Abstract
Sec1p/Munc18 (SM) proteins are essential for SNARE-mediated membrane trafficking. The formulation of unifying hypotheses for the function of the SM protein family has been hampered by the observation that two of its members bind their cognate syntaxins (Sxs) in strikingly different ways. The SM protein Vps45p binds its Sx Tlg2p in a manner analogous to that captured by the Sly1p–Sed5p crystal structure, whereby the NH2-terminal peptide of the Sx inserts into a hydrophobic pocket on the outer face of domain I of the SM protein. In this study, we report that although this mode of interaction is critical for the binding of Vps45p to Tlg2p, the SM protein also binds Tlg2p-containing SNARE complexes via a second mode that involves neither the NH2 terminus of Tlg2p nor the region of Vps45p that facilitates this interaction. Our findings point to the possibility that SM proteins interact with their cognate SNARE proteins through distinct mechanisms at different stages in the SNARE assembly/disassembly cycle.
Introduction
Intracellular compartmentalization into discrete membrane-bound organelles is a defining feature of eukaryotic cells. Communication between organelles is achieved through membrane fusion, which is a highly conserved and regulated process. Central to the membrane fusion machinery are the SNARE family of proteins, which are characterized by a helical motif preceding their COOH-terminal transmembrane domain (Jahn and Sudhof, 1999). A SNARE motif from one membrane assembles into a four-helix bundle with three SNARE motifs on another membrane, including one contributed by a syntaxin (Sx) family member (Jahn and Sudhof, 1999). The neuronal Sx, Sx1A, adopts two distinct conformations (Dulubova et al., 1999): in the closed conformation, the autonomously folded NH2-terminal Habc domain folds back onto the SNARE motif, rendering it unavailable for core complex formation; in the open conformation, the Habc domain moves away from the SNARE motif, leaving it free to participate in the core complex.
Although SNARE proteins are sufficient to drive bilayer fusion in vitro (Weber et al., 1998), other factors control membrane fusion in vivo. The Sec1p/Munc18 (SM) family of proteins are essential for SNARE-mediated traffic, but their mode of action is far from understood (Gallwitz and Jahn, 2003; Toonen and Verhage, 2003). Unlike SNARE proteins, which are common only in their shared SNARE motif, SM proteins are conserved across their entire length (600–700 residues; Jahn and Sudhof, 1999).
Munc18a was originally identified as a Sx1A-binding protein (Hata et al., 1993) whose binding to Sx1A prevents SNARE complex formation in vitro (Pevsner et al., 1994). The crystal structure of the Munc18a–Sx1A complex also supports a model in which Munc18a acts as a negative regulator of SNARE complex assembly, revealing that the SM protein is an arch-shaped molecule with three distinct domains forming a central cavity that cradles the Sx in its closed conformation (Misura et al., 2000). Consistent with this, no binding of Munc18a to Sx1A–SNARE complexes containing Sx1A in the open conformation is observed (Yang et al., 2000). In marked contrast, other SM proteins bind to assembled SNARE complexes (Gallwitz and Jahn, 2003); indeed, the yeast plasma membrane SM protein Sec1p was originally thought to bind only to the assembled SNARE complex and not to uncomplexed Sso1p (Carr et al., 1999), although this has recently been disputed (Scott et al., 2004).
Consistent with the crystal structure of the Munc18a–Sx1A complex, which shows contacts of the central cavity of the SM protein with both the SNARE and Habc domains of the Sx (Misura et al., 2000), most of the cytosolic portion of Sx1A is required to bind the SM protein (Kee et al., 1995; Misura et al., 2000). Therefore, it was surprising that short NH2-terminal sequences of Sx5 and 16 are both necessary and sufficient to capture their respective cognate SM proteins Sly1p and Vps45p (Dulubova et al., 2002, 2003). The crystal structure of Sly1p in complex with the NH2-terminal 45 residues of Sed5p, the yeast homologue of Sx5, reveals that the structure of Sly1p is very similar to that of Munc18a, with three domains arranged in an arch shape, forming a central cavity of ∼15 Å (Bracher and Weissenhorn, 2002). As described above, the central cavity of Munc18a cradles Sx1A in its closed conformation, with contacts made between the Sx and domains I and IIIa of the SM protein (Misura et al., 2000). The interaction between Sed5p and Sly1p is strikingly different from this, with the NH2-terminal peptide of the Sx buried in a hydrophobic pocket on the outer face of domain I (Bracher and Weissenhorn, 2002). This binding has been disrupted through mutations in both Sly1p and Sed5p and is not essential for membrane traffic (Peng and Gallwitz, 2004). As both proteins are essential for membrane traffic, it is therefore likely that their essential functions involve interactions other than that revealed by the crystal structure of the Sly1p–Sed5p complex (Peng and Gallwitz, 2004).
The differences between the interactions of SM proteins with their cognate Sxs have made understanding the action of this family of proteins difficult. We have previously shown that the SM protein Vps45p controls the entry of its cognate Sx Tlg2p into functional SNARE complexes (Bryant and James, 2001) and that it interacts with both uncomplexed Tlg2p and cis-SNARE complexes in vivo (Bryant and James, 2003). Tlg2p binds to Vps45p via its NH2-terminal region in a manner analogous to that captured by the Sly1p–Sed5p crystal structure (Dulubova et al., 2002). In this study, we disrupt this binding and demonstrate that membrane traffic is not perturbed. We also report a dominant-negative version of Vps45p, which has revealed that the SM protein interacts with the Tlg2p–SNARE complex through a mechanism distinct from the Sed5p–Sly1p hydrophobic pocket mode of binding. This is the first demonstration of an SM protein interacting with SNARE proteins via two distinct modes and is a finding that will enhance the formulation of a unifying hypothesis for the mechanism of action of this protein family.
Results
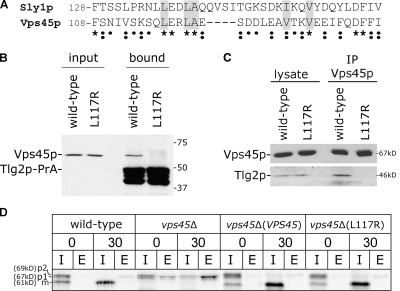
Mutation of L117 of Vps45p disrupts high-affinity binding to Tlg2p
Vps45p binds an NH2-terminal peptide of its cognate Sx Tlg2p in a manner analogous to the high-affinity interaction between Sed5p and a hydrophobic pocket on the outer face of domain I of Sly1p (Dulubova et al., 2002). This hydrophobic pocket is formed by five residues of Sly1p (L137, L140, A141, I153, and V156) that surround F10 of Sed5p (Bracher and Weissenhorn, 2002). Sequence alignment of Vps45p and Sly1p indicates that four of these residues are conserved (Fig. 1 A), with the fifth residue, I153, being replaced by a valine in Vps45p. Although this position is occupied by an isoleucine residue in Sly1p from Saccharomyces cerevisiae, a valine is found here in Sly1p sequences from other species, including humans and Drosophila melanogaster (Peng and Gallwitz, 2004).
Figure 1.
Abrogation of Vps45p binding to Tlg2p does not perturb membrane traffic. (A) Sly1p and Vps45p sequences from S. cerevisiae were aligned using Clustalw software. A portion of the alignment containing the hydrophobic pocket of Sly1p into which the NH2-terminal peptide of Sed5p inserts is shown (residues 128–165 of Sly1p and residues 108–141 of Vps45p). The five residues that form the hydrophobic pocket of Sly1p are shaded. Asterisks indicate residues that are identical between the two proteins. Double dots indicate a strong similarity between amino acid residues, and a single dot indicates similarity. (B) Tlg2p-PrA immobilized on IgG-Sepharose was incubated with cell lysates prepared from vps45Δ cells (NOzY1) harboring plasmids encoding either HA-Vps45p (pCOG070) or HA-Vps45pL117R (pCOG071). Bound material was subject to immunoblot analysis using HA antibodies (10% of the lysate used was included; input). (C) Vps45p and bound proteins were immunoprecipitated from vps45Δ cells (NozY1) harboring plasmids encoding either HA-Vps45p (pCOG070) or HA-Vps45pL117R (pCOG071). The amount of Vps45p and Tlg2p present in the immunoprecipitates was assessed by immunoblotting (a sample of the lysate was also included in this analysis; 10% on the Vps45p blot and 0.5% on the Tlg2p blot). (D) CPY sorting was followed in wild-type (RPY10), vps45Δ (NozY2), and vps45Δ cells carrying a centromeric plasmid expressing either wild-type VPS45 (pNB706) or a version of the gene encoding Vps45pL117R (pNB708). Cells were pulse labeled for 5 min with [35S]methionine/cysteine. CPY was immunoprecipitated from both intracellular (I) and extracellular (E) fractions at the beginning (0) and end (30) of a 30-min chase period.
