Abstract
Systemic lupus erythematosus (SLE) is a complex systemic autoimmune disease associated with multiple immunologic abnormalities. Prominent among these is upregulation of type I interferon (IFN)—a powerful immune adjuvant. IFN is, in part, produced in SLE in response to autoantigens in the form of self-nucleic acids and their associated nuclear proteins. Sources of these autoantigens include apoptotic and necrotic cells as well as neutrophils undergoing a specific form of cell death called NETosis. Although plasmacytoid dendritic cells are the main producers of IFN-a, other cells are important regulators of this process. Both genetic and environmental risk factors play a role in the development and pathogenesis of SLE. Further highlighting the importance of IFN, candidate gene and genome-wide association studies have identified a number of genes involved in type I IFN pathways associated with SLE. In this review, 3 monogenic deficiencies that result in lupus-like phenotypes and several polygenic variants that have been consistently associated with SLE are highlighted, and the relationship of these genes to IFN-a production is discussed. Clinical associations of the type I IFN pathway and the use of IFN-blocking agents as therapeutic agents in SLE are also reviewed.
Systemic Lupus Erythematosus
Systemic lupus erythematosus (SLE or lupus) is a complex autoimmune disease with diverse clinical manifestations ranging from mild rash and arthralgia to severe or life-threatening forms of the disease affecting the kidneys and central nervous system (Rahman and Isenberg 2008). Multiple abnormalities contribute to the pathogenesis of SLE. These include abnormal clearance of apoptotic cells and immune complexes (ICs) and low thresholds of activation of B and T lymphocytes leading to loss of self-tolerance and autoantibody production. These autoantibodies are directed against nucleic acids (DNA) and associated nuclear proteins as well as ribonuclear proteins (RNP) such as Ro, La, and Sm (Tan 1989). Tissue damage is mediated by deposition of pathogenic autoantibodies and ICs in the affected organs, followed by activation of downstream inflammatory pathways mediated by complement and FcR engagement of innate immune cells (Rahman and Isenberg 2008).
Both environmental and genetic risk factors are important in the development of SLE. The female to male ratio is 9:1, suggesting that hormones may be important in the development of the disease. Exposure to UV light is known to trigger SLE, and several viruses and bacterial infections have also been implicated in disease pathogenesis and development. Drug-induced lupus is a well-described syndrome in which lupus-like symptoms develop after exposure to certain medications, including hydralazine, procainamide, isoniozide, and interferon-alpha (IFN-a) (Sarzi-Puttini and others 2005). Twin studies reveal a 25% to 40% concordance rate among monozygotic twins and a 2% concordance rate among dizogotic twins (Harley and others 2009), emphasizing the strong genetic component in SLE. HLA associations and complement deficiencies, especially of early complement components C1q, C2, and C4 have long been known to be associated with SLE. To date, 3 monogenic deficiencies—C1q, 3 prime repair exonuclease 1 (TREX1), and tartrate-resistant acid phosphatase (TRAP)—have been identified that result in clinical phenotypes consistent with lupus. In addition, more than 100 common genetic variants (only 8 consistently replicated) that confer increased risk of SLE susceptibility but with small effect have been identified by genome-wide association studies (GWAS) (Harley and others 2009). Of note, all 3 monogenic syndromes and many of the genetic variants identified by GWAS are involved in the type I IFN pathway, further emphasizing the importance of this pathway in SLE (Deng and Tsao 2010; Sestak and others 2011).
IFNs: Types and Effect on Immune Function
There are 3 types of IFNs: type I (alpha, beta, omega, and other less common subtypes), type II (IFN-gamma), and type III (IFN-lambda) [interleukin 28 and 29 (IL-28 and IL-29)] (reviewed in Theofilopoulos and others 2005). Of the type I IFNs, IFN-b can be produced by almost any cell and release of this cytokine serves to prime or amplify type I IFN by other cells, especially plasmacytoid dendritic cells (pDCs) that are the main producers of IFN-a (reviewed in Swiecki and Colonna 2010). The 13 type I IFNs, each encoded by a separate gene on chromosome 9, as well as IFN-b signal through a common heterodimeric receptor, IFN-alpha receptor (IFNAR). The IFNAR is composed of 2 chains (IFNAR1 and IFNAR2) that signal through Jak1-Tyk2 and signal transducer and activator of transcription 1 protein (STAT1) (Uze and others 2007). The type I IFNs are important for protection against viral infections with powerful effects on both the innate and adaptive immune responses, but also have roles in cell growth regulation (Fitzgerald-Bocarsly and others 2008). Acting as an immune adjuvant, IFN-a increases the survival and activation of DCs and induces upregulation of the major histocompatibility complex (MHC) and costimulatory molecules such as CD40 and CD86. IFN also induces DCs to promote the secretion of BLyS and APRIL, which, in turn, lead to increased B-cell survival, differentiation, and isotype class-switching. Type I IFNs directly affect T cells by increasing survival, activation, and cross-priming as well as promoting Th1 differentiation (Reviewed in Baccala and others 2007).
Type I IFN: Cells, Sensors, and Pathways
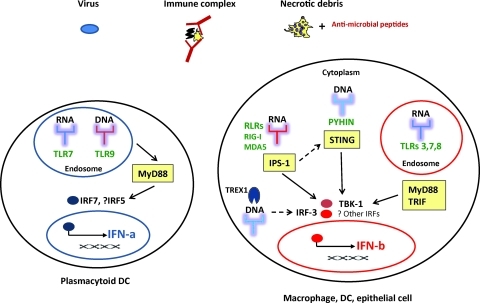
As the dominant alarm signal responding to virus infection, type I IFN (mostly IFN-b) can be produced by most cells, including macrophages, DCs, and epithelial cells (Fig. 1). In addition to virus infection, IFNs (mostly IFN-a) may be triggered by ICs containing nucleoproteins and, in some cases, by necrotic debris as discussed below. In most cells, a variety of cytosolic receptors or sensors exist to detect (foreign) nucleic acids. These sensors include Toll-like receptors (TLRs), retinoic acid-inducible gene I (RIG-I)-like receptors (RLR) comprising RIG-I, and the melanoma differentiation-associated gene 5 (MDA-5) helicases and pyrin and HIN200 domain-containing protein receptors (reviewed in Swiecki and Colonna 2010; Barber 2011). Cytoplasmic DNA-sensing receptors are not yet fully characterized, but activation of stimulator of IFN gene appears to be a common adapter in the non-TLR IFN pathway stimulated by DNA, and is also implicated in the RNA-stimulated RIG-I pathway (Barber 2011) (Fig. 1).
FIG. 1.
Innate signaling pathways leading to type I interferon (IFN) production. Various triggers—viruses, nucleic acid-containing immune complexes, and necrotic debris associated with antimicrobial peptides—are sensed by pattern recognition receptors in cells, including endosomal Toll-like receptors (TLRs) and cytoplasmic RIG-I-like receptors (RLRs) and pyrin and HIN200 domain-containing protein (PYHIN) receptors and lead to IFN production. TREX1 negatively regulates interferon stimulation by cytoplasmic DNA. Yellow boxes represent adapter molecules necessary for downstream signal transduction. IPS-1, interferon-beta promotor stimulator 1; IRF, interferon regulatory factor; MyD88, myeloid differentiation factor 88; STING, stimulator of interferon gene; TRIF, Toll-receptor associated activator of interferon; TBK-1, TANK binding kinase1.
