Summary
A human genome-wide linkage scan for obesity identified a linkage peak on chromosome 5q13–15. Positional cloning revealed an association of a rare haplotype to high body-mass index (BMI) in males but not females. The risk locus contains a single gene, “arrestin domain containing 3” (ARRDC3), an uncharacterized α-arrestin. Inactivating Arrdc3 in mice led to a striking resistance to obesity, with greater impact on male mice. Mice with decreased ARRDC3 levels were protected from obesity due to increased energy expenditure through increased activity levels and increased thermogenesis of both brown and white adipose tissues. ARRDC3 interacted directly with β-adrenergic receptors, and loss of ARRDC3 increased the response to β-adrenergic stimulation in isolated adipose tissue. These results demonstrate that ARRDC3 is a gender-sensitive regulator of obesity and energy expenditure and reveal a surprising diversity for arrestin family protein functions.
Introduction
There is a significant genetic component to obesity, with heritability estimated at 0.5–0.8 in twin studies (Stunkard et al., 1986). However, relatively few common genetic variants have been linked to obesity by genome-wide association studies and much of the genetic basis of obesity remains unexplained. (Walley et al., 2009; O’Rahilly, 2009).
The β-arrestins, prototypical regulators of cell receptor signaling and turnover, also have physiological roles in metabolism. In particular, β-arrestin 2 regulates insulin receptor signaling and may play a central role in diabetes (Luan et al., 2009). The β- and visual arrestins are members of a larger and more ancient family of arrestin-fold proteins that share structural similarity (Shi et al., 2006; Lin et al., 2008). A sub-family of these arrestin-fold proteins has been termed the “α-arrestins” (Alvarez, 2008), although as the structures of these proteins are not known, definitive classification is lacking (Aubry et al., 2009).
Mice and humans have at least five α-arrestins: ARRDC1–4 and TXNIP. Thioredoxin-interacting protein (TXNIP), first identified as an inhibitor of thioredoxin, is known to regulate metabolism and insulin responsiveness by inhibiting glucose uptake into muscle and by shifting glucose usage from glycolytic to aerobic pathways (Parikh et al., 2007; Chen et al., 2008; Chutkow et al., 2010). However, TXNIP inhibits glucose uptake independent of its interaction with thioredoxin, and this is a conserved function of at least one other α-arrestin (Patwari et al., 2009). The α-arrestins may therefore have conserved roles in metabolism, but their functions remain unclear.
We report here a male-specific association of ARRDC3 with high body mass index in humans and show that Arrdc3 deficiency in mice protects against obesity. These results reveal the α-arrestins as a family of proteins with roles in regulation of metabolism and the development of obesity.
Results
Genome-wide linkage and locus-wide association to male overweight/obesity at 5q13–15
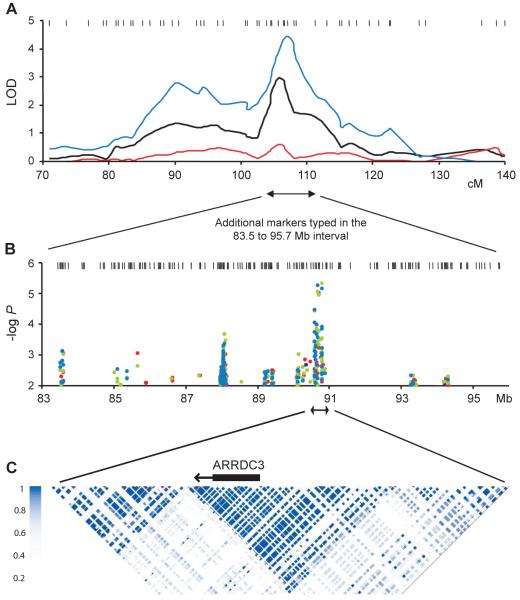
A genome-wide linkage scan (GWLS) at an average inter-marker distance (marker density) of 4cM, was performed on an Icelandic population as described in detail in the Supplemental Results and Methods, yielding two loci of interest in the GWLS for male obesity. In brief, individuals who exceeded a BMI of 27 were classified as “affecteds” in the linkage analysis and clustered into families with a genealogical database covering the entire nation, with the criterion that each affected was related to at least one other affected by six or fewer meiotic events. We report the results of our non-parametric affected-only linkage analysis, together with gender-specific analyses for dichotomous BMI (i.e. inclusive phenotypic groups of BMI ≥ 27, BMI ≥ 30, BMI ≥ 31…BMI ≥ 40) presenting a propensity for overweight, mild obesity, severe obesity and morbid obesity, in a Caucasian population of 3893 males and 4445 females. Top linkage scores were observed for BMI> 33, in male affecteds only, at 5q13–15 with distinct doublet peaks at 89 cM and 107 cM, and a second locus was identified on 12q24 at 134 cM (Suppl. Figure S1A).
Multiple investigators have previously reported linkages to obesity and/or type II diabetes overlapping 5q12-15 (Hager et al., 1998; Bell et al., 2004; Cheng et al., 2009). A noteworthy candidate gene, PCSK1, is present at 5q14 (Kilpeläinen et al., 2009), but the sequence variation responsible for the linkage at this locus has not been definitively identified. Further support for the linkage of the doublet peaks at 5q13–q15 in humans came from comparative mapping of major mouse quantitative trait loci (QTL), which revealed a close synteny between the human linkage peak and a mouse doublet QTL in chromosome 13 (Suppl. Figure S2B–D). This mouse QTL was observed for multiple traits including fat mass and plasma levels of leptin and insulin. We therefore genotyped an additional 27 markers in the interval between 79 and 122 cM at 5q, bringing the information content in the peak region up to 95%. This increased the peak LOD score at marker D5S618 in the male-only analysis to 4.44 (Figure 1A).
Figure 1.
Positional cloning identifies a linkage between obesity and ARRDC3 in males. (A) Linkage region on chromosome 5 showing strength of association to severe obesity (BMI > 33) in males (blue line), females (red line), and all subjects (black line). The male-specific locus on 5q13–q15 (LOD 4.6) was identified using 51 pedigrees containing 124 affected males and 257 relatives. The female-only scan included 97 families with 245 affected females and 612 relatives, and the unrestricted analysis (all subjects) was based on 168 families including 520 affecteds and 883 relatives. (B) Strength of association after additional marker genotyping in the 2-LOD drop for extremely obese males compared to population controls. Results are shown for single-marker association (black dots) and two-, three- and four-marker haplotypes (blue, green, and red dots respectively). (C) The genomic region of the peak haplotype association contains a single RefSeq gene, ARRDC3, in a discrete block of linkage disequilibrium. See also Figure S1.
Given the strength of the LOD score at 5q14–q15 and the syntenic match of the doublet peaks in mice and man, we carried out a locus-wide association study across a 13 Mb region (83–96 Mb in NCBI Build 34) that corresponds roughly to a drop of 2 in LOD score under the larger peak at 5q14–15. In addition to the seven markers in this region used for the linkage analysis, 171 microsatellite markers were genotyped, reaching an average intermarker density of 78 kb. This set of markers was genotyped in an independent group of 435 severely obese males (BMI ≥ 35) and 586 control subjects, all from the same population that was used for the GWLS. In addition to testing the alleles of each marker individually, we also tested haplotypes based on these markers for association to male obesity. The distribution of the markers across the locus and the result of the association analysis are shown in Figure 1B. The resulting at-risk haplotype (0 DG5S1387; 2 DG5S740; 0 DG5S1851; 4 DG5S744) was found at a frequency of 5.6% in affected subjects and 1.5% in controls (P = 0.0000056; adjusted P = 0.042; odds ratio = 3.8), and spanned the region 90.7 to 90.92 Mb. This region coincides with a single RefSeq gene in a discrete linkage disequilibrium block, arrestin domain containing 3 (ARRDC3), located at 90.76 Mb (Figure 1C).
Further analysis of the linkage conditional on the identified at-risk haplotype revealed that noncarriers of the rare at-risk haplotype contribute the majority of the linkage signal at 5q13–15. The inability to reveal a strong contribution of the mapped risk locus to the linkage for obesity is nevertheless consistent with the results for other gene linkages to common diseases identified by positional cloning. For example, the well-confirmed association of TCF7L2 alleles to type 2 diabetes showed no contribution of the discovered at-risk alleles to the corresponding LOD score (Grant et al., 2006). In light of these results, three possibilities remain to explain the linkage signal to male obesity at 5q13–15: the at-risk haplotype is correlated with an unidentified causal variant with substantially higher risk, other at-risk haplotypes across ARRDC3 exist, or there are other genes in the region harboring risk to obesity in males.