The mutation of Sly1p L137 to an arginine residue abolishes the binding of the SM protein to Sed5p without affecting membrane transport (Peng and Gallwitz, 2004). We found that the mutation of L117 of Vps45p to arginine similarly disrupts the binding of Vps45p to Tlg2p. A protein A (PrA) fusion protein containing the cytosolic domain of Tlg2p (Tlg2p-PrA) efficiently binds tagged Vps45p (HA-Vps45p) from a cell lysate (Fig. 1 B). However, Tlg2p-PrA is unable to bind a version of HA-Vps45p harboring the L117R mutation (Fig. 1 B). Similarly, immunoprecipitation of HA-Vps45pL117R from a cell lysate fails to coprecipitate Tlg2p, unlike its wild-type counterpart (Fig. 1 C). Despite its lack of binding to Tlg2p, HA-Vps45pL117R is able to function in membrane traffic (Fig. 1 D). vps45Δ mutant cells are unable to correctly sort the vacuolar hydrolase carboxypeptidase Y (CPY), and, instead, a precursor form of the protease is secreted from the cell (Piper et al., 1994). As shown in Fig. 1 D, the introduction of a centromeric plasmid driving the expression of either wild-type HA-Vps45p or HA-Vps45pL117R restores this defect, demonstrating that the L117R mutation, which abolishes the binding of Vps45p to Tlg2p, does not impair SM protein function in membrane trafficking. These data are consistent with the finding that abolition of the high-affinity binding of Sly1p to Sed5p does not perturb membrane trafficking in yeast (Peng and Gallwitz, 2004). However, studies using mammalian cells indicate that this binding is required for transport through the early secretory pathway (Yamaguchi et al., 2002; Dulubova et al., 2003; Williams et al., 2004). Any unifying hypothesis for the role of SM proteins needs to take account of these seemingly paradoxical observations. One possibility is that the mammalian cell systems used are more sensitive to perturbation than are the yeast systems. It may be that a requirement for the hydrophobic pocket binding between Sly1p and Vps45p and their respective Sxs could be uncovered under certain physiological conditions in yeast.
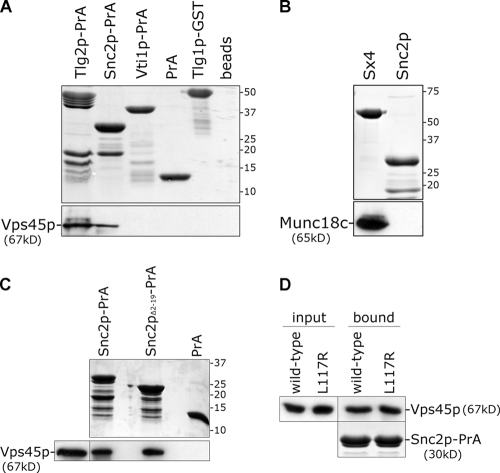
Vps45p binds to the v-SNARE Snc2p
Peng and Gallwitz (2004) reported the novel finding that an SM protein interacts directly with non-Sx SNAREs, leading them to propose a model for Sly1p function whereby the SM protein forms a bridge between t-SNAREs and their cognate v-SNAREs. We investigated whether Vps45p physically interacts with the non-Sx SNAREs present in the Tlg2p–SNARE complex. We expressed the cytosolic domains of Snc2p and Vti1p as PrA fusion proteins. Attempts to express a similar fusion protein containing the cytosolic domain of Tlg1p were unsuccessful, so we used a GST fusion protein (Tlg1p-GST; Coe et al., 1999). Fig. 2 A demonstrates that His6-Vps45p interacts directly with the cytosolic domains of both Tlg2p and the v-SNARE Snc2p (Snc2p-PrA). We found no evidence for an interaction of Vps45p with the cytosolic domains of either Tlg1p or Vti1p (Fig. 2 A). Fig. 2 B demonstrates that the binding between the cytosolic domain of Snc2p and Vps45p is specific, as Snc2p-PrA does not bind to another SM protein family member, Munc18c. Munc18c shares 50% similarity with Vps45p and binds specifically to its cognate Sx (Sx4; Fig. 2 B; Tellam et al., 1997).
Figure 2.
Vps45p binds directly to the v-SNARE Snc2p. Recombinant proteins purified from E. coli were immobilized on glutathione-agarose or IgG-Sepharose before incubation with either His6-tagged Vps45p or Munc18c purified from E. coli. After extensive washing, bound proteins were subject to SDS-PAGE followed by Coomassie blue staining (top) or immunoblotting with Vps45p or Munc18c antibodies. (A) Proteins comprising the cytosolic domains of Tlg2p, Snc2p, Vti1p, and Tlg1p fused to either PrA or GST were tested for their ability to bind His6-Vps45p. (B) The binding of Munc18c to Snc2p-PrA and GST-tagged syntaxin 4 (Sx4) was assessed. (C) The ability of Snc2p-PrA, Snc2pΔ2–19PrA, and the PrA moiety alone to bind His6-Vps45p was analyzed. (D) The ability of Snc2p-PrA to bind either His6-tagged Vps45p (wild type) or the same harboring the L117R mutation purified from bacteria was assessed and run next to 5% of the lysate used (input).
Unlike Sx t-SNAREs, the v-SNARE Snc2p has a short NH2-terminal region of 19 residues preceding the SNARE motif. A version of Snc2p-PrA lacking these residues (Snc2pΔ2–19-PrA) efficiently bound Vps45p (Fig. 2 C), demonstrating that the SNARE motif accounts for the Vps45p–Snc2p interaction. This is consistent with the binding of Sly1p to the v-SNAREs Bet1p and Sft1p, which is mediated through the SNARE motif of these v-SNAREs (Peng and Gallwitz, 2004). Snc2p does not bind Vps45p through the same mechanism as Tlg2p because HA-Vps45pL117R, which is unable to bind Tlg2p-PrA (Fig. 1 B), binds to Snc2p-PrA (Fig. 2 D).
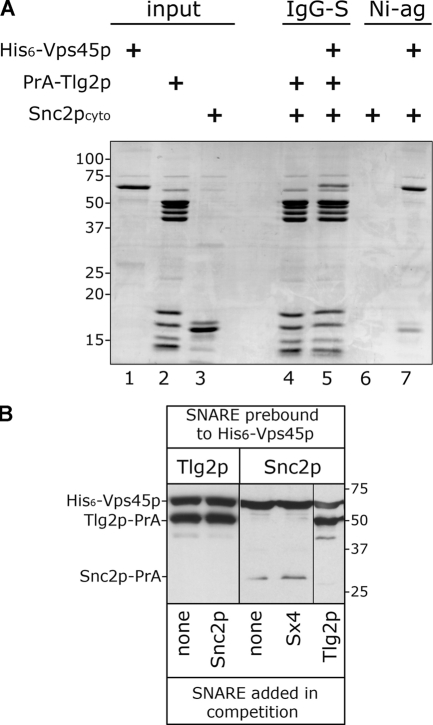
The finding that Tlg2p and Snc2p use different mechanisms to bind to Vps45p (Fig. 2 D) supports the bridging hypothesis of Peng and Gallwitz (2004). However, we were unable to observe Tlg2p and Snc2p binding to Vps45p simultaneously (Fig. 3 A). Tlg2p-PrA immobilized on IgG-Sepharose is unable to bind to the cytosolic domain of Snc2p (Snc2pcyto), which was produced using thrombin cleavage to remove the PrA tag from Snc2p-PrA (Fig. 3 A, lane 4). Attempts to bridge the two SNARE proteins by the addition of purified His6-Vps45p proved unsuccessful (Fig. 3 A, lane 5), with only His6-Vps45p and Tlg2p-PrA binding to IgG-Sepharose. Note that His6-Vps45p and Snc2pcyto are able to bind under these conditions, as Snc2pcyto can be bound to Ni2+–nitrilotriacetic acid–agarose in the presence of His6-Vps45p (Fig. 3 A, lane 7).
