Abstract
The conformations of 2,6-bis(2-anilinoethynyl)pyridine-based urea receptors were studied by single crystal X-ray diffraction methods and revealed a rich conformational flexibility influenced by solvents. Whereas receptor L1 in DMSO prefers an “S” conformation, receptor L1 crystallizes in an “O” conformation from DMSO/CH3OH binary solvent system, and a “W” conformation in the ternary solvent mixture DMSO/toluene/1,4-dioxane. In the case of L2, the molecule adopts an “S” conformation where water molecules are sandwiched between two molecules of L2 to form a dimer. Similar to L2, L3 also forms a dimer where water molecules are sandwiched between L3 molecules, which are capped with two molecules of DMSO. Such a capping DMSO solvate is lacking in the case of L2. Taken together, these results demonstrate that the conformation of 2,6-bis(2-anilinoethynyl) pyridine-based urea receptors can be dramatically manipulated and tuned by the choice of crystallization solvents.
Keywords: Conformations, Self-Assembly, Supramolecular chemistry, Host-guest chemistry
INTRODUCTION
Structure and function go hand in hand when describing chemical/biological systems. While the covalent structure of molecules is essential for proper function, the non-covalent intra- and intermolecular interactions that make up their higher ordered three-dimensional structure are equally important. A classic example showing the importance of three-dimensional structure is RNA.1 Despite the fact that all types of RNA are composed of four nucleic acids, its diverse range of three-dimensional structures makes RNA capable of a wide range of biological functions, ranging from protein translation to catalysis.2 The ability to control, manipulate or predict this higher-order structure is essential when trying to understand or emulate complex biological systems.1
One efficient strategy to manipulate the higher order structure of organic molecules involves varying the solvent. The properties of a given solvent system (e.g., dielectric constant, polarity, hydrogen bonding capacity) can greatly influence the stability of a specific conformer or assembly, and in turn can affect not only the properties of a molecule but also the function.3 Understanding how to control this conformational and structural polymorphism through the use of solvent manipulation could prove valuable in helping to preorganize a molecule into a desired conformation and assist in functional tunability.
One area of active research investigating the role of solvents in structural complexity is supramolecular chemistry. For example, in many host-guest systems the binding site of a host is rigidified in an attempt to maximize non-covalent interactions;4 however, by completely rigidifying a molecule one loses conformational switchability and the “induced fit” that could be gained by employing a more flexible system.5 Ideally, systems that mimic the designs found in nature could use the positive aspect of both a flexible and rigid system, using solvent to help enforce structural integrity without completely eliminating the conformational switchability that results from a host-guest interaction.6 In that scenario, the solvent acts as a conformational or structural switch to create a robust binding pocket for a guest. In a more elaborate design, the solvent might even induce formation of a tertiary structure in the host to create the binding pocket.
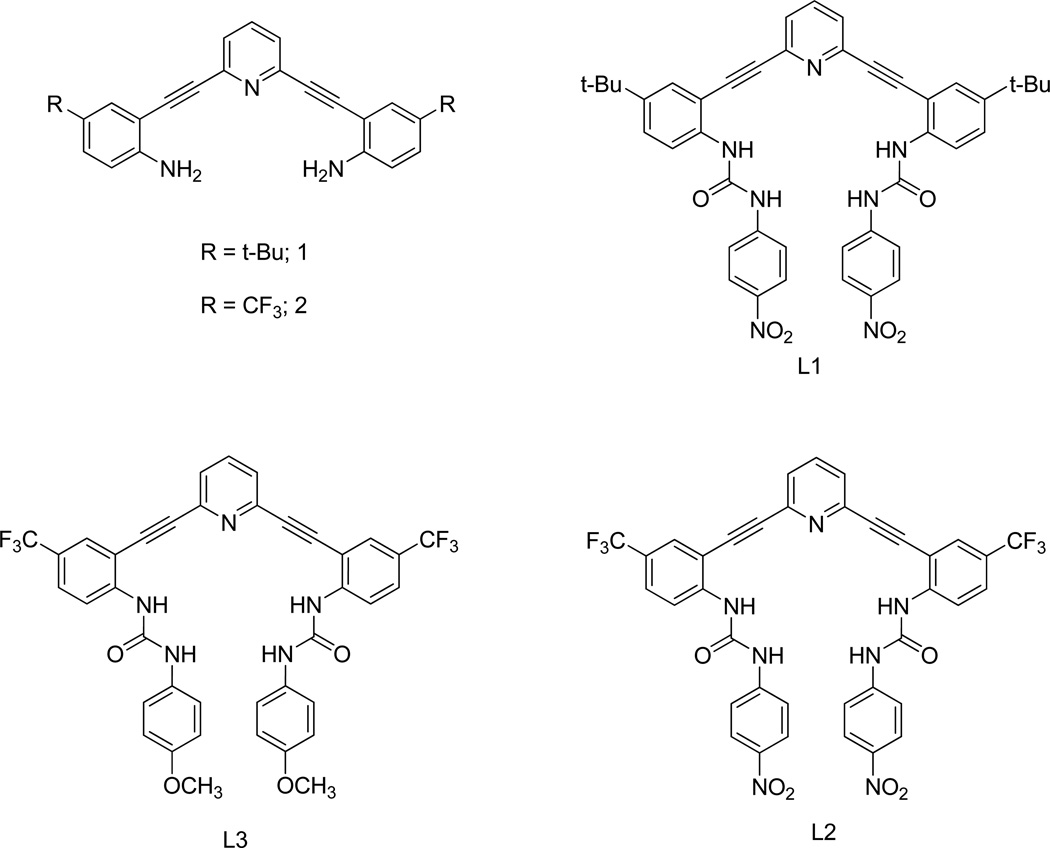
Meta-linked diethynylbenzene and diethynylpyridine skeletons have been widely employed for constructing various supramolecular complexes, because of their well-defined geometry as well as shape-persistent nature.7 In addition, the assembly of these molecules via relatively mild Sonogashira cross-coupling conditions permits access to a wide variety of artificial supramolecular complexes. We recently initiated a supramolecular project based on fluxional chromophoric scaffolds containing a central 2,6-bis(2-anilinoethynyl)pyridine moiety (e.g., 1 and 2) with two pendant phenyl sulfonamides8 or phenylureas.9 On the basis of the low rotation barrier about alkyne bonds, one can assume that these molecules exist as a mixture of rapidly interconverting conformers in solution, as well as multiple possible conformers in the solid state. We disclose herein the control of phenylurea-substituted 2,6-bis(2-anilinoethynyl)pyridine conformational switches L1, L2 and L3 by subtle differences in crystallization solvents. The effects on the preferred conformations of various external factors—particularly hydrogen bonding solvents as competitors for the urea groups in L1, L2 and L3—are investigated by X-ray crystallography, which reveals that the preferred conformation depends markedly on the choice of the solvents of crystallization. In two cases, solvent induces dimerization of the host molecules to create a binding pocket for two guests, suggesting a potential design strategy for guest-induced molecular switches. Given this receptor class is also inherently fluorescent, this scaffold may hold future promise in photochemically-driven switching as well.9a
RESULTS AND DISCUSSION
Crystallizations of L1, L2 and L3
Bisureas L1, L2 and L3 were prepared in moderate yields by the reaction of dianilines 1 and 2 with the appropriate phenyl isocyanate (Scheme 1).8–10 Single crystals of complexes L1•2DMSO, L1•2DMSO•CH3OH, L1•3DMF, L2•H2O•0.5DMSO, L3•H2O•DMSO (I), and L3•H2O•DMSO (II) suitable for X-ray analysis were obtained by crystallization from DMSO, DMSO/CH3OH, DMF/toluene/1,4-dioxane, DMSO, DMSO and DMSO/HCl, respectively (see ESI for structural parameters, Table S1).
