGE, transferred to nitrocellulose membranes, and probed with antifimbrillin antibodies. The arrow denotes the fimbrillin band at approximately 45 kDa.
Role of pg2131 in fimA expression.
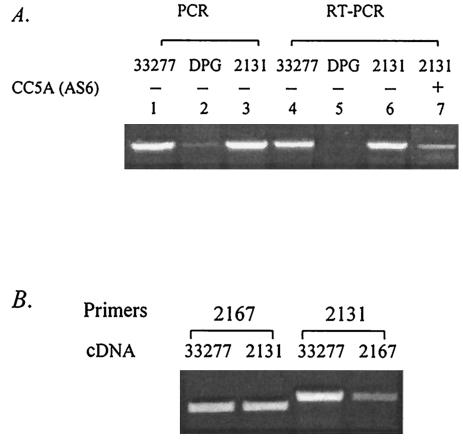
To analyze the role of pg2131 in fimA expression, mutant strain P. gingivalis 2131 was generated by insertional inactivation of pg2131, which was confirmed by PCR with primers pg2131for2 and pg2131rev2. The gene encoding PG2131 is adjacent to fimA (PG2132), and hence PCR was performed to ensure that the fimA gene was not disrupted by the mutation. As shown in Fig. 5, the fimA gene was amplified from wild-type 33277 and mutant 2131 by PCR, indicating that fimA is not disrupted in P. gingivalis 2131. As a negative control, P. gingivalis DPG3 with an insertional mutation in the fimA gene was also used, and as expected, a fimA PCR product could not be detected in this strain.
FIG. 5.
Expression of the differentially displayed genes in P. gingivalis mutants. (A) DNAs and total RNAs were extracted from P. gingivalis strains 33277, DPG3, and 2131. Bacteria were grown either under standard conditions (lanes 1, 2, 3, 4, 5, 6) or in the presence of CC5A fraction AS6 (lane 7). PCR and RT-PCR were performed with FE1 and FE2 to yield a 1,024-bp fimA fragment. (B) Total RNAs were extracted from wild-type 33277 and mutants 2131 and 2167. mRNAs were transcribed with RNase H reverse transcriptase. The 2167 primers pg2167for and pg2167rev2 were used to create an 800-bp pg2167 fragment. The 2131 primers pg2131for and pg2131rev2 were used to create an 1,100-bp pg2131 fragment.
Reverse transcription-PCR was performed to measure the mRNA levels of fimA in mutant strain 2131 and wild-type 33277. The results from RT-PCR showed that mutation of pg2131 did not affect fimA transcription under standard growth conditions (Fig. 5A). Similar to mutant 2167, P. gingivalis 2131 shut down transcription of fimA in response to elevated temperature (not shown), but unlike P. gingivalis 2167, mutation in pg2131 had no impact on the ability of P. gingivalis to communicate with S. cristatus CC5A (Fig. 5A), since the mutant, like the wild-type strain, reduced production of fimA mRNA in the presence of CC5A (AS6). These results imply that the pg2131 gene may not be directly involved in P. gingivalis signal transduction. Interestingly, the results from Western analysis and ELISA showed that the level of fimbrillin in surface extracts was significantly lower in P. gingivalis 2131 (Fig. 4).
Since it is possible that the low level of fimbrillin in the surface extract of mutant 2131 results from fimbrillin accumulation inside the cells, and pg2131 is responsible for fimbrillin translocation, we performed Western analyses and ELISA after lysing the bacterial cells with lysozyme. Negligible amounts of fimbrillin were detected in P. gingivalis strains DPG3 and 2131 (data not shown). These data suggest that the product of pg2131 is required for fimA expression posttranscriptionally.
To better understand the connectivity of the differentially displayed genes in the regulation of fimA, pg2131 expression was examined in P. gingivalis mutant 2167 with RT-PCR (Fig. 5B). Interestingly mRNA levels of pg2131 were significantly lower in mutant 2167 compared wild-type 33277. In contrast, the mRNA levels of pg2167 were similar in wild-type 33277 and mutant 2131. These results indicate that pg2131 expression may be controlled by the product of pg2167. Overexpression of pg2131 in the presence of CC5A proteins may be an effect of upregulation of pg2167 or downregulation of fimA. Therefore, pg2131 may not be a component of the transcriptional regulation pathways.
Role of PG2167 and PG2131 in binding CC5A signals.