Figure 3.
Tlg2p and Snc2p do not bind simultaneously to Vps45p. (A) ∼1 nmol Tlg2p-PrA immobilized on IgG-Sepharose was incubated with Snc2cyto alone (lane 4) or in addition to purified His6-Vps45p (lane 5). Protein mixtures were incubated at 4°C for ∼16 h, after which time proteins were eluted from IgG-Sepharose (IgG-S) after extensive washing with PBS. Eluted proteins were resolved by 12% SDS-PAGE and stained with Coomassie blue. Snc2cyto was incubated under the same conditions either with or without purified His6-Vps45p (lanes 6 and 7). An excess of Ni2+-agarose resin (Ni-ag) was added for the last hour of these incubations, after which time proteins that bound to the resin were detected by Coomassie blue staining after SDS-PAGE. Proportional samples of the input proteins were included on the same gel (lanes 1–3). Note that both Snc2cyto and His6-Vps45p were added in excess to Tlg2p-PrA (approximately five- and twofold, respectively). (B) ∼1 nmol His6-Vps45p was immobilized on Ni2+-agarose resin and incubated with either ∼1 nmol Tlg2p-PrA or Snc2p-PrA at 4°C for ∼16 h to prebind the SNARE. After extensive washing in PBS, either 50 nmol Snc2p-PrA, Tlg2p-PrA, or GST-Sx4 or no SNARE was added in a total volume of 1 ml. After incubation at 4°C for a further 16 h, material bound to the Ni2+-agarose resin was assessed by immunoblotting.
Although our inability to observe the two SNARE proteins binding to the SM protein does not disprove the bridging hypothesis of Peng and Gallwitz (2004), as such a bridging interaction is likely to be tightly regulated and may require a transient conformation of the SM protein that is not accessed in our experimental system, it does indicate that the Tlg2p- and Snc2p-binding sites on Vps45p may overlap or influence each other. Consistent with this, we found that Tlg2p-PrA is able to displace Snc2p-PrA from His6-Vps45p (Fig. 3 B). In contrast, Snc2p-PrA is not able to displace Tlg2p-PrA from the SM protein (Fig. 3 B).
A dominant-negative version of Vps45p binds SNARE complexes through a mechanism that is distinct from the Sly1p–Sed5p hydrophobic pocket interaction
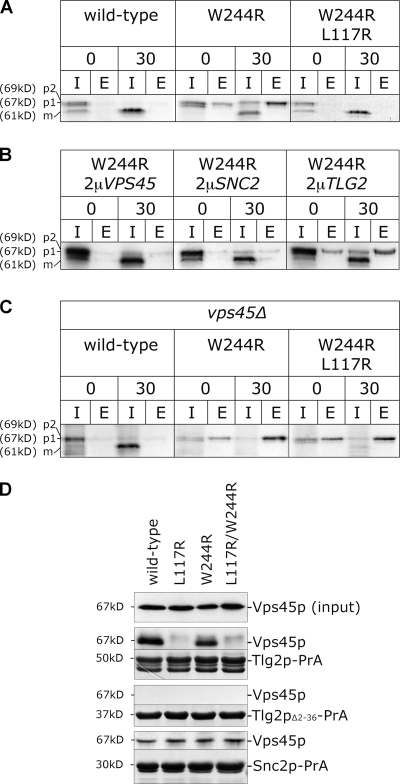
With the aim of gaining mechanistic insight into Vps45p function, we sought to isolate alleles of VPS45 that were dominant negative for CPY sorting (see Materials and methods). Fig. 4 A demonstrates that a version of Vps45p carrying a single amino acid substitution (W244R) exerts a dominant-negative effect on the sorting of CPY. Wild-type cells expressing Vps45pW244R from a multicopy (2μ based) plasmid missort ∼60% of their newly synthesized CPY. The wild-type gene expressed at the same level caused no defects in CPY sorting (Fig. 4 A).
Figure 4.
Tlg2p binding is required for the efficacy of a dominant-negative version of Vps45p. (A) CPY sorting was followed (as in Fig. 1) in wild-type cells (RPY10) harboring the following plasmids: wild-type (HA-Vps45p; pCOG070), W244R (HA-Vps45pW244; pCOG072), and L117R/W244R (HA-Vps45pL117R/W244R; pCOG073). (B) As in A, but with wild-type cells (RPY10) producing HA-Vps45pW244 from pCOG065 and also harboring the following plasmids: 2μVPS45 (pCOG070), 2μSNC2 (pCOG054), and 2μTLG2 (pHA-TLG2). (C) vps45Δ cells (NOzY2) producing HA-Vps45p and mutants thereof from the same plasmids as in A. (D) The binding of HA-Vps45p and indicated mutants from yeast cell lysates prepared from vps45Δ (NOzY1) cells transformed with pCOG070 (wild type), pCOG072 (W244R), pCOG071 (L117R), or pCOG073 (L117R/W244R) to Tlg2p-PrA, Tlg2pΔ2–36-PrA, and Snc2p-PrA was assessed as described in Fig. 1. Gels were also stained with Coomassie blue. The amount of Vps45p in 5% of each lysate used is also shown (input). I, intracellular; E, extracellular.
Introduction of a second multicopy plasmid carrying wild-type VPS45 suppressed the CPY missorting phenotype of this strain, indicating that Vsp45pW244R acts as a competitive antagonist of Vps45p function (Fig. 3 B). This is consistent with the finding that when Vsp45pW244R was introduced into a wild-type strain on a single-copy (CEN based) plasmid, no CPY secretion was observed (unpublished data). Thus, we conclude that the mutant is a dosage-dependent inhibitor of normal Vps45p function (an antimorph). The requirement for the mutant to be in excess relative to the normal protein may indicate that it competes for interaction with a target protein or proteins with which it forms a nonproductive complex. Indeed, it is clear that Vsp45pW244R is not able to perform all of the functions of its wild-type counterpart because cells in which the mutant gene is the only source of Vps45p are unable to sort CPY correctly (Fig. 4 C).
Our finding that the disruption of Vps45p binding to Tlg2p through L117 of the SM protein had no effect on CPY sorting (Fig. 1 D) and the finding that the analogous binding of Sly1p to Sed5p is not required for membrane traffic (Peng and Gallwitz, 2004) implies that the hydrophobic pocket mode of interaction between SM proteins and their Sxs is not critical. To further characterize the Vps45pW244R mutant, we combined the L117R mutation with the W244R mutation. Intriguingly, a mutant protein carrying both changes, Vps45pL117R/W244R, is not dominant negative for CPY secretion (Fig. 4 A). Fig. 4 D demonstrates that, like Vps45pL117R, Vps45pL117R/W244R does not bind to the cytosolic domain of Tlg2p, whereas Vps45pW244R does. This binding is facilitated through the NH2-terminal peptide of Tlg2p, as Vps45pW244R did not bind to a version of Tlg2p-PrA lacking the first 36 residues (Tlg2pΔ2–36-PrA; Fig. 4 D). These data reveal that the binding of Vps45p to the NH2-terminal region of Tlg2p via a mode analogous to that captured by the Sly1p–Sed5p crystal structure (Bracher and Weissenhorn, 2002) is required for Vps45pW244R to exert its dominant-negative effect. This is an interesting finding, as it provides support for a model in which this mode of binding plays a role in vivo (Yamaguchi et al., 2002; Dulubova et al., 2003; Williams et al., 2004) despite the fact that it is dispensable under normal circumstances in yeast (Fig. 1; Peng and Gallwitz, 2004).