In summary, we describe the clean identification of a single gene in the linkage disequilibrium region; however, given a lack of common SNPs that correlated with the relatively rare at-risk microsatellite haplotype, including expression (e) SNPs, and the inability to detect a strong contribution of the at-risk haplotype to the linkage signal, the human linkage analysis suggested an association between ARRDC3 and male obesity but did not fully prove a causal association. We therefore proceeded to test for a causal connection of ARRDC3 to obesity with gene expression studies in humans and knockout studies in mice.
ARRDC3 expression in human omental adipose tissue correlates with BMI in males
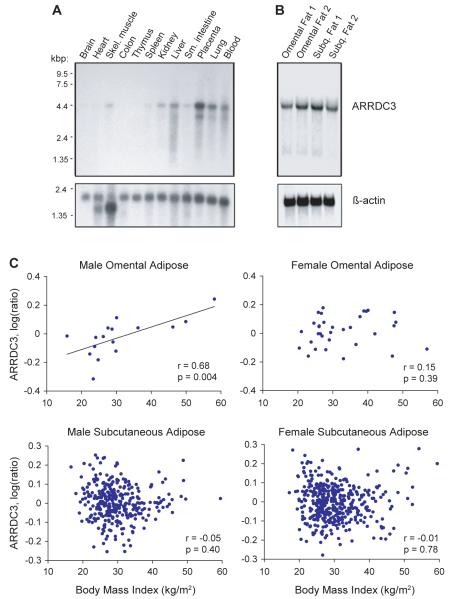
To define the distribution of ARRDC3 in human tissues, we performed northern analysis. A single transcript of ~4.4 kb was detected in placenta, skeletal muscle, spleen, lungs, and liver; ARRDC3 transcripts were not detected in human brain (Figure 2A). These results are consistent with a previous report describing Arrdc3 expression in mice, which also identified prominent expression in skeletal muscle, placenta, and kidney but relatively low expression in brain and spinal cord (Oka et al., 2006). The low neural expression of ARRDC3 was notable since few validated genes linked to obesity have been identified with prominent roles in peripheral tissues, especially adipose tissue (O’Rahilly, 2009; Thorleifsson et al., 2009). We therefore performed additional northern analysis in human adipose samples and detected expression of ARRDC3 in both omental and subcutaneous adipose (Figure 2B).
Figure 2.
Analysis of ARRDC3 gene expression. (A) Northern analysis of ARRDC3 expression in multiple human tissues. (B) Northern analysis of human omental fat and subcutaneous fat from two donors. (C) Correlation of body mass index with ARRDC3 expression levels in omental and subcutaneous adipose tissue measured by microarray analysis (for omental adipose, n = 16 males and 34 females; for subcutaneous adipose, n = 325 males and 417 females). See also Figure S2.
Given the robust expression of ARRDC3 in adipose tissues, we next tested whether ARRDC3 expression in adipose tissue samples were associated with BMI. Omental fat tissue biopsies were collected from 50 volunteers who were either obese individuals undergoing gastric bypass surgery or normal weight individuals undergoing elective abdominal surgery. RNA was isolated from the tissues, and microarray analyses were performed. Interestingly, ARRDC3 expression in omental adipose was significantly correlated with BMI in males, whereas there was no significant correlation in females (Figure 2C). The correlation of BMI with ARRDC3 expression in males remained statistically significant (P < 0.05) after removing the individual contributing most to the correlation. In the less metabolically active subcutaneous adipose tissue, ARRDC3 expression was not correlated with BMI (Figure 2C). The association of ARRDC3 expression and BMI in males, together with the male-specific linkage data, further suggest but do not establish ARRDC3 as the gene underlying obesity at this locus.
To define potential roles for ARRDC3 that might promote obesity, we explored the expression of Arrdc3 in adipose tissue in mice. We found no differences in Arrdc3 expression in isolated adipocytes compared to the stromal-vascular compartments in either subcutaneous or visceral fat depots (Suppl. Fig. S2A). It has been reported that Arrdc3 expression increased in human adipocytes relative to pre-adipocytes (Oka et al., 2006), so we examined ARRDC3 protein levels during differentiation of 3T3-L1 pre-adipocytes. On stimulation with differentiation medium, we observed a rapid but transient increase in Arrdc3 mRNA (data not shown). We then analyzed cell lysates by Western blotting with a new monoclonal antibody to ARRDC3 that does not cross-react with TXNIP or ARRDC4, detects an epitope in the C-terminal tail of ARRDC3, and detects endogenous ARRDC3 in 3T3-L1 cell lysates as a doublet band (Suppl. Fig. S2B-D). Western analysis confirmed the transient increase and demonstrated correspondence of Arrdc3 mRNA to ARRDC3 protein levels during differentiation (Suppl. Fig. S2E). However, ARRDC3 does not appear to directly promote adipogenesis, as overexpression of ARRDC3 inhibited differentiation of 3T3-L1s (Suppl. Fig. S2F).
ARRDC3 is induced by fasting in humans and mice
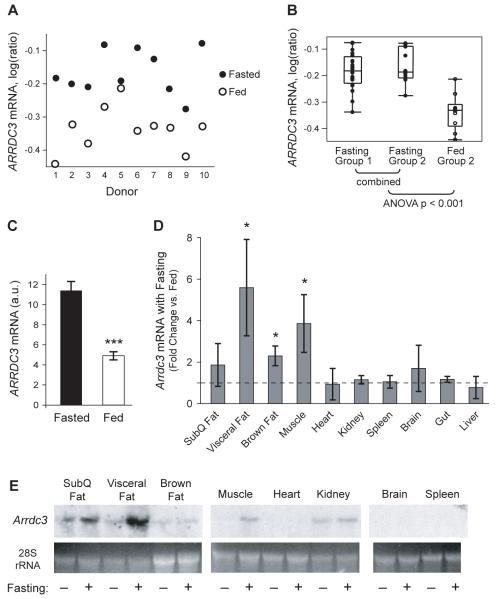
The fed–fasting transition plays an important role in regulating metabolic activity (Muoio and Newgard, 2006). We therefore examined whether ARRDC3 is implicated in the fed or fasting gene-expression signature in human adipose tissue. Microarray analysis of subcutaneous fat biopsy tissues from fasted and fed volunteers identified 402 genes differentially regulated between the fasted and fed state (P < 0.01 as the threshold for significance) (Suppl. Figure S3A). ARRDC3 was among the 10 most responsive genes to food intake, with ARRDC3 transcript levels higher during fasting (Figure 3A–B). This finding was technically confirmed by real-time PCR analysis for ARRDC3 (2.3-fold lower with feeding, P < 0.001) (Figure 3C). Of note, TXNIP was another responder gene (P = 0.0007) with higher expression during fasting (Suppl. Figure S3B). Given the similar responses to feeding, we tested whether expressions of ARRDC3 and TXNIP were correlated in the larger sample of subcutaneous adipose tissues described previously (Emilsson et al., 2008) and found a significant positive correlation (r = 0.44, P < 0.001) (Suppl. Figure S3C). These data support the notion that ARRDC3 is a fasting-induced gene involved in regulating metabolism and that these functions may be shared with other human α-arrestins such as TXNIP.
Figure 3.
ARRDC3 is upregulated by fasting in humans and mice. (A) ARRDC3 expression in human subcutaneous adipose tissue biopsies from a microarray analysis. A randomized cross-over design was used to obtain paired fed and fasted data for each donor (P < 0.001; n = 10). (B) The data in panel A were combined with results of microarray analysis from a second group of 10 donors who underwent fasting on both occasions (combined P < 0.001; n = 20 fasted and 10 fed). (C) Confirmation of the effect of feeding on Arrdc3 expression by real-time PCR (***P < 0.001; n = 10/group). (D) Arrdc3 expression in multiple mouse tissues in response to a 14-hour fast (relative to mice fed ad libitum) (*P < 0.05; n = 3–4/group). (E) Northern analysis of tissues from fed and 14-hour fasting mice. Muscle, heart, kidney, brain, and spleen represent discontiguous lanes from a single blot. Error bars represent mean ± SEM. See also Figure S3.
To determine whether the response to fasting is conserved across mammals, we measured Arrdc3 expression levels in multiple tissues of mice fed ad libitum or fasted 14 h. Real-time PCR analysis showed that fasting induced Arrdc3 expression most prominently in visceral (epididymal) fat (5.6-fold), with significant induction in brown fat (2.3-fold) and skeletal muscle (3.9-fold) as well (Figure 3D). We then performed northern analysis and confirmed a striking induction of Arrdc3 by fasting in visceral fat and skeletal muscle (Figure 3E).
