Abstract
Leptospira interrogans causes a variety of clinical syndromes in animals and humans. Although much information has accumulated on the importance of leptospiral lipopolysaccharide in protective antibody responses, relatively little is known about proteins that participate in immune responses. Identification of those proteins induced only in the host is particularly difficult. Using a novel double-antibody screen designed to identify clones in a gene library of L. interrogans serovar Pomona expressing host-inducible proteins, we have characterized a gene (lk75.3) encoding a sphingomyelinase-like preprotein of 648 amino acids with cytotoxic activity for equine pulmonary endothelial cells and weak hemolytic activity for equine and rabbit erythrocytes. lk73.5 was found as a single gene copy in all serovars of L. interrogans but not in other Leptospira spp. except L. inadai. The open reading frame (ORF) for Lk73.5 is followed by another partially homologous sequence containing an ORF (sph-like 2) for a 28.7-kDa peptide. Lk73.5 and Sph-like 2 share 95.1 and 97.7% amino acid identity with putative sphingomyelinases Sph2 and Sph1 (N terminus) from L. interrogans serovar Lai (S.-X. Ren, G. Fu, X.-G. Jiangk, R. Zeng, Y.-G. Miao, H. Xu, Y.-X. Zhang, H. Xiong, G. Lu, L.-F. Lu, H.-Q. Jiang, J. Jia, Y.-F. Tu, J.-X. Jiang, W.-Y. Gu, Y.-Q. Zhang, Z. Cai, H.-H. Sheng, H.-F. Yin, Y. Zhang, G.-F. Zhu, M. Wank, H.-L. Huangk, Z. Qian, S.-Y. Wang, Wei Ma, Z.-J. Yao, Y. Shen, B.-Q. Qiang, Q.-C. Xia, X.-K. Guo, A. Danchinq, I. S. Girons, R. L. Somerville, Y.-M. Wen, M.-H. Shik, Z. Chen, J.-G. Xuk, and G.-P. Zhao, Nature 422:88-893, 2003). Substantial homologies to sphingomyelinases from other leptospiras and other bacteria are also present. Lk73.5 was not detected in leptospiras cultured at 30 or 37°C. The recombinant protein reacted strongly with sera from recently infected mares but not with sera from horses vaccinated with commercial pentavalent bacterin. The host-inducible immunogenic Lk73.5 should have value in distinguishing vaccine from infection immune response.
Leptospirosis, a worldwide disease, is caused by several different Leptospira spp. that are invasive for a wide range of mammalian hosts, including humans, horses, dogs, pigs, cattle, and wildlife. Symptoms of infection vary from subclinical to potentially fatal with multiorgan involvement (30, 38, 50). Immune-based uveitis leading to blindness (12, 26, 40, 51) is a devastating sequela in humans and horses. Survival of the organism in water and persistent urinary shedding by wildlife, rodents, and livestock complicate control of the disease (30, 50). Increased economic and recreational activity in pristine areas as well as flooding associated with climatic change have increased human exposure to leptospiras. Indeed, several large outbreaks have occurred in Nicaragua (49, 52), Brazil (27), the United States (8), and Malaysia (9), and leptospirosis is now considered a reemerging infectious disease (30).
Knowledge of the epizootiology of leptospirosis has been important in the design of effective preventive strategies. Control by vaccination has been less effective, and leptospira biologicals are generally unsatisfactory in terms of efficacy, spectrum of cross protection, and duration of immunity. Leptospira vaccines, in general, stimulate protective immunity which is inferior to that developed following recovery from either acute or subclinical leptospirosis (3-5). Hypothetical antigens specifically induced during in vivo growth but not expressed by in vitro cultures may play an important role in stimulating protective immune responses. Moreover, recent findings have indicated that vaccine efficacy may be related to as-yet-unknown protein antigens with the ability to elicit Th1 immune responses (34, 35).
Over the past 5 years, novel methods such as in vivo expression technology, signature tag mutagenesis, and differential fluorescence induction have been used to identify host-induced antigens (21). The lack of a methodology for genetic manipulation of the leptospira genome limits application of these approaches in the study of the antigens in the pathogenic Leptospira spp. However, the pioneering work of Saint Girons et al. (13) provides an opportunity to express and investigate the activity of genes of pathogenic leptospiras in saprophytic host strains and suggests that genetic manipulation of pathogenic leptospiras may soon be possible.
Our approach to the identification of leptospiral host-inducible antigens has been to use a modification of in vivo-induced antigen technology (20), in which a gene library of Leptospira interrogans is screened with a convalescent-phase serum pool followed by a second screen with antiserum to bacterin of in vitro-cultured leptospira to identify clones positive for genes expressed only in vivo. Using this approach, we have identified a novel, in vivo-induced immunogenic sphingomyelinase-like protein of L. interrogans, determined its amino acid sequence, and studied its lytic and cytotoxic effects on erythrocytes and pulmonary endothelial cells.
MATERIALS AND METHODS
Bacterial strains and media.
L. interrogans serovars Pomona type kennewicki (JEN 4), Pomona (Pomona) Copenhageni (M 20), Canicola (Hond Utrech IV), Grippotyphosa (Andaman), Hardjo (Hardjoprajitno), and Bratislava (Jez Bratislava) were kindly provided by Mike Donahue (Livestock Disease Diagnostic Center, University of Kentucky, Lexington). The nonpathogenic Leptospira biflexa serovar Biflexa (Codice) was obtained from The National Veterinary Services Laboratories, Ames, Iowa. Leptospiras were cultivated in Johnson-Harris-bovine serum albumin-Tween 80 medium (24) (Bovuminar PLM-5 Microbiological Media; Intergen, Purchase, N.Y.) at 30°C unless otherwise indicated. Escherichia coli SOLR and XL1-Blue MRF′ (Stratagene, La Jolla, Calif.) were hosts for phage manipulation and plasmid excision. E. coli NovaBlue and BL21(DE3) (Novagen, Madison, Wis.) were used for cloning and expression of recombinant proteins and were routinely grown in Luria-Bertani broth or on Luria-Bertani agar.
Library screening.