In contrast to the universality of IFN-b production, the main IFN-a producing cell among human leukocytes is the pDC. Although pDCs are present in very low frequency in human peripheral blood, accounting for only 0.2%–0.5% of human peripheral blood mononuclear cells (PBMC), they have the ability to produce 10–100 times more type I IFNs than other cell types. pDCs are unique in that they constitutively express high levels of the endosomal TLRs 7 and 9 as well as IFN regulatory factors (IRFs) 5 and 7, allowing them to rapidly produce type I IFNs in response to viral infection. Activation of TLR7 or 9 induces signaling through the myeloid differentiation factor 88 (MyD88) pathway leading to phosphorylation of IRFs 5 and 7 (Fig. 1), which in turn translocate to the nucleus and stimulate expression of IFNs (reviewed in Cao 2009).
Type I IFNs in SLE Pathogenesis
In has been known for some time that there are elevated levels of IFNs in the serum of patients with SLE, and that this elevation correlated with disease activity (Hooks and others 1979; Ytterberg and Schnitzer 1982). Additional evidence that IFNs may have a pathogenic role in the development of SLE came from the observation that some patients who were treated with type I IFNs for either malignancy or chronic infections developed a lupus-like syndrome that was indistinguishable from spontaneous SLE (Ronnblom and others 1991; Sanchez Roman and others 1994). More recently, several groups have shown through microarray analysis that peripheral blood mononuclear cells (PBMCs) from SLE patients overexpress a large number of IFN-stimulated genes (ISGs) (Baechler and others 2003; Bennett and others 2003). This discovery dramatically focused attention on type I IFN in SLE.
Sources of Autoantigens in SLE and Regulation of Type I IFN Production by pDC
Apoptosis and SLE
One mechanism by which nuclear material may become a source of autoantigens and lead to systemic autoimmunity is through defective clearance of dead and dying cells. In healthy individuals, apoptotic cells are rapidly removed by macrophages—a process that is inherently anti-inflammatory in nature. In many mouse models and in human SLE, there is evidence of defective clearance of apoptotic cells (Munoz and others 2010), resulting in a transition to a necrotic form of cell death. It is proposed that release of (modified) nuclear antigens exposes self-antigens (particularly nucleic acids) to intracellular sensors that respond by producing inflammatory cytokines, including IFN-a (Navratil and others 2006), although whether and how this material enters responding cells remains to be determined. The adjuvant effects of IFN promote loss of self-tolerance, production of autoantibodies against nuclear material, and IC formation (Martin and Elkon 2005). The process of IC formation generated by SLE autoantibodies and antigen derived from dead and dying cells can be replicated in vitro using apoptotic or necrotic debris. These ICs bind to low affinity FcgRIIa receptors expressed on the surface of pDCs and are endocytosed. In the endosome, RNA or DNA present in these complexes activate TLR7 or 9, respectively, and induce IFN-a production by pDCs (Martin and Elkon 2005; Ronnblom and others 2006). The ability of SLE sera to induce IFN-a production appears to be well correlated with the presence of antibodies against small nuclear ribonuclearprotein particles (RNPs) such as Sm or U1RNP (Vollmer and others 2005). In addition, the presence of antibodies specific for Sm, U1RNP, Ro, and dsDNA in serum has been associated with higher expression of ISGs in SLE patients, supporting the clinical significance of these observations (Kirou and others 2005).
Neutrophils and SLE
In addition to the IFN gene signature observed in microarrays when whole blood was analyzed, upregulation of granulopoiesis-related genes was also observed in the arrays obtained from SLE patients with active disease (Bennett and others 2003). These granulopoiesis-related genes include enzymes and enzyme inhibitors as well as bactericidal proteins. It has also been noted that SLE patients have increased numbers of apoptotic neutrophils in peripheral blood that correlated with anti-DNA antibodies and disease activity (Courtney and others 1999). Two recent studies have provided evidence that activation and/or apoptosis of neutrophils may directly contribute to the IFN signature in SLE.
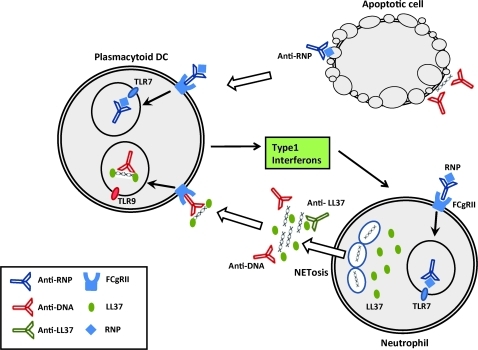
As part of their antimicrobial functions, activated neutrophils release web-like structures called neutrophil extracellular traps (NETs), composed of large amounts of nuclear DNA in a specific type of cell death termed NETosis (Brinkmann and others 2004). Of note, IFN-a can prime mature neutrophils in vitro to form NETs (Martinelli and others 2004). Lande and others (2011) reported that SLE serum contains ICs composed of neutrophil-derived antimicrobial peptides and self-DNA. Neutrophil-specific antimicrobial peptides protect DNA from nuclease degradation in vitro and are required for pDC activation and IFN production. Peptide/DNA complexes were released by neutrophils undergoing NETosis and were able to trigger IFN-a production in pDCs through activation of TLR9 (Lande and others 2011). Significantly, SLE serum contains antibodies against the antimicrobial peptide LL37 and self-DNA, which can induce NET formation by neutrophils isolated from healthy donors. Interestingly, neutrophils from SLE patients release more NET DNA than healthy donors, even in the absence of in vitro stimulation (Lande and others 2011). In the study by Garcia-Romo and others (2011) neutrophils isolated from SLE patients were shown to undergo accelerated cell death in vitro and express genes involved in type I IFN and TLR signaling pathways. This same expression pattern was seen in neutrophils from healthy donors after exposure to SLE serum in vitro. Neutrophils from patients with SLE, but not healthy donors, underwent NETosis upon exposure to anti-RNP antibodies and caused IFN-a production by pDCs in an FcgRII, NADPH, and TLR7-dependent manner (Garcia-Romo and others 2011). SLE NETs released large amounts of extracellular DNA, LL37 (an antimicrobial peptide), and HMGB1 (a protein that leads to type I IFN secretion by binding to DNA and targeting cytosolic DNA receptors). Healthy neutrophils exposed to IFN-a are susceptible to NETosis induced by anti-RNP antibodies in vitro (Garcia-Romo and others 2011). Together, these data identify a pathogenetically important link between neutrophils, pDCs, IFN-a production, and autoantibodies in SLE, and describe a cycle of chronic inflammation that may be important in disease pathogenesis (Fig. 2).
FIG. 2.
Stimulation of type I interferon by cell debris and immune complexes. Immune complexes (ICs) containing ribonucleoproteins (RNP) can be generated by the binding of autoantibodies to RNP containing antigens released by apoptotic or necrotic cells (top right). In addition, ICs containing deoxyribonucleoproteins (DNP) can be generated by the binding of autoantibodies to DNA or antimicrobial peptides, such as LL37, after NETosis of neutrophils (bottom right). Ultimately, these ICs are internalized by FcgR on plasmacytoid dendritic cells (pDC) and activate TLR7 or 9 resulting in the production of type I interferon in systemic lupus erythematosus (SLE; top left).