Generation of Arrdc3-deficient mice
Arrdc3-deficient mice were generated from a BayGenomics gene-trap clone containing an insertional mutation between exons 1 and 2 of Arrdc3 (Suppl. Figure S4A). Intercrossing heterozygous mice yielded homozygous offspring at the expected Mendelian ratio; however, there was significant perinatal mortality in homozygous pups. Of 631 mice surviving to weaning, 187 were wild-type (30%), 399 (63%) were heterozygotes, and 45 (7.1%) were homozygotes. Of the 45 homozygotes, 19 (42%) were male and 26 (58%) were female (P = 0.29 for difference from 50%). Transmission of the insertional mutation in Arrdc3 was confirmed by Southern analysis of genomic DNA digested with two different restriction enzymes (Suppl. Figure S4B); the loss of Arrdc3 expression was documented by northern analysis (Suppl. Figure S4C).
Arrdc3-deficient mice are resistant to age-induced obesity
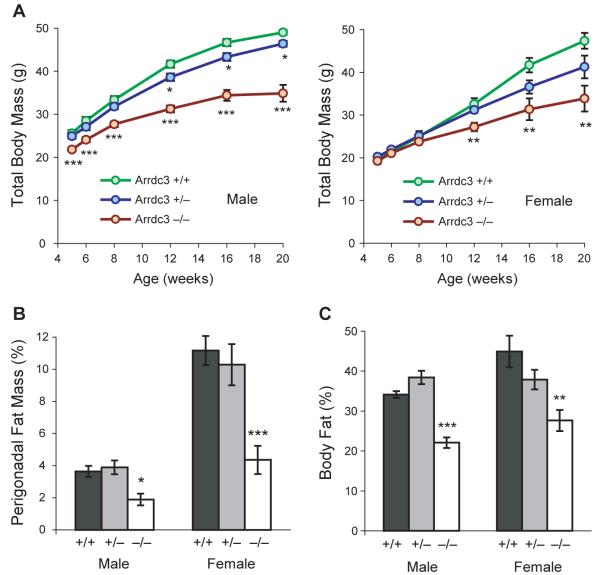
We tested whether Arrdc3-deficient male and female mice are resistant to obesity with age compared to wild-type littermate controls. C57BL/6 mice are prone to increased adiposity and insulin resistance on a chow diet by 18 weeks of age (Biddinger et al., 2005). We therefore weighed wild-type (+/+) mice as well as heterozygous (+/−) and homozygous (−/−) Arrdc3 knockout mice from weaning through 20 weeks of age (Figure 4A). Total body mass of male Arrdc3-null mice was lower than wild-type littermates at 5 weeks of age (15% lower, P < 0.001) and at 20 weeks of age (29% lower, P < 0.001). The effect of Arrdc3 deficiency on body weight was dependent on gene dosage; the weights of heterozygotes were lower than wild-type mice at 12 weeks of age (7.3% less, P < 0.05).
Figure 4.
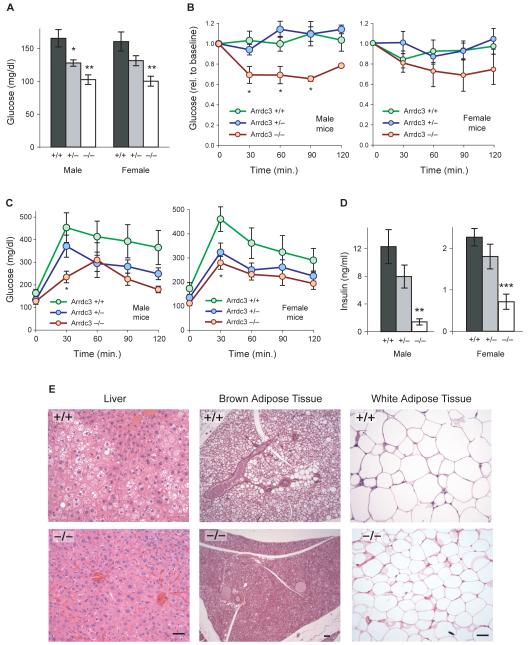
Arrdc3-deficient mice are resistant to age-induced obesity. (A) Total body mass of littermate wild-type (+/+), heterozygous (+/−), and homozygous (−/−) Arrdc3 knockout mice from 5 to 20 weeks of age. Male n = 10–18, 15–33, 6–11; female n = 9–10, 11–30, 7–14 for +/+, +/−, and −/− genotypes respectively. (B) Perigonadal fat mass expressed as a percentage of total body mass at 5–-7 months of age. Male n = 7, 6, 3; female n = 6, 9, 5. (C) Adiposity of male and female littermates measured by DEXA scanning at 8–12 weeks of age. Male n = 7, 6, 4; female n = 6, 7, 5. Error bars represent mean ± SEM; *P < 0.05; **P < 0.01; ***P < 0.001 vs. wild-type. See also Figure S4.
The effect of Arrdc3 deficiency in female mice was not as strong as in male mice, with little effect in young female mice. Total body mass of Arrdc3-null females was not different from wild-type littermates at 5 weeks of age (−/−, 19.3 ± 0.5 g; +/+, 19.3 ± 0.8 g). However, Arrdc3-null females were resistant to weight gain with age: at 20 weeks, body weight in Arrdc3-null mice was 29% lower than in wild-type mice (33.9 ± 3.0 g vs. 47.4 ± 1.9 g; P < 0.01). Heterozygous female mice also trended towards lower body mass than wild-type mice at 20 weeks of age (41.3 ± 2.6 g vs. 47.4 ± 1.9 g; P = 0.08).
Since Arrdc3-null mice were shorter than wild-type mice, we directly tested whether adiposity as a percentage of total body mass was different in Arrdc3-null mice. First, we dissected out and weighed fat pads of the older mice (5–7 months old) and calculated fat mass as a percentage of total body mass. Perigonadal fat pads were smaller in Arrdc3-null mice compared to wild-type mice in both males (1.9 ± 0.4% vs. 3.6 ± 0.3% of total body mass, P < 0.05) and females (4.4 ± 0.9% vs. 11.2 ± 0.9% of total body mass, P < 0.001) (Figure 4B). We then confirmed by DEXA scanning that differences in whole body adiposity are present just as differences in body mass are emerging, at 2–3 months of age (Figure 4C). Percentage body fat was lower in Arrdc3-null mice compared to wildtype mice in males (22.1 ± 1.3% vs. 34.1 ± 0.8%, P < 0.001) and females (27.6 ± 2.6 % vs. 44.9 ± 4.0%, P < 0.01).
Arrdc3-deficient mice retain normal glucose homeostasis
In general, mouse models and human disease states with low adiposity can be accompanied either by normal glucose homeostasis or by a lipodystrophy-like insulin resistance syndrome (Huang-Doran et al., 2010). We therefore tested the glucose tolerance and insulin response of male and female Arrdc3-deficient mice compared to wild-type littermates at 5–6 months of age.
Prior to injection of insulin, mice were fasted for 4 h and baseline blood glucose levels were measured (Figure 5A). Fasting glucose levels were elevated in wild-type mice and reduced in a gene dosage–dependent fashion in both male and female Arrdc3-deficient mice, consistent with protection from the hyperglycemia and insulin resistance that develops in the wild-type C57BL/6 mice with age (Biddinger et al., 2005). Intraperitoneal injection of a low dose (0.25 mU/g) of insulin had little effect on glucose levels in wild-type and heterozygous mice (Figure 5B). In contrast, Arrdc3-null mice retained a strong and sustained response to a low dose of insulin, with significantly lower glucose levels at 30, 60, and 90 min after injection compared to wild-type littermates. Male and female Arrdc3-null mice also cleared an intraperitoneal glucose load more quickly, with significantly lower glucose levels 30 min after injection (Figure 5C). Furthermore, we measured insulin levels in 3-month-old mice fed ad libitum (Figure 5D). Wild-type mice had much higher insulin levels than the Arrdc3-null mice, which retained insulin levels in the normal range (Andrikopoulos et al., 2008). These data demonstrate that Arrdc3 deficiency protects against obesity while maintaining normal insulin response and glucose homeostasis.
Figure 5.