A λZAPII library containing 3- to 5-kb random fragments of L. interrogans serovar Pomona type kennewicki chromosomal DNA (25) was screened to identify phage that expressed gene products reactive with a pool of sera from mares that had recently aborted due to L. interrogans serovar Pomona type kennewicki infection. Reactive plaques were picked, replated, and rescreened until all gave a positive signal with the antiserum. A secondary screen was performed with a pool of antisera from ponies vaccinated with bacterin prepared from L. interrogans serovar Pomona type kennewicki cultured at 30°C (36) to identify plaques reactive with sera for mares that had aborted but not with antiserum against the bacterin. Negative clones were selected as potential producers of in vivo-induced antigens. These clones were analyzed by immunoblotting to confirm expression and to estimate sizes of proteins.
Gel electrophoresis and immunoblotting.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed in an X-Cell SureLock Mini-Cell (Invitrogen, Carlsbad, Calif.) for 2 h at 125 V in Tris-glycine running buffer (25 mM Tris, 192 mM glycine, 0.1% SDS [pH 8.3]). Samples for electrophoresis were mixed with an equal volume of 2× gel loading buffer (100 mM Tris-Cl [pH 6.8], 4% SDS, 20% glycerol, 200 mM dithiothreitol, and 0.1% bromophenol blue) and boiled for 5 min before loading. The gels were rinsed twice in water and stained with Simply Blue Safe Stain (Invitrogen) for visualization of protein bands. Proteins were also transferred to Protran nitrocellulose membranes (0.2-μm pore size; Schleicher & Schuell, Keene, N.H.) and blocked with 3% gelatin in Tris-buffered saline (20 mM Tris, 150 mM NaCl, 0.05% Tween 20 [pH 7.5]). The membranes were incubated with antisera, followed by incubation with protein G conjugated to horseradish peroxidase (Zymed, San Francisco, Calif.). Bound conjugate was detected by using 12 mg of 4-chloro-1-naphthol (Sigma, St. Louis, Mo.), dissolved in 5 ml of methanol-25 ml of Tris-buffered saline-30 μl of 30% hydrogen peroxide, for approximately 10 min.
DNA sequencing and analysis.
Plasmids excised from selected recombinant phages by using the ExAssist helper phage and E. coli SOLR (Stratagene) were isolated by using the QIAprep spin miniprep kit (Qiagen, Valencia, Calif.) and sequenced by using standard T7 and M13 reverse and custom design primers. Sequencing was performed in a commercial DNA sequencing facility (Davis Sequencing LLC, Davis, Calif.), and editing was with Chromas 1.61 (Technelysium Pty Ltd., Queensland, Australia). Nucleotide sequences were aligned and connected by using DNASIS (Hitachi Software Engineering America, Ltd., San Diego, Calif.). Analyses of nucleotide sequence and deduced amino acid sequences were performed with DNASIS and the Genetics Computer Group package of programs (Wisconsin Package version 10.0; Genetics Computer Group, Madison, Wis.) Protein structure was also predicted by using PSORT (http://psort.nibb.ac.jp/), SignalP, TMHMM (http://www.cbs.dtu.dk/), and COILS (http://www.ch.embnet.org/index.html). Homologies were identified by a BLAST search with the National Center for Biotechnology Information server (http://www.ncbi.nlm.nih.gov/BLAST/).
Protein expression.
Primers expF and expR (Table 1), including an XhoI restriction enzyme site, were designed by using Primer 2 (Scientific & Educational Software, 1991). The forward primer was designed so that the recombinant His tag fusion would not include the predicted signal peptide. The sequence encoding mature Lk73.5 was PCR amplified from genomic DNA of L. interrogans serovar Pomona type kennewicki, which was denatured at 92°C for 2 min before 25 cycles of 92°C at 1 min, 56°C at 1 min, and 72°C for 3 min followed by digestion with XhoI and ligation into pET-15b (Novagen). The resulting construct was transformed into E. coli BL21(DE3) (Novagen). Expression of Lk73.5 was induced with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) when the culture reached an optical density of 0.6 at 600 nm, and cells were harvested after 3 h. Recombinant His6-Lk73.5 was isolated by using TALON metal affinity resin (Clontech Laboratories, Inc., Palo Alto, Calif.) under denaturation conditions. Fractions containing His6-Lk73.5 were combined and dialyzed against 20 mM Tris-50 mM NaCl buffer (pH 7.5). This preparation (recLk73.5) was used for all downstream applications.
TABLE 1.
Oligonucleotide primers used in this work
| Primer | Sequence (5′→3′)a | Location relative to lk73.5 start codon |
|---|---|---|
| expF | gcgctcgagTTACCCGAAAAAGAATCCTC | 72 |
| expR | gcgctcgagTCCGGATTTAAGAGGCCAGG | 1954 |
| LeN3 | CCATCGTGGAAGATTTGTGG | 571 |
Boldface indicates an XhoI site; lowercase indicates primer extension.
Sera.
Ponies were inoculated subcutaneously with 500 μg of recLk73.5 adsorbed to 30% aluminum hydroxide (Alhydrogel; Accurate Chemical & Scientific Corp., Westbury, N.Y.) in a volume of 2 ml and boosted 2 weeks later with the same dose. Serum was harvested by jugular venipuncture 5 weeks after the initial immunization. Sera from mares that had recently aborted due to naturally acquired infection with pathogenic Leptospira were kindly provided by Barbara Smith (Livestock Disease Diagnostic Center, University of Kentucky, Lexington). Sera from horses vaccinated with commercial leptospira pentavalent bacterin were obtained from horses in a riding stable in north Georgia. Sera from horses infected with Borrelia burgdorferi (10) and Ehrlichia sp. (11) were kindly provided by Yung-Fu Chang (Cornell University, Ithaca, NY). Negative control sera were obtained from ponies on the University of Kentucky North Farm with no known exposure to Leptospira.
Cytotoxicity of recLk73.5 for EEC.