Important components of signal transduction are the cell surface receptors that sense and transmit the signals. As PG2167 and PG2131 possess a theoretical signal sequence and can be predicted to be on the cell surface, we examined the possibility that these molecules could serve as receptors for binding to CC5A molecules. The AS6 fraction of S. cristatus CC5A was incubated with P. gingivalis 33277, 2131, 2167, and DPG3 for 1 h at room temperature in the presence of the protease inhibitors pepstatin (30 μM), phenylmethylsulfonyl fluoride (2 mM), benzamidine (2 mM,) leupeptin (215 μM), aprotinin (10 μM), and Nα-p-tosyl-l-lysine chloromethyl ketone (2 mM). Unbound molecules of CC5A were then collected by centrifugation and analyzed by SDS-PAGE and for inhibitory activity.
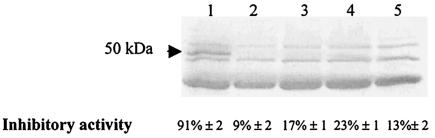
One band of ≈50 kDa, which is close to what we had estimated by chromatographic separation (21), was missing from the supernatant after interaction with P. gingivalis 33277 and with mutants 2167, 2131, and DPG3 (Fig. 6). As a control, E. coli DH5α was also tested in the binding assay. The protein profile of AS6 remained the same after AS6 was exposed to E. coli (data not shown), suggesting specific recognition between P. gingivalis and S. cristatus. In addition, the decrease in the 50-kDa protein level was accompanied by a decrease in the inhibitory activity of the supernatants after reaction with all the P. gingivalis strains. The inhibition of fimA expression was 5- to 10-fold lower (P < 0.05) than that with the AS6 fraction (Fig. 6). These data suggest that the inhibitory molecule may interact directly with the surface molecules of P. gingivalis and that fimbrillin, PG2131, and PG2167 do not act as the receptors for CC5A signaling.
FIG. 6.
Interaction of CC5A surface protein and P. gingivalis cells. The supernatants obtained from a 1-h reaction of CC5A AS6 with P. gingivalis strains were subjected to SDS-PAGE and stained with Coomassie blue. Lane 1, AS6 of CC5A; lanes 2, 3, 4, and 5, AS6 supernatants after incubation with P. gingivalis 33277, 2131, 2167, and DPG3, respectively. The arrow indicates the band missing from the supernatants exposed to P. gingivalis cells. The inhibitory activity of the supernatants on fimA expression was determined by β-galactosidase assay.
DISCUSSION
Dental plaque is a complex multispecies biofilm. While cell-to-cell communication is widely considered to be important in the formation and development of the oral bacterial community, there are few documented examples of interbacterial signaling. Our previous observations demonstrated that fimA expression is significantly inhibited in P. gingivalis in the presence of S. cristatus but is not modulated by other oral streptococci (21). Furthermore, P. gingivalis biofilm formation does not occur on a substratum of S. cristatus. Differential display PCR was used to identify the genes regulated in P. gingivalis in the presence of S. cristatus surface proteins. A number of genes were selectively regulated, indicating the existence of an extensive communication network between P. gingivalis and S. cristatus.
In this study, P. gingivalis genes that were differentially expressed in response to S. cristatus were further investigated. Mutant strain 2167, with an insertional mutation in pg2167, did not reduce fimA expression when exposed to S. cristatus proteins. Indeed, this P. gingivalis mutant produced more fimbrillin than the wild-type strain. The data suggest that the gene product of pg2167 is upregulated in response to the S. cristatus signal and then negatively regulates the expression of FimA at the transcriptional level. Independent studies have reported that P. gingivalis fimA expression is modulated by growth temperature (1, 23). The transcriptional activity of fimA is higher at a lower growth temperature (34°C) than at an elevated growth temperature (39°C). fimA expression in the pg2167 mutant strain responded similarly to growth temperature. Therefore, regulation through PG2167 would appear to be independent of the temperature-dependent control of fimA expression.
Mutational analysis of a second gene, pg2131, showed that pg2131 was required for optimal fimbrillin production. In fact, disruption of the pg2131 gene showed a result similar to that with the fimA mutation with regard to fimbrillin production. However, a mutation in pg2131 did not lead to a decrease of fimA transcription. We speculate that pg2131 is involved in posttranscriptional regulation of fimA expression. Posttranscriptional control has been observed in bacterial flagella and type III secretion systems (2, 3). It is believed that the additional layers of posttranscriptional control could increase the efficiency of gene expression. Although the posttranscriptional regulation mechanisms of the fimA gene in P. gingivalis are unknown, our data demonstrate that pg2131 transcription is under the (direct or indirect) control of pg2167, as a mutation in pg2167 causes reduced transcription of pg2131. It is possible that the observation of elevated pg2131 transcription in differential display PCR is due to an increase of pg2167 expression or a reduction of fimA transcription. In either event, pg2131 does not appear to be responsible for regulating the expression of fimA in response to S. cristatus.