Regulation of IFN-a production by pDC
A recent study by Eloranta and others (2009) explored the regulation of IFN-a production by RNA-containing IC-stimulated pDCs. PBMCs or pDCs primed with IFN-a2b and granulocyte-macrophage colony-stimulating factor before stimulation increased subsequent IFN-a production—suggesting that pDCs themselves may enhance their own production of IFN-a. Monocytes inhibited IFN-a production in PBMC cultures, but had no effect on IFN-a production in isolated pDC cultures. CD56+ natural killer (NK) cells enhanced IFN-a production by pDCs, an effect that is lost with addition of monocytes to the cultures. Interestingly, monocytes isolated from SLE patients were less effective at inhibiting RNA IC-stimulated pDCs. The inhibitory effects of monocytes on IFN-a production by RNA-IC-stimulated pDCs could be mediated through production of prostaglandin E2, tumor necrosis factor alpha (TNF-a), or reactive oxygen species (Eloranta and others 2009). The mechanism by which CD56+ NK cells promoted IFN-a production by pDC after RNA-containing IC was subsequently shown to be via secretion of macrophage inflammatory protein 1 beta (MIP-1b) and cell–cell contact through lymphocyte function-associated antigen 1. Perhaps counter-intuitively, NK cells from SLE patients showed lower capacity to promote IFN-a production by RNA-containing IC-stimulated pDCs, although capacity was fully restored with the addition of exogenous IL-12 and IL-18 (Hagberg and others 2011).
Genetics, IFN, and SLE
SLE has a strong genetic predisposition as evidenced by the high concordance rates in mono- and dizygotic twins (Harley and others 2009). Although many mouse models have been generated by manipulation of single genes, the lupus phenotype in many cases depended upon background lupus prone genes present in C57BL/6×Sv129 mice (Carlucci and others 2007). Similarly, single-gene disorders predisposing to SLE in the human population are very uncommon—the majority of patients have multiple genetic variants, each of which has a small effect that combine to contribute to disease (Harley and others 2009). Below we will discuss the only 3 known, highly penetrant, monogenic deficiencies involving the first component of complement, C1q; an intracellular DNA exonuclease, TREX-1; or an intracellular acid phosphatase, TRAP. Provocatively, all of these syndromes are associated with alterations in type I IFN production. In addition, data from increasingly powerful GWAS have identified variants in >100 genes that have been reported to confer increased risk for the development of disease through several different pathways. In the following sections, we briefly highlight mono- and polygenic alterations that involve the type I IFN pathway.
Monogenic Deficiencies Associated with SLE
C1q deficiency
An extremely strong genetic risk for SLE is conferred by defects in the early complement components (C1q, C4, and, to a lesser extent, C2). To date, deficiency in C1q, the first component of the classical pathway, is the strongest known susceptibility factor identified in humans. More than 90% of individuals who have a homozygous deficiency of C1q develop SLE, usually with more severe disease manifestations (Pickering and others 2000). Although complete C1q deficiency is very rare, reduced production, increased consumption, and autoantibodies to C1q may contribute to acquired lowering of C1q levels (Trendelenburg 2005). C1q not only has a role in clearance of apoptotic cells, but can also affect cytokine production by other immune cells (Fraser and others 2009).
Kirou and others (2005) reported that low complement levels are associated with the activation of the type I IFN pathway. Although these observations could be explained by increased disease activity, our group (Santer and others 2010) and Lood and others (2009) recently observed that C1q itself plays a direct role in the regulation of IFN-a stimulation by SLE ICs. The mechanisms whereby C1q protects against IFN-a production differed in these 2 studies. Whereas Lood and others reported that IC containing C1q were less stimulatory on isolated pDC, we observed that the presence of C1q in ICs diverted ICs away from pDC and favored binding to monocytes (that do not make IFN-a). Regardless of the mechanism involved, we demonstrated the clinical significance of these observations in that patients with C1q deficiency had increased serum and, in one case, CSF concentrations of IFN-a.
Our observation that C1q deficiency leads to defective suppression of IFN-a in response to nucleoprotein containing ICs raises the question of how IFN-a production is initiated in individuals with low C1q. There is good evidence to suggest that normal individuals are exposed to circulating ICs presumably after minor injury or infection and that one function of natural antibodies is to help facilitate the removal of tissue debris (Elkon and Casali 2008; Madi and others 2009). We propose that in C1q-sufficient individuals, monocytes and macrophages rapidly clear small amounts of ICs containing self-antigens. However, in C1q-deficient patients, these complexes would be less efficiently cleared by monocytes and would be more likely to engage pDCs with resulting induction of IFN-a. Once pDCs release IFN-a, this cytokine not only primes other cells of the immune system, but also renders pDCs more resistant to C1q-mediated inhibition (Santer and others 2010). Combined, these effects lead to a positive feedback loop that sustains the autoimmune response, as also evidenced by the reports of SLE induced by type I IFN therapy discussed above. Whether a similar series of events occurs in patients with antibodies to C1q remains to be determined.
Deficiency of TREX1 and other intracellular nucleases
Mutations of any 1 of 3 genes encoding the intracellular nucleases, TREX1 (the most abundant 3′ to 5′ DNA exonuclease), RNase H2 (digests DNA:RNA hybrids), and SAMHD1 (a putative nuclease), cause the Aicardi Goutieres syndrome (AGS) in children (Crow and others 2006a, 2006b). This syndrome is characterized by mental retardation, cerebral atrophy (white matter destruction), and calcification of the basal ganglia, and is associated with CSF lymphocyte pleocytosis. Elevated levels of IFN-a is seen in the CSF of patients with AGS, and some patients also develop lupus-like features with hypocomplementemia and antinuclear autoantibodies (Ramantani and others 2010). Of note, TREX1 mutations were found in 0.5%–2.7% of SLE patients (Lee-Kirsch and others 2007; Namjou and others 2011). In a recent analysis of over 8,000 multi-ancestral lupus patients (Namjou and others 2011), a TREX1 risk allele was associated with neurologic manifestations, especially seizures in patients of European descent. In addition, a strong association between a TREX1 single-nucleotide polymorphism and anti-nRNP antibodies was seen (Namjou and others 2011).
TREX1 is known to preferentially bind single-stranded DNA, and its homology to other editing enzymes suggested that it plays a role in DNA repair. However, TREX1 is most strongly implicated in interaction with an endoplasmic reticulum-associated DNA repair complex (the SET complex) that is involved in the caspase-independent, granzyme A-mediated form of apoptotic cell death (Chowdhury and others 2006). TREX1-knockout mice develop a lethal inflammatory myositis (Morita and others 2004). This finding was explained in a study by Stetson and others (2008), where TREX1 was shown to function as a negative regulator the cytosolic IFN stimulatory DNA response (Fig. 1). Mice deficient in TREX1 had an increase in IRF3-mediated type I IFN production that caused heart inflammation. TREX1-deficient mice lacking the type I IFN receptor were protected from disease and also did not develop autoantibodies (Stetson and others 2008). These data emphasize the pathogenic contribution of IFN-a in TREX1-mediated systemic autoimmunity.
TRAP deficiency
Although the immuno-osseous dysplasia spondyloenchondrodysplasia (SPENCD) has been regarded primarily as a skeletal dysplasia, SPENCD patients may also show immune dysfunction, with a particular susceptibility to the development of SLE. Two groups (Briggs and others 2011; Lausch and others 2011) have recently confirmed a high frequency of autoimmune phenotypes, including lupus, in SPENCD patients. Both studies identified loss-of-function mutations in the ACP5 gene, encoding TRAP, as causative of the disease. Increased type I IFN activity and the presence of an IFN signature provided a pivotal link between this mutation and lupus—but why?