Cytotoxicity of recLk73.5 was determined by measuring lactate dehydrogenase (LDH) released from equine pulmonary endothelial cells (EEC) (22, 33) (a gift of James MacLachlan, University of California, Davis), using a CytoTox96 cytotoxity kit (Promega, Madison, Wis.). Cells from a subconfluent culture in modified Eagle's medium were plated in triplicate on a 96-well plate (Nalge Nunc International, Rochester, N.Y.) and incubated for 12 to 15 h at 37°C in 5% CO2. A confluent monolayer of EEC was washed twice with phosphate-buffered saline, and serial dilutions of recLk73.5 from 0.16 to 50 μg/ml were added. Phospholipase C from Clostridium perfringens (25 μg/ml) and recQ1p42 (50 μg/ml) (36) were used as positive and negative controls, respectively. Following incubation for 1 h at 37°C in 5% CO2, the plate was centrifuged at 800 × g for 5 min, and 50 μl of supernatant from each well was transferred to another plate containing substrate solution and incubated at room temperature for 30 min. The absorbance was measured at 490 nm after addition of 50 μl of stop solution. Cytotoxicity was calculated by using the formula % cytotoxicity = 100 × (Asample − Abackground)/(Atotal − Abackground), where Asample was the absorbance at 490 nm of reaction mixture from treated cells and Abackground and Atotal were the absorbances of reaction mixtures from untreated cells and from cells lysed with lytic buffer, respectively. The experiment was repeated three times.
Hemolytic activity of recLk73.5.
Different concentrations of recLk73.5 and recQ1p42 (80 μg/ml), as a negative control, were added to a 10% suspension of rabbit erythrocytes in 10 mM Tris-100 mM NaCl (pH 7.5), followed by incubation at 37°C for 1 h. The suspensions were then immediately cooled in ice and centrifuged, and the absorbance of the supernatants was measured at 440 nm. Complete lysis was obtained by resuspension of erythrocytes in water.
Detection of lk73.3 in genomes of other leptospiras.
Genomic DNAs of L. interrogans serovars Bratislava, Canicola, Hardjo, Grippotyphosa, Copenhageni, and Pomona and of L. biflexa were isolated from 10 ml of stationary-phase cultures. Cells were resuspended in 400 μl of TE buffer (10 mM Tris-Cl [pH 8.0], 1 mM EDTA) containing 0.5% SDS and 100 μg of proteinase K per ml and incubated for 1 h at 37°C. NaCl was added to a final concentration of 1 M. DNA was precipitated from the aqueous phase with 2 volumes of 95% ethanol following protein extraction with phenol-chloroform and chloroform. Chromosomal DNA of B. burgdorferi was kindly provided by Yung-Fu Chang (Cornell University). DNAs of Leptospira borgpetersenii serovar Hardjo, Leptospira santarosiae serovar Tropica, Leptospira inadai serovar Malaya, Leptospira weilii serovar Coxi, Leptospira noguchi serovar Fortbragg, and Leptospira kirschneri serovar Gryppotyphosa were kindly provided by Richard Zuerner (National Animal Disease Center, Ames, Iowa). Leptospiral DNA was digested to completion with HindIII, and separated on a 0.8% agarose gel, and transferred to a Hybond-N nylon membrane (Amersham, Piscataway, N.J.) according to the manufacturer's protocol. A hybridization probe was produced by amplifying the 5′ end of lk73.5 with the expF and LeN3 primers and randomly labeling the amplicon with biotin by using the NEBlot phototype kit (New England BioLabs, Inc., Beverly, Mass.). DNA-DNA hybridization was performed overnight at 42°C in buffer containing 4× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 50% formamide, 5× Denhardt solution, 10% dextran sulfate, 0.5% SDS, and 100 μg of denatured salmon sperm DNA per ml. The membrane was washed twice at room temperature in 2× SSC with 0.1% SDS and twice at 60°C in 0.1× SSC with 0.1% SDS. Hybridization was detected with the Phototope-Star detection kit (New England BioLabs) according to the manufacturer's instructions.
Nucleotide sequence accession number.
The nucleotide sequence has been deposited in the GenBank database under accession number AF320511.
RESULTS
Isolation and characterization of the gene encoding Lk73.5.
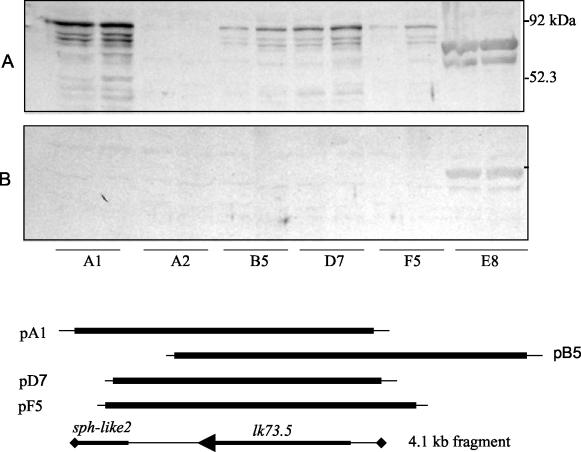
Eleven clones reactive with sera of mares that had aborted but not with antiserum to cultured leptospiras were identified. Six of these clones expressed an 80-kDa protein. Figure 1 shows the reactivities of sera used to screen the library with E. coli containing plasmids rescued from four of these recombinant phages. A series of lower-molecular-mass bands reactive only with sera from mares that had aborted were evident in each clone expressing the 80-kDa protein but not in an E. coli clone that did not express this protein. It was therefore concluded that the multiple bands are the result of proteolytic digestion. Restriction mapping of plasmids indicated that the inserts overlapped. Sequence analysis confirmed this and revealed two open reading frames in a 4.1-kb overlapping fragment (Fig. 1). The first encodes a protein with a calculated molecular mass of 73.5 kDa that was designated Lk73.5 (Fig. 2). The probable Shine-Dalgarno sequence (GGAGA) was located 9 nucleotides upstream of the ATG start codon. Several potential E. coli σ70 promoters were predicted for lk73.5. One consisted of the sequences TTcAtt and TAaaAT in the −35 and −10 regions (matches to the consensus sequence is indicated by uppercase) separated by 19 nucleotides. Three pairs of direct repeats, O1, O2, and O3, separated by 94, 126, and 26 nucleotides, respectively, flanked the potential promoter area. Immediately downstream of lk73.5, a stem-and-loop structure (ΔG = −22.3 kcal/mol) followed by five thymines resembled a ρ-independent transcriptional terminator. The amino terminus of Lk73.5 appears to consist of a 23-amino-acid signal peptide sequence with a lipobox-like (LIR↓C) structure at its carboxy terminus. The proceeding amino acid sequence contained four 25-mer repeats spanning residues 54 to 154. Two of these repeats, R2 and R3, are perfectly identical; the others are partially divergent. A second open reading frame located downstream of lk73.5 (Fig. 1) was designated sph-like 2 and encoded a 28.7-kDa peptide with partial homology to the amino terminus of Lk73.5.