Crystal Structure of L1•2DMSO
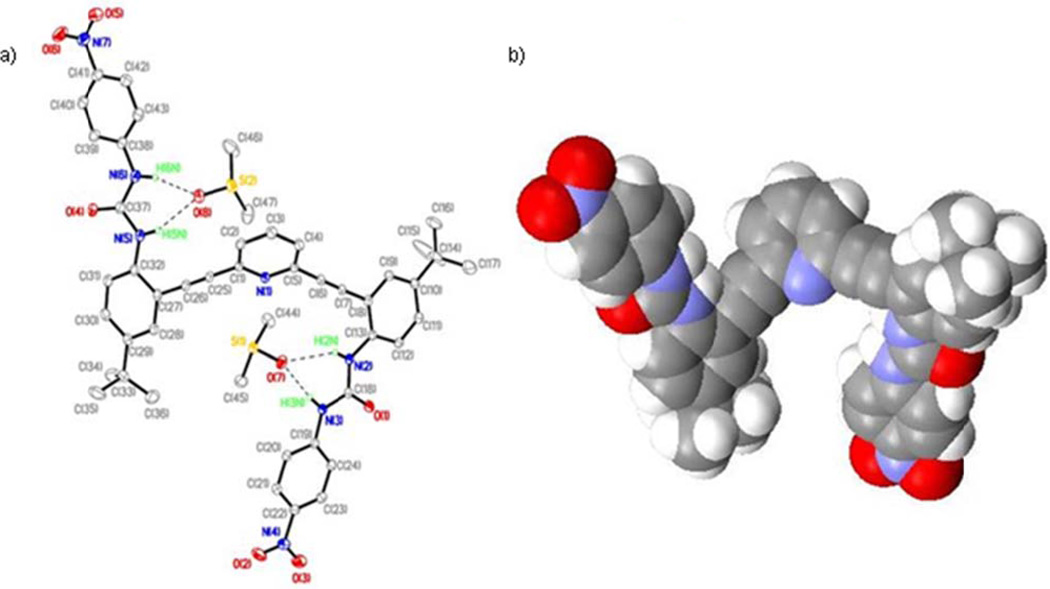
The neutral bis-urea ligand L1 crystallizes by slow evaporation from DMSO in monoclinic space group P21/c (Table S1) with two molecules of DMSO (L1•2DMSO). Figure 1a shows an ORTEP diagram of the asymmetric unit with atom numbering schemes. The crystal structure of L1•2DMSO shows strong intermolecular hydrogen-bonding interactions between urea N-H groups and solvent DMSO molecules (Table S2). Each of the two DMSO solvent molecules forms hydrogen bonds with each urea arm. Further, one of the DMSO molecules also makes a short contact with the central pyridine nitrogen (corresponding S⋯N contact is 3.133 Å). Surprisingly, there are no inter- or intramolecular NH⋯(O)C hydrogen-bonds present, which is unusual for such urea and amide based receptors.11,12 In the crystal structure of L1•2DMSO, the oxygen atom of the DMSO solvent acts as an acceptor and is involved in N-H⋯(O)S interactions with the donor urea hydrogen atoms H6N and H5N (Figure 1a). Similar hydrogen-bonding interactions are observed between another DMSO oxygen and H2N and H3N hydrogen atoms. Details of these hydrogen bonding interactions are shown in Table S2).
Figure 1.
(a) ORTEP diagram of L1•2DMSO with 30% thermal ellipsoids showing H-bonds between L1 and solvent molecules (dashed lines). Hydrogen atoms not involved in H-bonds are omitted for clarity. The O(7)⋯N(2), O(7)⋯N(3) and O(8)⋯N(5), O(8)⋯N(6) distances are 3.122(2), 2.757(2) and 2.935(3),2.790(3)Å, respectively;(b) space-filling model of the backward “S” conformation of L1 in L1•2DMSO.
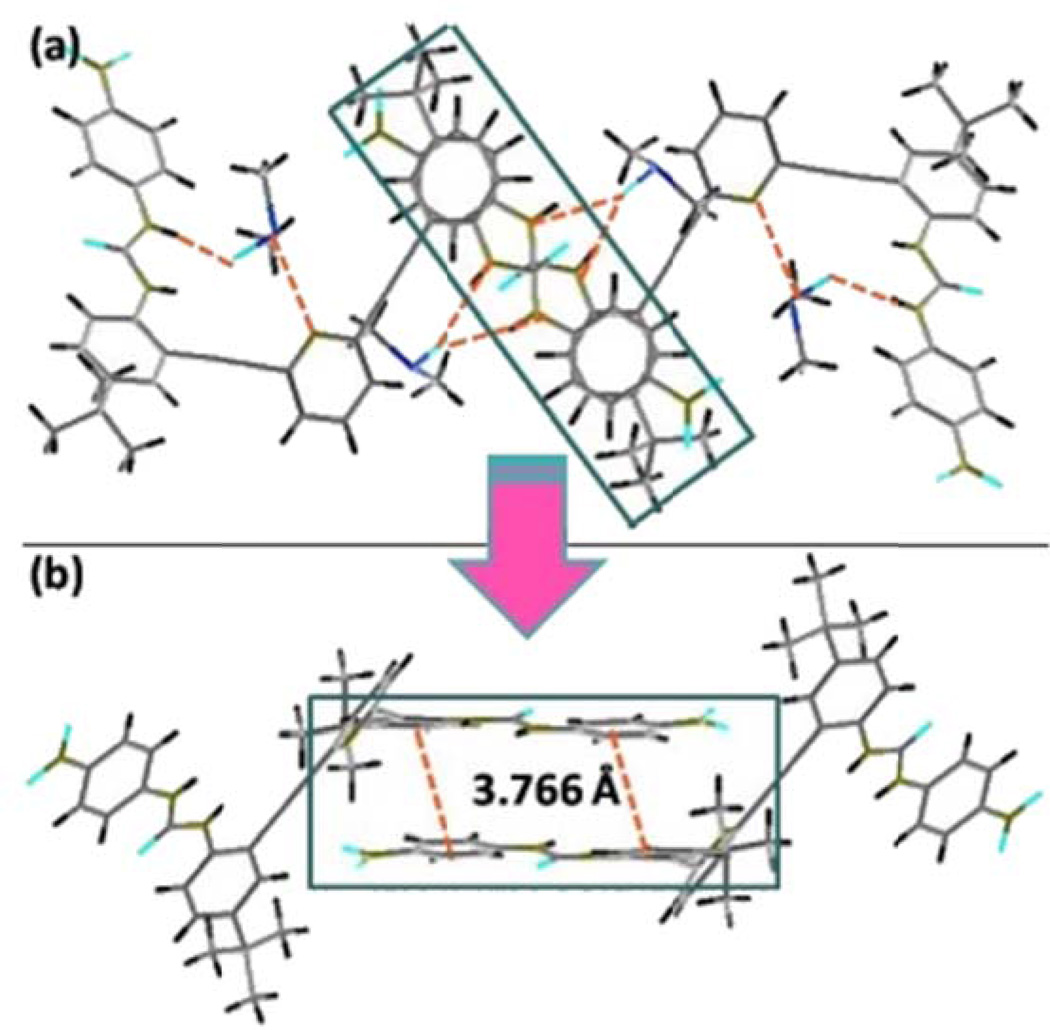
Interestingly, the solid state structure reveals that L1•2DMSO adopts a backward “S” conformation to bind the two molecules of DMSO. This is quite different from the previously reported crystal structure of the parent phenyl urea analog of L1 where the molecules crystallize as a tetramer with two different conformations: a backward “S” and a “W” which stack in an “SWWS” fashion.7 Unlike this “SWWS” motif, only the backward “S” conformation is observed in L1•2DMSO (Figure 1b). In addition to strong hydrogen-bonding interactions between solvent molecules (DMSO) and L1, the 3-D host network is propagated through several nonbonding interactions. For example, Figure 2 shows that both types of phenyl moieties participate in weak intermolecular π⋯π contacts. The centroid⋯centroid distance between the phenyl rings is 3.766 Å.
Figure 2.
Wireframe representation of the top (a) and side (b) views of the π⋯π interactions present in the extended structure of complex L12DMSO.