Arrdc3-deficient mice are resistant to the metabolic complications of obesity. (A) Fasting blood glucose levels of littermate wild-type (+/+), heterozygous (+/−) and homozygous (−/−) Arrdc3 knockout mice at 5–7 months of age. Male n = 6, 11, 3; female n = 7, 9, 6 for +/+, +/−, and −/− genotypes, respectively. (B) The same mice were then injected with low-dose (0.25 mU/g body mass) insulin into the peritoneum. Response of blood glucose levels to insulin is expressed relative to the baseline levels shown in panel A. (C) Glucose tolerance testing of male and female littermate mice at 5-6 months of age. Blood glucose levels were measured after an intraperitoneal injection of 2 g glucose/kg body mass. Male n = 6, 10, 3; female n = 5, 11, 7. (D) Insulin levels in fed mice at 3 months of age. Male n = 8, 10, 3; female n = 5, 13, 7. (E) Histological analysis of a 9-month-old male wild-type mouse and an Arrdc3 −/− littermate. Images are representative of those observed in 3 mice of each genotype. Scale bars, 10 μm. Error bars represent mean ± SEM; *P < 0.05; **P < 0.01 vs. wild-type.
Obesity with insulin resistance is often accompanied by increased lipid stores in the liver (Unger et al., 2010). Histological analysis of a 9-month-old wild-type mouse showed evidence of hepatic steatosis (Figure 5E), whereas the liver of an Arrdc3-null littermate lacked overt lipid accumulation. Similarly, wild-type interscapular brown fat showed lipid-laden cells, whereas Arrdc3-null brown fat showed minimal lipid deposition. Finally, histology of adjacent subcutaneous white fat was consistent with smaller adipocyte size in the Arrdc3-null mice.
Arrdc3-deficient mice have increased energy expenditure with thermogenic activation of brown and white adipose tissue
We next sought to identify potential mechanisms for resistance to obesity in Arrdc3-null mice. However, male Arrdc3-null mice weighed less than wild-type mice even at the time of weaning, raising the possibility that observed differences in outcomes could be confounded by differences in growth and development. Therefore, we focused on analyzing Arrdc3 deficiency in female mice.
Serum chemistry and hormone levels of 12–15-week-old female mice (Suppl. Table S1) showed that Arrdc3-null mice had lower leptin levels, consistent with the lower adiposity, as well as lower triglyceride levels and higher beta-hydroxybutyrate levels. The markedly lower leptin and insulin levels provide additional support for Arrdc3 as the gene underlying the mouse QTLs for insulin and leptin levels on chromosome 13 (Suppl. Figure S1B). However, there was no evidence for altered hormone levels as a primary cause for resistance to obesity.
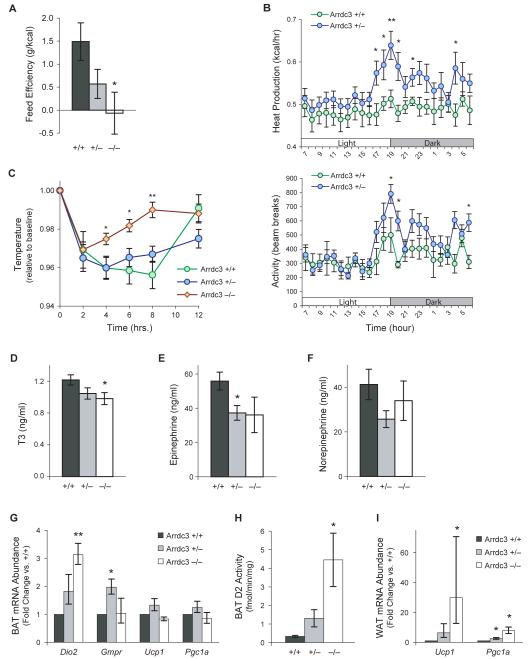
To investigate energy balance, we measured food intake in female mice just as differences in body mass were appearing among genotypes (at 8–10 weeks of age). Food intake was not significantly different among genotypes (Suppl. Figure S5A-C). In contrast, feed efficiency (change in body weight per calorie consumed) was lower in the Arrdc3-null mice compared to wild-type littermates (−/−, −0.7 ± 4.5 mg/kcal; +/−, 5.7 ± 3.1 mg/kcal; +/+: 14.9 ± 4.1 mg/kcal; P < 0.05 for −/− vs. +/+; P = 0.09 for +/− vs. +/+) (Figure 6A). While not excluding a contribution from reduced appetite, the markedly low feed efficiency suggested that Arrdc3-null mice resist obesity through either decreased nutrient absorption or increased energy expenditure.
Figure 6.
Arrdc3-deficient mice have increased energy expenditure and adipose tissue activation. (A) Food intake of 8–10-week-old female mice was measured over one week and feed efficiency calculated as the ratio of weight gain to energy intake. n = 11, 17, 6 for +/+, +/−, and −/− genotypes, respectively. (B) Metabolic cage analysis of Arrdc3 haploinsufficient and wildtype mice (n = 8 and 9 for +/+ and +/− genotypes, respectively). (C) Brown fat thermogenic function was assessed by housing mice at 4 °C. Rectal temperature was measured and expressed relative to baseline temperature (n = 6, 9, and 5). (D) Serum T3 levels in 3-month-old female mice (n = 5, 13, 6). (E-F) Serum catecholamine levels in 3-month-old female mice (n = 5, 14, 7). (G) Real-time PCR analysis of catecholamine-stimulated genes in brown adipose tissue of 3-month-old mice (n = 4, 5, 3). (H) Type 2 deiodinase activity in brown adipose tissue lysates (n = 5, 9, 5). (I) Real-time PCR analysis of thermogenic genes in visceral white adipose tissue (n = 4, 5, 3). Error bars represent mean ± SEM; *P < 0.05; **P < 0.01 vs. wild-type. See also Figure S5 and Table S1.
To test directly whether decreased Arrdc3 levels cause increased energy expenditure, gas exchange and activity levels were monitored by housing mice in metabolic cages. Interpretation of causality in metabolic cage analysis is most clear in groups of mice with the same body mass and composition (Butler and Kozak, 2010), yet even female Arrdc3-null mice trend towards lower body mass by 8–12 weeks of age. We therefore tested whether we could detect a difference between Arrdc3 wild-type and Arrdc3 heterozygous female mice in the metabolic cage analysis, since these mice have similar body mass and composition at 8–12 weeks of age. The analysis showed that Arrdc3 +/− mice had significantly higher oxygen uptake (Suppl. Fig. S5D) and higher calculated heat production than wild-type littermates, particularly during the dark (active) cycle (Figure 6B), demonstrating that decreased levels of ARRDC3 lead to an increase in energy expenditure. The respiratory exchange ratio (Suppl. Fig. S5E) did not show significant differences in fuel source usage. Activity levels were also significantly increased in Arrdc3 +/− mice, primarily at the light-to-dark feeding transition (Figure 6B), suggesting that increased energy expenditure in the mice is attributable to both increased basal metabolic rate and increased activity.
Given that Arrdc3 is regulated by feeding status in visceral white adipose and brown adipose tissues, we hypothesized that adipose tissue might be a source for the increased energy expenditure. Since brown fat is a major site of energy expenditure in mice (Foster and Frydman, 1979), we first examined whether Arrdc3-null female mice have increased brown fat thermogenic function. At room temperature, we observed a trend towards lower body temperature in the Arrdc3-deficient mice (−/−, 37.3 ± 0.4 °C; +/−, 37.6 ± 0.2 °C; +/+, 37.7 ± 0.2 °C; P = 0.40 for −/− vs. +/+; P = 0.92 for +/− vs. +/+), which could reflect increased heat loss from either lower subcutaneous adiposity or decreased sympathetic activation (Thomas and Palmiter, 1997). However, when housed at 4 °C, Arrdc3-null mice recovered their baseline body temperature more rapidly. Compared to wild-type littermates, Arrdc3-null mice were significantly warmer 4, 6, and 8 h after being housed at 4 °C (Figure 6C). After 12 h at 4 °C, all mice had returned to similar body temperatures relative to baseline. This indicates that Arrdc3 does not affect maximal levels of thermogenesis. Instead, the time course of early recovery suggests that Arrdc3-null mice have baseline differences that prime them to respond more rapidly to the cold.