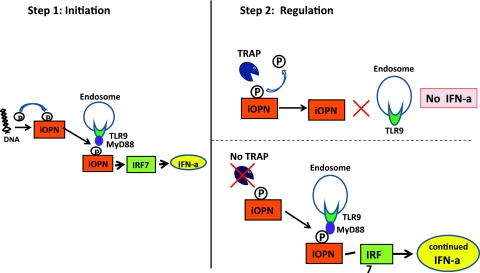
Osteopontin (OPN) is a glycoprotein that exists as both a secreted OPN and intracellular OPN (iOPN) protein. Post-translational modification and proteolytic cleavage indicate that OPN has a diversity of biological functions. In mouse pDC, iOPN has been described to regulate IFN-a secretion via MyD88 and TLR9 pathways. Since OPN is a recognized substrate for osteoclast-derived TRAP in vivo (Andersson and others 2003), it is possible that in the absence of TRAP in pDC, OPN would remain phosphorylated and maintain persistent stimulation of IFN-a through TLR9/MyD88. Lausch and others (2011) studied OPN and demonstrated that compared with controls, SPENCD patients have higher urinary levels of phosphorylated OPN. These results suggest that TRAP is responsible for dephosphorylating OPN and that this function is defective in patients with SPENCD. In summary, extracellular phosphorylated OPN results in osteoclast overactivation and the bony phenotype of SPENCD, whereas increased intracellular phosphorylated OPN enhances IFN-a levels, via TLR9 and MyD88 in pDCs (Fig. 3). Interestingly, variants in the OPN gene have been associated with SLE, and patients who carry risk alleles for this gene have been found to have elevated IFN-a activity (Kariuki and others 2009b).
FIG. 3.
Hypothetical 2-step model depicting how tartrate-resistant acid phosphatase (TRAP) deficiency may regulate IFN-a production in pDCs. In step 1, the presence of cytoplasmic DNA leads to phosphorylation of intracellular osteopontin (iOPN). This in turn facilitates binding to MyD88/TLR9, activation of IRF7, and IFN-a production. In step 2, in normal cells (top right) TRAP dephosphorylates iOPN leading to reduction in IFN-a production (negative regulation). In the absence of TRAP (bottom right), iOPN remains phosphorylated, resulting in persistent activation of the TLR9/MyD88 pathway and continued IFN-a production.
Polygenic Variants in SLE
Candidate gene studies and GWAS in SLE
The importance of the MHC region in SLE susceptibility has long been known. Among class II genes, HLA-DR2, HLA-DR3 (common in Europeans and Asians), and HLA-DR8 (more common in Hispanics) were identified. Important Class III genes include TNF, C2, C4A, and C4B (Deng and Tsao 2010). Since the discovery of the IFN gene signature in SLE, there have been a number of candidate gene studies performed analyzing genes involved in the type I IFN pathway. In addition, there have been 7 GWAS performed on lupus patients (5 European and 2 Asian) who have confirmed the association of many candidate genes and have also identified a number of associations with new genetic variants. These studies are comprehensively reviewed in (Deng and Tsao 2010; Lee and Bae 2010; Sestak and others 2011). Here, we focus on genes and pathways relevant to IFN-a stimulation or signaling.
IRF5
IRF5 is the gene outside the HLA locus that is most strongly and consistently associated with SLE (reviewed in Kyogoku and Tsuchiya 2007). The IRF family is comprised of 9 genes (IRF1-9) that recognize a DNA sequence called the IRF enhancer, which is frequently found in the promoter region upstream of a number of ISGs. IRF5 and IRF7 are activated upon TLR7 and 9 signaling, resulting in type I IFN production (Kyogoku and Tsuchiya 2007). Sigurdsson and others (2005) first observed an association with IRF5 and TYK2 (tyrosine kinase 2) in a linkage study of SLE patients from Sweden, Finland, and Iceland. Since then, IRF5 associations with SLE have been reproduced by many other groups, mostly in European populations, but also to a lesser extent in Hispanic, Asian, and African American patients (Lee and Bae 2010; Sestak and others 2011). To date, 4 functional variants of IRF5 have been identified that define both risk and protective haplotypes in SLE (Graham and others 2006, 2007). In European populations, the variants of IRF5 with the strongest probabilities of being causal in SLE are an SNP (rs10488631) location 3′ of IRF5, and a CGGGG insertion-deletion polymorphism located 64 base pairs upstream of the first untranslated exon of IRF5 (Sigurdsson and others 2008). Several groups reported that SLE patients carrying risk haplotypes for IRF5 have higher serum IFN-a activity (Niewold and others 2007; Rullo and others 2010), and this effect may be most prominent in patients with either anti-RNA binding protein (RBP) or anti-dsDNA antibodies. It should be noted that IRF5 is also implicated in B-lymphocyte activation and production of IL-12 by cells of the innate immune system.
Signal transducer and activator of transcription 4 protein
A strong association between SLE and STAT4, which encodes for STAT4, has been found in both candidate-gene and GWAS studies (Deng and Tsao 2010). The main action of STAT4 is on helper T (Th) cells—it is induced by IL-12, IL-23, and IFN-a, and promotes Th1 as well as Th17 responses (Korman and others 2008). In SLE, the risk variant for STAT4 is associated with both lower serum IFN-a levels and higher IFN-induced gene expression in PBMCs (Kariuki and others 2009a). IRF5 and STAT4 risk alleles confer additive risk for SLE (Sigurdsson and others 2008).
Tyrosine-protein phosphatase nonreceptor type 22
Tyrosine-protein phosphatase nonreceptor type 22 (PTPN22) is a phosphatase that modulates antigen receptor signal transduction in both T and B lymphocytes (Cohen and others 1999). Two functional variants of the PTPN22 gene have been identified—one confers a greater risk of SLE, whereas the other is protective (Kyogoku and others 2004; Orru and others 2009). One study has reported that SLE patients who carry the risk allele for PTPN22 had higher serum IFN-a activity and lower serum TNF levels (Kariuki and others 2008), suggesting that, in addition to effects on lymphocyte signaling, PTPN22 variants may also be involved in type IFN regulation.
IFN: Clinical Aspects
IFN: disease activity and manifestations
Upregulation of ISGs was reported to correlate with disease activity, the incidence of nephritis, and the presence of certain autoantibodies, especially those against RNPs (Baechler and others 2003; Kirou and others 2005). Some longitudinal studies suggest that the IFN signature correlates with disease activity, but there are conflicting results (see below). A pathologic role for IFN-a has also been suggested in neuropsychiatric lupus (NPSLE). Cerebral spinal fluid (CSF) from patients with NPSLE induced higher IFN-a production in in vitro bioassays compared with other disease controls (multiple sclerosis and other autoimmune diseases). The interferogenic activity of the CSF is dependent of the presence of autoantibodies, FcgRII engagement, as well as endosomal transport of predominantly ribonucleoprotein antigens (Santer and others 2009).