The CPY sorting defect that Vps45pW244R bestows upon wild-type cells is suppressed by the overexpression of SNC2 (Fig. 4 B). Overproduction of other SNARE proteins, including Tlg2p, did not suppress the phenotype of Vps45pW244R (Fig. 4 B). This suggests that Vps45pW244R blocks traffic through the endosomal system by titrating out the v-SNARE Snc2p. Therefore, we looked at the interaction of Vps45pW244R with the cytosolic domain of Snc2p. Fig. 4 D demonstrates that Vps45pW244R binds to Snc2p-PrA in a manner analogous to the wild-type SM protein and Vps45pL117. Fig. 4 D also demonstrates that Vps45pL117R/W244R similarly binds to Snc2p-PrA but not to Tlg2p-PrA, confirming that Vps45p binds to Snc2p through a mode distinct from that used for its interaction with Tlg2p (Fig. 2 D).
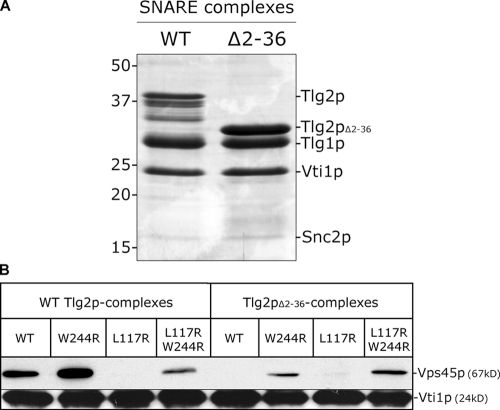
In addition to interacting with monomeric Tlg2p, Vps45p binds to cis-SNARE complexes (Bryant and James, 2003). Given that we found no differences between the binding of Vps45pW244R and its wild-type counterpart to either Tlg2p or Snc2p, we went on to investigate their binding to assembled SNARE complexes. Fig. 5 B demonstrates that Vps45p binds to SNARE complexes formed in vitro, whereas Vps45pL117R does not. Fig. 5 B also shows that Vps45pW244R binds to these complexes, as does the double mutant Vps45pL117R/W244R, indicating that the interaction between Vps45pW244R and SNARE complexes is not mediated through the NH2 terminus of Tlg2p and a hydrophobic pocket of Vps45p. To further investigate this interaction, we prepared SNARE complexes using the version of Tlg2p-PrA lacking the first 36 residues of the Sx (Tlg2pΔ2–36-PrA; Fig. 5 B). These complexes are unable to bind either wild-type Vps45p or Vps45pL117R, but, importantly, both Vps45pW244R and the double mutant Vps45pL117R/W244R were able to bind these SNARE complexes. These data suggest that the W244R mutation locks Vps45p in a form that interacts with the assembled SNARE complex and that this interaction does not require the NH2 terminus of Tlg2p.
Figure 5.
The NH2-terminal peptide of Tlg2p is not required for Vps45p to bind assembled SNARE complexes. (A) SNARE complexes containing either the entire cytosolic domain of Tlg2p (WT) or lacking residues 2–36 (Δ2–36) bound to Ni2+-agarose were resolved by SDS-PAGE on a 14% gel and were visualized by Coomassie staining. Note that the relatively weak band for Snc2p is caused by its poor Coomassie staining (McNew et al., 2000). The bands migrating between Tlg2p and Tlg1p in the wild-type lane are Tlg2p degradation products (confirmed by immunoblotting; not depicted). (B) The binding of HA-Vps45p and indicated mutants from yeast cell lysates described in Fig. 4 to the complexes represented in A was assessed as described in Materials and methods. Bound material was analyzed through immunoblotting with anti-HA antibodies and anti-Vti1p (to ensure that binding to equivalent amounts of complex was being assessed).
The interaction between the Vps45pL117R/W244R double mutant and the SNARE complex is weaker than the binding of wild-type Vps45p to the same (Fig. 5 B). This likely reflects the fact that the two versions of the SM protein are binding to the SNARE complexes via different mechanisms. Wild-type Vps45p binds to the assembled SNARE complex through the NH2-terminal peptide of Tlg2p (in a manner analogous to the binding captured by the Sly1p–Sed5p crystal structure). This is evident from the observation that wild-type Vps45p does not bind to complexes formed using Tlg2p that lacks its first 36 residues (Tlg2pΔ2–36; Fig. 5 B) and also by the observation that Vps45pL117R does not bind to SNARE complexes (containing full-length Tlg2p; Fig. 5 B). We speculate that the W244R mutation results in a protein that is locked in a conformation that binds to the assembled SNARE complex. It binds to this complex via a mode distinct from what is analogous to the Sly1p–Sed5p interaction (through the NH2 terminus of Tlg2p and the putative hydrophobic pocket in domain I of Vps45p). This is supported by the observation that the Vps45pW244R mutant binds to Tlg2pΔ2–36-containing SNARE complexes and also by the finding that the Vps45pL117R/W244R double mutant binds to SNARE complexes (Fig. 5 B). The Vps45pL117R/W244R double mutant binds less well to SNARE complexes (containing wild-type Tlg2p) than Vps45pW244R, presumably because the latter is able to bind to these complexes both through the Sly1p–Sed5p-type interaction and also via the binding mode enabled by the W244R mutation. This conclusion is supported by the reduced amount of Vps45pW244R binding to SNARE complexes that were formed using Tlg2pΔ2–36 (Fig. 5 B).
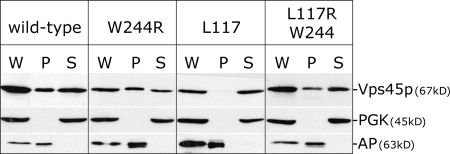
Vps45p is a peripheral membrane protein that attaches to membranes through its interaction with Tlg2p (Bryant and James, 2001). Like endogenous Vps45p (Bryant and James, 2001), HA-Vps45p is distributed between membrane and cytosol fractions obtained from wild-type cells, as is the dominant-negative mutant Vps45pW244R (Fig. 6). Consistent with its inability to bind to the cytosolic domain of Tlg2p, Vps45pL117R does not associate with membranes but instead localizes to the cytosol (Fig. 6). However, the double mutant Vps45pL117R/W244R is found attached to membranes despite its inability to bind monomeric Tlg2p (Fig. 4 D). Collectively with the data presented in Fig. 5, this indicates that Vps45pW244R binds to SNARE complexes in a way that does not require an interaction with Tlg2p akin to that captured by the Sly1p–Sed5p crystal structure. We suggest that this mode of interaction is not observed for wild-type Vps45p (or the L117R mutant; Fig. 5), as the W244R mutation is required to lock the molecule in a conformation capable of binding the SNARE complex in this way. However, the observation that the dominant-negative effect of Vps45pW244R is suppressed by an increased dose of the wild-type protein (Fig. 4 B) indicates that Vps45p does adopt this conformation during the course of its normal functional cycle. If the W244R mutation caused Vps45p to fold in such a way that it binds to the SNARE complex nonspecifically, it would be a neomorphic mutant (where the mutant protein has gained a new or significantly altered function with respect to the wild type). This is ruled out by our finding that an increased dose of the wild-type gene suppresses the dominant-negative effect of Vps45pW244R (Fig. 4 B).
Figure 6.
Vps45p associates with membranes through a manner distinct from the binding represented by the Sly1p–Sed5p crystal structure. vps45Δ (NOzY1) cells expressing HA-tagged versions of wild-type Vps45p (pNB706), W244R (pNB707), L117R (pNB708), or L117R/W244R (pNB709) from centromeric plasmids were fractionated into membrane pellet (P) and cytosol (S) fractions (W, whole cell extract). The amount of Vps45p in each of these fractions was analyzed through immunoblotting. Phosphoglycerate kinase (PGK) and AP were included in this analysis as markers for membranes and cytosol, respectively.