The initial nonshivering thermogenic response to cold is largely due to a hypothalamus-driven increase in sympathetic tone (Thomas and Palmiter, 1997) that stimulates β-adrenergic receptors in the brown fat, inducing cAMP, lipolysis, local thyroid hormone signaling, and UCP1 levels (Jimenez et al., 2002; de Jesus et al., 2001). However, at baseline, serum concentrations of 3,5,3′-triiodothyronine (T3), thyroxine (T4), epinephrine, and norepinephrine in 3-month-old Arrdc3-deficient female mice were either not significantly different, or were lower, than those of wild-type littermates (Figure 6D–F and Suppl. Table S1). The normal-to-low systemic thyroid and catecholamine levels, together with the expression pattern of Arrdc3, suggested that a primary effect of Arrdc3 is downstream of systemic hormones and at the level of the effector tissues such as brown fat.
We therefore harvested brown adipose tissue of the female mice (3 months old) and performed real-time PCR analysis of the catecholamine-induced genes Dio2 (Silva and Larsen, 1983), Gmpr (Salvatore et al., 1998), Ucp1 (Mory et al., 1984), and Pgc1a (Puigserver et al., 1998) (Figure 6G). Arrdc3 deficiency increased baseline levels of Dio2 transcripts in a gene dosage-dependent manner relative to wild-type (+/−, 1.8-fold increase; −/−, 3.1-fold increase; P = 0.14 for +/− vs. +/+, P < 0.01 for −/− vs. +/+), whereas Ucp1 and Pgc1a transcript levels were unchanged. Dio2 encodes the type 2 iodothyronine deiodinase (D2), which converts thyroxine to active T3 within tissues and is required for cold-induced nonshivering thermogenesis (de Jesus et al., 2001). We measured D2 activity in the brown fat and confirmed a ~10-fold increase in Arrdc3-null brown fat compared with wild-type brown fat (+/+, 0.34 ± 0.05; +/−, 1.3 ± 0.5; −/−, 4.5 ± 1.4 fmol/min/mg total protein; P = 0.07 for +/− vs. +/+, P < 0.05 for −/− vs. +/+) (Figure 6H).
Although we observed differences in brown fat activation, Arrdc3 is most strongly regulated in the visceral adipose tissue depot. While the interscapular brown fat depot is a major site of thermogenesis in rodents, it has recently been demonstrated that the distinction between white and brown fat is not absolute: white adipose tissue is interspersed with varying proportions of Ucp1-expressing cells resembling brown adipocytes (Ishibashi and Seale, 2010), and the relative numbers of these cells are increased by β-adrenergic signaling (Cousin et al., 1992). Therefore, we tested whether visceral white fat could also contribute to increased baseline thermogenesis through “browning” of the white fat. Analysis of mRNA extracted from visceral fat revealed a striking 30-fold upregulation in Ucp1 levels in Arrdc3-null mice relative to wild-type mice (Figure 6I). Expression of Pgc1a, a master transcriptional regulator of thermogenesis (Puigserver et al., 1998), was also significantly increased in Arrdc3-null and Arrdc3 heterozygous mice relative to wild-type mice (7.7-fold and 2.6-fold increases, respectively).
Taken together, these data show that Arrdc3-null mice have increased thermogenic capacity due in part to upregulation of the thermogenic program in both brown and white fat depots. This occurs in the absence of increased stimulation by systemic catecholamines and thyroid hormone, consistent with regulation of local intracellular signaling by Arrdc3.
ARRDC3 regulates adipose tissue response to catecholamines through interaction with β-adrenergic receptors
Although the molecular functions of the α-arrestins are currently unclear in mice and humans, several studies in yeast have shown that regulation of receptor turnover and endocytosis is an ancient function of the arrestin-fold proteins (Lin et al., 2008; Herrador et al., 2010). Recently, Nabhan and colleagues have reported that ARRDC3 interacts with the β2-adrenoceptor resulting in receptor ubiquitination in vitro (Nabhan et al., 2010), suggesting the hypothesis that loss of ARRDC3 leads to enhanced energy expenditure through increased intracellular β-adrenergic signaling.
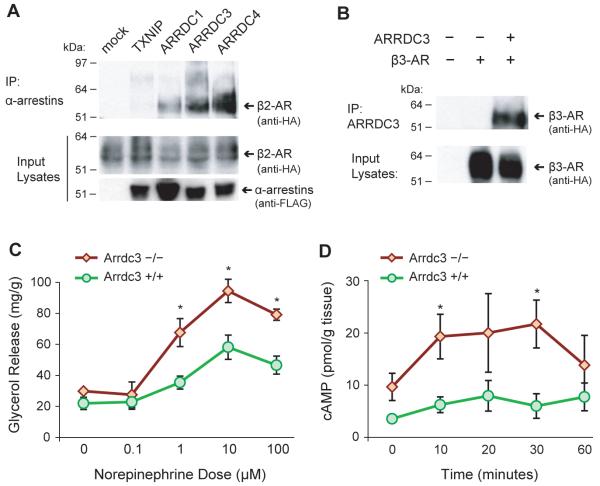
We first tested whether interaction with the β-adrenoceptor, a defining characteristic of the β-arrestins, is also a property of the α-arrestins. Four human α-arrestins and the β2-adrenergic receptor were overexpressed in 293 cells, and the interaction tested by co-immunoprecipitation (Figure 7A). No interaction between the β2-adrenoceptor and TXNIP was detected, and a weak interaction was observed with ARRDC1. In contrast, a robust interaction was detected with both ARRDC3 and ARRDC4, suggesting that interaction with the β2-adrenoceptor is at least partially conserved among α-arrestins. Since catecholamine stimulation induces the brown adipocyte thermogenic program largely through the β3-adrenoceptor, we next tested whether ARRDC3 also interacts with this receptor (Figure 7B). Immunoprecipitation of ARRDC3 revealed a strong interaction with the β3-adrenoceptor, supporting the hypothesis that ARRDC3 generally regulates β-adrenergic signaling.
Figure 7.
ARRDC3 interacts with β-adrenergic receptors to regulate signaling in adipose tissue. (A) Co-immunoprecipitation of HA-tagged β2-adrenergic receptor with FLAG-tagged α-arrestins. (B) Co-immunoprecipitation of β3-adrenergic receptor with ARRDC3. (C) Glycerol release from cultured white adipose tissue in response to norepinephrine (n = 3 mice/genotype). (D) Time course of cyclic AMP levels in white adipose tissue in response to norepinephrine (n = 3 mice/genotype). Error bars represent mean ± SEM; *P < 0.05 vs. wild-type.
Finally, we tested whether the in vitro interaction of ARRDC3 and the β-adrenoceptors is accompanied by a significant effect on adipose tissue function. We used a well-established functional assay, adrenergic stimulation of lipolysis in cultured adipose tissue (Björntorp, 1966), to assess the response to catecholamines in the absence of ARRDC3. Stimulation of excised visceral white adipose tissue with norepinephrine produced significantly higher glycerol release from Arrdc3-null animals compared to wild-type animals (Figure 7C). Since β-adrenergic signaling is largely mediated by cAMP, we also examined cAMP levels after stimulation by norepinephrine and found significantly higher cAMP levels in adipose tissue from Arrdc3-null mice compared to tissue from wild-type mice (Figure 7D).
Discussion
In the current study, we describe an association of high BMI in males to a rare haplotype across a single gene, ARRDC3, encoding a member of the arrestin superfamily of proteins. In addition, higher expression of ARRDC3 in visceral adipose tissue was associated with obesity in males. Motivated by the suggestion of a role for ARRDC3 in human obesity, we generated Arrdc3-deficient mice and demonstrated that Arrdc3 regulates obesity in mice.
Arrdc3-deficient mice were resistant to obesity in a gene dosage-dependent manner in both genders but with greater effect in males than in females. Arrdc3 deficiency protects from obesity through increased energy expenditure in part through enhanced thermogenesis in brown and white adipose tissue. Finally we show that ARRDC3 interacts directly with β2- and β3-adrenoceptors, resulting in increased intrinsic adipose tissue β-adrenergic signaling.
Our work identifies ARRDC3 as the second α-arrestin that regulates metabolic processes in vivo. Despite the shared ability to regulate metabolism, ARRDC3 and its homologous α-arrestins appear to operate by distinct molecular mechanisms. In vitro, TXNIP and ARRDC4 strongly inhibit glucose uptake into cells but ARRDC3 does not (Patwari et al., 2009). This is mirrored by differences in the phenotype of the Txnip-null and Arrdc3-null mice. Txnip-null mice have a profound fasting hypoglycemia with downregulated insulin levels due to increased peripheral tissue glucose uptake (Hui et al., 2008; Chutkow et al., 2010), whereas Arrdc3-deficient mice maintain glucose and insulin levels within the normal range. Most strikingly, Txnip-null mice are one of the few animal models that decouple the development of obesity from the development of insulin resistance: they are strongly protected against the development of diabetes despite increased adiposity (Chen et al., 2008; Chutkow et al., 2010), reminiscent of PPAR-gamma agonist therapy and adiponectin-overexpressing mice (Kim et al., 2007). In contrast, Arrdc3-null mice show no decoupling of adiposity and insulin resistance: they are protected against both obesity and dysregulated glucose homeostasis.