An important consideration is what happens to the IFN gene signature with therapy. Glucocorticoids remain one the mainstays of treatment in SLE, and it has been reported that the ISGs tend to normalize in lupus patients treated with corticosteroids (Bennett and others 2003; Guiducci and others 2010), suggesting that they track with disease activity. The primary anti-inflammatory mechanism of glucocorticoids is not through direct down-regulation of IFN-a, but instead is thought to be through nuclear factor kappa B (NF-kB) inhibition (De Bosscher and others 2000). Glucocorticoids have also been shown to induce apoptosis in many cell types, including pDCs—a response that is abrogated in the presence of TLR7 and 9 activation (Boor and others 2006). As has been discussed above, one of the main pathogenic mechanisms of IFN production in SLE is through activation of TLR7 and 9 signaling, which also activates the NF-kB pathway necessary for pDC survival. The effect of glucocoricoids on TLR7 and 9 activation of pDCs and subsecquent IFN-a production was analyzed in a recent study by Guiducci and others (2010). In addition to decreasing the amount of apoptotic cell death, TLR7 and 9 stimulation was shown to restore IFN-a production in pDCs exposed to glucocorticoids. In SLE patients (and 2 lupus-prone mouse strains), the inhibitory effects of glucocorticoids on the IFN pathway were reduced. Further, glucocorticoids did not affect NF-kB activation in TLR7 or 9-stimulated pDCs, thus preventing glucocorticoid-induced pDC death, with the consequence that systemic IFN-a levels were maintained (Guiducci and others 2010). Therapeutic approaches that attenuate either IFN-a production or its downstream effects are currently under investigation.
Therapy
Three different monoclonal antibodies targeting IFN-a, namely, MeDi-545 (sifalimumab), rhuMab iFn-α (rontalizumab), and nnC 0152–0000-0001, are in clinical trials. Results have been published from the first phase 1 study of sifalimumab (Yao and others 2009). Pairwise comparisons of patients treated with sifalimumab or placebo revealed that treatment caused a significant (P<0.05) dose-dependent inhibition of the IFN signature that was observed for the 3, 10, and 30 mg/kg doses of anti-IFN-a antibody. There was a similar trend of reduction in ISGs in the skin lesions of a small number of SLE patients analyzed. No serious adverse events were reported. Preliminary results of the phase 2a trial have been reported in abstract form (Merrill and others 2011). In this 14-week trial, although ISGs declined by an average of 40%, significant changes in disease activity measures were not observed. Longer term studies are eagerly awaited.
ISGs as biomarkers
Because of the reported association with disease activity, there has been interest in evaluating the IFN gene signature or ISG protein products as biomarkers of disease activity in lupus. Although longitudinal data are limited, 2 recent studies found that while a high IFN gene signature is associated with more active disease at a single point in time, its utility as a possible biomarker is limited (Landolt-Marticorena and others 2009; Petri and others 2009). However, different approaches for evaluating the IFN response are still under investigation. For example, Chaussabel and others (2008) have developed a unique method of analyzing blood transcriptional profiles by grouping gene transcripts with common biological functions into modules. They observed that modular profiles can discriminate between several different disease states—healthy, Streptococcus pneumonia infection, melanoma, post-transplant, and lupus. In SLE, the ISG module, among others, correlated with disease activity. Assays for protein products encoded by ISGs have also been used to evaluate disease activity. For example, it was reported that the combined measurement of serum levels of CXCL10 (IFNgamma-inducible 10-kd protein), CCL2 (monocyte chemotactic protein 1), and CCL19 (MIP-3b) was predictive of a lupus flare over the ensuing year (Bauer and others 2009).
Summary and Conclusions
Although long implicated in the pathogenesis of SLE, the pre-eminence of ISGs in microarrays has focused attention on the genesis and pathological role of this cytokine in SLE and related systemic autoimmune disorders. In a remarkably short space of time, this has led to clinical trials that attempt to block the adjuvant and inflammatory properties of this cytokine. Since IFN-a is also thought to be vital to immune defense against viruses, other strategies to interrupt either IFN-a induction or downstream effects will no doubt also be tested.
What is less clear is whether and how IFN-a is generated early in disease—before production of high titer IgG antibodies targeted against nucleoprotein antigens. More exciting discoveries will no doubt follow.
Acknowledgments
The authors acknowledge support from the NIH (RO1 AR48796 and R01 NS065933 to KBE) and NIH (T32AR 007108 to VS). Vivian Stone is also supported by a physician scientist development award from the American College of Rheumatology.
Author Disclosure Statement
No competing financial interests exist.
References
- Andersson G. Ek-Rylander B. Hollberg K. Ljusberg-Sjolander J. Lang P. Norgard M. Wang Y. Zhang SJ. TRACP as an osteopontin phosphatase. J Bone Miner Res. 2003;18(10):1912–1915. doi: 10.1359/jbmr.2003.18.10.1912. [DOI] [PubMed] [Google Scholar]
- Baccala R. Hoebe K. Kono DH. Beutler B. Theofilopoulos AN. TLR-dependent and TLR-independent pathways of type I interferon induction in systemic autoimmunity. Nat Med. 2007;13(5):543–551. doi: 10.1038/nm1590. [DOI] [PubMed] [Google Scholar]
- Baechler EC. Batliwalla FM. Karypis G. Gaffney PM. Ortmann WA. Espe KJ. Shark KB. Grande WJ. Hughes KM. Kapur V. Interferon-inducible gene expression signature in peripheral blood cells of patients with severe lupus. Proc Natl Acad Sci U S A. 2003;100(5):2610–2615. doi: 10.1073/pnas.0337679100. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber GN. Innate immune DNA sensing pathways: STING, AIMII and the regulation of interferon production and inflammatory responses. Curr Opin Immunol. 2011;23(1):10–20. doi: 10.1016/j.coi.2010.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer JW. Petri M. Batliwalla FM. Koeuth T. Wilson J. Slattery C. Panoskaltsis-Mortari A. Gregersen PK. Behrens TW. Baechler EC. Interferon-regulated chemokines as biomarkers of systemic lupus erythematosus disease activity: a validation study. Arthritis Rheum. 2009;60(10):3098–3107. doi: 10.1002/art.24803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett L. Palucka AK. Arce E. Cantrell V. Borvak J. Banchereau J. Pascual V. Interferon and granulopoiesis signatures in systemic lupus erythematosus blood. J Exp Med. 2003;197(6):711–723. doi: 10.1084/jem.20021553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boor PP. Metselaar HJ. Mancham S. Tilanus HW. Kusters JG. Kwekkeboom J. Prednisolone suppresses the function and promotes apoptosis of plasmacytoid dendritic cells. Am J Transplant. 2006;6(10):2332–2341. doi: 10.1111/j.1600-6143.2006.01476.x. [DOI] [PubMed] [Google Scholar]