Discussion
We set out to investigate the modes of interaction between Vps45p and the components of the Tlg2p–SNARE complex (Tlg2p, Tlg1p, Snc2p, and Vti1p). Building upon the findings of Dulubova et al. (2002) and Peng and Gallwitz (2004), we created a mutant version of Vps45p (L117R) in which the interaction between Vps45p and the NH2-terminal region of Tlg2p is abolished. This confirmed that Vps45p and Tlg2p bind to each other in a manner analogous to the interaction revealed by the Sly1p–Sed5p crystal structure (Bracher and Weissenhorn, 2002, Dulubova et al., 2002). We also identified a mutation in Vps45p, W244R, that creates a dominant-negative phenotype. Peng and Gallwitz (2004) proposed that the hydrophobic pocket mode of binding might serve to recruit the SM protein to sites of trans-SNARE complex formation. Our finding that the L117R mutation abolishes the membrane association of Vps45p is consistent with this (Fig. 6). We have demonstrated that as well as binding to Tlg2p in this mode, Vps45p binds to SNARE complexes via an unrelated mechanism. The SM protein likely goes through an ordered sequence of these interactions as the SNARE complex assembly/disassembly cycle proceeds with ongoing rounds of membrane fusion. These interactions are likely to be carefully orchestrated within the cell, with the SM protein switching between binding modes. Vps45pW244R could subvert this orderly progression by acting in the two modes simultaneously, and this may explain the deleterious effects of this mutant. Our studies of the Vps45pW244R mutant have revealed that as well as binding directly to the NH2-terminal segment of Tlg2p, Vps45p binds to SNARE complexes via an unrelated mechanism. The SM protein likely goes through an ordered sequence of these interactions as the SNARE complex assembly/disassembly cycle proceeds with ongoing rounds of membrane fusion. These interactions are likely to be carefully orchestrated within the cell, with the SM protein switching between binding modes. Vps45pW244R could subvert this orderly progression by acting in the two modes simultaneously, and this may explain the deleterious effects of this mutant.
Sequence alignments reveal that W244 of S. cerevisiae Vps45p is conserved in all Vps45p and Sly1p homologues; in most Sec1p homologues (including Munc18a and squid Sec1 [sSec1]), there is a leucine residue at this position. Structures of Munc18a (Misura et al., 2000), sSec1 (Bracher et al., 2000), and yeast Sly1 (Bracher and Weissenhorn, 2002) show that these residues (Sly1W278, nSec1L247, and sSec1L244) project into the protein interior to form part of a conserved hydrophobic cluster that includes F336 and L411 (in Sly1p; I296/Y358 in nSec1 and I293/Y356 in sSec1). In all three cases, this pair of residues lies at the internal ends of the two helices that form the helical hairpin of domain II; thus, the conserved hydrophobic cluster interfaces between the projecting helical hairpin and the rest of domain II. Insertion of a polar residue here may cause a conformational change that opens the arch, allowing unregulated binding to the SNARE complex. An open structure of this type is seen in sSec1, and it has been suggested that movements of the helical hairpin could affect the release of Sx1A during SNARE complex formation (Bracher et al., 2000).
The conserved hydrophobic cluster also interfaces with (1) the five-helix helical repeat of domain II, which forms part of the molecular surface at the top of the arch; and (2) the surface loop insertion in Sly1p, a motif that is well positioned to regulate access to the surface of the helical hairpin and that includes the site of the sly1-20 mutation, which renders Sly1p independent of the rab protein Ypt1 (Bracher and Weissenhorn, 2002). Thus, the W244R mutation could disrupt two different surface regions of the protein and affect binding sites for regulatory factors.
In conclusion, this study demonstrates that the SM protein Vps45p not only binds to its cognate Sx Tlg2p but also to the v-SNARE Snc2p, which participates in SNARE complexes with Tlg2p. We have also demonstrated that Vps45p uses two distinct binding modes to interact with Tlg2p and Tlg2p-containing SNARE complexes. This is an important finding, as one of the major hindrances to the formulation of a unifying hypothesis of SM protein function has been the observed differences in the interactions of members of this family with their cognate Sx–SNARE complexes. Our finding uncovers the possibility that SM proteins interact with their cognate SNARE proteins through distinct mechanisms at different stages in the SNARE assembly/disassembly cycle. Determining the functional significance of these two different modes of interaction between Vps45p and its cognate SNARE proteins will require a detailed analysis of the conformational state of Vps45pW244R that is bound to the assembled SNARE complex.
Materials and methods
Reagents
Antibodies against Vps45p, Vti1p, AP, phosphoglycerate kinase, and Munc18c have been described previously (Piper et al., 1994; Tellam et al., 1997; Coe et al., 1999; Bryant and James, 2001). The monoclonal antibody for the influenza HA epitope 3F10 was obtained from Roche. Yeast strains and plasmids used in this study are listed in Tables I and II.
Table I. Yeast strains used in this study.
| Strain | Genotype | Reference |
|---|---|---|
| RPY10 | MATα ura3-52 leu2-3 112 his4-519 ade6 gal2 | Piper et al., 1994 |
| SF838-9D | MATα ura3-52 leu2-3 112 his4-519 ade6 gal2 pep4-3 | Rothman and Stevens, 1986 |
| NOzY1 | MATα ura3-52 leu2-3 112 his4-519 ade6 gal2 pep4-3 vps45Δ∷Kanr | Bryant and James, 2001 |
| NOzY2 | MATα ura3-52 leu2-3 112 his4-519 ade6 gal2 vps45Δ∷Kanr | Bryant and James, 2001 |
All strains are congenic to RPY10.
Table II. Plasmids used in this study.
| Plasmid | Description |
|---|---|
| pCOG070 | Yeast expression (2μ URA3) plasmid encoding wild-type HA-Vps45p |
| pCOG071 | As pCOG070 encoding HA-Vps45pL117R |
| pCOG072 | As pCOG070 encoding HA-Vps45pW244R |
| pCOG073 | As pCOG070 encoding HA-Vps45pL117R/W244R |
| pCOG065 | Yeast expression (2μ LEU2) plasmid encoding HA-Vps45pW244R |
| pNB706 | Yeast expression (CEN, URA3) plasmid encoding wild-type HA-Vps45p |
| pNB707 | As pNB706 encoding HA-Vps45pW244R |
| pNB708 | As pNB706 encoding HA-Vps45pL117R |
| pNB709 | As pNB706 encoding HA-Vps45pL117R/W244R |
| pNB710 | E. coli expression vector encoding His6-Vps45p |
| pCOG067 | E. coli expression vector encoding His6-Vps45pL117R |
| pCOG022 | E. coli expression vector encoding two IgG-binding domains of S. aureus PrA |
| pCOG025 | E. coli expression vector encoding Tlg2p-PrA |
| pCOG026 | E. coli expression vector encoding Tlg2pΔ2–36-PrA |
| pCOG045 | E. coli expression vector encoding Snc2p-PrA |
| pCOG046 | E. coli expression vector encoding Snc2pΔ2–19-PrA |
| pCOG048 |
E .coli expression vector encoding Vti1p-PrA (PrA fused COOH terminally to the entire cytosolic domain [residues 1–187] of Vti1p) |
| Tlg1p-GST | E. coli expression vector encoding the cytosolic domain of Tlg1p harboring a GST taga |
| pCOG038 |
E. coli expression vector for the coexpression of the cytosolic domain of Tlg1p (residues 1–205) and the cytosolic domain of Snc2p harboring a COOH-terminal His6 tag |
| pCOG032 |
E. coli expression vector for the coexpression of the cytosolic domain of Vti1p (residues 1–187) and Tlg2p-PrA |
| pCOG033 | As pCOG032, but with the cytosolic domain of Tlg2p lacking residues 2–36 |
| pCOG054 | Yeast expression (2μ URA3) plasmid driving the overproduction of Snc2p |
| pHA-TLG2 | Yeast expression (2μ URA3) plasmid driving the overproduction of Tlg2pb |
All plasmids were constructed during the course of this study unless otherwise referenced. A detailed account of plasmid construction can be found in Materials and methods.