Arrdc3 is expressed in multiple non-neural tissues and may have more than one site of action. Furthermore, both increased metabolic rate and increased activity levels contributed to increased energy expenditure in the Arrdc3-deficient mice, and either one alone may be causal due to the interwoven control of these processes (Xu et al., 2011). However, we identify here that increased energy expenditure in Arrdc3-deficient mice is accompanied by more rapid cold-induced thermogenesis and activation of the thermogenic program in brown and white adipose tissue. In brown fat, we found upregulated tissue iodothyronine deiodinase (D2) activity. Increased conversion of T4 to active T3 locally via D2 has been shown to confer resistance to diet-induced obesity (Watanabe et al., 2006). Surprisingly, we saw no increase in Ucp1 expression in brown adipose tissue of Arrdc3-deficient mice. Consistent with these results, UCP1 is not required for increased oxygen consumption when mice are placed on a high-fat diet (Anunciado-Koza et al., 2008). On the other hand, Ucp1 and Pgc1a were strongly induced in white fat, and this “browning” of the white fat is now known to be a mechanism for upregulating thermogenesis and resisting obesity (Ishibashi and Seale, 2010).
Whereas multiple human studies consistently found that ARRDC3 is associated with obesity only in males, and Arrdc3-null male mice were more strongly affected, Arrdc3-null mice of both genders were resistant to obesity. This may be attributable to species differences such as the well known divergences in regulation of energy expenditure, hormone response, and steroid hormone levels between mice and humans (van Weerden et al., 1992; Farooqi et al., 1999). On the other hand, complete loss of Arrdc3 in mice is likely not directly comparable to the naturally occurring variation in ARRDC3 that was studied in humans.
It is possible that the observed results for the Arrdc3-null mice are biased by perinatal mortality. However, this is unlikely to affect the ultimate conclusion that ARRDC3 regulates obesity for several reasons. First, newborn Arrdc3-null pups are born at the expected Mendelian ratios, are able to breathe and feed, and have no detectable developmental defects. Second, increased perinatal mortality would be expected to bias against our hypothesis, since surviving pups are likely to have more robust growth than a hypothetical average that would include pups that did not survive. Finally, and most importantly, there was no increase in perinatal mortality for Arrdc3 +/− mice, yet most of the observed phenotypes show an Arrdc3 gene dose-dependent relationship.
Our data, together with two recent reports (Nabhan et al., 2010; Draheim et al., 2010), suggest the hypothesis that ARRDC3 retains a role in down-regulating receptor signaling, a defining feature of the classical arrestins. However, a precise delineation of ARRDC3 function in receptor endocytosis is still needed. Although recruitment of ARRDC3 to the β2-adrenoceptor is increased on stimulation by ligand (Nabhan et al., 2010), it is not clear whether binding of ARRDC3 is as specific for phosphorylated receptors as the arrestins. The residues in the arrestin polar core which are critical for this binding specificity are not conserved in the alpha-arrestins. In addition, the β3-adrenoceptor lacks the phosphorylation sites that lead to rapid downregulation in the β1 and β2 adrenoceptors (Nantel et al., 1993; Liggett et al., 1993), so investigation of the interaction and effects on endocytosis with endogenous ARRDC3 and β-adrenoceptors will be required to define the significance of the interaction.
In summary, we demonstrate that Arrdc3-deficiency protects against obesity through increased energy expenditure and suggest that ARRDC3 acts by increasing β-adrenergic signaling to activate thermogenesis in brown and white adipose tissues.
Experimental Procedures
Subjects
Ethical approval for the study was granted by the National Bioethics Committee (NBC 01-033) and the Icelandic Data Protection Authority. All participants in the study signed informed consent. The study is based on a subset of a large cohort of 8,338 Icelandic individuals who have participated in genetic studies of cardiovascular diseases and related comorbidities at deCODE Genetics. Most of the cohort did not have a history of cardiovascular disease but had been recruited as unaffected relatives of cardiovascular probands or as controls.
Adipose tissue isolation and gene expression analysis
Abdominal subcutaneous fat samples were obtained as described previously (Emilsson et al., 2008) from a population-based cohort with a distribution of BMI typical for a developed Western country. Visceral fat removal from 50 subjects was performed as follows: 10–20 g of adipose tissue samples from the greater omentum who were either obese individuals undergoing gastric bypass surgery or normal weight individuals undergoing elective abdominal surgery. Fat samples were removed with an ultrascissor (or by diathermy) and then placed in a plastic bag and moved through a 12-mm port. Before surgery, all subjects were fasted overnight and only saline and the necessary premedication and anaesthetics were infused intravenously until adipose tissue was removed. All samples were removed under general anesthesia. The samples were immediately frozen in liquid nitrogen and stored at −70 °C.
For technical replication, ARRDC3 expression was quantified by real-time PCR with Taqman assays spanning exons 7–8 of ARRDC3 and normalized to β-actin. Each sample was run in quadruplicate on an ABI Prism 7900HT machine. For northern blot analysis of visceral and subcutaneous adipose tissue, 5 μg of RNA was loaded per well where quantification of total RNA was carried out using the comparative CT method according to the manufacturer’s recommendations (User Bulletin #2, ABI Prism 7700 Sequence Detection System). For analysis of other human tissues, multiple tissue northern blots were obtained from Clontech (Human MTN Blot Cat. 7760-1). Membranes were probed with 32P-labeled full-length human ARRDC3 cDNA and a commercially available probe for the house-keeping gene β-actin.
Fed-fasting transcriptional analysis in subcutaneous fat
Subcutaneous fat biopsies were obtained sequentially (one week apart) from 20 human donors (14 females, 6 males). One group of ten subjects was assigned to fasting on both occasions while another group of ten entered a two-arm randomized cross-over study, in which each subject participated in both a fasting and a feeding arm of the study. All individuals fasted beginning at 9 pm on the day before the biopsy. Individuals were randomly assigned to one of two groups: the fasted group, in which the individual fasted throughout the morning until noon, or the fed group, in which the individual consumed a meal between 9 and 10 am. For each arm of the study, subcutaneous adipose tissue was collected 2 h after the meal or a matching time after overnight fasting. RNA was isolated from each of the samples and microarray analysis performed for 23,720 genes as detailed previously (Emilsson et al., 2008).
For mouse experiments, male C57BL/6Tac mice were either fasted overnight for 14 h or fed ad libitum before tissue harvest. Tissues were disrupted and RNA extracted in TriReagent (Molecular Research Center, Cincinnati, OH). RNA was reverse transcribed and Arrdc3 mRNA quantified by real-time PCR analysis with primers spanning exons 1–2.
Arrdc3-deficient mice
The BayGenomics mouse ES cell line CSE151 (129/OlaHsd strain) containing the insertion of a promoterless gene-trap cassette (pGT0lxf) between exons 1–2 of Arrdc3 was identified by 5′-RACE and PCR analysis (Stryke et al., 2003). Mouse chimeras generated from the targeted clone were bred with C57BL/6 mice, and heterozygous offspring were bred to yield Arrdc3-null and wild-type littermates that were used for all experiments in this study.
Mouse studies
All experiments were approved by the Harvard Medical Area Standing Committee on Animals. Mice were fed a breeding diet (PicoLab mouse diet 20) with an energy content of 23% protein, 21% fat, and 55% carbohydrates and housed in microisolator cages. Insulin tolerance tests in mice were performed by injecting 0.25 mUnits/g regular insulin intraperitoneally (IP) after 4 h of fasting. Glucose tolerance tests were performed by IP injection of 2 g/kg glucose after 6 h of fasting. Blood was collected from the tail vein at 30-min intervals and measured by glucometer (Contour, Bayer). Food intake was measured by weighing food remaining in cages of mice housed individually for one week. For cold tolerance testing, mice were housed at 4 °C in microisolator cages with ad libitum access to food and water. For northern analysis, 10 μg of total RNA was loaded per lane and membranes probed with 32P-labeled full-length Arrdc3 DNA. Realtime PCR analysis was performed with primers as described in the Supplemental Experimental Procedures and quantified by the comparative CT method relative to expression of Tbp (Lossos et al., 2003).