Crystal Structure of L1•2DMSO•CH3OH
The neutral ligand L1 crystallizes in triclinic space group P-1 (Table S1) with a molecule of MeOH and two molecules of DMSO when crystallized out of DMSO/MeOH mixtures (L1•2DMSO•CH3OH). Figure 3a shows the ORTEP diagram of the ligand and solvent guests with atom numbering scheme. The crystal structure of L1•2DMSO•CH3OH reveals hydrogen bonding interactions with bond methanol and DMSO solvent guests through NH⋯O interactions. The oxygen atoms of both solvent molecules form hydrogen bonds with both ureido NH groups (Figure 3a). One of the DMSO molecules sits in the binding pocket of L1 and forms H-bonds to the amide NH group. Another disordered DMSO is present in the lattice and is not involved in any hydrogen bonding. Details of these hydrogen bonding interactions are shown in Table S3.
Figure 3.
(a) ORTEP diagram of L1•2DMSO•CH3OH with 30% thermal ellipsoids showing H-bonds between L1 and solvent guest molecules; a disordered DMSO molecule and hydrogen atoms not involved in H-bonds are omitted for clarity. The O(1S)⋯N(1), O(1S)⋯N(2), O(1S)⋯N(3) and O(2S)⋯N(5), O(2S)⋯N(6) distances are 2.731(5), 3.202(5), 2.786(5) and 3.229(6), 2.767(6) Å, respectively;(b) Space-filling model of L1 found in a L1•2DMSO•CH3OH showing the “O” conformation in the solid state.
Unlike L1•2DMSO, L1•2DMSO•CH3OH adopts an “O” conformation in the crystalline state where both the nitrophenyl urea units are folded over each other to complete the “O” allowing both solvent guests to be enclosed in the circular cavity. This affords a “foldamer-type” conformation13 with hydrogen bonding of the urea protons to both CH3OH/DMSO solvent guest molecules featuring concomitant hydrogen bonding from CH3OH to the central pyridine nitrogen.
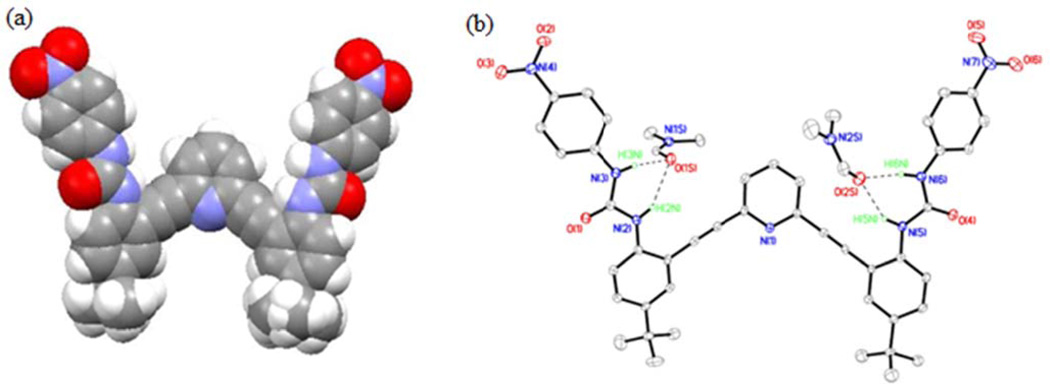
Crystal Structure of L1•3DMF
The neutral bisurea L1 also crystallizes out of a mixture of DMF/toluene/1,4-dioxane in triclinic space group P-1 (Table S1) with three molecules of DMF (L1•3DMF). Figure 4a shows the ORTEP diagram of the ligand moiety with atom numbering scheme. The crystal structure of L1•3DMF reveals strong intermolecular hydrogen-bonding interactions between the urea moieties and two of the DMF solvent molecules. Each of the two DMF “guests” form N-H⋯(O)CDMF hydrogen bonds with adjacent urea arms of L1 (H2N, H3N, H5N, H6N, Figure 4a). One additional DMF molecule does not form hydrogen bonds with any of the urea moieties (omitted for clarity). Details of the hydrogen bonding interactions are summarized in Table S4.
Figure 4.
(a) Space-filling view of L1 in the “W” conformation found in L1•3DMF; (b) ORTEP diagram of L1•3DMF with 30% thermal ellipsoids showing hydrogen bonding interactions between L1 and the DMF solvent molecules. One molecule of DMF and other hydrogen atoms which are not participating in hydrogen bonding with L1 are omitted for clarity. The O(1S)⋯N(2), O(1S)⋯N(3) and O(2S)⋯N(5), O(2S)⋯N(6) distances are 3.137(4), 2.851(4) and 3.052(4)Å, respectively.
A unique feature of L1•3DMF is that it adopts a “W” conformation in the crystalline state to create two identical urea binding sites for each of the two DMF solvent guests (Figure 4b). The conformation and hydrogen bonding interactions of L1 present in L1•3DMF differ dramatically from the backward “S” and “O” conformations observed in L1•2DMSO and L1•2DMSO•CH3OH, respectively (Figures 1 and 3) The L1 molecules are oriented in the crystal structure in a zigzag fashion through a variety of interactions, including NO2⋯CH3(tert-butyl), CH⋯C≡C, CH-π, CHsolvent⋯Ocarbonyl and CHsolvent- π interactions (Figure S1).
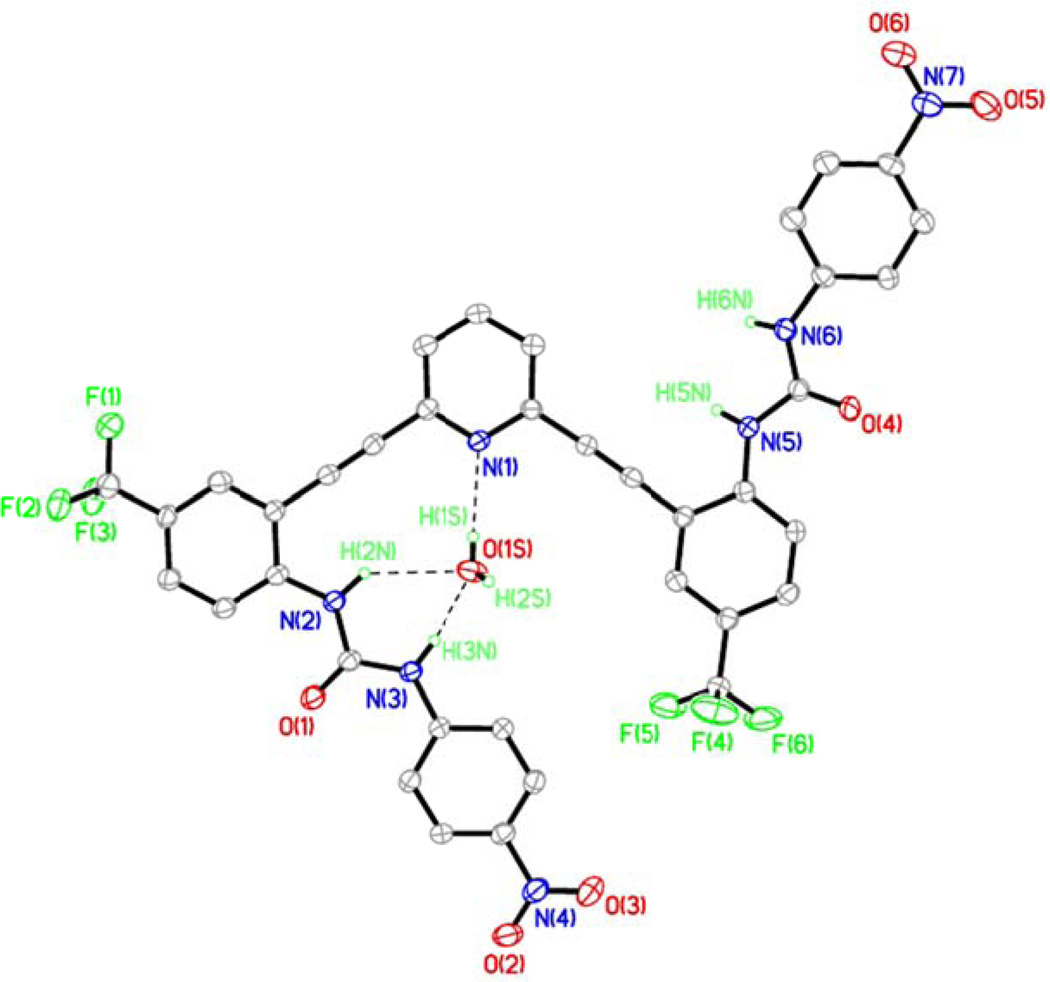
Crystal Structure of L2•H2O•0.5DMSO
X-ray crystallographic studies reveal that L2 crystallizes out of wet DMSO in monoclinic space group P2/n (Table S1). The asymmetric unit contains one L2 molecule, one water molecule and a molecule of DMSO disordered over two positions related by a two-fold axis. The ORTEP diagram of the ligand with atom numbering scheme is depicted in Figure 5. As shown in Figure 5 one of the urea groups points into the binding pocket to donate two hydrogen bonds to a water molecule. The water molecule is further hydrogen bonded to a urea carbonyl oxygen of an adjacent L2 molecule. This water molecule also donates a hydrogen bond to the pyridine nitrogen of the core. The details of these interactions are provided in Table S5.