FIG. 1.
Leptospiral proteins expressed by E. coli SOLR that contain plasmids rescued from phage clones. Mid-log-phase cultures were subjected to SDS-PAGE and immunoblotted with pools of sera from mares that had aborted (A) and from horses immunized with cultured leptospiras (B). Clones A1, B5, D7, and F5 produced an 80-kDa that was antigen reactive only with sera from mares that had aborted. A2 was an E. coli clone that did not express the antigen. Clone E8 expressed a leptospira protein that was reactive with both pools of sera. Proteins from each clone were loaded in duplicate. The lines represent alignment of the inserts from clones A1, B5, D7, and F5 and the 4.1-kb nucleotide sequence submitted to GenBank. lk73.5 and sph-like 2 are shown on the lower line. The positions of inserts of clones 2 and 8 were not determined.
FIG. 2.
Nucleotide sequence of the leptospiral gene encoding Lk73.5. The sequence of the 4.1-kb fragment is shown from position 271 through 2610. Probable −10 and −35 promoter sequences, ribosomal binding sites (RBS), and amino acid repeats R1 to R4 are underlined. The putative signal sequences are boxed, and a putative ρ-independent transcriptional terminator is indicated by dashed arrows. The direct nucleic acid repeats are shown in boldface.
Homology to other proteins.
The Lk73.5 protein showed 95.1% amino acid identity with a putative sphingomyelinase, Sph2, from L. interrogans serovar Lai (41). Excluding the amino-terminal repetitive sequence, 63.9 and 54.9% homology was shared with sphingomyelinase C from L. borgpetersenii serovar Hardjo (42) and with hemolytic protein of L. interrogans serovar Lai (28), respectively. The central part of Lk73.5 (180 to 462 amino acid residues) corresponding to the exo/endo/phos conservative domain (pfam03372.5) also demonstrated substantial homology to phospholipases from Staphylococcus aureus (49.8%) (39), Bacillus cereus (47.6%) (23), Listeria ivanovii (46.9%) (14), and Pseudomonas spp. (41.9%) (46).
Expression of Lk73.5 by cultured leptospiras.
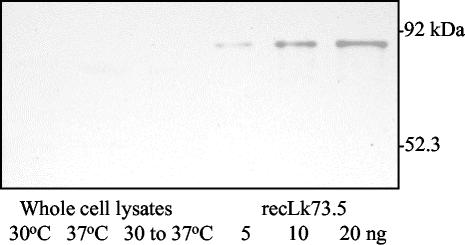
Western blot analysis of whole-cell lysates of L. interrogans serovar Pomona type kennewicki cultured at 30°C, at 37°C, and after a temperature shift from 30 to 37°C (Fig. 3) did not show detectable levels of Lk73.5 expression. Lk73.5 was not detected by Western blotting and enzyme-linked immunosorbent assay in supernatants of all leptospira cultures (data not shown).
FIG. 3.
Immunoblot of L. interrogans serovar Pomona type kennewicki cultured at 30°C, cultured at 30°C and shifted to 37°C, and cultured at 37°C. Whole-cell lysates containing 20 μg of total protein and 5, 10 and 20 ng of recLk73.5 were separated by SDS-PAGE, transferred to nitrocellulose, and probed with horse antiserum against recLk73.5.
Reactivity of recLk73.5 with horse sera.
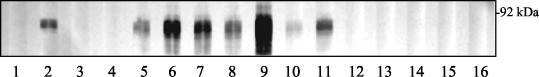
All sera from mares that had recently aborted showed strong reactivity with recLk73.5 (Fig. 4). Negative control sera included sera from horses infected with B. burgdorferi or human granulocytic ehrlichiosis agent or from horses with no exposure to these agents or leptospiras. Sera from horses immunized with L. interrogans serovar Pomona type kennewicki grown at 30 or 37°C or vaccinated with commercial pentavalent leptospira vaccine were unreactive.
FIG. 4.
Reactivity of recLk73.5 with horse sera. The protein was purified by metal affinity chromatography, subjected to SDS-PAGE separation, and probed with normal horse serum (lane 1), horse serum specific for recLk73.5 (lane 2), equine Lyme disease-positive serum (lane 3), Ehrlichia sp.-positive serum (lane 4), post-leptospiral abortion sera (lanes 5 to 11), horse antisera to bacterins prepared from L. interrogans serovar Pomona type kennewicki cultured at 30 and 37°C (lanes 12 and 13, respectively), and sera from horses immunized with pentavalent commercial bacterin (lanes 14 to 16).
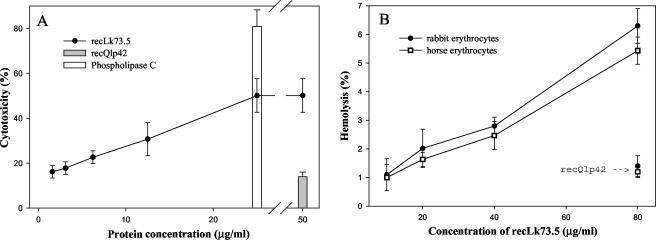
Hemolytic and cytotoxic activities of recLk73.5.
EEC exposed to serial dilutions of Lk73.5 released LDH in a concentration-dependent manner that reached a plateau at 25 μg/ml. The effect was less than but comparable to that for phospholipase C from C. perfringens, which was used as a positive control. Leptospira outer membrane lipoprotein Q1p42 (36), expressed and purified under the same conditions as Lk73.5, had almost no effect on release of LDH (Fig. 5A). recLk73.3 was only mildly hemolytic for rabbit and horse erythrocytes (Fig. 5B), and this activity was not increased in the presence of well-known phospholipase activators, such as Ca, Mg, and Mn (28, 46), at concentrations of 2 to 10 mM (data not shown).