Construction of plasmids made during the course of this study
Escherichia coli expression vectors.
pCOG022 was constructed by replacing the ORF encoding the S tag in pETDuet-1 (Novagen) with a sequence encoding two IgG-binding domains of Staphylococcus aureus PrA amplified from pZZ-HIS5 (Rayner and Munro, 1998) with oligonucleotide primers 27 (PacIPROTEINA; 5′-TTAATTAATCACGAATTCGCGTCTACTTTCGG-3′) and 47 (XhoIPROTEINA; 5′-CTCGAGCTGGTTCCGCGTGGATCCGTAGACAACAAATTCAACAAAG-3′). This plasmid drives the production of PrA with polylinker sites upstream of the sequence encoding PrA to facilitate the NH2-terminal tagging of proteins with PrA (pCOG022 also has polylinker sites downstream of the sequence encoding six His residues under T7 promoter control to facilitate the NH2-terminal His6 tagging of proteins).
pCOG025 was constructed by amplifying the sequence encoding the cytosolic domain of Tlg2p (residues 1–318) from pYCG-YOL018c (Seron et al., 1998) using oligonucleotides 44 (NdeITLG2; 5′-CATATGTTTAGAGATAGAACTAATTTA-3′) and 2 (XhoIstop TLG2; 5′-CTCGAGGTAGTGTGTTGCTTTATTCAAC-3′). The resulting 927-bp PCR product was subcloned into the NdeI–XhoI sites of pCOG022. This plasmid drives the production of Tlg2p-PrA from the T7 promoter. pCOG026 was constructed by amplifying the sequence encoding the cytosolic domain of Tlg2p lacking residues 2–36 (i.e., residues 37–318) from the template plasmid pYCG-YOL018c (Seron et al., 1998) using oligonucleotides 45 (NdeITLG2; 5′-CATATGGGTACATACCCGATGATG-3′) and 2 (XhoIstop TLG2; 5′-CTCGAGGTAGTGTGTTGCTTTATTCAAC-3′). The resulting 819-bp PCR product was subcloned into the NdeI–XhoI sites of pCOG022. This plasmid drives the production of Tlg2pΔ2–36-PrA from the T7 promoter. pCOG032 was constructed by amplifying the sequence encoding the cytosolic domain of Vti1p (residues 1–187) from pFvM29 (von Mollard et al., 1997) using oligonucleotides 7 (NcoIVTI1; 5′-CCATGGATGAGTTCCCTATTAATATCATAC-3′) and 8 (EcoRIstopstop VTI1; 5′-GAATTCTTATTAAGTCATTGTTTTTAGTGTCTTTAT-3′). The resulting 560-bp PCR product was subcloned into the NcoI–EcoRI sites of pCOG025. This plasmid drives the simultaneous production of Tlg2p-PrA and the cytosolic domain of Vti1p (as an untagged protein), both from the T7 promoter. pCOG033, which drives the simultaneous production of Tlg2pΔ2–36-PrA and the cytosolic domain of Vti1p (as an untagged protein) from the T7 promoter, was similarly constructed by subcloning the 560-bp PCR product formed using the aforementioned oligonucleotides 7 and 8 into the NcoI–EcoRI sites of pCOG026.
pCOG038, which drives the simultaneous production of the cytosolic domain of Snc2p carrying a COOH-terminal His6 tag, and the cytosolic domain of Tlg1p (as an untagged protein), both from the T7 promoter, was constructed in two stages. First, oligonucleotides 60 (NdeITLG1; 5′-CATATGAACAACAGTGAAGATCCG-3′) and 6 (XhoIstopstop TLG1; 5′-CTCGAGTTATTAACAATCGTCGTATTTTTCTTTA-3′) were used to amplify a 615-bp product encoding the cytosolic domain of Tlg1p (residues 1–205) from yeast genomic DNA. The resulting PCR product was subcloned into the NdeI–XhoI sites of pACYC-Duet (Novagen), generating pCOG037. Oligonucleotides 3 (EcoRIinframe SNC2; 5′-GAATTCGATGTCGTCATCAGTGCCATAGC-3′) and 4 (HindIIIstopstop SNC2; 5′-AAGCTTTTATTAATCTTTCCACCACATTTGCT-3′) were used to amplify a 270-bp product encoding the cytosolic domain of Snc2p (residues 1–88) from yeast genomic DNA. This was subcloned into the EcoRI–HindIII sites of pCOG037 to generate pCOG038. pCOG045 was created by amplifying the sequence encoding the cytosolic domain of Snc2p (residues 1–88) from yeast genomic DNA using oligonucleotides 127 (KpnIstart SNC2; 5′-GGTACCATGTCGTCATCAGTGCCATACG-3′) and 128 (XhoISNC2; 5′-CTCGAGATCTTTCCACCACATTTGCTTTC-3′). The resulting 270-bp fragment was subcloned into the KpnI–XhoI sites of pCOG022, yielding pCOG045. This plasmid drives the production of Snc2p-PrA from the T7 promoter. pCOG046 was constructed in a similar manner by subcloning a 210-bp PCR product obtained using oligonucleotides 128 (described above) and 129 (KpnIstart SNC2; 5′-GGTACCATGGCAAACCCAAATTCCCAAAAC-3′) to amplify the cytosolic domain of Snc2p lacking residues 2–19 (i.e., residues 20–88). This plasmid drives the production of Snc2pΔ2–19-PrA from the T7 promoter. pCOG048 was constructed by amplifying the sequence encoding the entire cytosolic domain of Vti1p (residues 1–187) from pCOG032 using oligonucleotides 133 (KpnIstart VTI1; 5′-GGTACCATGAGTTCCCTATTAATATCATAC-3′) and 134 (XhoIVTI1; 5′-CTCGAGAGTCATTGTTTTTAGTGTCTTTAT-3′) and subcloning the resulting 560-bp PCR product into the KpnI–XhoI sites of pCOG022. This plasmid drives the production of PrA fused COOH-terminally to the entire cytosolic domain of Vti1p (Vti1p-PrA).
pNB710, which drives the production of NH2-terminally His6-tagged Vps45p from the T7 promoter, was created by using oligonucleotides 9 (BamHIinframe VPS45; 5′-GGATTCGATGAACCTTTTTGATGTGGCTG-3′) and 10 (PstIadditionalstop VPS45; 5′-CTCGAGTTATTATTTTGCAGATCTAATAGAATC-3′) to amplify the entire coding region of VPS45 from yeast genomic DNA, generating pNB705. The resultant ∼1.7-kb product was subcloned first into pCR2.1 (Invitrogen) and subsequently as a BamHI–NotI fragment into pET-Duet (Novagen). pCOG067, which drives the expression of NH2-terminally His6-tagged Vps45pL117R from the T7 promoter, was created through site-directed mutagenesis of pNB710 using the oligonucleotides 202 (L117R; 5′-ATTGTCTCTAAATCTCAAAGAGAACGGCTAGCTGAATC-3′) and 203 (5′-GATTCAGCTAGCCGTTCTCTTTGAGATTTAGAGACAAT-3′).
Yeast expression vectors.
pCOG070, driving the overexpression of Vps45p harboring an HA tag at its COOH terminus, was constructed by first subcloning a 3.7-kb BamHI–SphI fragment from pTS18 (Piper et al., 1994) into YEplac195 (Gietz and Sugino, 1988) to generate YEpVPS45. The sequence encoding the HA tag was added using a sequential 4 primer PCR reaction (Horton et al., 1990) with the following oligonucleotide primers: oligonucleotide A (5′-GGTTCCTATCCATATGACGTTCCAGATTACGCTGCTCAGTGCTGAATAAGGATTATCTTATTCTAAAATTCTATTTTATATATGAGGC-3′; 45 bases encoding HA tag + stop codon and 43 bases of sequence immediately downstream of the VPS45 stop codon); oligonucleotide B (5′-GTTAGCTCACTCATTAGGC-3′; flanking primer in the vector lying outside the SphI site); oligonucleotide C (5′-CATGACAAGGACGATATATTAACC-3′; VPS45 primer approximately 200 bp upstream of the Bsu361 site); and oligonucleotide D (5′-TCAGCACTGAGCAGCGTAATCTGGAACGTCATATGGATAGGAACCTTTTGCAGATCTAATAGAATCCATATATTC-3′; HA tag + stop codon; same as in primer A [but complementary] plus last 10 codons of VPS45 ORF [without the stop codon]). The final 719-bp PCR product (obtained using oligonucleotides B and C as primers) was cloned as a Bsu361–SphI fragment into YEpVPS45. pCOG070 thus constructed encodes a Vps45p with the following COOH-terminal sequence: …YMDSIRSAKG SYPYDVPDYA AQC* (italicized sequence is the extreme COOH terminus of unmodified Vps45p, and the underlined sequence is the HA epitope).