Figure 5.
ORTEP diagram of L2•H2O•0.5DMSO with 30% thermal ellipsoids showing by hydrogen bonding interactions between L2 and H2O solvent molecule. A disordered DMSO molecule and H atoms not involved in H-bonds are omitted for clarity. The O(1S)⋯N(1), O(1S)⋯N(2) and O(1S)⋯N(3) are 2.867(4), 3.229(4) and 2.795(4) Å, respectively.
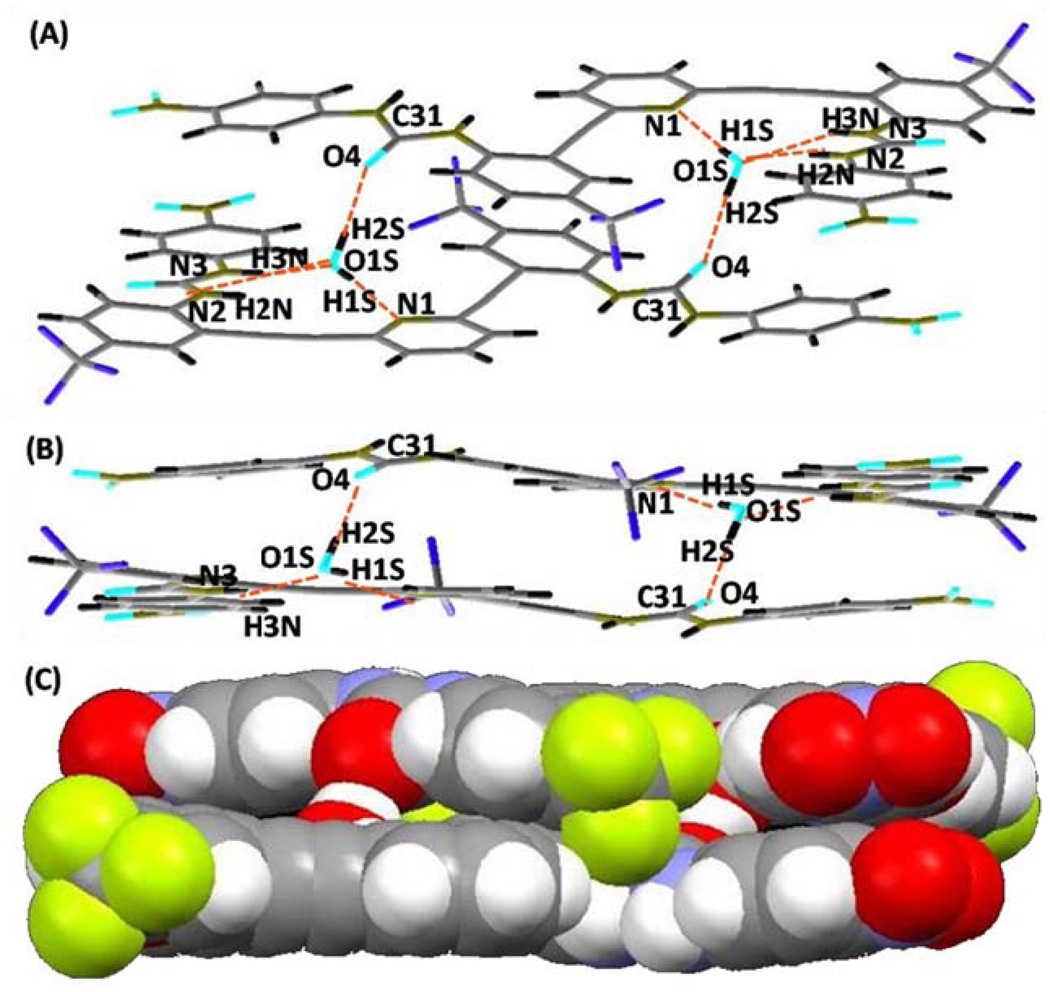
As observed in L1•2DMSO, L2 also adopts a backwards “S” conformation in the crystal structure of L2•H2O•0.5DMSO. However, unlike in L1•DMSO2, the presence of four water molecules per unit cell provides a bridge between adjacent “S” conformers to form a “SS” dimer in the crystal structure of L2•H2O•0.5DMSO (Figure 6). The 2+2 dimer structure is held together by two urea-water hydrogen bonds, two pyridine-water hydrogen bonds, and two urea carbonyl-water hydrogen bonds (Figure 6b). Since four L2 molecules are present per unit cell, there are two dimers per cell, each residing on an inversion center (view along inversion center shown in Figure 6b). A space-filling model of the dimer (Figure 6c) reveals how the two water molecules bridge the two L2 molecules of the dimer. The dimers stack along the b-axis forming channels to house the disordered DMSO solvent molecules (Figure S2).
Figure 6.
(a,b) Crystal structure of L2•H2O•0.5DMSO illustrating the 2+2 dimer formed with water molecules through inter and intramolecular hydrogen bonding. (c) Space-filling model showing the two water molecules bridging between two L2 molecules. A disordered DMSO molecule is omitted for clarity. The O(1S)⋯O(4) distances is 2.821(4) Å.