FIG. 5.
Cytotoxic (A) and hemolytic (B) activities of recLk73.5. Cytotoxicity was assayed by measuring LDH released from EEC after 1 h of incubation with different concentrations of recLk73.5. Phospholipase C from C. perfringens (25 μg/ml) and recQ1p42 (50 μg/ml) (36), expressed and purified under the same conditions as Lk73.5, were used as positive and negative controls, respectively. Hemolytic assays were performed on a 10% suspension of rabbit or horse erythrocytes in Tris-buffered saline at 37°C for 1 h with different concentrations of recLk73.5. RecQ1p42 (80 μg/ml) was used as a negative control. Hemolysis was expressed as the percentage of hemoglobin released from the same volume of erythrocytes in distilled water minus background values. Results represent the means from three experiments ± standard deviations.
lk73.5 in other leptospiras.
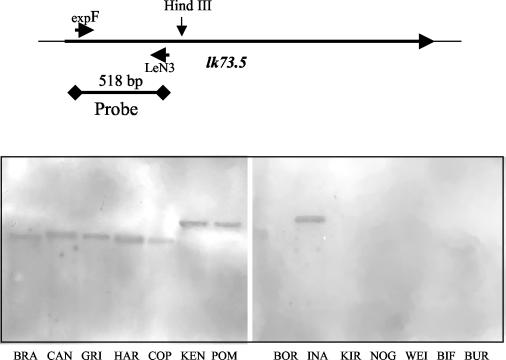
Southern blot hybridization with a probe consisting of the specific 5′ end of lk73.5 revealed only a single gene copy in all L. interrogans serovars. With the exception of L. inadai, other Leptospira spp. did not contain the gene. Bands that hybridized were of similar size (2.1 kb) for L. interrogans serovar Pomona type kennewicki and L. interrogans serovar Pomona, whereas for other leptospiras they varied from 1.7 to 2.5 kb, indicating differences in upstream sequences (Fig. 6). PCR amplification with primers expF and expR, specific for sequence encoding mature Lk73.5, produced bands that were of the predicted size for L. interrogans serovars Pomona and Canicola but slightly smaller for other leptospiras. No signals were detected for L. borgpetersenii, L. santarosiae, L. weilii, L. noguchi, or L. kirschneri (data not shown). Subsequent sequencing of the 5′ ends of amplicons showed that the differences are due to deletion of 75 nucleotides from the sequence encoding repeat R3, one of the two perfect repeats located in the N terminus of Lk73.5 (Fig. 7).
FIG. 6.
Southern blot analysis of genomic DNAs from selected strains of Leptospira spp. DNA of B. burgdorferi, the genomic DNA of which has no sequence homologous to lk73.5, was used as a negative control. Genomic DNAs were digested with HindIII and probed with a biotin-labeled fragment of the 5′ terminus of lk73.5, which has less than 40% overall homology with other leptospiral sphingomyelinase genes. Lanes: BRA, L. interrogans serovar Bratislava; CAN, L. interrogans serovar Canicola; GRI, L. interrogans serovar Grippotyphosa; HAR, L. interrogans serovar Hardjo; COP, L. interrogans serovar Copenhageni; KEN, L. interrogans serovar Pomona type kennewicki; POM, L. interrogans serovar Pomona; BOR, L. borgpetersenii; INA, L. inadai; KIR, L. kirschneri; NOG, L. noguchi; WEI, L. weilii; BIF, L. biflexa; BUR, B. burgdorferi. The schematic on the top shows the positions of the single HindIII site and of the sequence used as a probe.
FIG. 7.
PCR amplification of DNAs from different leptospiras and B. burgdorferi with primers expF and expR and comparison of the amino acid sequences predicted for the 5′ ends of amplicons. Lanes: BRA, L. interrogans serovar Bratislava: CAN, L. interrogans serovar Canicola; GRI, L. interrogans serovar Grippotyphosa; HAR, L. interrogans serovar Hardjo; COP, L. interrogans serovar Copenhageni; POM, L. interrogans serovar Pomona; KEN, L. interrogans serovar Pomona type kennewicki; BIF, L. biflexa; IN, L. inadai; BUR, B. burgdorferi; M, 1-kb DNA ladder.
DISCUSSION
Currently used leptospiral vaccines provide protection by inducing production of antibodies against lipopolysaccharides, serogroup-specific antigens found on the bacterial surface. These vaccines induce high levels of lipopolysaccharide-specific antibodies that confer short-term protection against acute infection by the corresponding serovars. A growing body of experimental data suggests that an effective cross-protective vaccine based on one or more leptospiral proteins is feasible (6, 19, 45). Because of their accessibility to host antibody, proteins located in the outer membrane are promising candidates for inclusion in future vaccines. Several such proteins, including LipL32 (18), LipL41 (44), and OmpL1 (17), for which an outer membrane location has been demonstrated experimentally, have been identified. This list might also include immunoreactive proteins such as hemolysin (28), GroEL (2), DnaK (1, 15), and LipL45 (32), the cellular location of which is not well defined. These proteins were identified either by screening chromosomal DNA libraries with rabbit antisera to in vitro-grown leptospira (LipL45) or by hybridization with oligonucleotide probes based on N-terminal amino acid sequences of particular proteins isolated from cultured leptospira (OmpL1, LipL41, and LipL32) and by PCR with degenerative primers (hemolysin). Thus, proteins involved in the infectious process and absent or poorly expressed on cultured leptospira might a priori have been excluded from further consideration. Our approach assumed that potential protective antigens, represented in the gene library with the same probability as other proteins, might be identified by sequential screening with convalescent-phase antibody and antibody specific for the antigens of in vitro (30°C)-cultured bacteria (bacterin).