Site-directed mutagenesis was used to construct pCOG071, pCOG073, and pCOG074 encoding versions of HA-Vps45p harboring L117R, W244R, and both L117R and W244R, respectively. pCOG065 was constructed by subcloning the ∼3.7-kb BamHI–SphI fragment containing the VPS45 ORF from pCOG072 into YEplac181 (2μ LEU2; Gietz and Sugino, 1988). pNB706, pNB707, pNB708, and pNB709 (CEN versions of pCOG070, pCOG071, pCOG072, and pCOG073) were created by subcloning the ∼3.7-kb BamHI–SphI fragment containing the VPS45 ORF from pCOG070, pCOG071, pCOG072, and pCOG073 into YCplac111 (CEN URA3; Gietz and Sugino, 1988).
pCOG054, a multicopy (URA3) plasmid driving the overproduction of an NH2-terminally tagged version of Snc2p, was constructed by using PCR to amplify a 1.1-kb fragment beginning ∼500 bp upstream of the SNC2 ATG and extending ∼300 bp downstream of the stop codon from yeast genomic DNA. The resulting PCR product was subcloned into XhoI–KpnI pRS426 (Sikorski and Hieter, 1989). Site-directed mutagenesis was then used to introduce a HpaI site immediately after the start codon. Self-annealing oligonucleotides encoding the HA epitope (YPYDVPDYA) were used to create pCOG054, a multicopy (URA3) plasmid driving the overproduction of an NH2-terminally tagged version of Snc2p.
Protein expression and purification
E. coli strain BL21-DE3 (Invitrogen) transformants were grown at 37°C to mid-log phase and induced for recombinant protein expression with 1 mM IPTG for 4 h at 37°C. Cells were harvested and lysed by sonication in PBS (25 ml/l of original culture). Clarified lysates (20,000 g for 20 min) were incubated with the appropriate resin (IgG-Sepharose, glutathione-agarose [GE Healthcare], or Ni2+–nitrilotriacetic acid–agarose [QIAGEN]). Note that resins were added such that the tagged protein was in excess of the available binding sites to saturate the resin (except in Fig. 3 as stated). After incubation (with mixing) at 4°C for 1 h, resins were washed extensively with PBS. Elution of proteins bound to IgG-Sepharose was achieved using 0.5 M HAc, pH 3.4. Thrombin cleavage of Snc2p-PrA to liberate Snc2pcyto was achieved by incubation of the protein immobilized on IgG-Sepharose with 10 U thrombin protease (Sigma-Aldrich) for 4 h at 25°C. Elution of His6-tagged proteins was achieved by incubation with PBS containing 250 mM imidazole followed by dialysis against PBS. His6-tagged Vps45p proteins were coexpressed with pTGroE (Yasukawa et al., 1995). Here, expression was induced for 16 h at 22°C. Expression and purification of GST-Sx4 and His6-tagged Munc18c was described previously (Tellam et al., 1997).
Identification of Vps45pW244R as conferring a dominant-negative phenotype for CPY sorting
Error-prone PCR was used to amplify the BamHI–SphI VPS45 fragment along with a 130-bp flanking sequence from the plasmid YEpVPS45. Mutagenized PCR product was transformed into wild-type yeast cells along with the parent plasmid of YEpVPS45 (YEplac195; Gietz and Sugino, 1988) that had been linearized by digestion with HindIII. Approximately 50,000 Ura+ transformants were screened for a Vps− phenotype using a CPY colony blot assay (Rothman and Stevens, 1986). Plasmids were rescued from six colonies that were found to strongly secrete CPY, and three of these resulted in CPY secretion upon retransformation into wild-type cells. Sequence analysis revealed that each of these plasmids carried multiple missense mutations within the VPS45 ORF. To identify the region of the ORF carrying the mutations leading to the dominant-negative phenotype, four zones were defined by the following restriction endonuclease sites: BstEII, position 825; NdeI, position 1030; and Bsu36I, position 1,570. These sites were used to create chimeras between the wild-type and mutated ORFs for each of the dominant-negative plasmids. These were tested for their ability to cause CPY secretion from wild-type yeast cells; in this way, a single mutation was identified as being responsible for causing a dominant-negative phenotype. Mutation of thymine 730 to a cytosine within the VPS45 ORF results in the replacement of W244 of Vps45p by an arginine residue (W244R).
In vitro SNARE complex formation and isolation
E. coli transformants were induced for the coexpression of His6-Snc2p, the untagged cytosolic domain of Tlg1p (both from pCOG038), the untagged cytosolic domain of Vti1p and Tlg2p-PrA (from pCOG032), or Tlg2pΔ2–36-PrA (from pCOG033) as in Protein expression and purification. IgG-Sepharose was added to cleared cell lysates as in Protein expression and purification. After binding for 1 h at 4°C and extensive washing with PBS, 25 U thrombin protease (Sigma-Aldrich) in 500 μl PBS was added to cleave the cytosolic domain of Tlg2p and bound material from the PrA tag during a 4-h incubation at 25°C. Ni2+-agarose resin was added to the supernatant containing the thrombin-cleaved complexes, and binding through His6-Snc2p was achieved at 4°C for 1 h followed by extensive washing with PBS containing 20 mM imidazole.
In vitro binding assays
The binding of Vps45p, mutants thereof, and Munc18c to fusion proteins and to assembled SNARE complexes was assessed by incubating the immobilized proteins with either a yeast cell lysate (prepared as previously described; Bryant and James, 2001) or purified His6-tagged Vps45p or Munc18c for 16 h at 4°C. After extensive washing with PBS, bound material was subjected to SDS-PAGE and Coomassie staining or immunoblot analysis.
Immunoprecipitation of Vps45p
HA-Vps45p and bound proteins were immunoprecipitated from yeast cell lysates as described previously (Bryant and James, 2003).
Pulse-chase radiolabeling and immunoprecipitation of CPY
The fate of newly synthesized CPY was followed by immunoprecipitation of the protein from intracellular and extracellular fractions of cells that had incorporated radiolabeled methionine into proteins synthesized during a 5-min time period as previously described (Piper et al., 1994).
Subcellular fractionation
Cells were fractionated using differential centrifugation after osmotic lysis of spheroplasts as previously described (Bryant and James, 2001), with the modification that the two membrane fractions were combined into one. In brief, 10-ml cultures were grown to OD600 = 1 and were incubated in 1 ml of 50 mM Tris-HCl, pH 8.0, and 1% 2-mercaptoethanol for 10 min at 30°C. Cells were converted to spheroplasts using 150 μg/ml zymolyase in 1 ml (1.2 M sorbitol, 50 mM potassium phosphate, pH 7.5, and 1 mM magnesium chloride) at 30°C for 45 min. Spheroplasts were washed with 1.2 M sorbitol before osmotic lysis in 1.2 ml of cold lysis buffer (0.2 M sorbitol, 50 mM Tris-HCl, pH 7.5, and 1 mM EDTA). Unbroken cells were removed by centrifugation for 5 min at 500 g (this and all subsequent centrifugation steps were performed at 4°C), yielding 1.2 ml of whole cell lysate. A 1.0-ml aliquot of whole cell lysate was subjected to centrifugation at 100,000 g for 30 min to yield membrane pellet and cytosolic/supernatant fractions. Membrane pellet was resuspended in 200 μl SDS sample buffer. Proteins were precipitated from the cytosolic/supernatant and 0.2 ml of whole cell lysate using 10% TCA and were resuspended in 200 and 40 μl SDS sample buffer, respectively.
Acknowledgments
We thank David James and Gwyn Gould for comments on this work and thank Alan Morgan for his patience.
This work was supported by grants from the Biotechnology and Biological Sciences Research Council (project grant to N.J. Bryant) and The Wellcome Trust (studentship to L.N. Carpp). Part of this work was carried out in Alan Morgan's laboratory (University of Liverpool). N.J. Bryant is a Prize Fellow of the Lister Institute of Preventive Medicine.
A. Boyd's present address is School of Biological Sciences, University of Liverpool, Liverpool L69 7ZB, UK.
Abbreviations used in this paper: CPY, carboxypeptidase Y; PrA, protein A; SM, Sec1p/Munc18; Sx, syntaxin.