- Briggs TA. Rice GI. Daly S. Urquhart J. Gornall H. Bader-Meunier B. Baskar K. Baskar S. Baudouin V. Beresford MW. Tartrate-resistant acid phosphatase deficiency causes a bone dysplasia with autoimmunity and a type I interferon expression signature. Nat Genet. 2011;43(2):127–131. doi: 10.1038/ng.748. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkmann V. Reichard U. Goosmann C. Fauler B. Uhlemann Y. Weiss DS. Weinrauch Y. Zychlinsky A. Neutrophil extracellular traps kill bacteria. Science. 2004;303(5663):1532–1535. doi: 10.1126/science.1092385. [DOI] [PubMed] [Google Scholar]
- Cao W. Molecular characterization of human plasmacytoid dendritic cells. J Clin Immunol. 2009;29(3):257–264. doi: 10.1007/s10875-009-9284-x. [DOI] [PubMed] [Google Scholar]
- Carlucci F. Cortes-Hernandez J. Fossati-Jimack L. Bygrave AE. Walport MJ. Vyse TJ. Cook HT. Botto M. Genetic dissection of spontaneous autoimmunity driven by 129-derived chromosome 1 Loci when expressed on C57BL/6 mice. J Immunol. 2007;178(4):2352–2360. doi: 10.4049/jimmunol.178.4.2352. [DOI] [PubMed] [Google Scholar]
- Chaussabel D. Quinn C. Shen J. Patel P. Glaser C. Baldwin N. Stichweh D. Blankenship D. Li L. Munagala I. A modular analysis framework for blood genomics studies: application to systemic lupus erythematosus. Immunity. 2008;29(1):150–164. doi: 10.1016/j.immuni.2008.05.012. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury D. Beresford PJ. Zhu P. Zhang D. Sung JS. Demple B. Perrino FW. Lieberman J. The exonuclease TREX1 is in the SET complex and acts in concert with NM23-H1 to degrade DNA during granzyme A-mediated cell death. Mol Cell. 2006;23(1):133–142. doi: 10.1016/j.molcel.2006.06.005. [DOI] [PubMed] [Google Scholar]
- Cohen S. Dadi H. Shaoul E. Sharfe N. Roifman CM. Cloning and characterization of a lymphoid-specific, inducible human protein tyrosine phosphatase, Lyp. Blood. 1999;93(6):2013–2024. [PubMed] [Google Scholar]
- Courtney PA. Crockard AD. Williamson K. Irvine AE. Kennedy RJ. Bell AL. Increased apoptotic peripheral blood neutrophils in systemic lupus erythematosus: relations with disease activity, antibodies to double stranded DNA, and neutropenia. Ann Rheum Dis. 1999;58(5):309–314. doi: 10.1136/ard.58.5.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crow YJ. Hayward BE. Parmar R. Robins P. Leitch A. Ali M. Black DN. van Bokhoven H. Brunner HG. Hamel BC. Mutations in the gene encoding the 3′-5′ DNA exonuclease TREX1 cause Aicardi-Goutieres syndrome at the AGS1 locus. Nat Genet. 2006a;38(8):917–920. doi: 10.1038/ng1845. others. [DOI] [PubMed] [Google Scholar]
- Crow YJ. Leitch A. Hayward BE. Garner A. Parmar R. Griffith E. Ali M. Semple C. Aicardi J. Babul-Hirji R. Mutations in genes encoding ribonuclease H2 subunits cause Aicardi-Goutieres syndrome and mimic congenital viral brain infection. Nat Genet. 2006b;38(8):910–916. doi: 10.1038/ng1842. others. [DOI] [PubMed] [Google Scholar]
- De Bosscher K. Vanden Berghe W. Haegeman G. Mechanisms of anti-inflammatory action and of immunosuppression by glucocorticoids: negative interference of activated glucocorticoid receptor with transcription factors. J Neuroimmunol. 2000;109(1):16–22. doi: 10.1016/s0165-5728(00)00297-6. [DOI] [PubMed] [Google Scholar]
- Deng Y. Tsao BP. Genetic susceptibility to systemic lupus erythematosus in the genomic era. Nat Rev Rheumatol. 2010;6(12):683–692. doi: 10.1038/nrrheum.2010.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkon K. Casali P. Nature and functions of autoantibodies. Nat Clin Pract Rheumatol. 2008;4(9):491–498. doi: 10.1038/ncprheum0895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eloranta ML. Lovgren T. Finke D. Mathsson L. Ronnelid J. Kastner B. Alm GV. Ronnblom L. Regulation of the interferon-alpha production induced by RNA-containing immune complexes in plasmacytoid dendritic cells. Arthritis Rheum. 2009;60(8):2418–2427. doi: 10.1002/art.24686. [DOI] [PubMed] [Google Scholar]
- Fitzgerald-Bocarsly P. Dai J. Singh S. Plasmacytoid dendritic cells and type I IFN: 50 years of convergent history. Cytokine Growth Factor Rev. 2008;19(1):3–19. doi: 10.1016/j.cytogfr.2007.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser DA. Laust AK. Nelson EL. Tenner AJ. C1q differentially modulates phagocytosis and cytokine responses during ingestion of apoptotic cells by human monocytes, macrophages, and dendritic cells. J Immunol. 2009;183(10):6175–6185. doi: 10.4049/jimmunol.0902232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Romo GS. Caielli S. Vega B. Connolly J. Allantaz F. Xu Z. Punaro M. Baisch J. Guiducci C. Coffman RL. Netting neutrophils are major inducers of type I IFN production in pediatric systemic lupus erythematosus. Sci Transl Med. 2011;3(73):73ra20. doi: 10.1126/scitranslmed.3001201. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham RR. Kozyrev SV. Baechler EC. Reddy MV. Plenge RM. Bauer JW. Ortmann WA. Koeuth T. Gonzalez Escribano MF. Pons-Estel B. A common haplotype of interferon regulatory factor 5 (IRF5) regulates splicing and expression and is associated with increased risk of systemic lupus erythematosus. Nat Genet. 2006;38(5):550–555. doi: 10.1038/ng1782. others. [DOI] [PubMed] [Google Scholar]
- Graham RR. Kyogoku C. Sigurdsson S. Vlasova IA. Davies LR. Baechler EC. Plenge RM. Koeuth T. Ortmann WA. Hom G. Three functional variants of IFN regulatory factor 5 (IRF5) define risk and protective haplotypes for human lupus. Proc Natl Acad Sci U S A. 2007;104(16):6758–6763. doi: 10.1073/pnas.0701266104. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guiducci C. Gong M. Xu Z. Gill M. Chaussabel D. Meeker T. Chan JH. Wright T. Punaro M. Bolland S. TLR recognition of self nucleic acids hampers glucocorticoid activity in lupus. Nature. 2010;465(7300):937–941. doi: 10.1038/nature09102. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagberg N. Berggren O. Leonard D. Weber G. Bryceson YT. Alm GV. Eloranta ML. Ronnblom L. IFN-{alpha} production by plasmacytoid dendritic cells stimulated with RNA-containing immune complexes is promoted by NK cells via MIP-1{beta} and LFA-1. J Immunol. 2011;186(9):5085–5094. doi: 10.4049/jimmunol.1003349. [DOI] [PubMed] [Google Scholar]