In previous work, a similar double-antibody screen of a chromosomal DNA library of L. interrogans serovar Pomona type kennewicki to detect proteins upregulated at 37°C led to discovery of the immunogenic lipoprotein Qlp42, expression of which is apparently induced by temperature shift during host invasion. Subsequently, LipL45, a homolog of this protein in L. kirschneri with 98% amino acid sequence identity, was described by Matsunaga et al. (32). Double-antibody screening potentially provides a means to detect leptospiral antigens that are expressed in response to cues produced during infection. Although several clones reactive with convalescent-phase but not with bacterin-specific antibody were isolated, the present study focused on Lk73.5, which was expressed by 6 of 11 clones remaining following the double-antibody screening. Lk73.5 consists of 648 amino acid residues, the first 23 of which are followed by cysteine, a possible lipoprotein signal sequence. Although arginine in the −1 position is quite unusual as a lipoprotein signal peptidase cleavage site (7, 16, 47, 48), limited data on the spirochetal lipobox suggest a less conservative definition. Experimental evidence will be required to confirm whether Lk73.5 has a lipoprotein mode of membrane anchoring.
Recombinant Lk73.5 was strongly reactive with sera from mares following leptospiral abortion but showed no reactivity with sera from horses immunized with bacterins prepared from L. interrogans serovar Pomona type kennewicki grown at 30 and 37°C or from horses vaccinated with a commercial bacterin containing inactivated cultures of L. interrogans serovars Canicola, Grippotyphosa, Hardjo, Icterohaemorrhagiae, and Pomona. In addition, no reactive band corresponding to Lk73.5 was detected by Western blot analysis of whole-cell lysates of L. interrogans serovar Pomona type kennewicki cultured at 30 or 37°C. These observations together suggest that Lk73.5 is produced only during infection. Repeats flanking the putative lk73.5 promoter may function in regulation of this gene activity in a manner similar to that described for the Gal repressosome of E. coli (31, 43).
The moderate cytotoxicity of Lk73.5 for primary EEC as shown by release of LDH supports a role of this protein in virulence. The vascular endothelium is an important site of pathology during leptospira infection, and resulting damage to blood vessels is responsible for the characteristic hemorrhages seen in the acute disease. The mechanism of action of Lk73.5 may involve enzymatic degradation of membrane components or pore formation as described for SphH of L. interrogans serovar Lai (29). It is possible that the typical vascular damage requires the cooperative effect of Lk73.5 and other sphingomyelinases.
In conclusion, our studies suggest that Lk73.5, as recently described for LigA, a protein of unknown significance in virulence (37), is a host-inducible antigen which elicits strong antibody responses in infected animals. Membrane damage to endothelial cells and erythrocytes indicates its potential as a virulence factor. Although its production by other leptospiras during host invasion has not yet been demonstrated, the presence of lk73.5 in all investigated L. interrogans strains and in at least one other species supports such a possibility. Thus, a body of direct and circumstantial evidence supports consideration of Lk73.5 as a component for inclusion in a subunit vaccine against leptospirosis. The protein should also have value in differentiation of antibody responses to vaccination with in vitro-grown leptospiras from those stimulated by natural infection.
Acknowledgments
This work was funded by the Keeneland Association. J. E. Nally was funded by a Maxwell and Muriel Gluck Fellowship in Equine Studies.
We thank Richard Zuerner for providing leptospiral DNAs and Mike Donahue, Barbara Smith, and Yung Fu Chang for antisera.
Editor: W. A. Petri, Jr.
REFERENCES
- 1.Ballard, S. A., M. Go, R. P. Segers, and B. Adler. 1998. Molecular analysis of the dnaK locus of Leptospira interrogans serovar Copenhageni. Gene 216:21-29. [DOI] [PubMed] [Google Scholar]
- 2.Ballard, S. A., R. P. Segers, N. Bleumink Pluym, J. Fyfe, S. Faine, and B. Adler. 1993. Molecular analysis of the hsp (groE) operon of Leptospira interrogans serovar copenhageni. Mol. Microbiol. 8:739-751. [DOI] [PubMed] [Google Scholar]
- 3.Bolin, C. A., and J. A. Cassells. 1990. Isolation of Leptospira interrogans serovar bratislava from stillborn and weak pigs in Iowa. J. Am. Vet. Med. Assoc. 196:1601-1604. [PubMed] [Google Scholar]
- 4.Bolin, C. A., J. A. Cassells, R. L. Zuerner, and G. Trueba. 1991. Effect of vaccination with a monovalent Leptospira interrogans serovar hardjo type hardjo-bovis vaccine on type hardjo-bovis infection of cattle. Am. J. Vet. Res. 52:1639-1643. [PubMed] [Google Scholar]
- 5.Bolin, C. A., R. L. Zuerner, and G. Trueba. 1989. Effect of vaccination with a pentavalent leptospiral vaccine containing Leptospira interrogans serovar hardjo type hardjo-bovis on type hardjo-bovis infection of cattle. Am. J. Vet. Res. 50:2004-2008. [PubMed] [Google Scholar]
- 6.Branger, C., C. Sonrier, B. Chatrenet, B. Klonjkowski, N. Ruvoen-Clouet, A. Aubert, G. Andre-Fontaine, and M. Eloit. 2001. Identification of the hemolysis-associated protein 1 as a cross-protective immunogen of Leptospira interrogans by adenovirus-mediated vaccination. Infect. Immun. 69:6831-6838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Braun, V., and H. C. Wu. 1994. Lipoproteins, structure, function, biosynthesis and model for protein export, p. 319-341. In J.-M. Ghuysen and R. Hakenbeck (ed.), New comprehensive biochemistry, vol. 27. Bacterial cell wall. Elsevier Science, Amsterdam, The Netherlands.