Statistical Analysis
All data is displayed as mean ± SEM. Inferences about differences between means were tested by 2-tailed Student’s t-test.
Supplementary Material
Highlights.
Genetic variation near ARRDC3 in humans is linked to high body-mass index in males but not females
Arrdc3-null mice resist obesity through increased energy expenditure.
Loss of Arrdc3 leads to increased brown fat activity, “browning” of the white fat, and lipolysis.
Alpha-arrestins interact with β-adrenergic receptors and loss of Arrdc3 leads to increased catecholamine response.
Acknowledgments
The authors wish to thank all the individuals who participated in this study, and the staff of the Clinical Research Center, deCODE Genetics, Reykjavik, Iceland, for their cooperation and assistance. We are also grateful for any consultation provided by Dr. Kari Stefansson and Dr. Gudmar Thorleifsson at deCode Genetics. Supported in part by NIH grants HL081523 (PP), HL048743 (RTL), HL103582 (RTL), HL090553 (SGY), DK36256 (PRL), DK076117 (AMZ), DK56626 (DEC) and DK48873 (DEC), and by a J. Ira and Nicki Harris Family research award (PP). VE and RD were employees and shareholders of Merck & Co at the time the research was performed. EES is an employee and shareholder of Pacific Biosciences.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alvarez CE. On the origins of arrestin and rhodopsin. BMC Evol Biol. 2008;8:222. doi: 10.1186/1471-2148-8-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrikopoulos S, Blair AR, Deluca N, Fam BC, Proietto J. Evaluating the glucose tolerance test in mice. Am. J. Physiol. Endocrinol. Metab. 2008;295:E1323–1332. doi: 10.1152/ajpendo.90617.2008. [DOI] [PubMed] [Google Scholar]
- Anunciado-Koza R, Ukropec J, Koza RA, Kozak LP. Inactivation of UCP1 and the glycerol phosphate cycle synergistically increases energy expenditure to resist diet-induced obesity. J. Biol. Chem. 2008;283:27688–27697. doi: 10.1074/jbc.M804268200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubry L, Guetta D, Klein G. The arrestin fold: variations on a theme. Curr. Genomics. 2009;10:133–142. doi: 10.2174/138920209787847014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell CG, Benzinou M, Siddiq A, Lecoeur C, Dina C, Lemainque A, Clément K, Basdevant A, Guy-Grand B, Mein CA, et al. Genome-wide linkage analysis for severe obesity in french caucasians finds significant susceptibility locus on chromosome 19q. Diabetes. 2004;53:1857–1865. doi: 10.2337/diabetes.53.7.1857. [DOI] [PubMed] [Google Scholar]
- Biddinger SB, Almind K, Miyazaki M, Kokkotou E, Ntambi JM, Kahn CR. Effects of diet and genetic background on sterol regulatory element-binding protein-1c, stearoyl-CoA desaturase 1, and the development of the metabolic syndrome. Diabetes. 2005;54:1314–1323. doi: 10.2337/diabetes.54.5.1314. [DOI] [PubMed] [Google Scholar]
- Björntorp P. Effect of ketone bodies on lipolysis in adipose tissue in vitro. J. Lipid Res. 1966;7:621–626. [PubMed] [Google Scholar]
- Butler AA, Kozak LP. A recurring problem with the analysis of energy expenditure in genetic models expressing lean and obese phenotypes. Diabetes. 2010;59:323–329. doi: 10.2337/db09-1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Hui ST, Couto FM, Mungrue IN, Davis DB, Attie AD, Lusis AJ, Davis RA, Shalev A. Thioredoxin-interacting protein deficiency induces Akt/Bcl-xL signaling and pancreatic beta-cell mass and protects against diabetes. FASEB J. 2008;22:3581–3594. doi: 10.1096/fj.08-111690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng C-Y, Kao WHL, Patterson N, Tandon A, Haiman CA, Harris TB, Xing C, John EM, Ambrosone CB, Brancati FL, et al. Admixture Mapping of 15,280 African Americans Identifies Obesity Susceptibility Loci on Chromosomes 5 and X. PLoS Genet. 2009:5. doi: 10.1371/journal.pgen.1000490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chutkow WA, Birkenfeld AL, Brown JD, Lee H-Y, Frederick DW, Yoshioka J, Patwari P, Kursawe R, Cushman SW, Plutzky J, et al. Deletion of the alpha-arrestin protein Txnip in mice promotes adiposity and adipogenesis while preserving insulin sensitivity. Diabetes. 2010;59:1424–1434. doi: 10.2337/db09-1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cousin B, Cinti S, Morroni M, Raimbault S, Ricquier D, Pénicaud L, Casteilla L. Occurrence of brown adipocytes in rat white adipose tissue: molecular and morphological characterization. J. Cell. Sci. 1992;103(Pt 4):931–942. doi: 10.1242/jcs.103.4.931. [DOI] [PubMed] [Google Scholar]
- Draheim KM, Chen H-B, Tao Q, Moore N, Roche M, Lyle S. ARRDC3 suppresses breast cancer progression by negatively regulating integrin beta4. Oncogene. 2010 doi: 10.1038/onc.2010.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emilsson V, Thorleifsson G, Zhang B, Leonardson AS, Zink F, Zhu J, Carlson S, Helgason A, Walters GB, Gunnarsdottir S, et al. Genetics of gene expression and its effect on disease. Nature. 2008;452:423–428. doi: 10.1038/nature06758. [DOI] [PubMed] [Google Scholar]
- Farooqi IS, Jebb SA, Langmack G, Lawrence E, Cheetham CH, Prentice AM, Hughes IA, McCamish MA, O’Rahilly S. Effects of recombinant leptin therapy in a child with congenital leptin deficiency. N. Engl. J. Med. 1999;341:879–884. doi: 10.1056/NEJM199909163411204. [DOI] [PubMed] [Google Scholar]
- Foster DO, Frydman ML. Tissue distribution of cold-induced thermogenesis in conscious warm- or cold-acclimated rats reevaluated from changes in tissue blood flow: the dominant role of brown adipose tissue in the replacement of shivering by nonshivering thermogenesis. Can. J. Physiol. Pharmacol. 1979;57:257–270. doi: 10.1139/y79-039. [DOI] [PubMed] [Google Scholar]
- Grant SFA, Thorleifsson G, Reynisdottir I, Benediktsson R, Manolescu A, Sainz J, Helgason A, Stefansson H, Emilsson V, Helgadottir A, et al. Variant of transcription factor 7-like 2 (TCF7L2) gene confers risk of type 2 diabetes. Nat. Genet. 2006;38:320–323. doi: 10.1038/ng1732. [DOI] [PubMed] [Google Scholar]
- Hager J, Dina C, Francke S, Dubois S, Houari M, Vatin V, Vaillant E, Lorentz N, Basdevant A, Clement K, et al. A genome-wide scan for human obesity genes reveals a major susceptibility locus on chromosome 10. Nat. Genet. 1998;20:304–308. doi: 10.1038/3123. [DOI] [PubMed] [Google Scholar]
- Herrador A, Herranz S, Lara D, Vincent O. Recruitment of the ESCRT machinery to a putative seven-transmembrane-domain receptor is mediated by an arrestin-related protein. Mol. Cell. Biol. 2010;30:897–907. doi: 10.1128/MCB.00132-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang-Doran I, Sleigh A, Rochford JJ, O’Rahilly S, Savage DB. Lipodystrophy: metabolic insights from a rare disorder. J. Endocrinol. 2010;207:245–255. doi: 10.1677/JOE-10-0272. [DOI] [PubMed] [Google Scholar]
- Hui STY, Andres AM, Miller AK, Spann NJ, Potter DW, Post NM, Chen AZ, Sachithanantham S, Jung DY, Kim JK, et al. Txnip balances metabolic and growth signaling via PTEN disulfide reduction. Proc. Natl. Acad. Sci. U.S.A. 2008;105:3921–6. doi: 10.1073/pnas.0800293105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishibashi J, Seale P. Medicine. Beige can be slimming. Science. 2010;328:1113–1114. doi: 10.1126/science.1190816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jesus LA, Carvalho SD, Ribeiro MO, Schneider M, Kim S-W, Harney JW, Larsen PR, Bianco AC. The type 2 iodothyronine deiodinase is essential for adaptive thermogenesis in brown adipose tissue. J. Clin. Invest. 2001;108:1379–1385. doi: 10.1172/JCI13803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez M, Léger B, Canola K, Lehr L, Arboit P, Seydoux J, Russell AP, Giacobino JP, Muzzin P, Preitner F. Beta(1)/beta(2)/beta(3)-adrenoceptor knockout mice are obese and cold-sensitive but have normal lipolytic responses to fasting. FEBS Lett. 2002;530:37–40. doi: 10.1016/s0014-5793(02)03387-2. [DOI] [PubMed] [Google Scholar]