- Harley IT. Kaufman KM. Langefeld CD. Harley JB. Kelly JA. Genetic susceptibility to SLE: new insights from fine mapping and genome-wide association studies. Nat Rev Genet. 2009;10(5):285–290. doi: 10.1038/nrg2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooks JJ. Moutsopoulos HM. Geis SA. Stahl NI. Decker JL. Notkins AL. Immune interferon in the circulation of patients with autoimmune disease. N Engl J Med. 1979;301(1):5–8. doi: 10.1056/NEJM197907053010102. [DOI] [PubMed] [Google Scholar]
- Kariuki SN. Crow MK. Niewold TB. The PTPN22 C1858T polymorphism is associated with skewing of cytokine profiles toward high interferon-alpha activity and low tumor necrosis factor alpha levels in patients with lupus. Arthritis Rheum. 2008;58(9):2818–2823. doi: 10.1002/art.23728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kariuki SN. Kirou KA. MacDermott EJ. Barillas-Arias L. Crow MK. Niewold TB. Cutting edge: autoimmune disease risk variant of STAT4 confers increased sensitivity to IFN-alpha in lupus patients in vivo. J Immunol. 2009a;182(1):34–38. doi: 10.4049/jimmunol.182.1.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kariuki SN. Moore JG. Kirou KA. Crow MK. Utset TO. Niewold TB. Age- and gender-specific modulation of serum osteopontin and interferon-alpha by osteopontin genotype in systemic lupus erythematosus. Genes Immun. 2009b;10(5):487–494. doi: 10.1038/gene.2009.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirou KA. Lee C. George S. Louca K. Peterson MG. Crow MK. Activation of the interferon-alpha pathway identifies a subgroup of systemic lupus erythematosus patients with distinct serologic features and active disease. Arthritis Rheum. 2005;52(5):1491–1503. doi: 10.1002/art.21031. [DOI] [PubMed] [Google Scholar]
- Korman BD. Kastner DL. Gregersen PK. Remmers EF. STAT4: genetics, mechanisms, and implications for autoimmunity. Curr Allergy Asthma Rep. 2008;8(5):398–403. doi: 10.1007/s11882-008-0077-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyogoku C. Langefeld CD. Ortmann WA. Lee A. Selby S. Carlton VE. Chang M. Ramos P. Baechler EC. Batliwalla FM. Genetic association of the R620W polymorphism of protein tyrosine phosphatase PTPN22 with human SLE. Am J Hum Genet. 2004;75(3):504–507. doi: 10.1086/423790. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyogoku C. Tsuchiya N. A compass that points to lupus: genetic studies on type I interferon pathway. Genes Immun. 2007;8(6):445–455. doi: 10.1038/sj.gene.6364409. [DOI] [PubMed] [Google Scholar]
- Lande R. Ganguly D. Facchinetti V. Frasca L. Conrad C. Gregorio J. Meller S. Chamilos G. Sebasigari R. Riccieri V. Neutrophils activate plasmacytoid dendritic cells by releasing self-DNA-peptide complexes in systemic lupus erythematosus. Sci Transl Med. 2011;3(73):73ra19. doi: 10.1126/scitranslmed.3001180. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landolt-Marticorena C. Bonventi G. Lubovich A. Ferguson C. Unnithan T. Su J. Gladman DD. Urowitz M. Fortin PR. Wither J. Lack of association between the interferon-alpha signature and longitudinal changes in disease activity in systemic lupus erythematosus. Ann Rheum Dis. 2009;68(9):1440–1446. doi: 10.1136/ard.2008.093146. [DOI] [PubMed] [Google Scholar]
- Lausch E. Janecke A. Bros M. Trojandt S. Alanay Y. De Laet C. Hubner CA. Meinecke P. Nishimura G. Matsuo M. Genetic deficiency of tartrate-resistant acid phosphatase associated with skeletal dysplasia, cerebral calcifications and autoimmunity. Nat Genet. 2011;43(2):132–137. doi: 10.1038/ng.749. others. [DOI] [PubMed] [Google Scholar]
- Lee-Kirsch MA. Gong M. Chowdhury D. Senenko L. Engel K. Lee YA. de Silva U. Bailey SL. Witte T. Vyse TJ. Mutations in the gene encoding the 3′-5′ DNA exonuclease TREX1 are associated with systemic lupus erythematosus. Nat Genet. 2007;39(9):1065–1067. doi: 10.1038/ng2091. others. [DOI] [PubMed] [Google Scholar]
- Lee HS. Bae SC. What can we learn from genetic studies of systemic lupus erythematosus? Implications of genetic heterogeneity among populations in SLE. Lupus. 2010;19(12):1452–1459. doi: 10.1177/0961203310370350. [DOI] [PubMed] [Google Scholar]
- Lood C. Gullstrand B. Truedsson L. Olin AI. Alm GV. Ronnblom L. Sturfelt G. Eloranta ML. Bengtsson AA. C1q inhibits immune complex-induced interferon-alpha production in plasmacytoid dendritic cells: a novel link between C1q deficiency and systemic lupus erythematosus pathogenesis. Arthritis Rheum. 2009;60(10):3081–3090. doi: 10.1002/art.24852. [DOI] [PubMed] [Google Scholar]
- Madi A. Hecht I. Bransburg-Zabary S. Merbl Y. Pick A. Zucker-Toledano M. Quintana FJ. Tauber AI. Cohen IR. Ben-Jacob E. Organization of the autoantibody repertoire in healthy newborns and adults revealed by system level informatics of antigen microarray data. Proc Natl Acad Sci U S A. 2009;106(34):14484–14489. doi: 10.1073/pnas.0901528106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin DA. Elkon KB. Autoantibodies make a U-turn: the toll hypothesis for autoantibody specificity. J Exp Med. 2005;202(11):1465–1469. doi: 10.1084/jem.20052228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinelli S. Urosevic M. Daryadel A. Oberholzer PA. Baumann C. Fey MF. Dummer R. Simon HU. Yousefi S. Induction of genes mediating interferon-dependent extracellular trap formation during neutrophil differentiation. J Biol Chem. 2004;279(42):44123–44132. doi: 10.1074/jbc.M405883200. [DOI] [PubMed] [Google Scholar]
- Merrill J. Chindalore V. Box J. Rothfield N. Fiechtner J. Sun J. Ethgen D. Results of a randomized, placebo-controlled, phase 2a study of sifalimumab, an anti-interferon-alpha monoclonal antibody, administered subcutaneously in subjects with systemic lupus erythematosus. EULAR. 2011. www.abstracts2view.com/eular/view.php?nu=EULAR11L_THU0411&terms= www.abstracts2view.com/eular/view.php?nu=EULAR11L_THU0411&terms=
- Morita M. Stamp G. Robins P. Dulic A. Rosewell I. Hrivnak G. Daly G. Lindahl T. Barnes DE. Gene-targeted mice lacking the Trex1 (DNase III) 3′→5′ DNA exonuclease develop inflammatory myocarditis. Mol Cell Biol. 2004;24(15):6719–6727. doi: 10.1128/MCB.24.15.6719-6727.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz LE. Janko C. Schulze C. Schorn C. Sarter K. Schett G. Herrmann M. Autoimmunity and chronic inflammation - two clearance-related steps in the etiopathogenesis of SLE. Autoimmun Rev. 2010;10(1):38–42. doi: 10.1016/j.autrev.2010.08.015. [DOI] [PubMed] [Google Scholar]
- Namjou B. Kothari PH. Kelly JA. Glenn SB. Ojwang JO. Adler A. Alarcon-Riquelme ME. Gallant CJ. Boackle SA. Criswell LA. Evaluation of the TREX1 gene in a large multi-ancestral lupus cohort. Genes Immun. 2011;12:270–279. doi: 10.1038/gene.2010.73. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navratil JS. Liu CC. Ahearn JM. Apoptosis and autoimmunity. Immunol Res. 2006;36(1–3):3–12. doi: 10.1385/IR:36:1:3. [DOI] [PubMed] [Google Scholar]