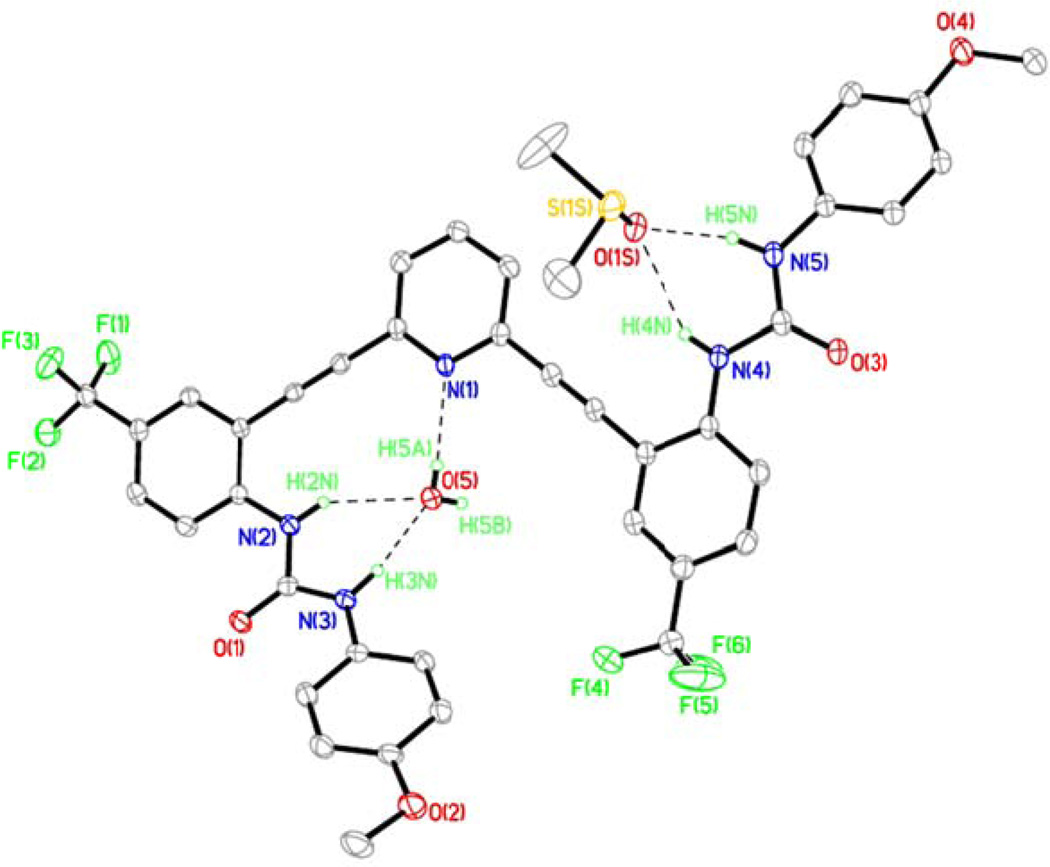
Crystal Structure of L3•H2O•DMSO (I)
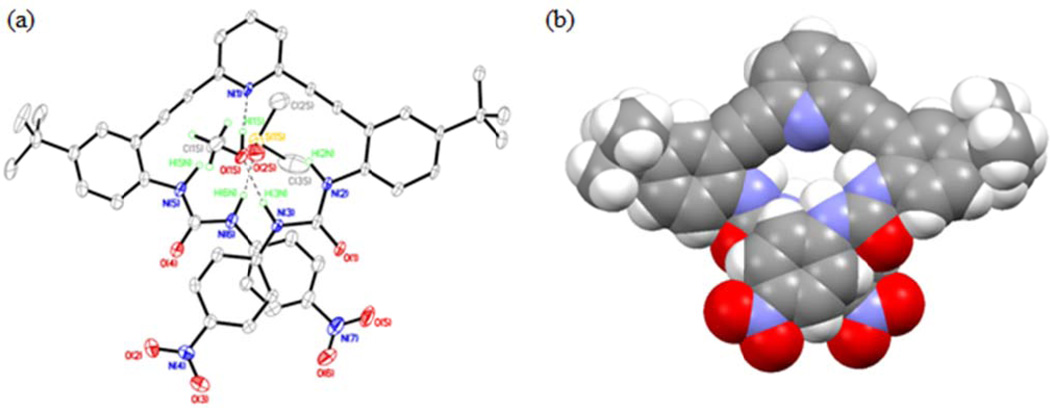
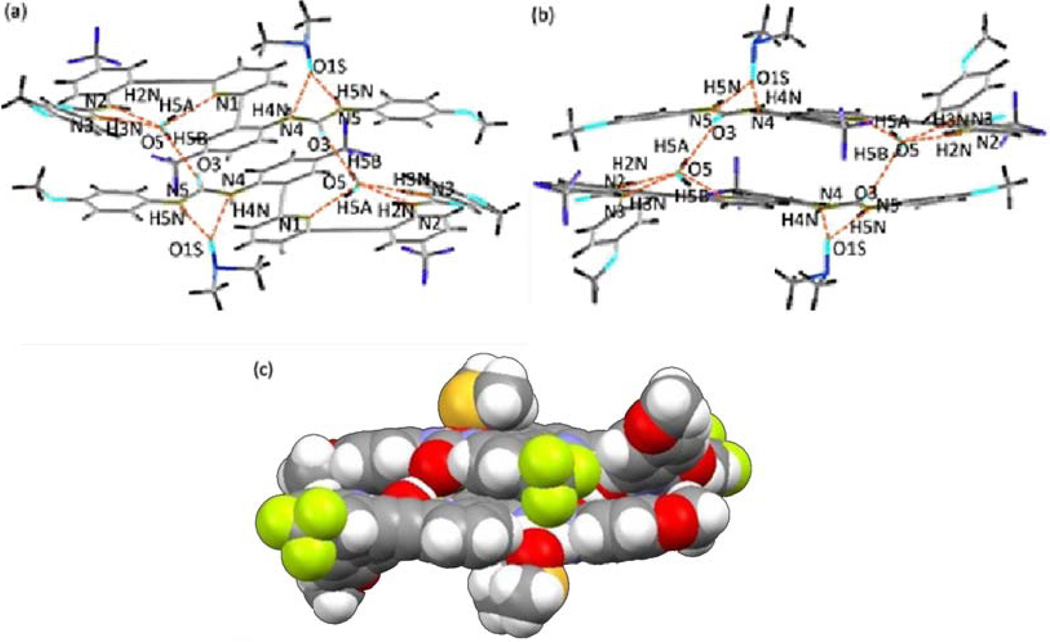
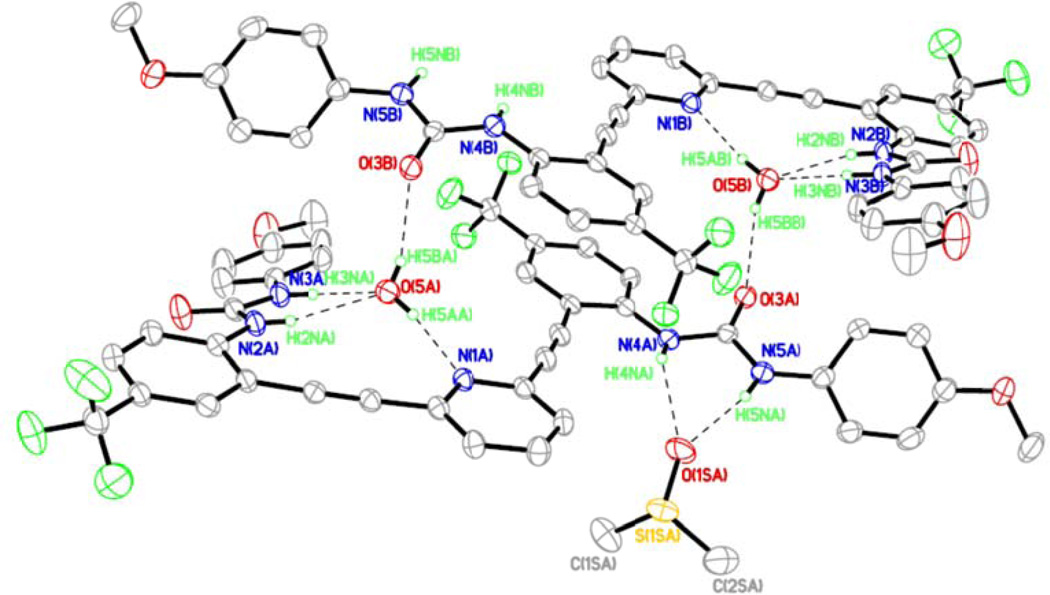
L3 crystallizes out of DMSO in triclinic space group P-1 (Table S1) with one molecule of DMSO and a water molecule (L3•H2O•DMSO). Figure 7 shows an ORTEP diagram of L3 and an atom numbering scheme. The crystal structure of L3•H2O•DMSO reveals strong intermolecular hydrogen-bonding interactions between the urea NH groups and bound solvent DMSO and water molecules. Despite having less acidic urea protons14 the hydrogen bond lengths to the water guest are shorter than in L2•H2O•0.5DMSO, presumably due to crystal packing. A water molecule occupies the binding pocket outlined by one urea arm and the pyridine core in L3 and forms hydrogen bonds with both the ureido NH groups and the pyridine nitrogen (Figures 7 and 8). The DMSo solvent molecule occupies the other urea arm by forming hydrogen bonds to the two ureido NH groups (H4N(A) and H5N(A)). Details of these hydrogen bonding interactions are shown in Table S6. The water molecule in the binding pocket also makes an intermolecular hydrogen bond with a urea oxygen of an adjacent molecule of L3 to form a stacked dimer similar to the one observed L2•H2O•2DMSO (Figure 8 a, b). L3 adopts a backward “S” conformation in L3•H2O•DMSO similar to the conformations found in L1•2DMSO and L2•H2O•2DMSO. Also similar to the structure observed in L2•H2O•2DMSO, L3•H2O•DMSO exists as a dimer held together by two bridging water molecules in the binding pocket formed by the pyridine core (Figure 8). The 2+2 dimer structure is held together by two urea-water hydrogen bonds, two pyridine-water hydrogen bonds, and two urea carbonyl-water hydrogen bonds (Figure 8a, b). A space-filling model of one of the dimers is depicted in Figure 8c, clearly showing how both water molecules are sandwiched by two units of the receptors through hydrogen bonding interactions. Two molecules of DMSO cap each side of the dimer.