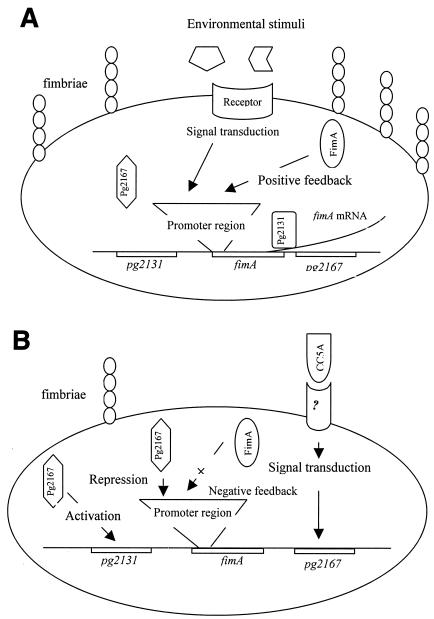
The presently available data suggest a model of fimbriation in P. gingivalis whereby communication between P. gingivalis and S. cristatus CC5A begins with recognition of the CC5A signal or signals by a P. gingivalis receptor (Fig. 7). Fimbrillin itself is not the receptor, as a fimA-null mutant binds CC5A proteins to the same extent as wild-type P. gingivalis. In response to the S. cristatus signal, P. gingivalis suppresses expression of the fimA gene. A component of this repression system, PG2167, is overexpressed under this condition. PG2131 is involved in posttranscriptional regulation of fimbrillin, although the role and regulation of PG2131 are as yet unclear. Overexpression of pg2131 may be a direct response to S. cristatus signal or a result of depletion of fimA expression. The latter could be a strategy used by P. gingivalis to compensate for decreasing production of fimbrillin. Our previous study suggests that FimA can positively regulate its own expression (20). Therefore, suppression of fimA transcription by PG2167 may further interrupt fimA expression due to a reduction of fimbrillin production. Recently, Hayashi et al. suggested that a two-component signal regulation system commonly found in the expression of bacterial virulence factors might also be involved in fimbrillin expression in P. gingivalis, based on the results of mutagenesis (7). The components responsible for fimbrillin regulation were a histidine protein kinase and a response regulator.
FIG. 7.
Model for fimA expression. (A) fimA expression is upregulated, which may be mediated through a two-component system. Expression of pg2167 is repressed without CC5A signal. (B) P. gingivalis interacts with an S. cristatus surface protein via an unknown receptor. Subsequent signal transduction induces pg2167 expression, and the latter may prevent fimA transcription and promote pg2131 transcription. The production of fimbrillin is decreased as a result of transcriptional downregulation of fimA, even though pg2131 is upregulated by pg2167. Reduced production of fimbrillin further reduces fimA transcription through negative feedback.
Collectively, these observations indicate that more than one mechanism of regulation controls the level of fimA expression. We expect that a more complete model of regulation of fimA will emerge from detailed genomic and proteomic studies.
Acknowledgments
The support of the NIDCR (DE00401, DE12505 and DE11111) is gratefully acknowledged.