- 8.Centers for Disease Control and Prevention. 1998. Update: leptospirosis and unexplained acute febrile illness among athletes participating in triathlons—Illinois and Wisconsin, 1998. JAMA 280:1474-1475. [PubMed] [Google Scholar]
- 9.Centers for Disease Control and Prevention. 2001. Update: outbreak of acute febrile illness among athletes participating in Eco-Challenge-Sabah 2000—Borneo, Malaysia, 2000. Morb. Mortal. Wkly. Rep. 50:21-24. [PubMed] [Google Scholar]
- 10.Chang, Y. F., V. Novosol, S. P. McDonough, C. F. Chang, R. H. Jacobson, T. Divers, F. W. Quimby, S. Shin, and D. H. Lein. 2000. Experimental infection of ponies with Borrelia burgdorferi by exposure to Ixodid ticks. Vet. Pathol. 37:68-76. [DOI] [PubMed] [Google Scholar]
- 11.Chang, Y.-F., V. Novosol, E. Dubovi, S. J. Wong, F. K. Chu, C.-F. Chang, F. D. Piero, S. Shin, and D. H. Lein. 1998. Experimental infection of the human granulocytic ehrlichiosis agent in horses. Vet. Parasitol. 78:137-245. [DOI] [PubMed] [Google Scholar]
- 12.Chu, K. M., R. Rathinam, P. Namperumalsamy, and D. Dean. 1998. Identification of Leptospira species in the pathogenesis of uveitis and determination of clinical ocular characteristics in south India. J. Infect. Dis. 177:1314-1321. [DOI] [PubMed] [Google Scholar]
- 13.Girons, I. S., P. Bourhy, C. Ottone, M. Picardeau, D. Yelton, R. W. Hendrix, P. Glaser, and N. Charon. 2000. The LE1 bacteriophage replicates as a plasmid within Leptospira biflexa: construction of an L. biflexa-Escherichia coli shuttle vector. J. Bacteriol. 182:5700-5705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gonzalez-Zorn, B., G. Dominguez-Bernal, M. Suarez, M. T. Ripio, Y. Vega, S. Novella, and J. A. Vazquez-Boland. 1999. The smcL gene of Listeria ivanovii encodes a sphingomyelinase C that mediates bacterial escape from the phagocytic vacuole. Mol. Microbiol. 33:510-523. [DOI] [PubMed] [Google Scholar]
- 15.Guerreiro, H., J. Croda, B. Flannery, M. Mazel, J. Matsunaga, M. Galvao Reis, P. N. Levett, A. I. Ko, and D. A. Haake. 2001. Leptospiral proteins recognized during the humoral immune response to leptospirosis in humans. Infect. Immun. 69:4958-4968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haake, D. A. 2000. Spirochaetal lipoproteins and pathogenesis. Microbiology 146:1491-1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haake, D. A., C. I. Champion, C. Martinich, E. S. Shang, D. R. Blanco, J. N. Miller, and M. A. Lovett. 1993. Molecular cloning and sequence analysis of the gene encoding OmpL1, a transmembrane outer membrane protein of pathogenic Leptospira spp. J. Bacteriol. 175:4225-4234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haake, D. A., G. Chao, R. L. Zuerner, J. K. Barnett, D. Barnett, M. Mazel, J. Matsunaga, P. N. Levett, and C. A. Bolin. 2000. The leptospiral major outer membrane protein LipL32 is a lipoprotein expressed during mammalian infection. Infect. Immun. 68:2276-2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haake, D. A., M. K. Mazel, A. M. McCoy, F. Milward, G. Chao, J. Matsunaga, and E. A. Wagar. 1999. Leptospiral outer membrane proteins OmpL1 and LipL41 exhibit synergistic immunoprotection. Infect. Immun. 67:6572-6582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Handfield, M., J. L. Brady, A. Progulske-Fox, and J. D. Hillman. 2000. IVIAT: a novel method to identify microbial genes expressed specifically during human infections. Trends Microbiol. 8:336-339. [DOI] [PubMed] [Google Scholar]
- 21.Handfield, M., and R. C. Levesque. 1999. Strategies for isolation of in vivo expressed genes from bacteria. FEMS Microbiol. Rev. 23:69-91. [DOI] [PubMed] [Google Scholar]
- 22.Hedges, J., C. Demaula, B. Moore, B. McLaughlin, S. Simon, and N. MacLachlan. 2001. Characterization of equine E-selectin. Immunology 103:498-504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johansen, T., F. B. Haugli, H. Ikezawa, and C. Little. 1988. Bacillus cereus strain SE-1: nucleotide sequence of the sphingomyelinase C gene. Nucleic Acids Res. 16:10370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnson, R. C., and V. G. Harris. 1967. Differentiation of pathogenic and saprophytic leptospires. I. Growth at low temperatures. J. Bacteriol. 94:27-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jusuf, S. D. 1997. Proteins of Leptospira interrogans serovar pomona type kennewicki reactive with serum antibodies of aborting mares and their fetuses. Doctoral dissertation. University of Kentucky, Lexington.
- 26.Kalsow, C. M., and A. E. Dwyer. 1998. Retinal immunopathology in horses with uveitis. Ocul. Immunol. Inflamm. 6:239-251. [DOI] [PubMed] [Google Scholar]
- 27.Ko, A. I., M. Galvao Reis, C. M. Ribeiro Dourado, W. D. Johnson, Jr., and L. W. Riley. 1999. Urban epidemic of severe leptospirosis in Brazil. Lancet 354:820-825. [DOI] [PubMed] [Google Scholar]
- 28.Lee, S. H., K. A. Kim, Y. G. Park, I. W. Seong, M. J. Kim, and Y. J. Lee. 2000. Identification and partial characterization of a novel hemolysin from Leptospira interrogans serovar lai. Gene 254:19-28. [DOI] [PubMed] [Google Scholar]
- 29.Lee, S. H., S. Kim, S. C. Park, and M. J. Kim. 2002. Cytotoxic activities of Leptospira interrogans hemolysin SphH as a pore-forming protein on mammalian cells. Infect. Immun. 70:315-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Levett, P. N. 2001. Leptospirosis. Clin. Microbiol. Rev. 14:296-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lewis, D. E. A., and S. Adhya. 2002. In vitro repression of the gal promoters by GalR and HU depends on the proper helical phasing of the two operators. J. Biol. Chem. 277:2498-2504. [DOI] [PubMed] [Google Scholar]