Figure 7.
ORTEP diagram of L3•H2O•DMSO (I) showing hydrogen bonding interaction between L3 and solvent guest DMSO and water molecules (30% thermal ellipsoids). The O(5A)⋯N(1A), O(5A)⋯N(2A), O(5A)⋯N(3a) and O(1SA)⋯N(4A), O(1SA)⋯N(5A) distances are 2.809, 3.030(3), 2.799(3) and 3.030(3), 3.660(3) Å respectively.
Figure 8.
(a and b) Crystal structure of L3•H2O•DMSO illustrating the 2+2 dimer formed with water molecules through inter- and intramolecular hydrogen bonding. The O(3)⋯O(5) distances is 2.804(3) Å. (c) Space-filling model of the complex showing the two water molecules sandwiched between adjacent L3 molecules.
Crystal Structure of L3•H2O•DMSO (II)
Attempts to crystallize L3 with mineral acids of various shapes and varying conjugate basicities such as HCl and HNO3 under similar crystallization conditions failed to produce a protonated L3 receptor (e.g., L3•HX). Instead, only crystal of the free receptor could be isolated. Interestingly, while failing to crystallize the protonated core pyridine, attempts to crystallize L3 in the presence of hydrochloric acid in DMSO produced single crystals of chemical formula L3•H2O•DMSO (II), but in which L3 is found in a slightly different conformation (Figure 9).
Figure 9.
ORTEP diagram L3•H2O•DMSO (II) with 30% thermal ellipsoids featuring the hydrogen bonding interactions between L3 and solvent molecules H2O and DMSO H atoms involved in H-bonds are shown for clarity. The O(5A)⋯N(1A), O(5A)⋯N(2A), O(5A)⋯N(3A), O(5A)⋯O(3B) and O(1SA)⋯N(4A), O(1SA)⋯N(5A) distances are 2.871(5), 3.039(5), 2.828(5),2.774(5) and 2.950(4), 2.795(5) Å, respectively.
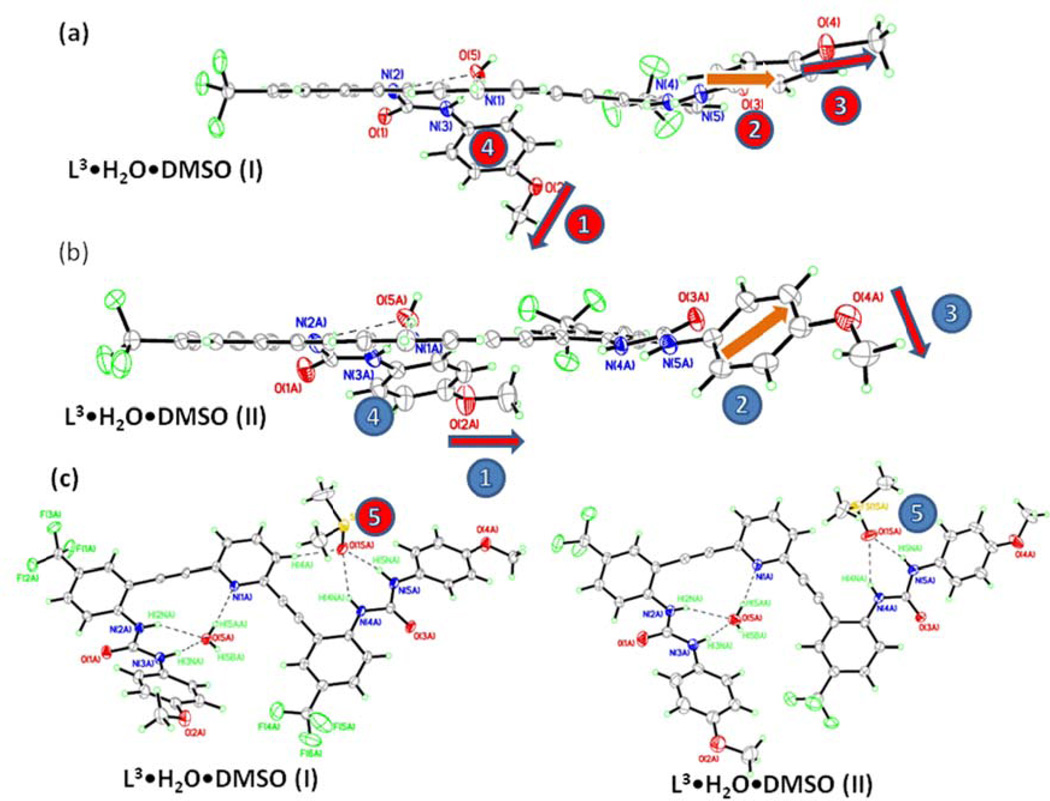
As in L3•H2O•DMSO (I), L3 again crystallizes in triclinic space group P-1 (Table S1) with one molecule of DMSO and a water molecule in L3•H2O•DMSO (II). An ORTEP diagram of the dimer showing an atom numbering scheme is provided in Figure 9. The crystal structure of L3•H2O•DMSO (II) shows strong intermolecular hydrogen-bonding interactions between the ureas and solvent DMSO and water molecules. Very similar hydrogen-bonding schemes are observed between complexes I and II, and the dimeric structures are quite similar. Details of these hydrogen bonding interactions are shown in Table S7. Although both complexes of L3 (I and II) adopt similar backward “S” conformations, there are a few differences apparent in their crystal structures (Figure 10). In complex II the methoxy group on the phenyl urea that is bound to the water guest is pointing nearly perpendicular to the plane of the bisethynylpyridine core (Figure 10a, labeled with a red 1). The other methoxy group (labeled with a red 3 in Figure 10a) is nearly parallel to the core plane. This trend is reversed in complex II: the methoxy group on the urea bound to the water guest is now parallel to the core (blue 1 in Figure 10b), whereas the other methoxy group is nearly perpendicular to the core (blue 3). Furthermore, the phenylurea groups are also oriented in different directions (Figure 10a,b labeled as 2 and 4). The DMSO guests are also oriented slightly differently in each complex (labeled as 5 in Figure 10): the orientation of the DMSO oxygen atom in I is more nearly perpendicular to the core, allowing an extra weak hydrogen bond with a phenyl CH group.
Figure 10.
ORTEP diagrams showing the differences between L3•H2O•DMSO complexes I (a, red labels) and II (b, blue labels). The numerical labels are: 1) methoxy groups on urea bound to water; 2&4) phenylurea orientation, 3) methoxy groups on urea bound to DMSO; 5) DMSO guest; c) ORTEP comparisons of I and II showing the different orientations of the DMSO guest (5).