- Kilpeläinen TO, Bingham SA, Khaw K-T, Wareham NJ, Loos RJF. Association of variants in the PCSK1 gene with obesity in the EPIC-Norfolk study. Hum. Mol. Genet. 2009;18:3496–3501. doi: 10.1093/hmg/ddp280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J-Y, van de Wall E, Laplante M, Azzara A, Trujillo ME, Hofmann SM, Schraw T, Durand JL, Li H, Li G, et al. Obesity-associated improvements in metabolic profile through expansion of adipose tissue. J Clin Invest. 2007;117:2621–2637. doi: 10.1172/JCI31021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liggett SB, Freedman NJ, Schwinn DA, Lefkowitz RJ. Structural basis for receptor subtype-specific regulation revealed by a chimeric beta 3/beta 2-adrenergic receptor. Proc. Natl. Acad. Sci. U.S.A. 1993;90:3665–3669. doi: 10.1073/pnas.90.8.3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CH, MacGurn JA, Chu T, Stefan CJ, Emr SD. Arrestin-related ubiquitin-ligase adaptors regulate endocytosis and protein turnover at the cell surface. Cell. 2008;135:714–25. doi: 10.1016/j.cell.2008.09.025. [DOI] [PubMed] [Google Scholar]
- Lossos IS, Czerwinski DK, Wechser MA, Levy R. Optimization of quantitative real-time RT-PCR parameters for the study of lymphoid malignancies. Leukemia. 2003;17:789–795. doi: 10.1038/sj.leu.2402880. [DOI] [PubMed] [Google Scholar]
- Luan B, Zhao J, Wu H, Duan B, Shu G, Wang X, Li D, Jia W, Kang J, Pei G. Deficiency of a beta-arrestin-2 signal complex contributes to insulin resistance. Nature. 2009;457:1146–1149. doi: 10.1038/nature07617. [DOI] [PubMed] [Google Scholar]
- Mory G, Bouillaud F, Combes-George M, Ricquier D. Noradrenaline controls the concentration of the uncoupling protein in brown adipose tissue. FEBS Lett. 1984;166:393–396. doi: 10.1016/0014-5793(84)80120-9. [DOI] [PubMed] [Google Scholar]
- Muoio DM, Newgard CB. Obesity-related Derangements in Metabolic Regulation. Annu. Rev. Biochem. 2006;75:367–401. doi: 10.1146/annurev.biochem.75.103004.142512. [DOI] [PubMed] [Google Scholar]
- Nabhan JF, Pan H, Lu Q. Arrestin domain-containing protein 3 recruits the NEDD4 E3 ligase to mediate ubiquitination of the beta2-adrenergic receptor. EMBO Rep. 2010;11:605–611. doi: 10.1038/embor.2010.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nantel F, Bonin H, Emorine LJ, Zilberfarb V, Strosberg AD, Bouvier M, Marullo S. The human beta 3-adrenergic receptor is resistant to short term agonist-promoted desensitization. Molecular Pharmacology. 1993;43:548–555. [PubMed] [Google Scholar]
- O’Rahilly S. Human genetics illuminates the paths to metabolic disease. Nature. 2009;462:307–314. doi: 10.1038/nature08532. [DOI] [PubMed] [Google Scholar]
- Oka S, Masutani H, Liu W, Horita H, Wang D, Kizaka-Kondoh S, Yodoi J. Thioredoxin-binding protein-2-like inducible membrane protein is a novel vitamin D3 and peroxisome proliferator-activated receptor (PPAR)gamma ligand target protein that regulates PPARgamma signaling. Endocrinology. 2006;147:733–743. doi: 10.1210/en.2005-0679. [DOI] [PubMed] [Google Scholar]
- Parikh H, Carlsson E, Chutkow WA, Johansson LE, Storgaard H, Poulsen P, Saxena R, Ladd C, Schulze PC, Mazzini MJ, et al. TXNIP regulates peripheral glucose metabolism in humans. PLoS Med. 2007;4:e158. doi: 10.1371/journal.pmed.0040158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patwari P, Chutkow WA, Cummings K, Verstraeten VLRM, Lammerding J, Schreiter ER, Lee RT. Thioredoxin-independent regulation of metabolism by the alpha-arrestin proteins. J. Biol. Chem. 2009;284:24996–25003. doi: 10.1074/jbc.M109.018093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puigserver P, Wu Z, Park CW, Graves R, Wright M, Spiegelman BM. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell. 1998;92:829–839. doi: 10.1016/s0092-8674(00)81410-5. [DOI] [PubMed] [Google Scholar]
- Salvatore D, Bartha T, Larsen PR. The Guanosine Monophosphate Reductase Gene Is Conserved in Rats and Its Expression Increases Rapidly in Brown Adipose Tissue during Cold Exposure. Journal of Biological Chemistry. 1998;273:31092–31096. doi: 10.1074/jbc.273.47.31092. [DOI] [PubMed] [Google Scholar]
- Shi H, Rojas R, Bonifacino JS, Hurley JH. The retromer subunit Vps26 has an arrestin fold and binds Vps35 through its C-terminal domain. Nat. Struct. Mol. Biol. 2006;13:540–548. doi: 10.1038/nsmb1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva JE, Larsen PR. Adrenergic activation of triiodothyronine production in brown adipose tissue. Nature. 1983;305:712–713. doi: 10.1038/305712a0. [DOI] [PubMed] [Google Scholar]
- Stryke D, Kawamoto M, Huang CC, Johns SJ, King LA, Harper CA, Meng EC, Lee RE, Yee A, L’Italien L, et al. BayGenomics: a resource of insertional mutations in mouse embryonic stem cells. Nucleic Acids Res. 2003;31:278–281. doi: 10.1093/nar/gkg064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stunkard AJ, Foch TT, Hrubec Z. A twin study of human obesity. JAMA. 1986;256:51–54. [PubMed] [Google Scholar]
- Thomas SA, Palmiter RD. Thermoregulatory and metabolic phenotypes of mice lacking noradrenaline and adrenaline. Nature. 1997;387:94–97. doi: 10.1038/387094a0. [DOI] [PubMed] [Google Scholar]
- Thorleifsson G, Walters GB, Gudbjartsson DF, Steinthorsdottir V, Sulem P, Helgadottir A, Styrkarsdottir U, Gretarsdottir S, Thorlacius S, Jonsdottir I, et al. Genome-wide association yields new sequence variants at seven loci that associate with measures of obesity. Nat. Genet. 2009;41:18–24. doi: 10.1038/ng.274. [DOI] [PubMed] [Google Scholar]
- Unger RH, Clark GO, Scherer PE, Orci L. Lipid homeostasis, lipotoxicity and the metabolic syndrome. Biochim. Biophys. Acta. 2010;1801:209–214. doi: 10.1016/j.bbalip.2009.10.006. [DOI] [PubMed] [Google Scholar]
- Walley AJ, Asher JE, Froguel P. The genetic contribution to non-syndromic human obesity. Nat. Rev. Genet. 2009;10:431–442. doi: 10.1038/nrg2594. [DOI] [PubMed] [Google Scholar]
- Watanabe M, Houten SM, Mataki C, Christoffolete MA, Kim BW, Sato H, Messaddeq N, Harney JW, Ezaki O, Kodama T, et al. Bile acids induce energy expenditure by promoting intracellular thyroid hormone activation. Nature. 2006;439:484–489. doi: 10.1038/nature04330. [DOI] [PubMed] [Google Scholar]
- van Weerden WM, Bierings HG, van Steenbrugge GJ, de Jong FH, Schröder FH. Adrenal glands of mouse and rat do not synthesize androgens. Life Sci. 1992;50:857–861. doi: 10.1016/0024-3205(92)90204-3. [DOI] [PubMed] [Google Scholar]
- Xu X, Ying Z, Cai M, Xu Z, Li Y, Jiang SY, Tzan K, Wang A, Parthasarathy S, He G, et al. Exercise ameliorates high-fat diet-induced metabolic and vascular dysfunction, and increases adipocyte progenitor cell population in brown adipose tissue. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2011;300:R1115–1125. doi: 10.1152/ajpregu.00806.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.