- 32.Matsunaga, J., T. A. Young, J. K. Barnett, D. Barnett, C. A. Bolin, and D. A. Haake. 2002. Novel 45-kilodalton leptospiral protein that is processed to a 31-kilodalton growth-phase-regulated peripheral membrane protein. Infect. Immun. 70:323-334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moore, B. D., U. B. R. Balasuriya, Jodi F. Hedges, and N. J. MacLachlan. 2002. Growth characteristics of a highly virulent, a moderately virulent, and an avirulent strain of equine arteritis virus in primary equine endothelial cells are predictive of their virulence to horses. Virology 298:39-44. [DOI] [PubMed] [Google Scholar]
- 34.Naiman, B. M., D. Alt, C. A. Bolin, R. Zuerner, and C. L. Baldwin. 2001. Protective killed Leptospira borgpetersenii vaccine induces potent Th1 immunity comprising responses by CD4 and γδ T lymphocytes. Infect. Immun. 69:7550-7558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Naiman, B. M., S. Blumerman, D. Alt, C. A. Bolin, R. Brown, R. Zuerner, and C. L. Baldwin. 2002. Evaluation of type 1 immune response in naive and vaccinated animals following challenge with Leptospira borgpetersenii serovar hardjo: involvement of WC1+γδ and CD4 T cells. Infect. Immun. 70:6147-6157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nally, J. E., S. Artiushin, and J. F. Timoney. 2001. Molecular characterization of thermoinduced immunogenic proteins Q1p42 and Hsp15 of Leptospira interrogans. Infect. Immun. 69:7616-7624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Palaniappan, R. U. M., Y.-F. Chang, S. S. D. Jusuf, S. Artiushin, J. F. Timoney, S. P. McDonough, S. C. Barr, T. J. Divers, K. W. Simpson, P. L. McDonough, and H. O. Mohammed. 2002. Cloning and molecular characterization of an immunogenic LigA protein of Leptospira interrogans. Infect. Immun. 70:5924-5930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Plank, R., and D. Dean. 2000. Overview of the epidemiology, microbiology, and pathogenesis of Leptospira spp. in humans. Microbes Infect. 2:1265-1276. [DOI] [PubMed] [Google Scholar]
- 39.Projan, S. J., K. J., B. Kreiswirth, S. L. Moghazeh, W. Eisner, and R. P. Novick. 1989. Nucleotide sequence: the beta-hemolysin gene of Staphylococcus aureus. Nucleic Acids Res. 17:3305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rathinam, S. R., S. Rathnam, S. Selvaraj, D. Dean, R. A. Nozik, and P. Namperumalsamy. 1997. Uveitis associated with an epidemic outbreak of leptospirosis. Am. J. Ophthalmol. 124:71-79. [DOI] [PubMed] [Google Scholar]
- 41.Ren, S.-X., G. Fu, X.-G. Jiangk, R. Zeng, Y.-G. Miao, H. Xu, Y.-X. Zhang, H. Xiong, G. Lu, L.-F. Lu, H.-Q. Jiang, J. Jia, Y.-F. Tu, J.-X. Jiang, W.-Y. Gu, Y.-Q. Zhang, Z. Cai, H.-H. Sheng, H.-F. Yin, Y. Zhang, G.-F. Zhu, M. Wank, H.-L. Huangk, Z. Qian, S.-Y. Wang, Wei Ma, Z.-J. Yao, Y. Shen, B.-Q. Qiang, Q.-C. Xia, X.-K. Guo, A. Danchinq, I. S. Girons, R. L. Somerville, Y.-M. Wen, M.-H. Shik, Z. Chen, J.-G. Xuk, and G.-P. Zhao. 2003. Unique physiological and pathogenic features of Leptospira interrogans revealed by whole-genome sequencing. Nature 422:88-893. [DOI] [PubMed] [Google Scholar]
- 42.Segers, R. P., A. van der Drift, A. de Nijs, P. Corcione, B. A. van der Zeijst, and W. Gaastra. 1990. Molecular analysis of a sphingomyelinase C gene from Leptospira interrogans serovar hardjo. Infect. Immun. 58:2177-2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Semsey, S., M. Geanacopoulos, D. E. A. Lewis, and S. Adhya. 2002. Operator-bound GalR dimers close DNA loops by direct interaction: tetramerization and inducer binding. EMBO J. 21:4349-4356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shang, E., T. Summers, and D. Haake. 1996. Molecular cloning and sequence analysis of the gene encoding LipL41, a surface-exposed lipoprotein of pathogenic Leptospira species. Infect. Immun. 64:2322-2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sonrier, C., C. Branger, V. Michel, N. Ruvoen-Clouet, J. P. Ganiere, and G. Andre-Fontaine. 2001. Evidence of cross-protection within Leptospira interrogans in an experimental model. Vaccine 19:86-94. [DOI] [PubMed] [Google Scholar]
- 46.Sueyoshi, N., K. Kita, N. Okino, K. Sakaguchi, T. Nakamura, and M. Ito. 2002. Molecular cloning and expression of Mn2+-dependent sphingomyelinase/hemolysin of an aquatic bacterium, Pseudomonas sp. strain TK4. J. Bacteriol. 184:540-546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sutcliffe, I., and R. Russell. 1995. Lipoproteins of gram-positive bacteria. J. Bacteriol. 177:1123-1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sutcliffe, I. C., and D. J. Harrington. 2002. Pattern searches for the identification of putative lipoprotein genes in Gram-positive bacterial genomes. Microbiology 148:2065-2077. [DOI] [PubMed] [Google Scholar]
- 49.Trevejo, R. T., J. G. Rigau Perez, D. A. Ashford, E. M. McClure, C. Jarquin Gonzalez, J. J. Amador, J. O. de los Reyes, A. Gonzalez, S. R. Zaki, W. J. Shieh, R. G. McLean, R. S. Nasci, R. S. Weyant, C. A. Bolin, S. L. Bragg, B. A. Perkins, and R. A. Spiegel. 1998. Epidemic leptospirosis associated with pulmonary hemorrhage—Nicaragua, 1995. J Infect. Dis. 178:1457-1463. [DOI] [PubMed] [Google Scholar]
- 50.Vinetz, J. M. 2001. Leptospirosis. Curr. Opin. Infect. Dis. 14:527-538. [DOI] [PubMed] [Google Scholar]
- 51.Wollanke, B., B. W. Rohrbach, and H. Gerhards. 2001. Serum and vitreous humor antibody titers in and isolation of Leptospira interrogans from horses with recurrent uveitis. J. Am. Vet. Med. Assoc. 219:795-800. [DOI] [PubMed] [Google Scholar]
- 52.Zaki, S. R., and W. J. Shieh. 1996. Leptospirosis associated with outbreak of acute febrile illness and pulmonary haemorrhage, Nicaragua, 1995. Lancet 347:535-536. [DOI] [PubMed] [Google Scholar]