Conclusions
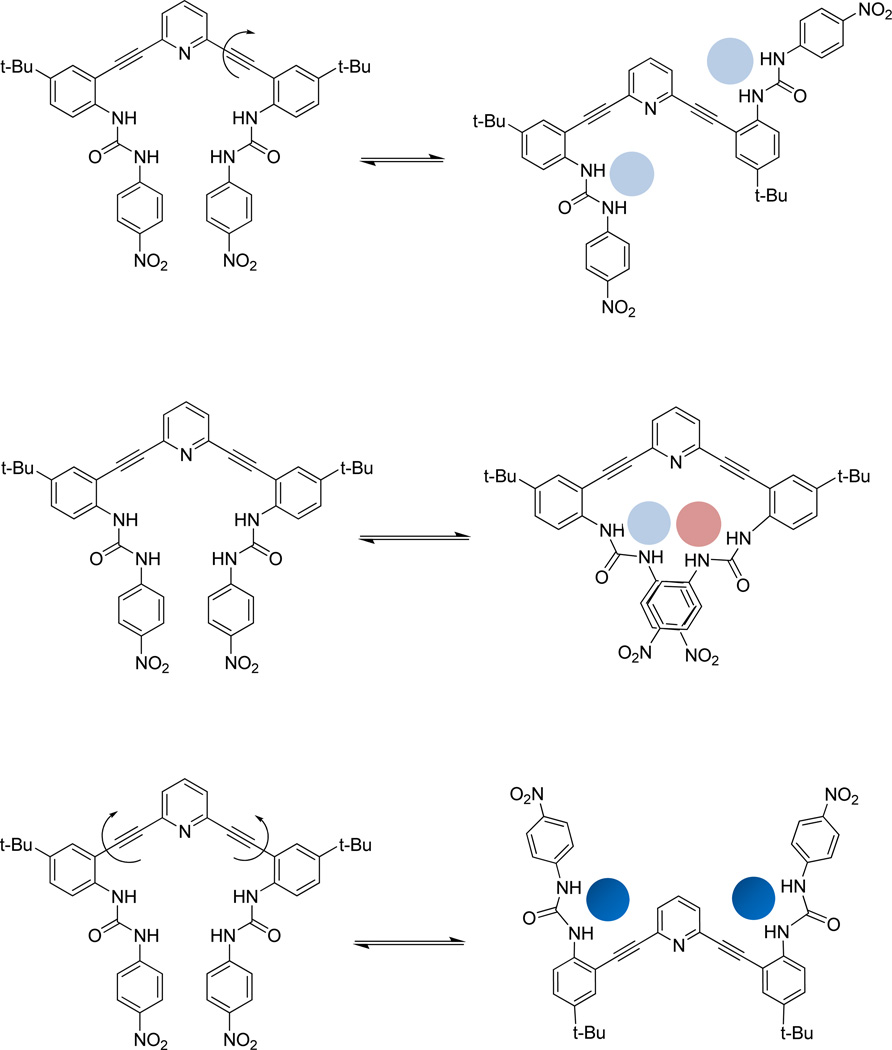
We have utilized 2,6-bis(2-anilinoethynyl)pyridylureas as receptors for neutral solvent guests. Three different conformations are observed about the bisethynyl cores: “S”, “O” and “W” in six different crystal structures depending on the solvents (guests) of crystallization. The receptors (L1, L2 and L3) each have two rotatable –C≡C–bond(s) that can potentially impart a variety of topological structures, and the interaction of the urea groups with potential guests dictate the conformations that are observed in the crystal structures (Chart 2). Remarkably, L1 exhibits each of the different conformations (backward “S”, “O” and “W”) depending on the choice of single, binary or ternary solvent systems for crystallization (DMSO, DMSO/CH3OH and DMF/toluene/1,4-dioxane, respectively). On the other hand, in the case of the structures formed from L2 and L3, aggregated structures are preferred to form as dimers that sandwich what appear to be strongly bound water guests. This receptor class exhibits complicated solution speciation, and the NMR spectra of the free base compounds are often broad and difficult to interpret.9 The solid state structures reported herein may suggest some of the aggregates that are present in these equilibrating solutions. We are attempting to understand the solution structures of these complex systems as well, and relate this conformational flexibility to the emergent fluorescent properties exhibited by this receptor class.8a
Chart 2.
Schematic showing interconversion between a starting “U” conformation into backwards “S” (top), “O” (middle) and “W” (bottom) conformations.
EXPERIMENTAL SECTION
General Procedure
All chemicals were of reagent grade, obtained from commercial sources, and used without further purification. Reagent grade solvents were used as provided by the supplier.
L1•2DMSO
Obtained by dissolving L1 (15 mg, 0.02 mmol) in 5 mL of DMSO. After addition of solvent, the clear solution was filtered and kept for crystallization at room temperature. Yellowish crystals suitable for X-ray analysis were obtained after 2 d by slow evaporation at room temperature.
L1•2DMSO•CH3OH
Obtained by dissolving L1 (15 mg, 0.02 mmol) in 5 mL of 1:1 DMSO/CH3OH binary solvent mixture. Yellowish crystals suitable for X-ray analysis were obtained after 3 d by slow evaporation at room temperature.
L1•3DMF
Obtained by dissolving L1 (15 mg, 0.02 mmol) in DMF/toluene/1,4-dioxane ternary solvent mixture. Crystals suitable for X-ray analysis were obtained after 2 d by slow evaporation at room temperature.
L2•H2O•0.5DMSO2
Obtained by dissolving L2 (15 mg, 0.02 mmol) in 5 ml of DMSO. Crystals suitable for X-ray analysis were obtained by slow evaporation at room temperature.
L3•H2O•DMSO (I)
Obtained by dissolving L2 (15 mg, ~0.02 mmol) in DMSO. Crystals suitable for X-ray analysis were obtained by slow evaporation at room temperature.
L3•H2O•DMSO (II)
Obtained by dissolving L3 (15 mg, ~0.02 mmol) in DMSO containing a few drops of HCl. Crystals suitable for X-ray analysis were obtained by slow evaporation at room temperature.
General X-ray Crystallography Experimental
Diffraction intensities for L1•2DMSO, L1•2DMSO•CH3OH, L1•3DMF, L2•H2O•0.5DMSO2, L3•H2O•DMSO (I), L3•H2O•DMSO (II) were collected at 173(2) K on a Bruker Apex CCD diffractometer using MoKα radiation λ= 0.71073 Å. Space groups were determined based on systematic absences (L2•H2O•0.5DMSO2 and L1•2DMSO) and intensity statistics (L1•2DMSO•CH3OH, L1•3DMF, L3•H2O•DMSO (I), L3•H2O•DMSO (II)). Absorption corrections were applied by SADABS.15 Structures were solved by direct methods and Fourier techniques and refined on F2 using full matrix least-squares procedures. All non-H atoms were refined with anisotropic thermal parameters except carbon atoms in the disordered t-Bu groups in L1•3DMF which were refined with isotropic thermal parameters. Hydrogen atoms in L2•H2O•0.5DMSO2, L3•H2O•DMSO (I), L1•2DMSO and L1•3DMF were refined in calculated positions in a rigid group model except those at N atoms involved in H-bonds which were found on the residual density maps and refined with isotropic thermal parameters without any restrictions. In L1•2DMSO•CH3OH and L3•H2O•DMSO (II) all H atoms were found from the residual density maps and refined with isotropic thermal parameters except H atoms in terminal Me groups in solvent DMSO molecule and t-Bu groups in L1•2DMSO•CH3OH, which were refined in calculated positions in a rigid group model. In L1•2DMSO•CH3OH there are two disordered solvent DMSO molecules. Me groups in one of them are disordered over two positions in a 1:1 ratio. The second DMSO solvent molecule is highly disordered and was treated by SQUEEZE.16 Correction of the X-ray data by SQUEEZE (89 electron/cell) was close to the required value (84 electron/cell) for two molecules in the full unit cell. The solvent DMSO molecule in L2•H2O•0.5DMSO2 is also disordered over two positions related by a two-fold axis. H atoms in this disordered solvent molecule were not taken into consideration. X-ray diffraction for all crystals investigated in this work was weak especially at high angles. Thus for all structures only reflections with θ ≤ 25 (θ ≤ 24 for L1•2DMSO•CH3OH) were included in the final refinements. The crystallographic data and details of data collections and refinements of the structures are given in the Supporting Information. All calculations were performed by the Bruker SHELXTL (v. 6.10) package.17
Supplementary Material
Chart 1.