References
- Bracher, A., and W. Weissenhorn. 2002. Structural basis for the Golgi membrane recruitment of Sly1p by Sed5p. EMBO J. 21:6114–6124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracher, A., A. Perrakis, T. Dresbach, H. Betz, and W. Weissenhorn. 2000. The X-ray crystal structure of neuronal Sec1 from squid sheds new light on the role of this protein in exocytosis. Structure. 8:685–894. [DOI] [PubMed] [Google Scholar]
- Bryant, N.J., and D.E. James. 2001. Vps45p stabilizes the syntaxin homologue Tlg2p and positively regulates SNARE complex formation. EMBO J. 20:3380–3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant, N.J., and D.E. James. 2003. The Sec1p/Munc18 (SM) protein, Vps45p, cycles on and off membranes during vesicle transport. J. Cell Biol. 161:691–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr, C.M., E. Grote, M. Munson, F.M. Hughson, and P.J. Novick. 1999. Sec1p binds to SNARE complexes and concentrates at sites of secretion. J. Cell Biol. 146:333–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coe, J.G., A.C. Lim, J. Xu, and W. Hong. 1999. A role for Tlg1p in the transport of proteins within the Golgi apparatus of Saccharomyces cerevisiae. Mol. Biol. Cell. 10:2407–2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulubova, I., S. Sugita, S. Hill, M. Hosaka, I. Fernandez, T.C. Sudhof, and J. Rizo. 1999. A conformational switch in syntaxin during exocytosis: role of munc18. EMBO J. 18:4372–4382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulubova, I., T. Yamaguchi, Y. Gao, S.W. Min, I. Huryeva, T.C. Sudhof, and J. Rizo. 2002. How Tlg2p/syntaxin 16 ‘snares’ Vps45. EMBO J. 21:3620–3631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulubova, I., T. Yamaguchi, D. Arac, H. Li, I. Huryeva, S.W. Min, J. Rizo, and T.C. Sudhof. 2003. Convergence and divergence in the mechanism of SNARE binding by Sec1/Munc18-like proteins. Proc. Natl. Acad. Sci. USA. 100:32–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallwitz, D., and R. Jahn. 2003. The riddle of the Sec1/Munc-18 proteins - new twists added to their interactions with SNAREs. Trends Biochem. Sci. 28:113–116. [DOI] [PubMed] [Google Scholar]
- Gietz, R.D., and A. Sugino. 1988. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 74:527–534. [DOI] [PubMed] [Google Scholar]
- Hata, Y., C.A. Slaughter, and T.C. Sudhof. 1993. Synaptic vesicle fusion complex contains unc-18 homologue bound to syntaxin. Nature. 366:347–351. [DOI] [PubMed] [Google Scholar]
- Horton, R.M., Z.L. Cai, S.N. Ho, and L.R. Pease. 1990. Gene splicing by overlap extension: tailor-made genes using the polymerase chain reaction. Biotechniques. 8:528–535. [PubMed] [Google Scholar]
- Jahn, R., and T.C. Sudhof. 1999. Membrane fusion and exocytosis. Annu. Rev. Biochem. 68:863–911. [DOI] [PubMed] [Google Scholar]
- Kee, Y., R.C. Lin, S.C. Hsu, and R.H. Scheller. 1995. Distinct domains of syntaxin are required for synaptic vesicle fusion complex formation and dissociation. Neuron. 14:991–998. [DOI] [PubMed] [Google Scholar]
- McNew, J.A., F. Parlati, R. Fukuda, R.J. Johnston, K. Paz, F. Paumet, T.H. Sollner, and J.E. Rothman. 2000. Compartmental specificity of cellular membrane fusion encoded in SNARE proteins. Nature. 407:153–159. [DOI] [PubMed] [Google Scholar]
- Misura, K.M., R.H. Scheller, and W.I. Weis. 2000. Three-dimensional structure of the neuronal-Sec1-syntaxin 1a complex. Nature. 404:355–362. [DOI] [PubMed] [Google Scholar]
- Peng, R., and D. Gallwitz. 2004. Multiple SNARE interactions of an SM protein: Sed5p/Sly1p binding is dispensable for transport. EMBO J. 23:3939–3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pevsner, J., S.C. Hsu, J.E. Braun, N. Calakos, A.E. Ting, M.K. Bennett, and R.H. Scheller. 1994. Specificity and regulation of a synaptic vesicle docking complex. Neuron. 13:353–361. [DOI] [PubMed] [Google Scholar]
- Piper, R.C., E.A. Whitters, and T.H. Stevens. 1994. Yeast Vps45p is a Sec1p-like protein required for the consumption of vacuole-targeted, post-Golgi transport vesicles. Eur. J. Cell Biol. 65:305–318. [PubMed] [Google Scholar]
- Rayner, J.C., and S. Munro. 1998. Identification of the MNN2 and MNN5 mannosyltransferases required for forming and extending the mannose branches of the outer chain mannans of Saccharomyces cerevisiae. J. Biol. Chem. 273:26836–26843. [DOI] [PubMed] [Google Scholar]
- Rothman, J.H., and T.H. Stevens. 1986. Protein sorting in yeast: mutants defective in vacuole biogenesis mislocalize vacuolar proteins into the late secretory pathway. Cell. 47:1041–1051. [DOI] [PubMed] [Google Scholar]
- Scott, B.L., J.S. Van Komen, H. Irshad, S. Liu, K.A. Wilson, and J.A. McNew. 2004. Sec1p directly stimulates SNARE-mediated membrane fusion in vitro. J. Cell Biol. 167:75–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seron, K., V. Tieaho, C. Prescianotto-Baschong, T. Aust, M.O. Blondel, P. Guillaud, G. Devilliers, O.W. Rossanese, B.S. Glick, H. Riezman, et al. 1998. A yeast t-SNARE involved in endocytosis. Mol. Biol. Cell. 9:2873–2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski, R.S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 122:19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tellam, J.T., S.L. Macaulay, S. McIntosh, D.R. Hewish, C.W. Ward, and D.E. James. 1997. Characterization of Munc-18c and syntaxin-4 in 3T3-L1 adipocytes. Putative role in insulin-dependent movement of GLUT-4. J. Biol. Chem. 272:6179–6186. [DOI] [PubMed] [Google Scholar]
- Toonen, R.F., and M. Verhage. 2003. Vesicle trafficking: pleasure and pain from SM genes. Trends Cell Biol. 13:177–186. [DOI] [PubMed] [Google Scholar]
- von Mollard, G.F., S.F. Nothwehr, and T.H. Stevens. 1997. The yeast v-SNARE Vti1p mediates two vesicle transport pathways through interactions with the t-SNAREs Sed5p and Pep12p. J. Cell Biol. 137:1511–1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber, T., B.V. Zemelman, J.A. McNew, B. Westermann, M. Gmachl, F. Parlati, T.H. Sollner, and J.E. Rothman. 1998. SNAREpins: minimal machinery for membrane fusion. Cell. 92:759–772. [DOI] [PubMed] [Google Scholar]
- Williams, A.L., S. Ehm, N.C. Jacobson, D. Xu, and J.C. Hay. 2004. rsly1 binding to syntaxin 5 is required for endoplasmic reticulum-to-Golgi transport but does not promote SNARE motif accessibility. Mol. Biol. Cell. 15:162–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi, T., I. Dulubova, S.W. Min, X. Chen, J. Rizo, and T.C. Sudhof. 2002. Sly1 binds to Golgi and ER syntaxins via a conserved N-terminal peptide motif. Dev. Cell. 2:295–305. [DOI] [PubMed] [Google Scholar]
- Yang, B., M. Steegmaier, L.C. Gonzalez Jr., and R.H. Scheller. 2000. nSec1 binds a closed conformation of syntaxin1A. J. Cell Biol. 148:247–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasukawa, T., C. Kanei-Ishii, T. Maekawa, J. Fujimoto, T. Yamamoto, and S. Ishii. 1995. Increase of solubility of foreign proteins in Escherichia coli by coproduction of the bacterial thioredoxin. J. Biol. Chem. 270:25328–25331. [DOI] [PubMed] [Google Scholar]