- Niewold TB. Hua J. Lehman TJ. Harley JB. Crow MK. High serum IFN-alpha activity is a heritable risk factor for systemic lupus erythematosus. Genes Immun. 2007;8(6):492–502. doi: 10.1038/sj.gene.6364408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orru V. Tsai SJ. Rueda B. Fiorillo E. Stanford SM. Dasgupta J. Hartiala J. Zhao L. Ortego-Centeno N. D'Alfonso S. A loss-of-function variant of PTPN22 is associated with reduced risk of systemic lupus erythematosus. Hum Mol Genet. 2009;18(3):569–579. doi: 10.1093/hmg/ddn363. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petri M. Singh S. Tesfasyone H. Dedrick R. Fry K. Lal P. Williams G. Bauer J. Gregersen P. Behrens T. Longitudinal expression of type I interferon responsive genes in systemic lupus erythematosus. Lupus. 2009;18(11):980–989. doi: 10.1177/0961203309105529. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickering MC. Botto M. Taylor PR. Lachmann PJ. Walport MJ. Systemic lupus erythematosus, complement deficiency, and apoptosis. Adv Immunol. 2000;76:227–324. doi: 10.1016/s0065-2776(01)76021-x. [DOI] [PubMed] [Google Scholar]
- Rahman A. Isenberg DA. Systemic lupus erythematosus. N Engl J Med. 2008;358(9):929–939. doi: 10.1056/NEJMra071297. [DOI] [PubMed] [Google Scholar]
- Ramantani G. Kohlhase J. Hertzberg C. Innes AM. Engel K. Hunger S. Borozdin W. Mah JK. Ungerath K. Walkenhorst H. Expanding the phenotypic spectrum of lupus erythematosus in Aicardi-Goutieres syndrome. Arthritis Rheum. 2010;62(5):1469–1477. doi: 10.1002/art.27367. others. [DOI] [PubMed] [Google Scholar]
- Ronnblom L. Eloranta ML. Alm GV. The type I interferon system in systemic lupus erythematosus. Arthritis Rheum. 2006;54(2):408–420. doi: 10.1002/art.21571. [DOI] [PubMed] [Google Scholar]
- Ronnblom LE. Alm GV. Oberg KE. Autoimmunity after alpha-interferon therapy for malignant carcinoid tumors. Ann Intern Med. 1991;115(3):178–183. doi: 10.7326/0003-4819-115-3-178. [DOI] [PubMed] [Google Scholar]
- Rullo OJ. Woo JM. Wu H. Hoftman AD. Maranian P. Brahn BA. McCurdy D. Cantor RM. Tsao BP. Association of IRF5 polymorphisms with activation of the interferon alpha pathway. Ann Rheum Dis. 2010;69(3):611–617. doi: 10.1136/ard.2009.118315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez Roman J. Castillo Palma MJ. Garcia Diaz E. Ferrer Ordinez JA. Systemic lupus erythematosus induced by recombinant alpha interferon treatment. Med Clin. 1994;102(5):198. [PubMed] [Google Scholar]
- Santer DM. Hall BE. George TC. Tangsombatvisit S. Liu CL. Arkwright PD. Elkon KB. C1q deficiency leads to the defective suppression of IFN-alpha in response to nucleoprotein containing immune complexes. J Immunol. 2010;185(8):4738–4749. doi: 10.4049/jimmunol.1001731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santer DM. Yoshio T. Minota S. Moller T. Elkon KB. Potent induction of IFN-alpha and chemokines by autoantibodies in the cerebrospinal fluid of patients with neuropsychiatric lupus. J Immunol. 2009;182(2):1192–1201. doi: 10.4049/jimmunol.182.2.1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarzi-Puttini P. Atzeni F. Iaccarino L. Doria A. Environment and systemic lupus erythematosus: an overview. Autoimmunity. 2005;38(7):465–472. doi: 10.1080/08916930500285394. [DOI] [PubMed] [Google Scholar]
- Sestak AL. Furnrohr BG. Harley JB. Merrill JT. Namjou B. The genetics of systemic lupus erythematosus and implications for targeted therapy. Ann Rheum Dis. 2011;70(Suppl. 1):i37–i43. doi: 10.1136/ard.2010.138057. [DOI] [PubMed] [Google Scholar]
- Sigurdsson S. Goring HH. Kristjansdottir G. Milani L. Nordmark G. Sandling JK. Eloranta ML. Feng D. Sangster-Guity N. Gunnarsson I. Comprehensive evaluation of the genetic variants of interferon regulatory factor 5 (IRF5) reveals a novel 5 bp length polymorphism as strong risk factor for systemic lupus erythematosus. Hum Mol Genet. 2008;17(6):872–881. doi: 10.1093/hmg/ddm359. others. [DOI] [PubMed] [Google Scholar]
- Sigurdsson S. Nordmark G. Goring HH. Lindroos K. Wiman AC. Sturfelt G. Jonsen A. Rantapaa-Dahlqvist S. Moller B. Kere J. Polymorphisms in the tyrosine kinase 2 and interferon regulatory factor 5 genes are associated with systemic lupus erythematosus. Am J Hum Genet. 2005;76(3):528–537. doi: 10.1086/428480. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stetson DB. Ko JS. Heidmann T. Medzhitov R. Trex1 prevents cell-intrinsic initiation of autoimmunity. Cell. 2008;134(4):587–598. doi: 10.1016/j.cell.2008.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiecki M. Colonna M. Unraveling the functions of plasmacytoid dendritic cells during viral infections, autoimmunity, and tolerance. Immunol Rev. 2010;234(1):142–162. doi: 10.1111/j.0105-2896.2009.00881.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan EM. Antinuclear antibodies: diagnostic markers for autoimmune diseases and probes for cell biology. Adv Immunol. 1989;44:93–151. doi: 10.1016/s0065-2776(08)60641-0. [DOI] [PubMed] [Google Scholar]
- Theofilopoulos AN. Baccala R. Beutler B. Kono DH. Type I interferons (alpha/beta) in immunity and autoimmunity. Annu Rev Immunol. 2005;23:307–336. doi: 10.1146/annurev.immunol.23.021704.115843. [DOI] [PubMed] [Google Scholar]
- Trendelenburg M. Antibodies against C1q in patients with systemic lupus erythematosus. Springer Semin Immunopathol. 2005;27(3):276–285. doi: 10.1007/s00281-005-0007-y. [DOI] [PubMed] [Google Scholar]
- Uze G. Schreiber G. Piehler J. Pellegrini S. The receptor of the type I interferon family. Curr Top Microbiol Immunol. 2007;316:71–95. doi: 10.1007/978-3-540-71329-6_5. [DOI] [PubMed] [Google Scholar]
- Vollmer J. Tluk S. Schmitz C. Hamm S. Jurk M. Forsbach A. Akira S. Kelly KM. Reeves WH. Bauer S. Immune stimulation mediated by autoantigen binding sites within small nuclear RNAs involves Toll-like receptors 7 and 8. J Exp Med. 2005;202(11):1575–1585. doi: 10.1084/jem.20051696. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Y. Richman L. Higgs BW. Morehouse CA. de los Reyes M. Brohawn P. Zhang J. White B. Coyle AJ. Kiener PA. Neutralization of interferon-alpha/beta-inducible genes and downstream effect in a phase I trial of an anti-interferon-alpha monoclonal antibody in systemic lupus erythematosus. Arthritis Rheum. 2009;60(6):1785–1796. doi: 10.1002/art.24557. others. [DOI] [PubMed] [Google Scholar]
- Ytterberg SR. Schnitzer TJ. Serum interferon levels in patients with systemic lupus erythematosus. Arthritis Rheum. 1982;25(4):401–406. doi: 10.1002/art.1780250407. [DOI] [PubMed] [Google Scholar]