Structures of scaffolds 1, 2 and 2,6-bis(2-anilinoethynyl)pyridine urea receptors L1, L2 and L3.
ACKNOWLEDGMENT
This research was supported by the NIH (GM087398) and the University of Oregon (UO). C.N.C. acknowledges the NSF for an Integrative Graduate Education and Research Traineeship (DGE-0549503).
Footnotes
ASSOCIATED CONTENT
Supporting Information. Six crystallographic files in CIF format of all structures, tables of crystallographic parameters, tables of hydrogen bonding distances and angles. This material is available free of charge via the Internet http://pubs.acs.org.
REFERENCES
- 1.Nelson DL, Cox MM. Lehninger Principles of Biochemistry. 5th ed. W. H. Freeman; 2008. [Google Scholar]
- 2.(a) Pley HW, Flaherty KM, McKay DB. Nature. 1994;372:68–74. doi: 10.1038/372068a0. [DOI] [PubMed] [Google Scholar]; (b) Dunkle JA, Wang L, Feldman MB, Pulk A, Chen VB, Kapral GJ, Noeske J, Richardson JS, Blanchard SC, Cate JHD. Science. 2011;332:981–984. doi: 10.1126/science.1202692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.(a) Maerz AK, Fowler DA, Mossine AV, Mistry M, Kumari H, Barnes CL, Deakyne CA, Atwood JL. New J. Chem. 2011;35:784. [Google Scholar]; (b) Scheiner S, Kar T. J. Phys. Chem. B. 2005;109:3681–3689. doi: 10.1021/jp0446736. [DOI] [PubMed] [Google Scholar]; (c) Prabhu N, Sharp K. Chem. Rev. 2006;106:1616–1623. doi: 10.1021/cr040437f. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Chopra D, Mohan TP, Rao KS, Row TNG. CrystEngComm. 2005;7:374. [Google Scholar]; (e) Ni X, Zhang Y, Song J, Zheng H. J. Cryst. Growth. 2007;299:365–368. [Google Scholar]; (f) Wong MW, Frisch MJ, Wiberg KB. J. Am. Chem. Soc. 1991;113:4776–4782. [Google Scholar]; (g) Schiel C, Hembury GA, Borovkov VV, Klaes M, Agena C, Wada T, Grimme S, Inoue Y, Mattay J. J. Org. Chem. 2006;71:976–982. doi: 10.1021/jo0518654. [DOI] [PubMed] [Google Scholar]
- 4.(a) Steed JW, Wallace KJ. In: Advances in Supramolecular Chemistry. Gokel GW, editor. Vol. 9. New York: Cerberus; 2003. [Google Scholar]; (b) Cram DJ. Angew. Chem. Int. Ed. 1986;25:1039–1057. [Google Scholar]; (c) Hettche F, Reiß P, Hoffmann RW. Chemistry - A European Journal. 2002;8:4946–4956. doi: 10.1002/1521-3765(20021104)8:21<4946::AID-CHEM4946>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]; (d) Steed JW, Atwood JL. Supramolecular Chemistry. John Wiley and Sons; 2009. [Google Scholar]
- 5.(a) Kondo S-ichi, Sato M. Tetrahedron. 2006;62:4844–4850. [Google Scholar]; (b) Lee DH, Im JH, Lee J-H, Hong J-I. Tetrahedron Lett. 2002;43:9637–9640. [Google Scholar]; (c) Lin Z-hua, Xie L-xia, Zhao Y-gang, Duan C-ying, Qu J-ping. Org. Biomol. Chem. 2007;5:3535. doi: 10.1039/b708983j. [DOI] [PubMed] [Google Scholar]
- 6.Swinburne AN, Paterson MJ, Beeby A, Steed JW. Chem. Eur. J. 2010;16:2714–2718. doi: 10.1002/chem.200903293. [DOI] [PubMed] [Google Scholar]
- 7.(a) Nelson JC, Saven JG, Moore JS, Wolynes PG. Science. 1997;277:1793–1796. doi: 10.1126/science.277.5333.1793. [DOI] [PubMed] [Google Scholar]; (b) Yang XW, Yuan LH, Yamamoto KA, Brown L, Feng W, Furukawa M, Zeng XC, Gong B. J. Am. Chem. Soc. 2004;126:3148–3162. doi: 10.1021/ja039416d. [DOI] [PubMed] [Google Scholar]; (c) Inouye M, Waki M, Abe H. J. Am. Chem. Soc. 2004;126:2022–2202. doi: 10.1021/ja039371g. [DOI] [PubMed] [Google Scholar]; (d) Kim T, Arnt L, Atkins E, Dew GN. Chem.-Eur. J. 2006;12:2423–2427. doi: 10.1002/chem.200501283. [DOI] [PubMed] [Google Scholar]
- 8.Berryman OB, Johnson CA, II, Zakharov LN, Haley MM, Johnson DW. Angew. Chem. Int. Ed. 2008;47:117–120. doi: 10.1002/anie.200703971. [DOI] [PubMed] [Google Scholar]
- 9.(a) Carroll CN, Coombs BA, McClintock SP, Johnson CA, II, Berryman OB, Johnson DW, Haley MM. Chem. Commun. 2011;47:5539–5541. doi: 10.1039/c1cc10947b. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Carroll CN, Berryman OB, Johnson CA, II, Zakharov LN, Haley MM, Johnson DW. Chem. Commun. 2009:2520–2522. doi: 10.1039/b901643k. [DOI] [PubMed] [Google Scholar]
- 10.Engle JM, Carroll CN, Zakharov LN, Johnson DW, Haley MM. manuscript in preparation. [Google Scholar]
- 11.Beer PD, Gale PA. Angew. Chem. Int. Ed. 2001;40:488–516. [PubMed] [Google Scholar]
- 12.(a) Lakshminarayanan PS, Ravikumar I, Suresh E, Ghosh P. Chem. Commun. 2007:5214–5216. doi: 10.1039/b713365k. [DOI] [PubMed] [Google Scholar]; (b) Ravikumar I, Lakshminarayanan PS, Arunachalam M, Suresh E, Ghosh P. Chem. Commun. 2007:5214–5216. doi: 10.1039/b713365k. [DOI] [PubMed] [Google Scholar]; (c) Lakshminarayanan PS, Suresh E, Ghosh P. Inorg. Chem. 2006;45:4372–4380. doi: 10.1021/ic052159o. [DOI] [PubMed] [Google Scholar]
- 13.(a) Matthew TS, Moore JS. Org. Lett. 2004;6:469–472. doi: 10.1021/ol036238k. [DOI] [PubMed] [Google Scholar]; (b) Smaldone RA, Moore JS. Chem. Commun. 2008:1011–1013. doi: 10.1039/b716122k. [DOI] [PubMed] [Google Scholar]; (c) Hill DJ, Moore JS. Proc. Nat. Acad. Sci. 2002;99:5053–5057. doi: 10.1073/pnas.072642799. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Prince RB, Barnes SA, Moore JS. J. Am. Chem. Soc. 2000;122:2758–2762. [Google Scholar]
- 14.Abraham MH, Grellier PL, Prior DV, Duce PP, Morris JJ, Taylor PJ. J. Chem. Soc., Perkin Trans. 1989;2:699–711. [Google Scholar]
- 15.Sheldrick GM. Bruker/Siemens Area Detector Absorption Correction Program. Madison, WI: Bruker AXS; 1998. [Google Scholar]
- 16.Van der Sluis P, Spek AL. Acta Cryst., Sect. A. 1990;A46:194–201. [Google Scholar]
- 17.SHELXTL-6.10 "Program for Structure Solution, Refinement and Presentation". 5465 East Cheryl Parkway, Madison, WI 53711-5373 USA: BRUKER AXS Inc; [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.