Editor: V. J. DiRita
REFERENCES
- 1.Amano, A., A. Sharma, H. T. Sojar, H. K. Kuramitsu, and R. J. Genco. 1994. Effects of temperature stress on expression of fimbriae and superoxide dismutase by Porphyromonas gingivalis. Infect. Immun. 62:4682-4685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bonifield, H. R., S. Yamaguchi, and K. T. Hughes. 2000. The flagellar hook protein, FlgE, of Salmonella enterica serovar Typhimurium is posttranscriptionally regulated in response to the stage of flagellar assembly. J. Bacteriol. 182:4044-4050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cambronne, E. D., and O. Schneewind. 2002. Yersinia enterocolitica type III secretion: yscM1 and yscM2 regulate yop gene expression by a posttranscriptional mechanism that targets the 5′ untranslated region of yop mRNA. J. Bacteriol. 184:5880-5893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Childs, W. C., and R. J. Gibbons. 1990. Selective modulation of bacterial attachment to oral epithelial cells by enzyme activities associated with poor oral hygiene. J. Periodontol. Res. 25:172-178. [DOI] [PubMed] [Google Scholar]
- 5.Hamada, S., T. Ogawa, H. Shimauchi, Y. Kusumoto. 1992. Induction of mucosal and serum immune responses to specific antigen of periodontal bacteria. Adv. Exp. Med. Biol. 327:71-81. [DOI] [PubMed] [Google Scholar]
- 6.Hase, C. C., and J. J. Mekalanos. 1998. TcpP protein is a positive regulator of virulence gene expression in Vibrio cholerae. Proc. Natl. Acad. Sci. 95:730-734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hayashi, J., K. Nishikawa, R. Hirano, T. Noguchi, and F. Yoshimura. 2000. Identification of a two-component signal transduction system involved in fimbriation of Porphyromonas gingivalis. Microbiol. Immunol. 44:279-282. [DOI] [PubMed] [Google Scholar]
- 8.Kolenbrander, P. E., R. N. Andersen, D. S. Blehert, P. G. Egland, J. S. Foster, and R. J. Palmer Jr. 2002. Communication among oral bacteria. Microbiol. Mol. Biol. Rev. 66:486-505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lamont, R. J., A. El-Sabaeny, Y. Park, G. S. Cook, J. W. Costerton, and D. R. Demuth. 2002. Role of the Streptococcus gordonii SspB protein in the development of Porphyromonas gingivalis biofilms on streptococcal substrates. Microbiology 148:1627-1636. [DOI] [PubMed] [Google Scholar]
- 10.Lamont, R. J., C. A. Bevan, S. Gil, R. E. Persson, and B. Rosan. 1993. Involvement of Porphyromonas gingivalis fimbriae in adherence to Streptococcus gordonii. Oral Microbiol. Immunol. 8:272-276. [DOI] [PubMed] [Google Scholar]
- 11.Lamont, R. J., S. G. Hersey, and B. Rosan. 1992. Characterization of the adherence of Porphyromonas gingivalis to oral streptococci. Oral Microbiol. Immunol. 7:193-197. [DOI] [PubMed] [Google Scholar]
- 12.Lee, J., H. Sojar, G. Bedi, and R. J. Genco. 1992. Synthetic peptides analogous to the fimbrillin sequence inhibit adherence of Porphyromonas gingivalis. Infect. Immun. 60:1662-1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee, S. W., J. D. Hillman, and A. Progulske-Fox. 1996. The hemagglutinin genes hagB and hagC of Porphyromonas gingivalis are transcribed in vivo as shown by use of a new expression vector. Infect. Immun. 64:4802-4810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Malek, R., J. G. Fisher, A. Caleca, M. Stinson, C. J. van Oss, J. Y. Lee, M. I. Cho, R. J. Genco, R. T. Evans, and D. W. Dyer. 1994. Inactivation of the Porphyromonas gingivalis fimA gene blocks periodontal damage in gnotobiotic rats. J. Bacteriol. 176:1052-1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miller, J. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 16.Ogawa, T., H. Uchida, and S. Hamada. 1994. Porphyromonas gingivalis fimbriae and their synthetic peptides induce proinflammatory cytokines in human peripheral blood monocyte cultures. FEMS Microbiol. Lett. 116:237-242. [DOI] [PubMed] [Google Scholar]
- 17.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 18.Socransky, S. S., C. Smith, and A. D. Haffajee. 2002. Subgingival microbial profiles in refractory periodontal disease. J. Clin. Periodontol. 29:260-268. [DOI] [PubMed] [Google Scholar]
- 19.Weinberg, A., C. M. Belton, Y. Park, and R. J. Lamont. 1997. Role of fimbriae in Porphyromonas gingivalis invasion of gingival epithelial cells. Infect. Immun. 65:313-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xie, H., W. O. Chung, Y. Park, and R. J. Lamont. 2000. Regulation of the Porphyromonas gingivalis fimA (fimbrillin) gene. Infect. Immun. 68:6574-6579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xie, H., G. Cook, J. W. Costerton, G. Bruce, T. M. Rose, and R. J. Lamont. 2000. Intergeneric communication in dental plaque biofilms. J. Bacteriol. 182:7067-7069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xie, H., and R. J. Lamont. 1999. Promoter architecture of the Porphyromonas gingivalis fimbrillin gene. Infect. Immun. 67:3227-3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xie. H., S. Cai, and R. J. Lamont. 1997. Environmental regulation of fimbrial gene expression in Porphyromonas gingivalis. Infect. Immun. 65:2265-2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ximenez-Fyvie, L. A., A. D. Haffajee, and S. S. Socransky. 2000. Microbial composition of supra- and subgingival plaque in subjects with adult periodontitis. J. Clin. Periodontol. 27:722-732. [DOI] [PubMed] [Google Scholar]